Abstract
Bcl-2 family proteins are essential regulators of cell death and exert their primary pro- or anti-apoptotic roles at the mitochondrial outer membrane. Previously, pro- and anti-apoptotic Bcl-2 proteins have been shown to interact with the voltage dependent anion channel (VDAC) of the outer mitochondrial membrane. VDAC is a 283-residue integral membrane protein that forms an aqueous pore in the outer mitochondrial membrane, through which metabolites and other small molecules pass between the cytosol and intermembrane space. The essential life-sustaining function of VDAC in metabolite trafficking is believed to be regulated by proteins of the Bcl-2 family. The protective role of anti-apoptotic Bcl-xL may be through its interaction with VDAC. Here, VDAC has been expressed, purified, and refolded into a functional form amenable to NMR studies. Various biophysical experiments indicate that micelle-bound VDAC is in intermediate exchange between monomer and trimer. Using NMR spectroscopy, gel filtration, and chemical cross-linking we obtained direct evidence for binding of Bcl-xL to VDAC in a detergent micelle system. The VDAC-interacting region of Bcl-xL was characterized by NMR with chemical shift perturbation and transferred cross saturation. The interaction region was mapped to a putative helical hairpin motif of Bcl-xL that was found to insert into detergent micelles. Our results suggest that Bcl-xL can bind to 1 or 2 VDAC molecules forming heterodimers and heterotrimers. Our characterization of the VDAC/Bcl-xL complex offer initial structural insight into the role of anti-apoptotic Bcl-xL in regulating apoptotic events in the mitochondrial outer membrane.
Keywords: VDAC, Bcl-xL, NMR, membrane protein, apoptosis
Apoptosis, or programmed cell death, is an essential cellular process strictly regulated by the Bcl-2 family of proteins. Three subfamilies of the Bcl-2 proteins are structurally homologous with different functions: the anti-apoptotic Bcl-2 proteins, e.g. Bcl-2, Bcl-xL, and Mcl-1, the multidomain pro-apoptotic proteins Bax and Bak, and the BH3-only pro-apoptotic proteins, Bid, Bad, Bim, etc. (1–7). BH3-only proteins are activated by various signals and transmit death signals by activating multidomain Bcl-2 proteins or occupying the binding pocket of anti-apoptotic Bcl-2 proteins, thus inhibiting their protective function (8, 9). Activation by pro-apoptotic BH3-only proteins initiates oligomerization of multidomain pro-apoptotic Bcl-2 proteins in the outer mitochondrial membrane (10, 11). Bax/Bak oligomerization leads to the disruption of membrane integrity and the release of certain apoptogenic factors, such as cytochrome c, which signal further events ultimately resulting in apoptotic cell death. Anti-apoptotic Bcl-xL is distributed between the cytoplasm and the outer mitochondrial membrane. In the cytoplasm anti-apoptotic proteins are believed to prevent cell death by binding and sequestering pro-apoptotic Bcl-2 proteins through their BH3 regions. High resolution structural studies of the Bcl-2 family proteins have only addressed their soluble, cytoplasmic forms (2–5). Therefore, crucial information about the behavior of Bcl-2 proteins in a membrane environment is currently unavailable. Bcl-2 proteins insert into mitochondrial membranes (12–14) and a complete understanding of Bcl-2 protein function in regulating apoptosis demands knowledge of their structure and interactions in a membrane setting. A particularly intriguing role for Bcl-xL is found in its interaction with the voltage dependent anion channel (VDAC1) (15). VDAC has been implicated in apoptosis through interactions with both pro- and anti-apoptotic members of the Bcl-2 family. The role of these interactions in apoptosis and normal cell function remains an issue of debate. An interaction of Bcl-xL with VDAC has been demonstrated biochemically (16) but its effect on the function of VDAC or role in regulating apoptosis is disputed. One report claims that Bcl-xL closes VDAC to prevent the formation of a permeability transition pore by maintaining mitochondrial membrane stability (16). A conflicting report suggests that Bcl-xL prevents cell death by maintaining VDAC in its active, open conformation to allow essential metabolite transport across the membrane (17).
VDAC is a 283-residue integral outer membrane protein of the mitochondrion involved in small molecule trafficking from the cytoplasm to the mitochondrion and forms the primary pathway for metabolite transport between the mitochondrion and cytoplasm (18, 19). It forms an aqueous pore, and is responsible for the translocation of ATP, ADP, and other metabolites across the outer mitochondrial membrane. VDAC is predicted to have a β-barrel structure, similar to bacterial outer membrane proteins. It remains debated whether VDAC is a functional monomer (20–22), dimer (23, 24), trimer (25) or in conformational equilibrium between different oligomeric states (26). Understanding the function of VDAC in metabolite transport and apoptosis on a structural level requires knowledge of its potential for oligomerization. VDAC has been proposed as a key component of the permeability transition pore through which apoptogenic factors, such as cytochrome c, pass during apoptosis. The predicted open pore β-barrel structure for VDAC has led to the idea that it might form a pathway for cytochrome c release. However, the estimated diameter of VDAC based on cryo-electron microscopy and electrophysiological studies is too narrow to allow passage of cytochrome c (27, 28). An oligomerization model of VDAC has been adopted, where multiple molecules of VDAC, possibly in complex with other proteins, would form a larger pore that could allow protein leakage across the outer mitochondrial membrane (29).
Our studies aim to clarify the biological role for the interaction between Bcl-xL and VDAC. Preparation of a functional form of the 32 kDa membrane protein VDAC is described. The oligomeric state of VDAC is addressed by chemical cross-linking and gel filtration. The binding interface between the 2 membrane proteins, Bcl-xL and VDAC, is described as determined by NMR spectroscopy. We use chemical shift perturbation and transferred cross saturation to characterize the VDAC-interacting region of Bcl-xL. The results of this study will lead to a better understanding of the role for the VDAC/Bcl-xL interaction in apoptosis and small molecule transport across the mitochondrion.
MATERIALS AND METHODS
VDAC Expression
The cDNA for human voltage-dependent anion channel 1 (VDAC1) with the corresponding GenBank accession #BC008482 was obtained as a full length clone from the Mammalian Gene Collection of OpenBiosystems (Huntsville, AL). A C-terminally 6x-His-tagged version of VDAC1 was cloned by standard PCR methods into the NdeI/XhoI sites of pET21a (Novagen). VDAC was expressed in BL21(DE3) cells by IPTG induction with 1 mM IPTG at 37°C for 3–5 hrs or with 200 µM IPTG at 20°C for 16–20 hrs.
Denaturing Purification of VDAC
VDAC was purified from inclusion bodies under denaturing conditions on a Ni-agarose resin (Qiagen) and eluted in Ni-elution buffer (7.5 M urea, 100 mM NaH2PO4, 10 mM Tris/HCl pH 8.0, and 250 mM imidazole). Cation exchange under denaturing conditions in 8 M urea was necessary as a next step in purification. Cation exchange chromatography required preparation of the denatured VDAC sample in a low salt-containing buffer prior to loading on the cation exchange column to remove any salt or contaminating materials which would inhibit binding of VDAC to the column. Purified VDAC in Ni-elution buffer was precipitated by dialysis against 4 L of 2 mM EDTA in a 12000 MWCO dialysis membrane. Precipitated VDAC was isolated by centrifugation at 18000 rpm in an SS-34 rotor. VDAC was resuspended in cation exchange start buffer (8 M urea, 25 mM sodium phosphate pH 6.5, 5 mM DTT) and sonicated to ensure efficient solubilization. VDAC in cation exchange start buffer was loaded onto a 5 mL HiTrap SP FF cation exchange column (GE Healthcare) at ~2 mL/min. VDAC was eluted from the cation exchange column in cation exchange start buffer + 100 mM NaCl. Further purification of the denatured VDAC sample was accomplished by gel filtration chromatography under denaturing conditions in denaturing gel filtration buffer (6 M guanidinium hydrochloride, 100 mM sodium phosphate pH 7.0, 100 mM NaCl, 1 mM EDTA, 5 mM DTT) at a flow rate of 0.5 mL/min.
VDAC Refolding
Purified VDAC in denaturing gel filtration buffer was refolded by dropwise dilution at 4°C with stirring into refold buffer (25mM sodium phosphate pH 7.0, 100 mM NaCl, 1 mM EDTA, 5 mM DTT, 1% lauryl dimethylamine oxide (LDAO)) to yield a final protein:detergent micelle ratio of ~1:20 (assuming an LDAO aggregation number of 76). After overnight stirring at 4°C the refolded VDAC sample was then dialyzed against phosphate buffer to remove guanidinium hydrochloride. Further purification on a Superdex-200 gel filtration column pre-equilibrated with LDAO GFC (gel filtration chromatography) buffer (25 mM sodium phosphate pH 7.0, 1 mM EDTA, 5 mM DTT, 0.1% LDAO) was employed to remove misfolded or aggregated VDAC and trace impurities. Analytical gel filtration on a Superose 12 10/300 GL (GE Healthcare) column was used to check the aggregation state of the purified sample. Pure VDAC samples were pooled and for NMR samples, concentrated to 0.7–1 mM. Detergent concentration is monitored by 1D proton NMR throughout the purification and a protein:micelle ratio is maintained at ~1:2–1:10. When necessary, excess detergent was removed from the sample by a series of dilution and concentration steps with a buffer lacking detergent.
