Abstract
Recent studies report that alternatively spliced exons tend to occur in longer introns, which is attributed to the length constraints for splice site pairing for the two major splicing mechanisms, intron definition versus exon definition. Using genome-wide studies of EST and microarray data from human and mouse, we have analyzed the distribution of various subsets of alternatively spliced exons, based on their inclusion level and evolutionary history, versus increasing intron length. Alternative exons may be included in either a major or minor fraction of all transcripts (known as major-form and minor-form exons, respectively). We find that major-form exons are seven- to eightfold more likely to be contained in short introns (<400 nt) than minor-form exons, which occur preferentially in longer introns. Since minor-form exons are more likely to be novel (∼75%), this implied that novel exons arise more frequently in longer introns. To test this hypothesis, we used whole genome alignments to classify exons according to their phylogenetic age. We find that older exons, i.e., exons that are conserved in all mammals, predominate at shorter intron lengths, for both major- and minor-form exons. In contrast, exons that arose recently during primate evolution are more prevalent at longer intron lengths (>1000 nt). This suggests that the observed correlation of longer intron lengths with alternatively spliced exons may be at least partly due to biases in the probability of exon creation, which is higher in long introns.
Keywords: evolution, intron length, alternative splicing, exon creation, inclusion level
INTRODUCTION
A central theme in the study of alternative splicing has been the elucidation of factors that affect the regulation of splicing, including splice sites, spliceosomal proteins, splicing factors, and their binding sites, such as exonic splicing enhancers (ESEs) and silencers (ESSs) (Black 2003; Wang et al. 2006). It has long been known that intron length and exon length were crucial factors for efficient splicing, because of the length constraints for splice site pairing for the two major splicing mechanisms, intron definition versus exon definition (Talerico and Berget 1994; Berget 1995; Sterner et al. 1996; Romfo et al. 2000).
Recently, several studies have suggested that intron length can play an important role in governing alternative splicing, both in Drosophila and in the human genome (Fox-Walsh et al. 2005; Dewey et al. 2006; Kim et al. 2007b). Specifically, recent bioinformatics analyses of genome databases showed that exons flanked by longer introns were much more likely to be alternatively spliced compared with exons flanked by short introns (Fox-Walsh et al. 2005; Kim et al. 2007b). In vitro splicing assays for Drosophila doublesex/fruitless gene constructs with varying intron lengths linked this effect to a switch from the intron definition mechanism to exon definition (Fox-Walsh et al. 2005). These experiments indicated that intron definition was much more efficient than exon definition at splicing exons with weak splice sites. Since the exon definition mechanism becomes dominant when the flanking introns are long, the investigators suggested that the probability of exon skipping would rise with increasing intron length, providing a straightforward explanation for the observed positive correlation between intron length and alternative splicing. These results suggested that intron length is an important causal factor that determines which exons are alternatively spliced rather than constitutively spliced.
Intron length can affect many processes in addition to the switch between intron definition versus exon definition (Moriyama et al. 1998; Deutsch and Long 1999; Marais et al. 2005). Thus, one question that bears consideration is whether some other mechanism might explain the correlation of intron length with increased alternative splicing. For example, since alternative splicing is strongly associated with recent exon creation events (Sorek et al. 2002; Boue et al. 2003; Lev-Maor et al. 2003; Modrek and Lee 2003; Artamonova and Gelfand 2004; Ast 2004; Lareau et al. 2004; Pan et al. 2004; Singer et al. 2004; Cusack and Wolfe 2005; Xing and Lee 2006; Alekseyenko et al. 2007), if novel exons are simply created more frequently in long introns than in short introns, that could also explain these results. For example, a recent study of exon creation in vertebrate genome evolution found that creation of novel splice sites via point mutations was one of the predominant mechanisms of exon creation (Alekseyenko et al. 2007). Since the probability of exon creation would therefore be expected to be proportional to the number of mutable sites in an intron, this model would predict that longer introns should have a higher probability of containing recently created exons.
This represents a very different interpretation of the observed effect. The original model attributed these results to the current functional constraints of splicing mechanisms—the length constraints of intron definition versus exon definition. In contrast, the evolutionary explanation would suggest that these patterns might not be functional, and may simply reflect biases in the probability of exon creation during genome evolution.
One way to investigate these contrasting models is provided by the observation that distinct types of alternative splicing can be distinguished by their inclusion level, defined as the fraction of transcripts of a gene that include a specific alternative exon. Specifically, many studies have shown that minor-form exons (defined as alternative exons with low inclusion levels that constitute a small fraction of the total transcripts from the gene) have quite different evolutionary characteristics than major-form exons (defined as alternative exons with high inclusion levels that constitute the majority of the total transcripts) (Modrek and Lee 2003; Lareau et al. 2004; Pan et al. 2004; Resch et al. 2004; Baek and Green 2005; Cusack and Wolfe 2005; Wang et al. 2005; Xing and Lee 2005, 2006; Malko et al. 2006; Alekseyenko et al. 2007; Irimia et al. 2007a). Whereas major-form exons and constitutive exons are generally ancient (>90% are older than 300 million years [Myr]) (Alekseyenko et al. 2007), minor-form exons have undergone a much higher rate of exon creation during vertebrate evolution, and >80% of current minor-form exons were created within the last 300 Myr of vertebrate evolution (Alekseyenko et al. 2007).
Thus, one obvious question is whether there is a significant difference in intron lengths between major- and minor-form exons, and to examine their possible evolutionary basis. In this paper, we analyzed this question using three independent datasets for measuring exon inclusion levels: (1) human EST data, (2) mouse EST data, and (3) microarray quantitation of exon skipping in mouse tissue samples (Pan et al. 2006). We further subdivided our analyses into the separate effects of upstream and downstream intron lengths, as well as exon length. Next, using large-scale genome alignments (e.g., the alignment of 17 vertebrate genomes from human to fish, obtained from the UCSC Genome Browser at http://genome.ucsc.edu/; Kuhn et al. 2007), we have analyzed in detail the evolutionary history of alternative exons, to distinguish those which were created recently (e.g., during the last 100 Myr) versus those that are older (i.e., conserved over a broader range of branches of the vertebrate phylogeny).