Circular Dichroism Spectroscopy
Far-UV CD spectroscopy was performed on an AVIV spectrometer Model 62DS at 18°C in a 0.1 cm pathlength cell. Spectra were recorded from 260-195 nm with 1 nm increments and 8 scans. Background correction was performed by subtracting a spectrum from buffer and linear smoothing was applied to the final spectrum. Secondary structure estimation was made against a 37-protein reference set with the program SELCON3 (30).
Bcl-xL Preparation
Human Bcl-xL lacking a long unstructured loop (corresponding to wild-type residues 45–84) and a putative transmembrane region (residues 210–233) was employed for all experiments in this study (31). 6x-His tagged Bcl-xL was expressed in BL21(DE3) cells. Bcl-xL was purified by standard Ni-affinity chromatography followed by size exclusion chromatography with a Superdex75 (Amersham) column in 25 mM sodium phosphate pH 7.0, 1 mM EDTA, and 5 mM DTT. Bcl-xL was concentrated to ~2 mM then LDAO from a 1 M stock was added to bring the protein:micelle ratio to approximately 1:6. 15N-Ala and 15N-Asp samples of Bcl-xL were grown in the CT16 auxotrophic strain provided by David Waugh (National Cancer Institute). Bcl-xL samples for transferred cross saturation require extensive deuteration to avoid spin diffusion through amides. Extensively deuterated Bcl-xL samples were grown in 99.8% D2O, 2H6-glucose (98%), and 15N-ammonium chloride (99%).
NMR Spectroscopy
NMR experiments were performed at 30°C on Varian INOVA 500 and 600 MHz, and Bruker 500, 600, and 750 MHz spectrometers, equipped with cryogenically cooled probes.
Resonance Assignments of Bcl-xL in LDAO Micelles
NMR chemical shift assignment was accomplished using a standard set of triple resonance experiments: HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HNCO, and HN(CA)CO. A 3D 15N-edited NOESY-HSQC and (1H, 15N)-HSQC spectra of Bcl-xL with specific 15N-labeling at Ala, Asp, Ile, Leu, Val, Lys, Phe, Thr, and Tyr were also used for the assignment. The non-uniformly sampled double-TROSY hNcaNH experiment was also employed for sequential amide correlation (32). The backbone chemical shift assignment of Bcl-xL in DPC micelles was also used as a guide in the assignment (Philippe Hajduk and Stephen Fesik, personal communication).
NMR Titrations/Chemical Shift Perturbation
NMR titrations of VDAC with metabolites were performed by addition of concentrated ATP, NADH, and NAD to [2H,15N]-VDAC. Metabolite was added at VDAC:metabolite ratios of 1:0.5, 1:1, 1:2, 1:5, 1:10, 1:20 and 1:30. 1H,15N-TROSY-HSQC spectra were recorded at 30°C on a Varian INOVA 600 MHz spectrometer equipped with a cryoprobe and monitored for changes in chemical shift.
Chemical shift perturbation experiments were performed by 15N-labeling either VDAC or Bcl-xL and adding the unlabeled partner at 50%, 100%, and 200%. Monitoring chemical shift changes of VDAC in the presence of Bcl-xL was performed by recording TROSY-HSQC spectra of a 2H,15N-labeled VDAC sample. Protonated, 15N-labeled Bcl-xL was used to record changes in the fast HSQC spectrum after adding unlabeled VDAC.
Transferred Cross Saturation
Transferred cross-saturation experiments were performed with perdeuterated, 15N-labeled Bcl-xL in detergent micelles in a buffer containing 80% D2O. Unlabeled VDAC was added at 20%. The final NMR sample contained 800 µM Bcl-xL, 160 µM VDAC, and 395 mM LDAO in 25 mM sodium phosphate (pH 7.0) buffer, 5 mM DTT, 1 mM EDTA, and 80/20 D2O/H2O. Experiments were performed at 30°C on a Bruker 750 MHz spectrometer with a cryoprobe. Transferred cross-saturation experiments were performed by applying a WURST-2 adiabatic pulse prior to a fast-HSQC experiment. In order to effectively suppress artifact signals from both water and detergent, an amide-selective REBURP is applied during the reverse INEPT period of the HSQC. Irradiation frequency of VDAC was chosen based on the 13C-HSQC of a methyl-ILV-labeled sample of VDAC. A 13C-HSQC of VDAC perdeuterated and selectively 13C,1H-labeled at ILV-methyl groups showed a well separated methyl peak at 0.34 ppm (251 Hz). The closest detergent signal is at 0.87 ppm (651 Hz). Therefore, on-resonance saturation with a 2.5 ms WURST adiabatic pulse having a bandwidth of 1000 Hz was performed at 0 Hz in order to irradiate this methyl group as well as nearby VDAC proton signals, sufficiently far from LDAO resonances so as not to inadvertently saturate the detergent. This narrow bandwidth avoided saturation of nearby detergent resonances as determined by 1D experiments. Experiments were performed with 1, 2, and 3 sec saturation durations.
Size Exclusion Chromatography
Size exclusion chromatography was carried out on a Superdex 200 16/60 column (GE Healthcare) in LDAO GFC buffer at 4°C with a flow rate of 1ml/min, while monitoring the absorbance at 280 nm. Analysis of Bcl-xL and VDAC binding by size exclusion chromatography was performed after mixing equimolar amounts of VDAC and Bcl-xL to give 10.5 µM in both and incubation for 1 hour. Analysis of VDAC cross-linking by size exclusion was performed after quenching the reaction with 1M Tris/HCl pH 8.0.
Chemical Cross-linking
Chemical cross-linking reactions were performed at room temperature with ethylene glycol bis[succinimidylsuccinate] EGS and glutaraldehyde. EGS was from Pierce Biotechnology (Rockford, Ill.) and 70% glutaraldehyde was from Sigma (St. Louis, MO). Cross-linking reactions of VDAC alone were performed with 1.1–1.2 mM VDAC and 1–1.5 mM glutaraldehyde or EGS. Cross-linking of VDAC to Bcl-xL was performed at 9 µM for both proteins and 18 or 180 µM glutaraldehyde. Reactions were quenched after 2 hrs either by adding 1 M Tris/HCl pH 8.0 before gel filtration or SDS-PAGE sample loading buffer prior to analysis by SDS-PAGE.
Western Blotting
Immunoblotting was performed with mouse monoclonal anti-VDAC antibody (Anti-Porin 31HL (Ab-1) (89–173/016) from Calbiochem, San Diego, CA) raised against the N-terminal region of human VDAC1 and mouse monoclonal anti- Bcl-xL antibody (2H12: sc23958 from Santa Cruz Biotechnology, Santa Cruz, CA). The goat anti-mouse IgG-HRP (sc-2005 from Santa Cruz Biotechnology, Santa Cruz, CA) was used as the secondary antibody. The HRP-conjugated secondary antibody was detected with the SuperSignal West Pico Chemiluminescene Kit (Pierce Biotechnology, Rockford, IL). His-tag detection by Western blotting was performed with an HRP-conjugated primary antibody recognizing the 6x His-tag (Roche, Indianapolis, IN).
RESULTS
VDAC Expression, Purification, and Refolding
Recombinantly expressed VDAC was directed to the insoluble fraction at up to 30 mg per liter culture. Optimal refolding efficiency of VDAC necessitated Ni-affinity, cation exchange, and gel filtration chromatography under denaturing conditions prior to refolding. SDS-PAGE analysis after Ni-affinity purification revealed an impurity at 15% abundance. Immunodetection with an anti-His antibody showed that the impurity contained a His-tag and was likely to be a truncation product of VDAC. Subsequent N-terminal sequencing confirmed the impurity as a truncation product of VDAC beginning at L144. Cation exchange under denaturing conditions was employed to remove the majority of the truncation product accomplished by thorough washing with cation exchange start buffer. VDAC was eluted from the cation exchange column in cation exchange start buffer + 100 mM NaCl. Cation exchange was followed by gel filtration under denaturing conditions. This step removed higher and lower molecular weight impurities and exchanged the buffer from urea to GdnHCl, which was necessary for refolding in LDAO; it was observed that refolding efficiency is higher from 6 M GdnHCl than from 8 M urea. Of the various detergents tested (LDAO, DPC, DHPC, β-OG, LM, C8E4, SDS), only LDAO resulted in a stable, folded form of VDAC suitable for NMR studies.