Using these data, we have sought to address two questions. First, is the effect of intron length reported by Fox-Walsh et al. (2005) a generic effect that applies equally to all types of alternatively spliced exons, or is it more specific? Our data indicate that major-form versus minor-form alternative exons differ significantly in intron length distribution, especially at short intron lengths. Second, does this pattern have a possible evolutionary explanation? Our results suggest that it does: This pattern corresponded directly to a strong bias favoring the creation of new exons in long introns as opposed to short introns. These findings indicate the need for further studies of the interplay between splicing mechanism constraints versus evolutionary biases in alternative splicing.
RESULTS
Effect of skip intron length on exon inclusion levels
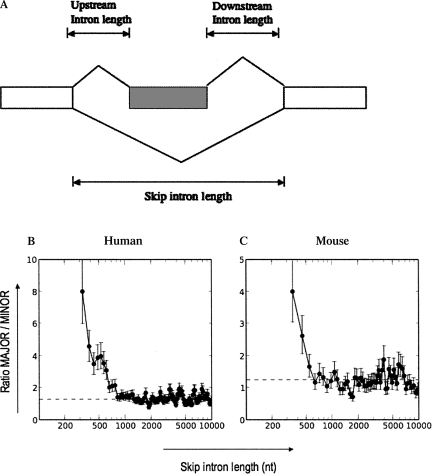
To test whether the distribution of major-form versus minor-form exons changes as a function of varying intron length, we have performed a genome-wide analysis of the effect of intron length on major-form versus minor-form alternative exons. Using a genome-wide analysis of EST data, we first classified alternatively spliced exons by their inclusion level: minor-form exons, defined as exons with an inclusion level of less than one-third of all transcripts, versus major-form exons, defined as exons with an inclusion level greater than two-thirds of all transcripts. We measured inclusion levels for 9799 alternative exons from human EST data, and for 3347 alternative exons from mouse EST data (see Materials and Methods for details). We first focused on the effect of the length of the intron containing the alternative exon (which we will refer to as the “skip intron length,” since splicing of this intron results in exon skipping; see Fig. 1A for illustration).
FIGURE 1.
Effect of skip intron length on exon inclusion levels. (A) Schematic illustration showing skipped exon (hatched box) and skip, upstream, and downstream introns. Skip intron length = length of left intron + length of skipped exon + length of right intron. (B,C) Effect of skip intron length on exon inclusion levels. (B) Human data set (Hg17); (C) mouse data set (mm7). (X-axis) Skip intron length, (y-axis) ratio of MAJOR/MINOR exons, (dotted line) ratio of MAJOR/MINOR exons over the entire population (1.29 in hg17; 1.25 in mm7). Error bars represent one standard deviation. The data were sorted by increasing intron length and plotted using a sliding window average; for details, see Materials and Methods.
These data showed a strong relationship between skip intron length and exon inclusion level (Fig. 1B,C). The ratio of major-form versus minor-form exons decreased markedly as a function of increasing skip intron length. Alternative exons contained in short introns (<400 nucleotides [nt]) were seven- to eightfold more likely to be major-form than minor-form. By contrast, alternative exons in longer introns (>800 nt) showed an approximately equal ratio of major-form versus minor-form exons (P-value of the difference is <2.2 × 10−16; see Table 1). The same patterns were observed independently in the human genome (Fig. 1B; using human EST data to measure inclusion levels) and in the mouse genome (Fig. 1C; using mouse EST data to measure inclusion levels). These two datasets made the transition to an approximately equal proportion of major-form and minor-form exons at an intron length of ∼600 nt.
TABLE 1.
Comparison of intron lengths between major- and minor-form exons in hg17
Effect of upstream and downstream intron length on exon inclusion levels
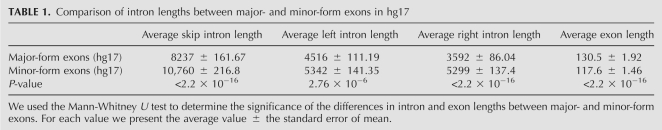
A previous study has indicated that upstream intron length has greater impact on whether an exon is likely to be alternatively spliced than downstream intron length does (Fox-Walsh et al. 2005). Since upstream intron length and downstream intron length are integral components of the skip intron length (see Fig. 1A), we decided to investigate this reported bias in more detail. Specifically, Fox-Walsh et al. (2005) reported that Drosophila and human exons with an upstream intron >4 kb were several-fold more likely to display exon skipping than exons with short upstream introns, but downstream intron length appeared to have relatively little effect.
To assess whether the quantitative inclusion level for each alternatively spliced exon displays a similar bias, we analyzed the effect of upstream versus downstream intron lengths on the ratio of major-form exons versus minor-form exons (Fig. 2). Both upstream intron length and downstream intron length displayed an effect similar to that of the skip intron length: At very short intron lengths (≤100 bp), major-form exons outnumbered minor-form exons by a factor of three or more, but this ratio declined rapidly with increasing intron length (Fig. 2; Table 1). This decay was slightly more gradual for upstream introns (up to ∼150 nt) than for downstream introns, although it is not clear whether this difference is significant. Again, we obtained reproducible results from the independent human EST (Fig. 2A,C) and mouse EST datasets (Fig. 2B,D). It is clear that the upstream and downstream intron length threshold for this decrease (∼100 nt) is much shorter than the length threshold (4000 nt) reported by Fox-Walsh et al. (2005) for increased likelihood that an exon will be alternatively spliced.
FIGURE 2.
Effect of upstream and downstream intron lengths on exon inclusion levels. Effect of upstream intron length (A,B) and downstream intron length (C,D) in the human dataset (A,C) and mouse dataset (B,D), respectively. (X-axis) Intron length, (y-axis) ratio of MAJOR/MINOR exons, (dotted line) ratio of MAJOR/MINOR exons over the entire population (1.29 in hg17; 1.25 in mm7). Error bars represent one standard deviation. The data were sorted by increasing intron length and plotted using a sliding window average; for details, see Materials and Methods.
Effect of exon length on exon inclusion levels
In light of the relatively consistent effects of skip intron length, upstream intron length, and downstream intron length, we decided to examine the effect of the final component, exon length (note: skip intron length = upstream intron length + exon length + downstream intron length). Surprisingly, this analysis showed that exon length appeared to have an opposite effect on inclusion (Fig. 3). Short exons (<50 nt) were much less likely to be major-form than minor-form, and the ratio of major-form to minor-form then increased by approximately four- to fivefold as a function of increasing exon length, up to ∼170 nt. The same effect was seen in the independent human (Fig. 3A) and mouse (Fig. 3B) datasets. The fact that this trend is opposite to that observed for skip intron length is surprising, given that the exon length contributes directly to the skip intron length (see Fig. 1). Since the exon length is part of the skip intron length, other things being equal, the skip intron length increases linearly with increasing exon length.
FIGURE 3.
Effect of exon length on exon inclusion levels. (A) Analysis of the human dataset (Hg17), (B) analysis of the mouse dataset (mm7). (X-axis) Exon length, (y-axis) ratio of MAJOR/MINOR exons, (dotted line) ratio of MAJOR/MINOR exons over the entire population (1.29 in hg17; 1.25 in mm7). Error bars represent one standard deviation. The data were sorted by increasing exon length and plotted using a sliding window average; for details, see Materials and Methods.
Independent analysis of intron length effects on inclusion level measured by quantitative microarrays
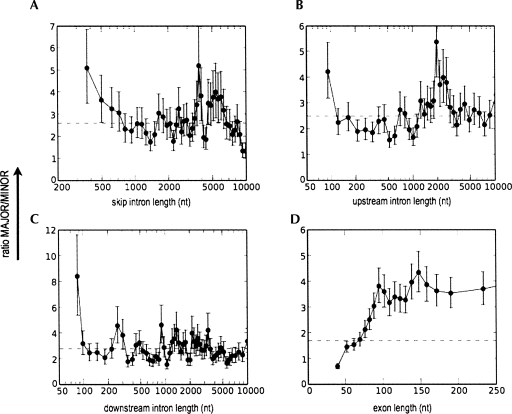
To assess the possibility that our results might be due to EST artifacts, we performed an independent analysis based on inclusion levels measured using a DNA microarray. We used data from a microarray designed by Blencowe and co-workers (Pan et al. 2004), including 3055 alternative exons in the mouse genome. Their inclusion level measurements for each exon have been carefully calibrated statistically and validated by quantitative RT-PCR (Pan et al. 2004). Since they measured inclusion levels for each exon in 10 different mouse tissues, we simply took the average inclusion level from all 10 samples (see Materials and Methods for details).
The microarray data showed the same trends as were observed in the human and mouse EST data (Fig. 4). The major-form/minor-form ratio decreased over twofold as a function of increasing skip intron length from 300 to 800 nt (Fig. 4A). In contrast, it showed a strong increase as a function of increasing exon length, up to ∼170 nt (Fig. 4D), again in agreement with the EST-based results. For both upstream and downstream intron length (Fig. 4B,C), it was maximum for very short introns (<100 nt), decreasing very rapidly (up to ∼120 nt) to become approximately constant. These data indicate that our results are broadly reproducible via different experimental approaches, and are not an artifact of EST biases or other errors.
FIGURE 4.
Independent analysis of intron and exon length effects on inclusion level measured in mouse by quantitative microarrays. Effects of skip intron length (A), upstream intron length (B), downstream intron length (C), and exon length (D) are shown. For all graphs, the x-axis represents intron or exon lengths and y-axis represents the ratio of MAJOR/MINOR exons. Error bars represent one standard deviation. (Dotted line) Ratio of MAJOR/MINOR exons over the entire population (2.59). The data were sorted by increasing intron or exon length and plotted using a sliding window average; for details, see Materials and Methods.
Effect of intron length on the phylogenetic age of exons
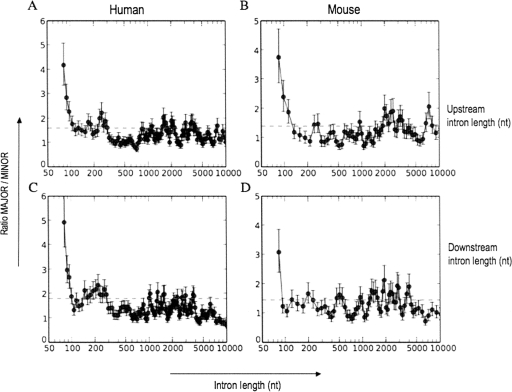
Since major-form and minor-form exons have been shown to have very different evolutionary histories and ages (for review, see Xing and Lee 2006), it is important to ask whether these intron length effects could arise from a strictly evolutionary model. In particular, Alekseyenko et al. (2007) reported that new minor-form exons were created during vertebrate evolution at a much higher rate than constitutive or major-form exons. Thus, the intron length effects described above could have a purely evolutionary explanation, if exon creation rates are themselves affected by intron length. Indeed, Alekseyenko and colleagues found that creation of novel splice sites by point mutations was the most commonly observed mechanism of minor-form exon creation. According to this model, therefore, the likelihood of creation of a novel minor-form exon should be proportional to the number of mutable sites in an intron, and thus should be higher in long introns than in short introns. To test this hypothesis, we analyzed the relative occurrence of evolutionarily “new” exons (i.e., exons created during recent primate evolution) versus “old” exons (i.e., exons conserved across a much longer range of mammalian evolution), as a function of intron length. For each exon in our databases, we analyzed its conservation across a phylogenetic tree consisting of 17 vertebrate genomes. Using multi-genome alignments from UCSC, we scored the exon as conserved or not conserved in each genome based on whether the exon and its splice sites were aligned in that genome and preserved as valid splice sites (see Materials and Methods for details).