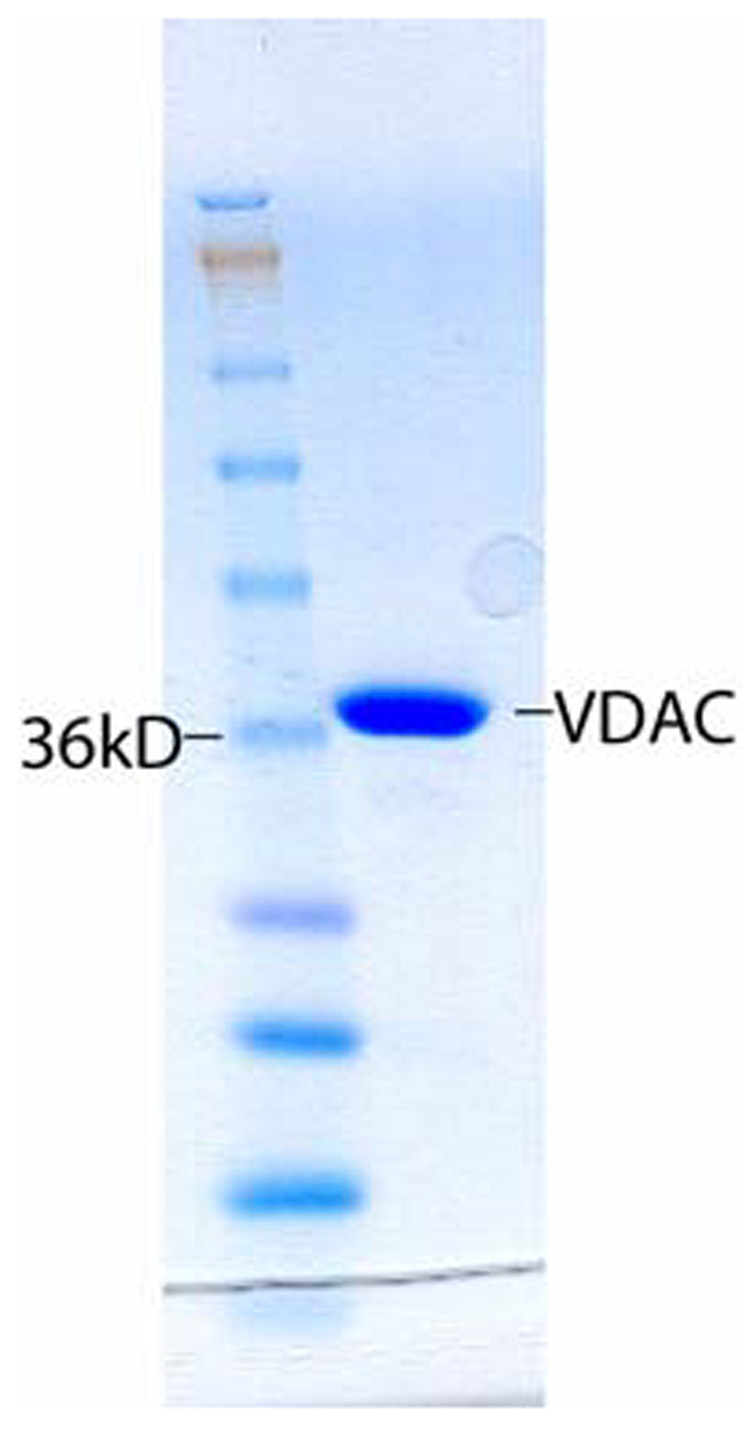
Purified VDAC in denaturing gel filtration buffer is refolded by dropwise dilution at 4°C with stirring into LDAO refold buffer (25 mM sodium phosphate pH 7.0, 100 mM NaCl, 1 mM EDTA, 5 mM DTT, 1% lauryl dimethylamine oxide (LDAO)) to yield a final protein:detergent micelle ratio of ~1:20 (assuming an LDAO aggregation number of 76). After refolding of VDAC into LDAO, preparative gel filtration showed a major species corresponding to pure, refolded VDAC and a minor species eluting in the void volume corresponding to aggregated or misfolded VDAC and other impurities. The pure VDAC fractions were pooled and concentrated when necessary for NMR samples. A SDS-PAGE gel of the final purified VDAC sample shows a major band close to the expected molecular weight of 32 kDa (Figure 1). NMR samples contained 0.7–1 mM VDAC and 200–700 mM LDAO.
FIGURE 1.
SDS-PAGE gel of purified VDAC. The calculated molecular weight is 32 kDa.
Circular Dichroism Spectroscopy
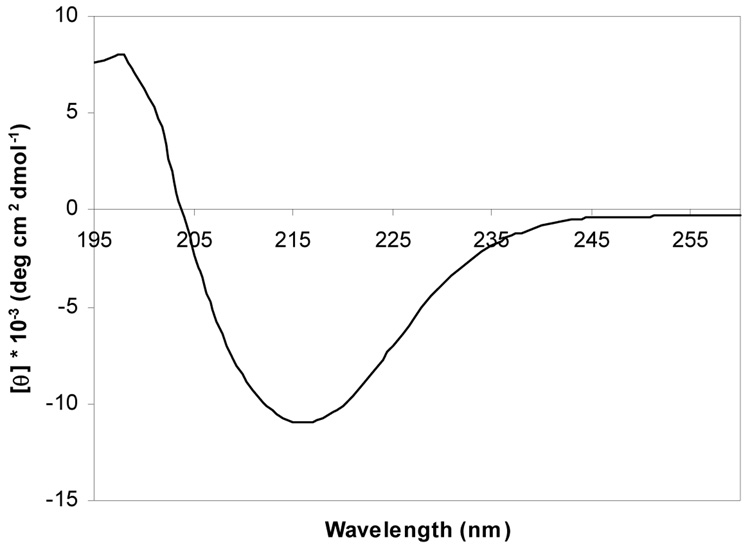
The CD spectrum of recombinantly expressed and refolded VDAC exhibits similar features as previous studies of mitochondria-purified and refolded VDAC from various sources: a minimum at 216 nm and a crossover to positive ellipticity at 206 nm (33–35) (Figure 2). Estimation of secondary structure gives 39.7% β-sheet, 17.5% α-helix, 13.3% β-turn, and 28.4% random coil. We may, therefore, conclude that refolded human VDAC adopts a similar folded conformation with high β-sheet content similar to previous studies of mitochondrial and refolded VDAC.
FIGURE 2.
Far-UV CD spectrum of refolded human VDAC in 0.1% LDAO at pH 7.
Titration of VDAC with Metabolites
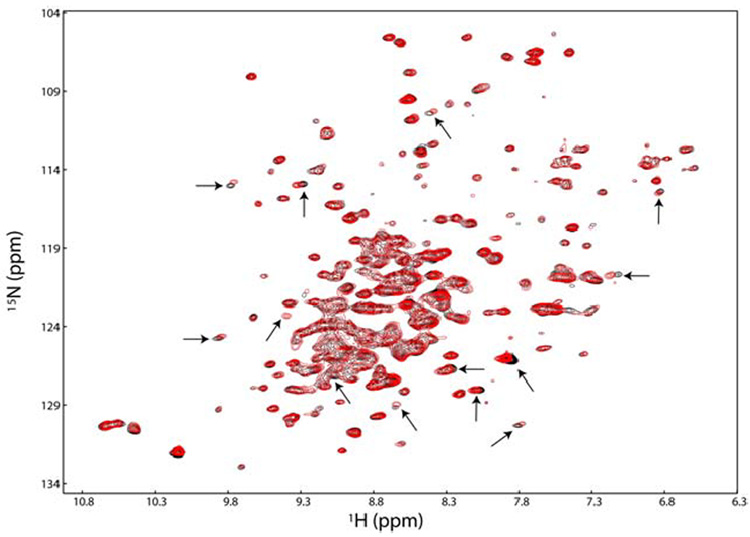
The TROSY-HSQC spectrum of VDAC shows dispersion indicative of a folded protein of high β-sheet content (Figure 3). Chemical shift perturbation experiments by titration of [2H,15N]-VDAC with the physiological substrates β-NADH, β-NAD, and ATP indicate an interaction between VDAC and the metabolites. Figure 3 shows representative spectra from the titrations. A large excess (≥10-fold) of metabolite was required to observe a noticeable change in the VDAC spectra. Significantly, a response from VDAC in the presence of these physiological substrates shows that the recombinantly expressed and reconstituted membrane protein is functional. Comparison of the three spectra reveals that NAD and NADH (not shown) induce shifts at positions in common with ATP and indicate a common interaction site.
FIGURE 3.
TROSY-HSQC spectra of 500 µM 2H,15N-VDAC in the presence (red) and absence (black) of 30-fold excess ATP. Peaks that undergo obvious changes are indicated with an arrow.
Additionally, the peaks that change after substrate/inhibitor binding are among the many, which diminish in intensity during D2O exchange experiments (data not shown), suggesting that the interaction between VDAC and these small molecules occurs at a solvent exposed region. Some of the same peaks are shifted for the three different metabolites. Shifts at the same peak positions show that the binding region on VDAC for the different metabolites is similar. The low number of affected peaks reflects the small size of the ligand compared to VDAC and indicates that metabolite binding does not cause a significant conformational change. The requirement of a large excess of metabolite reflects the low binding affinity of VDAC for these molecules (36–38). A low binding affinity is consistent with the reported function of VDAC as a transporter of metabolites: efficient transport is expected to require a low binding affinity of the transporter for the ligand. VDAC is a transporter of small metabolites, so it is impractical and unlikely that tight binding would occur. VDAC may act as a sensor to metabolite substrates and the peaks which are perturbed may be the signature of this sensor region.
Chemical Shift Perturbation of VDAC and Bcl-xL
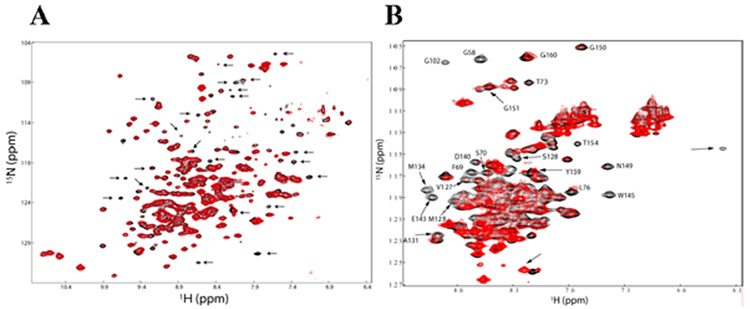
To investigate the reported interaction between Bcl-xL and VDAC, the effects of Bcl-xL on the TROSY-HSQC spectra of VDAC were analyzed. Addition of Bcl-xL to 2H,15N-VDAC caused chemical shift changes and differential line broadening for a subset of peaks (Figure 4A), thus, confirming a specific interaction between the two membrane proteins. Observation of chemical shift changes for VDAC upon addition of Bcl-xL is further evidence for a functional form of VDAC and demonstrates that a significant direct interaction between VDAC and Bcl-xL takes place.
FIGURE 4.
(A) Chemical shift perturbation of 2H, 15N-VDAC in the presence of unlabeled Bcl-xL. The TROSY-HSQC spectrum of VDAC in the absence (black) and presence of 50% Bcl-xL (red). Arrows indicate some of the major changes in the VDAC spectrum. (B) HSQC spectra of Bcl-xL in LDAO micelles in the absence of VDAC (black) and in the presence of VDAC at 1:1 ratio (red). Sample conditions: 300 µM 15N-Bcl-xL, 300 µM VDAC, 64 mM LDAO, 6% D2O. The assignment of some of the shifted peaks is indicated.
To further probe the interaction between VDAC and Bcl-xL, the inverse was performed by monitoring changes in the HSQC spectrum of 15N-labeled Bcl-xL in the presence of unlabeled VDAC. HSQC spectra of 15N-Bcl-xL/VDAC1 at ratios of 1:0.5, 1:1 (Figure 4B), and 1:2 were recorded. VDAC induces dramatic changes (chemical shift changes and/or differential line broadening) for a subset of Bcl-xL peaks in the HSQC spectra. Approximately 25% of Bcl-xL peaks are affected by the presence of VDAC, while 75% remain unchanged. In order to map the interaction interface, residues of Bcl-xL corresponding to peaks shifted by VDAC were identified by employing a standard NMR assignment strategy.
Assignment and Secondary Structure of Bcl-xL in LDAO Micelles
Because the dispersion of NMR resonances for Bcl-xL in LDAO is very poor, it was necessary to utilize an extensive set of 15N-specifically labeled samples for assignment by residue type. The recently described double-TROSY-hNcaNH experiment, which correlates sequentially connected backbone amides, also helped to establish new connectivities and verify previous assignment obtained through other NMR experiments (32). The backbone assignment of Bcl-xL in DPC micelles served as a guide for assignment in LDAO micelles since many of the carbon chemical shifts were similar for Bcl-xL in the two detergents. Partial backbone assignment of Bcl-xL was achieved to only 59%, due in large part to the crowding and overlapping of resonances.
Only limited structural information has been obtainable for Bcl-xL in LDAO micelles. Comparison of Bcl-xL spectra obtained in the absence (data not shown) and presence of detergent micelles indicates that a dramatic structural change occurs. The overall appearance of the 1H,15N-HSQC spectrum of Bcl-xL in LDAO and carbon chemical shifts are very similar to Bcl-xL in DPC ((39) and personal communication). Based on the similarity between the chemical shifts for Bcl-xL in LDAO to those in DPC, it is reasonable that Bcl-xL would adopt a similar structure in both micelle-mimicking environments. The solution state NMR characterization of Bcl-xL in DPC micelles (39), a solid state study in lipid bilayers (40), and biochemical approaches (41, 42) propose a model for Bcl-xL in which 2 central helices insert into membranes via a helical hairpin, while the rest of the protein becomes loosely ordered compared to the globular structure in aqueous buffer (2) but retains alpha-helical secondary structural elements. The secondary structure of Bcl-xL in LDAO micelles was calculated for assigned residues with the program TALOS (43) and helices 5, 6, and 7 were predicted to have been retained for Bcl-xL in LDAO, consistent with the helical hairpin model of Bcl-xL in membranes.
NMR Characterization of the VDAC-interacting Region of Bcl-xL
We could identify changes in approximately 48 peaks in the Bcl-xL 1H,15N-HSQC spectrum. Of these, over 80% (38/48) could be assigned, despite the overall low level of assignment for Bcl-xL in LDAO (60%). Numerous changes occur throughout the Bcl-xL spectrum but the majority of changes are located within and directly adjacent to the putative membrane anchor formed by helices 5 and 6 (Figure 4B). The majority of Bcl-xL residues affected by VDAC are clustered in one region but spectral changes also occur in regions distant in the primary sequence from the helical hairpin (Figure 5).
FIGURE 5.
Primary sequence of Bcl-xL. The regions predicted by TALOS to be helical are highlighted gray. Secondary structure calculation by TALOS was only possible for assigned residues. Helices reported for Bcl-xL in DPC micelles are indicated with a black bar (α1-α7). The Bcl-2 homology regions are indicated (BH1-BH4). Residues in red indicate a significant effect in chemical shift perturbation experiments, those in green experienced a significant saturation effect, and those in blue experience a significant effect in both experiments.
Spectral changes from chemical shift perturbation studies can arise from either direct interaction or from allosteric effects due to structural changes transmitted through distant interaction sites. We wanted to address the possibility of ambiguity from chemical shift perturbation by obtaining direct structural information about the binding region of Bcl-xL on VDAC. For strongly interacting, tight protein complexes conventional NOESY spectroscopy can be employed to establish the binding interface. However, weak interactions exhibiting fast to intermediate exchange behavior on the NMR timescale are not tractable to conventional NOESY because of weak NOEs for the fast exchange case and unobservable signals in the intermediate exchange regime. Most of the spectral changes of Bcl-xL induced by VDAC at stoichiometric concentration are extensive peak broadening, suggesting binding kinetics in the intermediate exchange regime, where conventional NOEs are not observable. Therefore, the recently introduced method of transferred cross saturation is well-suited to investigate this system (44–46).
Saturation transfer effects are readily observed if the concentration of the binding partner is kept sufficiently low to effectively create a fast exchange situation so that peaks are not broadened and peak positions are not shifted relative to the reference spectrum. Saturation of the protonated binding partner is transferred to the perdeuterated, 15N-labeled protein. Spin diffusion transfers the saturation throughout the irradiated, protonated protein, while at the same time transferring saturation to the 15N-labeled protein also by spin diffusion. The saturation is transferred through the interface of the complex and decreases the population difference predicted by the Boltzmann distribution for spins of the labeled binding partner. Spin diffusion in the 15N-labeled protein is limited by preparing an extensively deuterated sample. The observed effect is that amide positions that are in proximity to the protonated protein will have a reduced intensity in HSQC spectra recorded after irradiation. Peak intensities in the spectrum acquired with saturation are compared with the intensities of a non-irradiated reference spectrum. Reduction in intensity indicates that the corresponding residue is in direct contact with the binding partner (47).
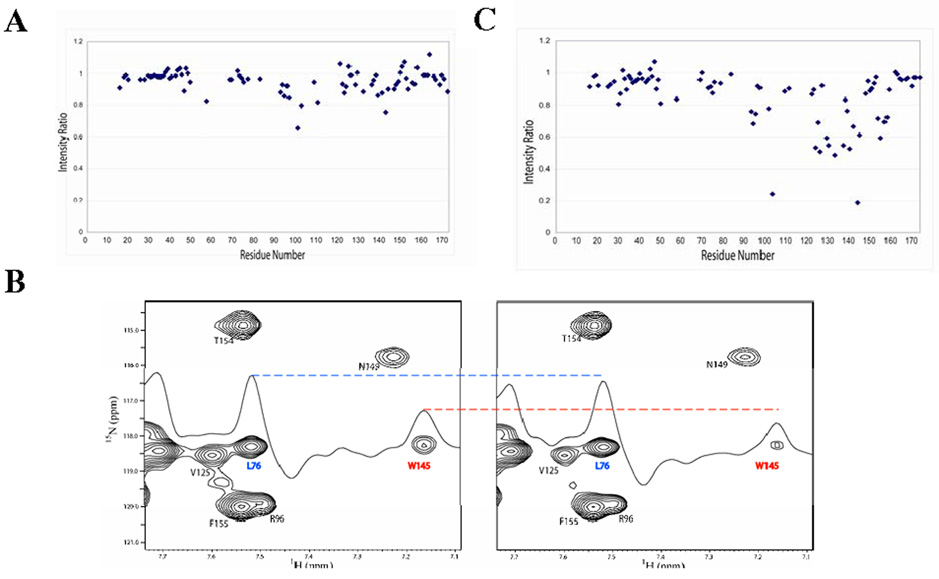
With transferred cross saturation we were successful in identifying Bcl-xL residues involved in binding to VDAC. In the cross saturation experiments a subset of Bcl-xL peaks experienced an intensity reduction of 5% or greater and were thus considered significant in forming the interaction interface with VDAC (Figure 6A, Table 1). The most pronounced saturation effects were observed for residues from L94 to V156, a region encompassing the helical hairpin. Transferred cross saturation confirmed the helical hairpin motif as forming the binding interface of the VDAC/ Bcl-xL complex. The results of the chemical shift perturbation studies and transferred cross saturation are summarized in Figure 5. Similar to chemical shift perturbation, cross saturation effects were observed for residues distant in primary sequence from the helical hairpin. Chemical shift perturbation was observed for residues S18, E46, S47, and A49, and cross saturation effects were felt at residues F16 and S47. Thus, cross saturation experiments confirmed these regions (F16-S18 and E46-A49), which are distant in primary sequence from the helical hairpin, in contributing to Bcl-xL -binding with VDAC. The utility of transferred cross saturation in verifying residues involved in direct interaction is further demonstrated by comparison with the results of the chemical shift perturbation experiment for Trp145 and Leu76. Chemical shift perturbation of Bcl-xL in the presence of VDAC shows a significant change in peak position for Leu76 and differential line broadening for Trp145 (Figure 4B). Trp145 is part of the putative helical hairpin region in helix 6, whereas Leu76 is located distant from the helical hairpin region, C-terminal to helix 3. The transferred cross saturation experiment shows a significant effect for W145, but a much smaller saturation effect for L76 (Figure 6B). Therefore, the method of transferred cross saturation was effective in differentiating between residues that are more directly involved in the formation of the VDAC/ Bcl-xL complex.