For minor-form exons (Fig. 5A,C), we defined “new” exons as those that were created during recent primate evolution, specifically, alternative exons that have valid splice sites only in human and chimpanzee, but not in rhesus macaque, mouse, and other species in the dataset. Such exons likely were created 5 Myr to 40 Myr ago, based on the evolutionary branch times of human versus chimp and macaque. We compared these with “old” exons, defined as those that are conserved (with valid splice sites) in all mammals (human, chimp, macaque, and mouse) but not in nonmammalian vertebrate species (chicken and zebrafish). Such exons likely were created 100 Myr to 250 Myr ago, based on the evolutionary branch times of mammals versus other vertebrate species.
FIGURE 5.
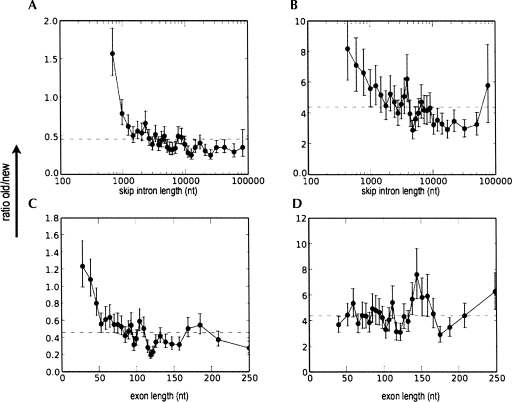
Effect of intron and exon lengths on the age of human exons. The ratio of old versus new exons is plotted against skip intron length (A,B) or exon length (C,D), for minor-form exons (A,C) and major-form exons (B,D). Exons were classified according to whether they were recently created during primate evolution (“new,” created during the last 25 Myr, subsequent to the divergence of macaque vs. human and chimp, as indicated by conservation of valid splice sites only in human and chimpanzee, but not in rhesus macaque, mouse, chicken, or fish), or not (“old,” >90 Myr old, as indicated by splice site conservation across mammals; see Materials and Methods for details). Dotted line shows ratio of old/new exons over the entire population (0.46 in minor, 4.4 in major). Error bars represent one standard deviation. The data were sorted by increasing intron or exon length and plotted using a sliding window average; for details, see Materials and Methods.
This analysis showed that skip intron length does indeed have a strong effect on the evolutionary age of minor-form exons (Fig. 5A): In short introns (≤1000 bp), old exons outnumbered new exons by nearly two to one, but this ratio declined rapidly with increasing intron length. For introns >10 kb, the ratio of old/new exons converged to ∼0.3. We observed this trend not only as a function of skip intron length, but also as functions of upstream or downstream intron length (data not shown).
We repeated this analysis for major-form exons (Fig. 5B). Since few major-form exons are conserved only in human and chimp, we extended the definition of “new” exons to include those that are conserved in primates (human, chimp, and macaque) but not in mouse. We compared these with “old” exons, defined as those conserved in both primates and other mammals (human, chimp, macaque, and mouse). Such exons likely are at least 100 Myr old. These results showed a similar trend: For introns ≤1 kb, old exons greatly outnumbered new exons by a factor of six, but this decreased with increasing intron length, to a ratio of about three.
To complete this picture, we also looked at the effect of exon length on phylogenetic age for both minor-form and major-form exons (Fig. 5C,D). For minor-form exons, we observed a similar trend as for intron length: At short exon lengths (≤50 bp), old exons outnumbered new exons, and this ratio decreased more than threefold with increasing exon length. At an exon length >150 bp, new exons predominated (Fig. 5C). In contrast, the length of major-form exons showed no correlation with their phylogenetic age. Novel major-form exons exhibited no significant length differences from old major-form exons (Fig. 5D).