FIGURE 6.
(A) Plot of the intensity ratios with and without saturation of VDAC mapped onto the primary sequence of Bcl-xL. Residues with missing data points have not been assigned. (B) Portion of HSQC spectrum of [2H,15N]-Bcl-xL without (left) and with (right) 2sec saturation of VDAC. A 1D slice is shown which intersects with peaks from L76 and W145, showing the effective saturation of W145 and significantly less saturation for L76. (C) Plot of intensity ratios with and without saturation of LDAO at 0.87 ppm mapped onto the primary sequence of Bcl-xL.
Table 1.
Intensity Ratio and Intensity Reduction from Transferred Cross Saturation of VDAC to Indicated Residues of Bcl-xL
| Residue | Intensity Ratio | % Intensity Reduction |
|---|---|---|
| 16 | 0.91 | 7.26 |
| 47 | 0.89 | 9.34 |
| 58 | 0.82 | 16.32 |
| 94 | 0.88 | 10.13 |
| 95 | 0.93 | 5.14 |
| 96 | 0.86 | 12.52 |
| 97 | 0.92 | 6.02 |
| 98 | 0.85 | 13.98 |
| 102 | 0.66 | 33.29 |
| 104 | 0.80 | 19.22 |
| 112 | 0.81 | 17.20 |
| 125 | 0.88 | 10.38 |
| 126 | 0.92 | 6.38 |
| 130 | 0.93 | 5.25 |
| 134 | 0.89 | 9.72 |
| 138 | 0.93 | 5.50 |
| 141 | 0.86 | 12.27 |
| 143 | 0.88 | 10.51 |
| 145 | 0.76 | 23.06 |
| 146 | 0.90 | 8.21 |
| 149 | 0.90 | 8.14 |
| 150 | 0.93 | 5.25 |
| 156 | 0.90 | 8.35 |
| 171 | 0.93 | 5.49 |
| 175 | 0.89 | 9.94 |
Not all of the residues identified as undergoing significant chemical shift changes experienced a significant cross saturation effect (Figure 5). This might be an indication of allosteric effects or may be a result of the inherent low sensitivity of the transferred cross saturation experiment compared to chemical shift changes. The combined results of the experiments, however, clearly show that the region of Bcl-xL primarily involved in the formation of the binding surface with VDAC is located within residues 94–156. This region encompasses the putative helical hairpin motif responsible for membrane anchoring (Figure 5). Saturation transfer was also performed by irradiating a detergent resonance to confirm that this region is, in fact, involved in membrane targeting.
The effect of saturation of the detergent was determined for Bcl-xL in the absence of VDAC to establish the region involved in micelle insertion. On-resonance saturation of the aliphatic methyl protons of LDAO at 0.87 ppm was performed in the absence of VDAC. Significant saturation effects are felt by the N-terminal part of helix 5, most of helix 6, and part of helix 7, thus confirming these regions as targeting Bcl-xL to membranes (Figure 6C). Importantly, the same regions involved in targeting of Bcl-xL to the micelle also contain the residues involved in interaction with VDAC.
Size Exclusion Chromatography of the VDAC/Bcl-xL Complex
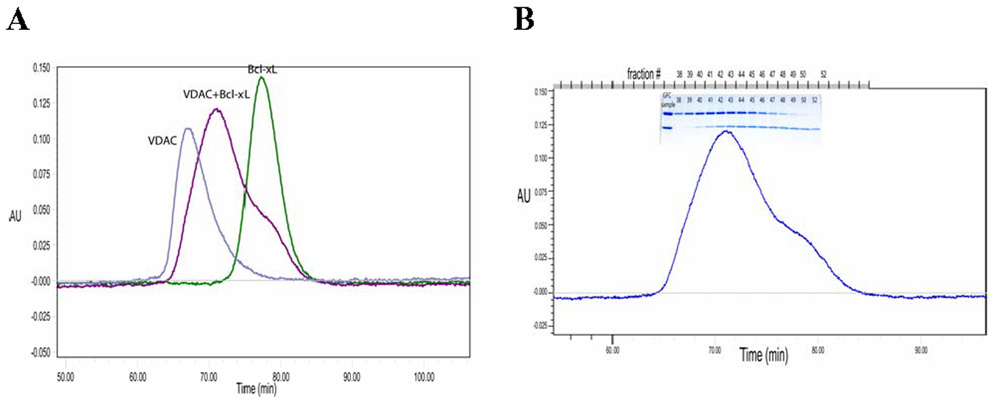
Gel filtration was used to determine whether Bcl-xL and VDAC eluted together as a complex or separately when mixed at equal ratios. The retention profile of the mixture was compared with that of the individual proteins with the same loading for each component. When the sample of Bcl-xL+VDAC, mixed at an equimolar ratio, was analyzed by size exclusion chromatography the major peak eluted at a time intermediate between the retention times of VDAC and Bcl-xL alone (Figure 7A).
FIGURE 7.
(A) Size exclusion chromatography demonstrates the formation of a complex between VDAC and Bcl-xL. The samples were VDAC alone (blue trace), Bcl-xL (green) and VDAC+ Bcl-xL (purple). (B) SDS-PAGE shows the altered elution profiles from size exclusion for VDAC and Bcl-xL when in complex compared to the isolated proteins. SDS-PAGE samples were taken for the gel filtration fractions as indicated above the lanes in the gel.
SDS-PAGE analysis of the gel filtration fractions shows that the maximum amount of Bcl-xL elutes earlier when in the presence of VDAC than for Bcl-xL alone (Figure 7B), and the maximum of VDAC in the complex elutes later than for VDAC alone. Co-elution of VDAC and Bcl-xL demonstrates the formation of a complex and supports a model of both proteins occupying the same micelle. Later elution of the complex than VDAC alone suggests that the VDAC/Bcl-xL complex is, in fact, smaller than VDAC alone. When fractions 41–44 of the complex were pooled and re-injected on the gel filtration column, the same elution profile is observed as for the original VDAC/Bcl-xL mixture in Figure 7B (not shown), suggesting that the profile is that of the stable VDAC/Bcl-xL complex.
Chemical Cross-linking of VDAC and Bcl-xL
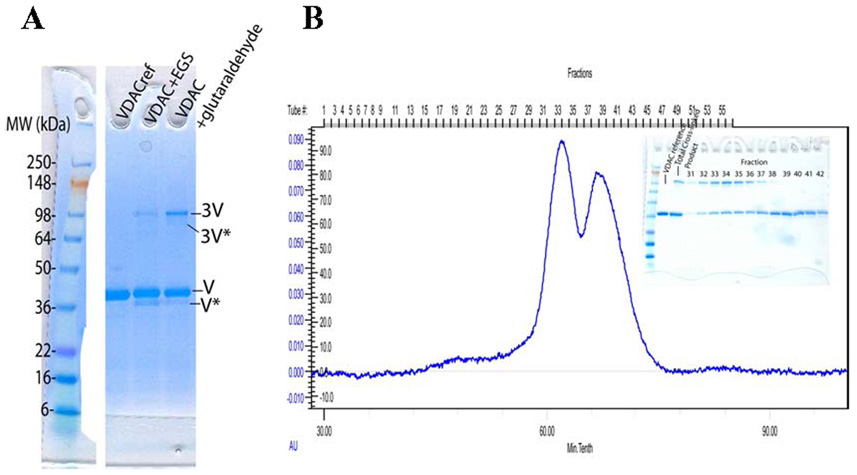
Chemical cross-linking was performed in order to investigate the reported self-association properties of VDAC. Cross-linking of VDAC was tested with 2 different reagents, EGS and glutaraldehyde. Test reactions were performed with 1.2 mM VDAC and 1 mM cross-linking reagent for 2 hr at room temperature. SDS-PAGE analysis reveals the predominant forms of cross-linked product to be a trimer (3V), an intramolecularly cross-linked monomer (V*), and an intramolecularly cross-linked trimer (3V*) (Figure 8A). In contrast to other cross-linking studies reporting self-association of VDAC as dimers, trimers, and tetramers (26) our study shows almost exclusive trimer formation.
FIGURE 8.
(A) SDS-PAGE gel of VDAC cross-linking reactions with EGS and glutaraldehyde. Left: molecular weight standard. Lane 2: VDAC without cross-linker, lane 3: VDAC + EGS, lane 4: VDAC + glutaraldehyde. (B) Cross-linked VDAC was purified by gel filtration chromatography and fractions were analyzed by SDS-PAGE (inset).
To address whether LDAO-purified VDAC was a tightly-associated trimer or in equilibrium between trimer and other forms, size exclusion chromatography was performed for a cross-linking reaction. The cross-linked product elutes much earlier than unreacted VDAC (62.1 min vs 67.0 min), indicating significantly different hydrodynamic properties: a difference in total apparent molecular weight and/or hydrodynamic radius (Figure 8B). The cross-linked trimer might behave as a larger species than a completely compact VDAC trimer if the affinity of monomer to form trimer is too low to stabilize a compact trimer on the timescale of the gel filtration run. This would cause the cross-linked trimer to be more extended than a compact VDAC trimer, resulting in earlier elution of the cross-linked species. This provides evidence that LDAO-purified VDAC is not a tightly-associated trimer but most likely in equilibrium between trimer and monomer.
Further support for exchange behavior between monomer and higher oligomer was provided by analytical gel filtration chromatography. A concentration-dependent elution profile was observed with increasing elution time for decreasing VDAC loading concentrations (data not shown).
Even further evidence for a concentration-dependent oligomerization was seen in TROSY-HSQC spectra at very low VDAC concentration. At low VDAC concentration a significant signal increase was observed for a subset of peaks, indicating an equilibrium between different multimeric forms (data not shown).
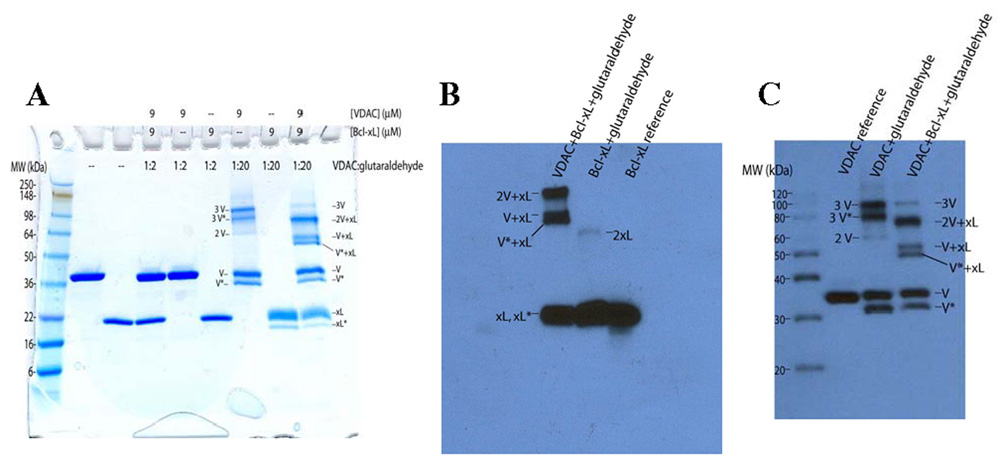
It was postulated that if VDAC was in exchange between monomer and trimer (or other oligomeric forms) that the interaction partner Bcl-xL might exert a negative effect on this self-association property of VDAC. Therefore, the effect of Bcl-xL on the self-association of VDAC was investigated by chemical cross-linking. A clear difference was observed in the cross-linking pattern when VDAC and Bcl-xL at equimolar ratios underwent cross-linking with both EGS (not shown) and glutaraldehyde (Figure 8A) compared with VDAC cross-linking alone. At a VDAC:glutaraldehyde ratio of 1:20, a difference is observed for VDAC in the presence of Bcl-xL. There is a dramatic decrease in the amount of VDAC trimer formed and new bands appear which do not show up in cross-linking reactions of VDAC and Bcl-xL alone (Figure 9A). In control reactions, no cross-linked product is formed for Bcl-xL alone and VDAC forms mostly trimers when cross-linked alone. A very intense band appears for the cross-linking of VDAC performed in the presence of Bcl-xL at ~90 kDa. There are also bands that appear at ~60 kDa and 55 kDa. The identities of the bands were determined by Western blotting of the reactions with antibodies against Bcl-xL and VDAC (Figure 9B, Figure 9C). The bands were identified as follows: 2VDAC+Bcl-xL (90 kDa), 1VDAC+Bcl-xL (60 kDa), and 1VDAC*+Bcl-xL* (55 kDa). (The lower shifted bands are interpreted as intramolecularly cross-linked species, VDAC* and Bcl-xL*.) The calculated molecular weights are consistent with 2:1 and 1:1 VDAC/Bcl-xL heterocomplexes: 2VDAC+1Bcl-xL (84.5 kDa), 1VDAC+1Bcl-xL (52.6 kDa). Thus, Bcl-xL caused a significant decrease in the amount of VDAC trimer formed and favored the formation of VDAC-VDAC-Bcl-xL and VDAC-Bcl-xL cross-linked species.
FIGURE 9.
(A) Glutaraldehyde cross-linking in the presence and absence of Bcl-xL. Cross-linking was performed with 2-fold (lanes 4–6) and 20-fold (7–9) excess of glutaraldehyde over VDAC. Lane 1 is the protein marker, lane 2 is uncross-linked VDAC, lane 3 is uncross-linked Bcl-xL. Immunostaining of Bcl-xL (B), and VDAC (C) verifies the identities of the cross-linked products as indicated here and described in the text.
DISCUSSION
The large 32 kDa human mitochondrial membrane protein VDAC has been prepared in a form suitable for NMR structural studies. Titration with metabolites provided evidence of an interaction with VDAC and supports the preparation of a functional form of VDAC. The similarity of the CD spectrum of refolded human VDAC to previous studies of mitochondrially-isolated VDAC lends further support that our preparation produced a functional form of VDAC.
An interaction between the anti-apoptotic protein Bcl-xL and VDAC was demonstrated by chemical shift changes in 1H,15N correlation experiments for both proteins upon addition of the binding partner. Specific changes observed for a small subset of peaks in the HSQC spectra of VDAC and Bcl-xL induced by the unlabeled membrane protein interaction partner is a strong indication of a specific complex between VDAC and Bcl-xL formed through a distinct binding interface. This is evidence for a significant interaction between Bcl-xL and VDAC in the membrane-mimicking environment of LDAO detergent micelles. The region of Bcl-xL involved in forming the interaction interface with VDAC was determined by chemical shift perturbation and confirmed by transferred cross saturation. Transferred cross saturation was complimentary to chemical shift perturbation in defining the interaction interface between Bcl-xL and VDAC.
The binding region involves mostly the C-terminal region of helix 4 through the putative helical hairpin motif of helices 5 and 6, and into helix 7 (Figure 10). Shimizu, et al. showed that the BH4 region of Bcl-xL, at the N-terminus encompassing amino acids 4–23, is necessary for its VDAC-inhibiting activity, but dispensable for direct binding to VDAC (48). However, if the BH4 can inhibit its activity it is probably involved in direct contact with VDAC. It may be that the BH4 does not contribute enough binding energy to be noticeable under the conditions of their pulldown assay. Fesik and co-workers showed that the BH4 region of Bcl-xL has reduced mobility, like the helical hairpin, and speculate that this region binds to the surface of the micelle (39). Our results are generally in agreement with these studies. Small effects are observed for residues in the BH4 region in the presence of VDAC. We observe a chemical shift change and a saturation effect in the BH4 region for residues S18 and F16, respectively. This implicates the BH4 in contributing partially to the interaction with VDAC. We do not observe strong saturation transfer effects on the regions in the BH4 upon irradiation of the detergent. This is consistent with the idea that the BH4 would interact only peripherally with the micelle as proposed by Fesik and co-workers. Our data could fit a model where the BH4 would bind on the surface of micelles and at the same time be involved in a weak interaction with VDAC. The BH4 region might reach over the top of VDAC without forming the major interface within the membrane. Our studies identified Bcl-xL residues 94–156 as forming the primary VDAC-interacting interface. We have shown that the putative helical hairpin motif of Bcl-xL comprised of helices 5 and 6 is responsible for membrane targeting and for complex formation with VDAC, while the BH4 region might also contribute to binding. We are also investigating the role of the putative transmembrane region at the C-terminus of Bcl-xL in membrane targeting and VDAC interaction.
FIGURE 10.