Analysis of GC content, splice site strength, and intron conservation
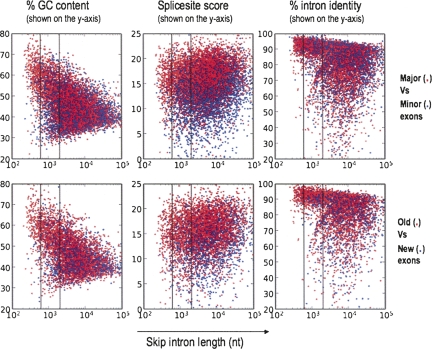
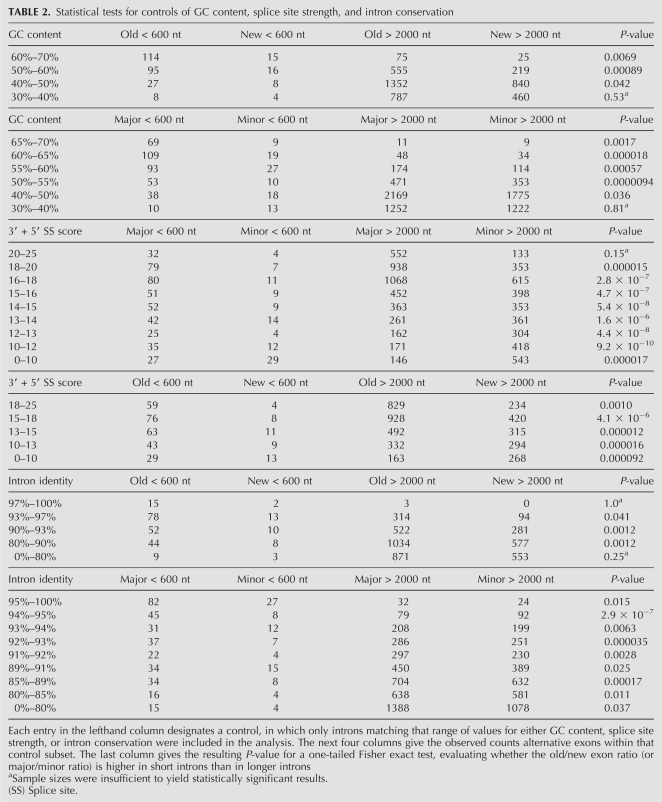
Since additional factors such as GC content have been observed to correlate with intron length (Duret et al. 1995; Chamary and Hurst 2004; Haddrill et al. 2005; Kondrashov et al. 2006; Gazave et al. 2007) and could affect our analysis, we analyzed the effect of these factors on our results. For example, GC content is on average higher in short introns than in long introns (Duret et al. 1995; Gazave et al. 2007). Is it possible then that GC content could explain our results? To test this hypothesis, we generated scatterplots of GC content versus intron length, for both evolutionarily old and new exons (Fig. 6). These data show that there is a correlation between GC content and intron length. However, they also show that for a given range of GC content (e.g., 50%–60%), short introns (<600 nt) have a markedly higher ratio of old versus new exons compared with long introns (>2000 nt). To control for GC content, we reanalyzed introns with high GC content (≥55%) and medium GC content (40%–55%) separately, and found that this relationship between intron length and old/new ratio was reproduced in both cases (Fig. 6). This relationship was statistically significant in every GC content range examined (40%–50%; 50%–60%; 60%–70%; see Table 2), except at the lower tail (0%–40%), where there were insufficient counts for the statistical test. Similarly, the relationship between intron length and major/minor ratio was also statistically significant after controlling for GC content (Table 2; Fig. 6).
FIGURE 6.
Distributions of GC content, splice site score, and intron conservation vs. intron length. (Upper plots) Each datapoint represents a single major-form exon (red) or minor-form exon (blue). (Lower plots) A single “old” exon (red; conserved in all mammals) or “new” exon (blue; created during the last 40 Myr of primate evolution). In all plots, the x-axis is the skip intron length (log-scale). In the lefthand plots, the y-axis is the percentage GC content within the intron. In the middle plots, the y-axis is the sum of the splice site scores for the 3′ and 5′ splice sites of the alternative exon. In the righthand plots, the y-axis is the combined percentage sequence identity for the upstream and downstream introns, measured between human and rhesus macaque (see Materials and Methods for details). The two vertical lines in each plot represent the skip intron length thresholds (“short introns” <600 nt; vs. “long introns” >2000 nt) used for statistical significance tests (see Table 2).
TABLE 2.
Statistical tests for controls of GC content, splice site strength, and intron conservation
We also examined whether our results could be explained by splice site strength, which has been previously reported to be lower in minor-form exons than in major-form and constitutive exons (Itoh et al. 2004; Baek and Green 2005; Zheng et al. 2005). The scatterplot of maxent splice site score versus intron length (Fig. 6) confirms that effect, but also shows that for any given range of splice site scores (e.g., 14–15), short introns (<600 nt) have a markedly higher major/minor ratio than long introns (>2000 nt). This relationship was statistically significant over all splice site score ranges examined (Table 2), except the upper tail (20–25), where there were insufficient counts.
Finally, we also tested the effect of intron conservation levels on our observed relationship between intron length and evolutionarily old versus new exons (Fig. 6; Table 2). Since it has been reported that exon creation mechanisms involve intronic point mutations (Sorek et al. 2002; Lev-Maor et al. 2003; Alekseyenko et al. 2007; Sorek 2007), it is possible that the frequency of exon creation events might correlate with increasing local mutation rates (or equivalently, reduced sequence conservation). To control for this effect, we computed intron sequence identity between human and rhesus macaque, the genome used as the outgroup for defining evolutionarily “new” exons in our analysis. In each range of intron sequence identity (e.g., 90%–93%), we found that short introns still had a much higher old/new ratio than long introns, except the upper and lower tails (97%–100%; 0%–80%), where there were too few counts to test the hypothesis.
DISCUSSION
One consistent conclusion from these results is that not only does intron length have important effects on alternative splicing, it also affects subsets of alternatively spliced exons differently, such as major-form exons versus minor-form exons, or recently created exons versus evolutionarily older exons. While these data are broadly consistent with the observation of Fox-Walsh et al. (2005) and Kim et al. (2007b) that exons are more likely to be alternatively spliced in long introns, our data show that such intron length effects are not uniform in how they affect different types of alternative exons.
It is interesting to note that Fox-Walsh et al. (2005) presented their bioinformatics analysis explicitly in the context of experimental studies of length constraints of the two major splicing mechanisms, intron definition versus exon definition. Using in vitro splicing assays with two Drosophila introns (from doublesex and fruitless), they showed that short introns of ≤200 nt yielded much more efficient splicing of a weak exon, apparently because such short introns are efficiently spliced by the intron definition mechanism, which cannot operate on long introns. They suggested that this effect might explain the tendency of alternative exons to be found preferentially in long introns: exons with weak splice sites might be rescued by this effect in short introns (spliced with 100% efficiency), but spliced with reduced efficiency in long introns (resulting in alternative splicing). This explanation attributes the association of alternative splicing with long introns to the current functional constraints of splicing mechanisms, specifically, to the restriction of the intron definition mechanism to short introns.