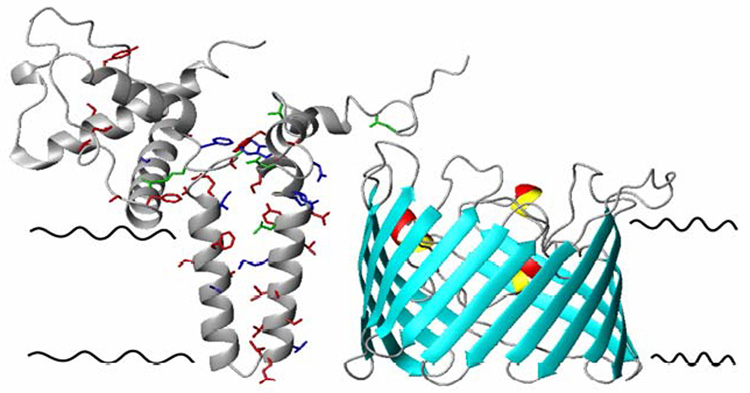
Left: Model of membrane-bound Bcl-xL illustrating the insertion of helices 5 and 6 into the membrane. Side chains of residues that undergo a significant change in chemical shift perturbation studies are red, transferred cross saturation are green, those which show a significant effect in both experiments are in blue.
Right: Homology model of VDAC1 based on alignment with 1PRN, generated with SWISSMODEL. This model illustrates 15–16 strands, as predicted for VDAC1.
We can rationalize a model of the interaction complex with the two membrane proteins oriented parallel to one another in the plane of the membrane. The helical hairpin would be oriented parallel to the barrel axis of VDAC, and the interaction interface between the two proteins would involve the entire length of the helical hairpin of Bcl-xL and a yet to be determined face of the β-barrel. A structural model of Bcl-xL in membranes was produced by incorporating previously published structural data (39–41) and our results from transferred cross saturation of detergent. The model of Bcl-xL presented (Figure 10) shows helices 5 and 6 anchored in the membrane and highlights the residues involved in binding to VDAC. A model of VDAC based on homology with the porin from Rhodopseudomonas blastica (PDB code: 1PRN) was created with SWISSMODEL and is presented in Figure 10. The interaction between the two membrane proteins is implied in this figure, and a complete structural characterization is still in progress.
Size exclusion chromatography and SDS-PAGE analysis of the fractions show that the maximum amount of Bcl-xL elutes earlier when in complex with VDAC than for Bcl-xL alone and is reliable evidence for an interaction. The maximum amount of VDAC in the complex elutes later than for VDAC alone and suggested that VDAC alone was larger than the VDAC/Bcl-xL complex. Further investigation by chemical cross-linking was performed and supported this hypothesis.
Chemical cross-linking studies with both EGS and glutaraldehyde reveal that VDAC forms multimers. The major cross-linked VDAC product is consistent with a trimer. Observation of VDAC cross-linking to itself is not necessarily proof that the native form of VDAC self-associates, because cross-linking studies are notorious for producing false positives, since the reagent may react with any available free amino group. However, the preferential formation of trimer over dimer is an indication that a specific interaction is taking place. Formation of almost exclusively trimers strongly suggests that VDAC is either a stable trimer or in equilibrium between monomer and trimer. This conclusion is supported by the work of Zalk, et al. where they show that VDAC in mitochondria and purified in Triton X-100 detergent has a tendency to form dimers, trimers, and tetramers (26). Trimeric VDAC has also been suggested from cryo-EM images of VDAC and from electrophysiological measurements showing multiple, rather than single, conductance states (25). VDAC also shares low but significant homology (~20% identity, and 40% similarity) with the bacterial membrane protein from Rhodopseudomonas blastica that forms a native trimer. Although the homology is not very high it is enough to support the hypothesis that VDAC may at least form transient trimers or oligomers possibly in a manner reminiscent of bacterial β-barrel membrane proteins. If a VDAC monomer is predicted to fold similarly as the trimeric bacterial porin, it is also reasonable to expect that trimerization through a homologous region occurs. Although from cross-linking we have evidence that LDAO-purified VDAC preferentially forms trimers, we acknowledge that VDAC could exist in other multimeric forms. Oligomerization state can be detergent-dependent (49), and since it has been reported that VDAC can adopt a dimeric form (23), or exist in a dynamic equilibrium between different forms (26) in Triton X-100, we accept that VDAC has the potential to take forms other than trimer.
Striking observations were made when VDAC cross-linking was performed in the presence of Bcl-xL. Cross-linking performed in the presence of Bcl-xL showed that self-association of VDAC is inhibited and association with Bcl-xL is promoted. A significant decrease in the formation of the major cross-linked VDAC trimer and intramolecularly cross-linked VDAC trimer were observed. Bcl-xL formed cross-links with VDAC as heterotrimers of 2 VDAC+1Bcl-xL, 1VDAC+1Bcl-xL, and 1 intramolecularly cross-linked VDAC+1Bcl-xL. Together, gel filtration and chemical cross-linking studies suggest that VDAC alone is a trimer, dimer, or multimer in dynamic equilibrium between states, although our data are most consistent with LDAO-purified VDAC existing as a trimer. In the presence of Bcl-xL VDAC elutes as a smaller molecular weight species than VDAC alone. Calculated molecular weights for a 2:1 VDAC:Bcl-xL complex and VDAC trimer are 84.5 and 95.5 kDa, respectively. Cross-linking of VDAC alone showed a product at ~100 kDa consistent with a VDAC homotrimer. Cross-linking of VDAC in the presence of Bcl-xL revealed a lower molecular weight VDAC:Bcl-xL product consistent with a 2:1 heterotrimer and a 1:1 heterodimer, consistent with the order of elution from the gel filtration column for the VDAC/Bcl-xL.
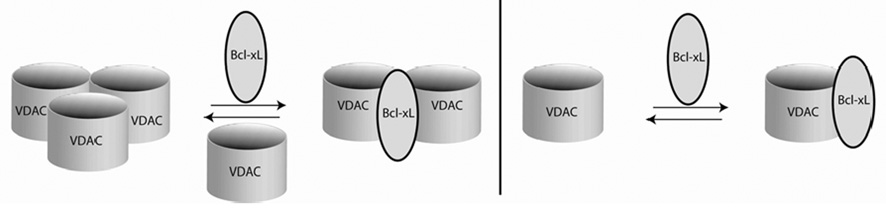
Our data show that Bcl-xL is capable of binding to VDAC monomers and dimers, but not to VDAC trimers, since no cross-linked products with molecular weights higher than the VDAC cross-linked trimer were observed. A model for the interaction could be that Bcl-xL displaces one VDAC monomer from the trimer when it binds, forming a heterotrimer of 2 VDAC molecules and one Bcl-xL. The availability of one VDAC monomer after displacement from the homotrimer facilitates cross-linking of one Bcl-xL to one VDAC monomer. Cross-linking of 1 Bcl-xL to 1 VDAC also indicates the relevance of heterodimer formation. In addition to demonstrating the physical binding between VDAC and Bcl-xL, this cross-linking study also shows that Bcl-xL interferes with VDAC trimerization, supporting the relevance of homotrimer formation for VDAC. The gel filtration and cross-linking data lead us to a model in which 1 Bcl-xL binds to 2 VDAC molecules and displaces one VDAC monomer. In cross-linking experiments Bcl-xL inhibits homooligomerization of VDAC, resulting in the predominance of hetero-cross-linked products. We propose a dynamic VDAC/Bcl-xL complex composed of 2 VDAC and 1 Bcl-xL as well as a 1:1 complex (Figure 11).
FIGURE 11.
Heterotrimeric (left) and heterodimeric (right) models for the VDAC/Bcl-xL complex as suggested from cross-linking results in this study.
Our results show that VDAC has a tendency to form trimers and that trimerization of VDAC is likely in equilibrium between monomeric or other multimeric forms. Our results argue against a simple monomeric species of VDAC. We also show direct binding of Bcl-xL to VDAC demonstrating the relevance of this interaction. The ability of VDAC to form multimers and the ability of Bcl-xL to interact with and in the process inhibit multimer formation, highlight the dynamic nature of VDAC. This dynamic structural property of VDAC may reflect its ability to form complexes with other proteins, lending support for its possible involvement in forming the so-called permeability transition pore (16) or a cytochrome c-permeant pore (29).
As regulators of cellular fate, the Bcl-2 family of proteins may serve multiple functions: by directly causing (pro-apoptotic) or preventing (anti-apoptotic) the disruption of the mitochondrial membrane, and by regulating essential metabolite flux between the mitochondrion and the cytoplasm via VDAC. Bcl-xL binding may serve to change VDAC conductive properties. Regulation of metabolite transfer across the outer membrane may be an additional mechanism by which Bcl-2 proteins exert their control over cellular fate. It remains unclear whether anti-apoptotic Bcl-2 proteins prevent cell death by inhibiting a VDAC-dependent permeability transition pore (15, 16, 50) or by holding VDAC in an open conformation to facilitate metabolite flux (17). Formation of the permeability transition pore by VDAC has been proposed to involve pro-apoptotic proteins tBid (51) and/or Bax (16, 52). However, there is controversy over the identity of the permeability transition pore (53). The results described here will direct future studies to better understand the role of VDAC and Bcl-2 proteins in apoptosis and normal mitochondrial function.
From this study the VDAC-binding interface of Bcl-xL was characterized by NMR. NMR and biochemical data implicate the helical hairpin region of Bcl-xL in mediating heterotrimer formation with two VDAC molecules as well as heterodimer formation. Through a combined strategy of chemical shift perturbation, transferred cross saturation, gel filtration, and chemical cross-linking we have characterized the binding of 2 large membrane proteins in micelles (Bcl-xL at 21 kDa, VDAC at 32 kDa, in a micelle of at least 20 kDa). This study underscores the suitability of NMR spectroscopy for the characterization of very large membrane protein complexes. Our results offer initial insights and lay the groundwork for further investigating models for VDAC and Bcl-2 protein function in apoptosis on a structural level.