Our data suggest the possibility of a different, evolutionary explanation for at least some of these intron length effects. The fact that long introns show a greatly increased occurrence of minor-form exons (as opposed to major-form exons) initially suggested this possibility. This proposal has two elements. First, evolutionary models of exon creation by point mutations yielding valid splice sites predict that the likelihood of creation of a new exon will be directly proportional to the number of mutable sites, and thus to the length of the enclosing intron. Second, many studies have shown that novel exon creation shows a strong bias to produce alternatively spliced exons, and particularly minor-form exons (Modrek and Lee 2003; Lareau et al. 2004; Pan et al. 2004; Baek and Green 2005; Wang et al. 2005; Malko et al. 2006; Xing and Lee 2006; Alekseyenko et al. 2007; Nurtdinov et al. 2007), at least in mammals and dipterans, but not in nematodes (Irimia et al. 2007a,b). Taken together, these ideas predict that the association of alternative splicing with long introns might simply be a consequence of the fact that new exons are more likely to be created in long introns. Our phylogenetic analysis of 17 vertebrate genomes confirmed this prediction: Newly created exons are indeed enriched in longer introns compared with short introns. While these data in no way rule out the explanation proposed by Fox-Walsh et al. (2005), they do suggest the possibility that this pattern might simply be an artifact of biases for where new exons are most likely to be created during evolution—in long introns. This question requires further study.
Our data also raise the question of whether major-form versus minor-form alternative splicing might differ somewhat in their splicing mechanisms. We observed an inverse correlation between intron length and exon inclusion levels. This implies that major-form exons may have shorter introns than minor-form exons. The differences in intron sizes suggest differences in splicing modes between major-form and minor-form exons. Our observations also suggest a positive correlation between exon length and inclusion levels. This is consistent with data suggesting that minor-form exons are shorter on average than major-form exons (Sorek et al. 2004; Baek and Green 2005; Lev-Maor et al. 2007). The restraints on exon length may reflect size limitations to facilitate bridging across the exon. Large exons are only a problem if paired with larger introns (Sterner et al. 1996). Thus, increasing exon lengths, in the presence of large introns, would result in failure of exon inclusion. It is also interesting to speculate that the observed positive correlation of major/minor ratio with increasing exon length might offer an explanation of the clear difference in slope for the decrease in major/minor ratio as a function of skip intron length (gradually descending, up to ∼700 nt; Fig. 1) versus as a function of upstream or downstream intron length (sharply descending, ∼100–150 nt; Fig. 2). Based on the upstream/downstream curves, one would expect the skip length curve to show a sharp decrease ∼400–500 nt; however, the observed effect of exon length would be expected to change this into a more gradual decay, as is observed.
These data are consistent with previous evidence that indicates a difference in splicing regulation between minor- and major-form exons. Minor-form exons show evidence of increased purifying selection on RNA sequence (Kaufmann et al. 2004; Baek and Green 2005; Xing and Lee 2005; Plass and Eyras 2006; Lev-Maor et al. 2007). This effect is especially pronounced at the exon flanks, where splicing regulatory sites are located (Plass and Eyras 2006; Xing et al. 2006). This requirement for sequence conservation suggests that splicing of minor-form exons is under tighter regulation. It also suggests the presence of splicing regulatory sequences such as ESEs. In this context, it is interesting that ESE density has been reported to increase with increasing intron length, at least up to intron sizes of 1.5 kb (Dewey et al. 2006). At intron lengths >1.5 kb, increasing intron lengths are associated with increased splice site strength, but show no correlation with ESE density (Dewey et al. 2006). ESEs may be important for the splicing of minor-form exons, although this remains a subject of ongoing research.
MATERIALS AND METHODS
EST, mRNA, and microarray data sources
The datasets analyzed were:
Human data: All exon skipping observations from the ASAP2 human splicing data (Jan 06; hg17; EST and mRNA data previously described in Kim et al. 2007a), along with information about the length of the flanking introns, inclusion level (see below), etc. The total number of high confidence exon skips included in this analysis was 9799.
Mouse data: All exon skipping observations are from the ASAP2 mouse splicing data (Jan 06; mm7; Kim et al. 2007a), along with information about the length of the flanking introns, etc. Total number of high confidence exon skips was 3347.
Microarray data: Obtained from Pan et al. (2004), kindly provided by Dr. Blencowe (Univ. of Toronto). Pan et al. (2004, 2006) used a custom array design to measure exon inclusion levels for more than 3000 alternatively spliced exons in 10 mouse tissues. Total number of exon skips analyzed was 3055.
Classification of exons according to inclusion level
For the human and mouse EST datasets
We calculated the exon inclusion level from EST data as previously described (Xu et al. 2002; Modrek and Lee 2003). Briefly, given EST counts for the skip intron, upstream intron, and downstream intron splicing events of Ns, Nu, and Nd respectively, we estimated the inclusion level:
 |
Exons were assigned to the MAJOR-form category if the inclusion level was >67%, and to the MINOR-form category if the inclusion level was <33%. If inclusion levels were between 33% and 67%, exons were assigned to the MEDIUM-form category. To ensure adequate EST coverage for distinguishing major-form from minor-form exons accurately, we required that the total number of EST observations for these splice events, (specifically, [Nu + Nd]/2 + NS) be ≥10 for the hg17 dataset, and ≥5 for the mm7 dataset. The number of major-form and minor-form exons were roughly equal in both human and mouse datasets (1.29 and 1.25, respectively; also see Table 3).
TABLE 3.
Total number of exon skips in each dataset and their inclusion levels
For microarray data
To obtain an average inclusion level for each exon, we simply took the average of its inclusion levels measured in each of the 10 mouse tissues analyzed by the Blencowe lab (Pan et al. 2004). The numbers of major-form and minor-form exons in this dataset are shown in Table 3.
Global alignment analyses of splice site conservation and phylogenetic classification of exons according to exon age
We performed our analyses using data from the Vertebrate Exon Evolution Database (VEEDB; available at http://bioinfo.mbi.ucla.edu/VEEDB/index.xml), a previously published analysis of exon and splice site conservation in 17 vertebrate genomes (Alekseyenko et al. 2007) based on multi-genome alignments from the UCSC genome browser database (Kuhn et al. 2007; available at http://genome.ucsc.edu/) and global alignment of introns using full dynamic programming. Briefly, VEEDB maps each exon in a given genome to the precise genomic location in each aligned genome, together with scoring information for whether the splice sites are conserved in each genome, as a criterion for functional exon conservation widely adopted in comparative genomics studies (Ovcharenko et al. 2004; Hsieh et al. 2006; Alekseyenko et al. 2007). If the target genome aligns to the exon sequence and its GT/AG splice sites are conserved, the exon is scored as conserved; if the splice sites are mutated, it is scored as non-exonic (in the absence of a valid splice site, the sequence is unlikely to be spliced as an exon in the mature mRNA).
Human exons from ASAP2 (Kim et al. 2007a) were classified based on whether the splice sites flanking the potential exonic sequence were conserved in the following species (shown in Table 4): Human (Homo sapiens), chimpanzee (Pan troglodytes), rhesus macaque (Macaca mulatta), mouse (Mus musculus), chicken (Gallus gallus), and zebrafish (Danio rerio). These representative species were chosen to provide broad coverage of genera (mammals, birds, fish) and evolutionary distance, and on the basis of genome assembly quality and coverage. We classified exons as “old” if they are conserved (with valid splice sites) across the mammals (human, chimp, macaque, and mouse) but not in non-mammalian vertebrate species (chicken and zebrafish). Such exons are at least 90 Myr old (created prior to the divergence of mouse and human). We classified exons as “new” (recent exon creation) if they had valid splice sites only in human and chimpanzee, but not in rhesus macaque, mouse, chicken, or fish. These “new” exons are thus alternative exons that were created during the last 25 Myr, subsequent to the divergence of macaque and primates.
TABLE 4.
Total number of exons classified by phylogenetic age and inclusion levels
Data analyses
To assess the effect of intron and exon lengths (see Fig. 1 for a schematic illustration of our nomenclature) on exon inclusion levels, we divided all skipped exons into two categories: MAJOR (major-form) or MINOR (minor-form) as described above and in Xu et al. (2002) and Modrek and Lee (2003). To plot the relationship between intron length versus the ratio of MAJOR/MINOR exons, we sorted the data by increasing intron length and used a sliding window (with a typical window size of 100 exons), to calculate the average intron length and ratio of MAJOR/MINOR exons within each window. We calculated confidence intervals for the ratios using the β probability distribution, which provides the posterior for the binomial based on observed counts of n1, n2 (where n1 = number of MAJOR exons and n2 = number of MINOR exons), using the Python Scipy package (Jones et al. 2001; http://www.scipy.org/).
Similarly, to assess the effect of intron and exon lengths on phylogenetic age of exons, we classified all exons as “old” or “new,” as described above. The ratio of old/new exons was plotted against average intron/exon length. Average intron/exon lengths and confidence intervals were calculated exactly as described above.
GC content was calculated for each skip intron, by computing the overall GC content of the combined upstream intron plus downstream intron (in other words, excluding the alternative exon from the calculation). Splice site strengths were calculated using MaxEntScan (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) (Eng et al. 2004; Yeo and Burge 2004), to calculate splice site scores of the 5′ SS + 3′ SS of all alternative exons in our dataset. Intron conservation was measured for each human skip intron by measuring the overall percent sequence identity for the upstream plus downstream introns (in other words, excluding the alternative exon from the calculation). Upstream or downstream introns <120 nt were excluded. For each upstream or downstream intron, the first and last 50 nt were excluded, since previous studies have shown that patterns of alternative splicing can strongly affect sequence conservation in these flanking intronic regions (Sorek and Ast 2003; Kaufmann et al. 2004; Baek and Green 2005; Xing and Lee 2005; Zheng et al. 2005; Sugnet et al. 2006). The clipped intron regions were then queried against the UCSC hg17_multiz17way multigenome alignment (using the software package Pygr, http://bioinformatics.ucla.edu/pygr) to obtain the aligned region in rhesus macaque. Sequence identities were counted between the aligned human versus macaque sequences, summed for the upstream and downstream intron pair, and divided by the total length of the upstream and downstream intron pair regions used for the analysis, to obtain the percentage sequence identity.
ACKNOWLEDGMENTS
We thank Qi Wang, Douglas Black, and Brent Graveley for helpful comments and discussions on the manuscript. This work was supported by grants from the NIH (U54 RR021813) and DOE (DE-FC02-02ER63421) and a Dreyfus Foundation Teacher-Scholar Award to C.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1024908.