ACKNOWLEDGEMENT
We thank Dr. David Waugh (National Cancer Institute) for the generous gift of the auxotrophic strain CT16. We thank Drs. Stephen Fesik and Philippe Hajduk (Abbott Laboratories) for kindly providing backbone chemical shift assignment for Bcl-xL in DPC.
Footnotes
This work was supported by NIH (Grant GM075879).
Abbreviations: 1D, one-dimensional; ATP, adenosine triphosphate; β-OG, beta-octylglucoside; CD, circular dichroism; C8E4, tetraethylene glycol monooctylether; DHPC, dihexanoylphosphocholine; DPC, dodecylphosphocholine; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; GdnHCl, guanidinium hydrochloride; GFC, gel filtration chromatography; HRP, horse raddish peroxidase; HSQC, heteronuclear single quantum correlation; LDAO, lauryl dimethylamine oxide; LM, lauryl maltoside; MWCO, molecular weight cutoff; NAD nicotinamide adenine dinucleotide; NOESY, nuclear Overhauser effect spectroscopy; SDS, sodium dodecyl sulfate; SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis; TROSY, transverse relaxation optimized spectroscopy; NMR, nuclear magnetic resonance spectroscopy.
REFERENCES
- 1.Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG. J Biol Chem. 2005;280:4738–4744. doi: 10.1074/jbc.M411434200. [DOI] [PubMed] [Google Scholar]
- 2.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong SL, Ng SL, Fesik SW. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 3.Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayoshi ED, Oltersdorf T, Fesik SW. Proc Natl Acad Sci U S A. 2001;98:3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 5.McDonnell JM, Fushman D, Milliman CL, Korsmeyer SJ, Cowburn D. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki M, Youle RJ, Tjandra N. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 7.Hinds MG, Lackmann M, Skea GL, Harrison PJ, Huang DC, Day CL. EMBO J. 2003;22:1497–1507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 10.Kelekar A, Thompson CB. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 11.Huang DC, Strasser A. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 12.Desagher S, Martinou JC. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 13.Hsu Y-T, Wolter KG, Youle RJ. PNAS. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinou JC, Green DR. Nat Rev Mol Cell Biol. 2001;2:63–67. doi: 10.1038/35048069. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu S, Matsuoka Y, Shinohara Y, Yoneda Y, Tsujimoto Y. J Cell Biol. 2001;152:237–250. doi: 10.1083/jcb.152.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu S, Narita M, Tsujimoto Y. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, Li XX, Gottleib E, Hill RB, Thompson CB, Colombini M. J Biol Chem. 2001;276:19414–19419. doi: 10.1074/jbc.M101590200. [DOI] [PubMed] [Google Scholar]
- 18.Rostovtseva T, Colombini M. Biophys J. 1997;72:1954–1962. doi: 10.1016/S0006-3495(97)78841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodge T, Colombini M. J Membr Biol. 1997;157:271–279. doi: 10.1007/s002329900235. [DOI] [PubMed] [Google Scholar]
- 20.Rostovtseva TK, Tan W, Colombini M. J Bioenerg Biomembr. 2005;37:129–142. doi: 10.1007/s10863-005-6566-8. [DOI] [PubMed] [Google Scholar]
- 21.Rostovtseva TK, Liu TT, Colombini M, Parsegian VA, Bezrukov SM. Proc Natl Acad Sci U S A. 2000;97:7819–7822. doi: 10.1073/pnas.140115397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng S, Blachly-Dyson E, Colombini M, Forte M. J Bioenerg Biomembr. 1992;24:27–31. doi: 10.1007/BF00769527. [DOI] [PubMed] [Google Scholar]
- 23.Linden M, Gellerfors P. Biochim Biophys Acta. 1983;736:125–129. doi: 10.1016/0005-2736(83)90177-3. [DOI] [PubMed] [Google Scholar]
- 24.Dolder M, Zeth K, Tittmann P, Gross H, Welte W, Wallimann T. J Struct Biol. 1999;127:64–71. doi: 10.1006/jsbi.1999.4141. [DOI] [PubMed] [Google Scholar]
- 25.Mannella CA, Colombini M, Frank J. Proc Natl Acad Sci U S A. 1983;80:2243–2247. doi: 10.1073/pnas.80.8.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zalk R, Israelson A, Garty E, Azoulay-Zohar H, Shoshan-Barmatz V. Biochem J. 2004;386:73–83. doi: 10.1042/BJ20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannella CA, Forte M, Colombini M. J Bioenerg Biomembr. 1992;24:7–19. doi: 10.1007/BF00769525. [DOI] [PubMed] [Google Scholar]
- 28.Song J, Midson C, Blachly-Dyson E, Forte M, Colombini M. J Biol Chem. 1998;273:24406–24413. doi: 10.1074/jbc.273.38.24406. [DOI] [PubMed] [Google Scholar]
- 29.Vander Heiden MG, Thompson CB. Nat Cell Biol. 1999;1:E209–E216. doi: 10.1038/70237. [DOI] [PubMed] [Google Scholar]
- 30.Sreerama N, Venyaminov SY, Woody RW. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 32.Frueh DP, Sun ZY, Vosburg DA, Walsh CT, Hoch JC, Wagner G. J Am Chem Soc. 2006;128:5757–5763. doi: 10.1021/ja0584222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koppel DA, Kinnally KW, Masters P, Forte M, Blachly-Dyson E, Mannella CA. J Biol Chem. 1998;273:13794–13800. doi: 10.1074/jbc.273.22.13794. [DOI] [PubMed] [Google Scholar]
- 34.Shao L, Kinnally KW, Mannella CA. Biophys J. 1996;71:778–786. doi: 10.1016/S0006-3495(96)79277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y, Jiang C, Chen Q, Tang H. Biochem Biophys Res Commun. 2003;303:475–482. doi: 10.1016/s0006-291x(03)00359-0. [DOI] [PubMed] [Google Scholar]
- 36.Colombini M. Mol Cell Biochem. 2004;256–257:107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 37.Lee AC, Zizi M, Colombini M. J Biol Chem. 1994;269:30974–30980. [PubMed] [Google Scholar]
- 38.Rostovtseva TK, Komarov A, Bezrukov SM, Colombini M. Biophys J. 2002;82:193–205. doi: 10.1016/S0006-3495(02)75386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Losonczi JA, Olejniczak ET, Betz SF, Harlan JE, Mack J, Fesik SW. Biochemistry. 2000;39:11024–11033. doi: 10.1021/bi000919v. [DOI] [PubMed] [Google Scholar]
- 40.Franzin CM, Choi J, Zhai D, Reed JC, Marassi FM. Magn Reson Chem. 2004;42:172–179. doi: 10.1002/mrc.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Saez AJ, Mingarro I, Perez-Paya E, Salgado J. Biochemistry. 2004;43:10930–10943. doi: 10.1021/bi036044c. [DOI] [PubMed] [Google Scholar]
- 42.Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW. Mol Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 43.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 44.Nishida N, Sumikawa H, Sakakura M, Shimba N, Takahashi H, Terasawa H, Suzuki EI, Shimada I. Nat Struct Biol. 2003;10:53–58. doi: 10.1038/nsb876. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi K, Yokogawa M, Matsuda T, Sugai M, Kawano S, Kohno T, Nakamura H, Takahashi H, Shimada I. Structure. 2003;11:1381–1392. doi: 10.1016/j.str.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Yokogawa M, Takeuchi K, Shimada I. J Am Chem Soc. 2005;127:12021–12027. doi: 10.1021/ja0511772. [DOI] [PubMed] [Google Scholar]
- 47.Nakanishi T, Miyazawa M, Sakakura M, Terasawa H, Takahashi H, Shimada I. J Mol Biol. 2002;318:245–249. doi: 10.1016/S0022-2836(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu S, Konishi A, Kodama T, Tsujimoto Y. Proc Natl Acad Sci U S A. 2000;97:3100–3105. doi: 10.1073/pnas.97.7.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pappert G, Schubert D. Biochim Biophys Acta. 1983;730:32–40. doi: 10.1016/0005-2736(83)90313-9. [DOI] [PubMed] [Google Scholar]
- 50.Tsujimoto Y, Shimizu S. Biochimie. 2002;84:187–193. doi: 10.1016/s0300-9084(02)01370-6. [DOI] [PubMed] [Google Scholar]
- 51.Rostovtseva TK, Antonsson B, Suzuki M, Youle RJ, Colombini M, Bezrukov SM. J Biol Chem. 2004;279:13575–13583. doi: 10.1074/jbc.M310593200. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu S, Shinohara Y, Tsujimoto Y. Oncogene. 2000;19:4309–4318. doi: 10.1038/sj.onc.1203788. [DOI] [PubMed] [Google Scholar]
- 53.Krauskopf A, Eriksson O, Craigen WJ, Forte MA, Bernardi P. Biochim Biophys Acta. 2006;1757:590–595. doi: 10.1016/j.bbabio.2006.02.007. [DOI] [PubMed] [Google Scholar]