REFERENCES
- Alekseyenko A., Kim N., Lee C. Global analysis of exon creation vs. loss, and the role of alternative splicing, in 17 vertebrate genomes. RNA. 2007;13:661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artamonova I.I., Gelfand M.S. Evolution of the exon–intron structure and alternative splicing of the MAGE-A family of cancer/testis antigens. J. Mol. Evol. 2004;59:620–631. doi: 10.1007/s00239-004-2654-3. [DOI] [PubMed] [Google Scholar]
- Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Baek D., Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boue S., Letunic I., Bork P. Alternative splicing and evolution. BioEssays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- Chamary J.V., Hurst L.D. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: Evidence for selectively driven codon usage. Mol. Biol. Evol. 2004;21:1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- Cusack B.P., Wolfe K.H. Changes in alternative splicing of human and mouse genes are accompanied by faster evolution of constitutive exons. Mol. Biol. Evol. 2005;22:2198–2208. doi: 10.1093/molbev/msi218. [DOI] [PubMed] [Google Scholar]
- Deutsch M., Long M. Intron–exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C.N., Rogozin I.B., Koonin E.V. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. BMC Genomics. 2006;7:311. doi: 10.1186/1471-2164-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J. Mol. Evol. 1995;40:308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- Eng L., Coutinho G., Nahas S., Yeo G., Tanouye R., Babaei M., Dork T., Burge C., Gatti R.A. Nonclassical splicing mutations in the coding and noncoding regions of the ATM gene: Maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh K.L., Dou Y., Lam B.J., Hung S.P., Baldi P.F., Hertel K.J. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E., Marques-Bonet T., Fernando O., Charlesworth B., Navarro A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biol. 2007;8:R21. doi: 10.1186/gb-2007-8-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P.R., Charlesworth B., Halligan D.L., Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S.J., Lin C.Y., Liu N.H., Chow W.Y., Tang C.Y. GeneAlign: A coding exon prediction tool based on phylogenetical comparisons. Nucleic Acids Res. 2006;34:W280–W284. doi: 10.1093/nar/gkl307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M., Rukov J.L., Penny D., Garcia-Fernandez J., Vinther J., Roy S.W. Widespread evolutionary conservation of alternatively spliced exons in Caenorhabditis . Mol. Biol. Evol. 2007a;25:375–382. doi: 10.1093/molbev/msm262. [DOI] [PubMed] [Google Scholar]
- Irimia M., Rukov J.L., Penny D., Roy S.W. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol. Biol. 2007b;7:188. doi: 10.1186/1471-2148-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Washio T., Tomita M. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA. 2004;10:1005–1018. doi: 10.1261/rna.5221604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E., Oliphant T., Peterson P. Scipy: Open source scientific tools for Python . 2001 http://www.scipy.org/
- Kaufmann D., Kenner O., Nurnberg P., Vogel W., Bartelt B. In NF1, CFTR, PER3, CARS and SYT7, alternatively included exons show higher conservation of surrounding intron sequences than constitutive exons. Eur. J. Hum. Genet. 2004;12:139–149. doi: 10.1038/sj.ejhg.5201098. [DOI] [PubMed] [Google Scholar]
- Kim N., Alekseyenko A., Roy M., Lee C. The ASAP II database: Analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007a;35:D93–D98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Magen A., Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007b;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F.A., Ogurtsov A.Y., Kondrashov A.S. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. 2006;240:616–626. doi: 10.1016/j.jtbi.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., et al. The UCSC genome browser database: Update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Green R.E., Bhatnagar R.S., Brenner S.E. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G., Goren A., Sela N., Kim E., Keren H., Doron-Faigenboim A., Leibman-Barak S., Pupko T., Ast G. The “alternative” choice of constitutive exons throughout evolution. PLoS Genet. 2007;3:e203. doi: 10.1371/journal.pgen.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malko D.B., Makeev V.J., Mironov A.A., Gelfand M.S. Evolution of exon–intron structure and alternative splicing in fruit flies and malarial mosquito genomes. Genome Res. 2006;16:505–509. doi: 10.1101/gr.4236606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G., Nouvellet P., Keightley P.D., Charlesworth B. Intron size and exon evolution in Drosophila . Genetics. 2005;170:481–485. doi: 10.1534/genetics.104.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B., Lee C. Alternative splicing in the human, mouse and rat genomes is associated with an increased rate of exon creation/loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Moriyama E.N., Petrov D.A., Hartl D.L. Genome size and intron size in Drosophila . Mol. Biol. Evol. 1998;15:770–773. doi: 10.1093/oxfordjournals.molbev.a025980. [DOI] [PubMed] [Google Scholar]
- Nurtdinov R.N., Neverov A.D., Favorov A.V., Mironov A.A., Gelfand M.S. Conserved and species-specific alternative splicing in mammalian genomes. BMC Evol. Biol. 2007;7:249. doi: 10.1186/1471-2148-7-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I., Boffelli D., Loots G.G. eShadow: A tool for comparing closely related sequences. Genome Res. 2004;14:1191–1198. doi: 10.1101/gr.1773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass M., Eyras E. Differentiated evolutionary rates in alternative exons and the implications for splicing regulation. BMC Evol. Biol. 2006;6:50. doi: 10.1186/1471-2148-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch A., Xing Y., Alekseyenko A., Modrek B., Lee C. Evidence for a sub-population of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romfo C.M., Alvarez C.J., van Heeckeren W.J., Webb C.J., Wise J.A. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe . Mol. Cell. Biol. 2000;20:7955–7970. doi: 10.1128/mcb.20.21.7955-7970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S.S., Mannel D.N., Hehlgans T., Brosius J., Schmitz J. From “junk” to gene: Curriculum vitae of a primate receptor isoform gene. J. Mol. Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- Sorek R. The birth of new exons: Mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Shemesh R., Cohen Y., Basechess O., Ast G., Shamir R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.A., Carlo T., Berget S.M. Architectural limits on split genes. Proc. Natl. Acad. Sci. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet C.W., Srinivasan K., Clark T.A., O'Brien G., Cline M.S., Wang H., Williams A., Kulp D., Blume J.E., Haussler D., et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S.M. Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zheng H., Yang S., Yu H., Li J., Jiang H., Su J., Yang L., Zhang J., McDermott J., et al. Origin and evolution of new exons in rodents. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xinshu X., Nostrand E.V., Burge C. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl. Acad. Sci. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C. Alternative splicing and RNA selection pressure—Evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Xing Y., Wang Q., Lee C. Evolutionary divergence of exon flanks: A dissection of mutability and selection. Genetics. 2006;173:1787–1791. doi: 10.1534/genetics.106.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Burge C. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zheng C.L., Fu X.D., Gribskov M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA. 2005;11:1777–1787. doi: 10.1261/rna.2660805. [DOI] [PMC free article] [PubMed] [Google Scholar]