Abstract
We have searched for a minimal interaction motif in τ protein that supports the aggregation into Alzheimer-like paired helical filaments. Digestion of the repeat domain with different proteases yields a GluC-induced fragment comprising 43 residues (termed PHF43), which represents the third repeat of τ plus some flanking residues. This fragment self assembles readily into thin filaments without a paired helical appearance, but these filaments are highly competent to nucleate bona fide PHFs from full-length τ. Probing the interactions of PHF43 with overlapping peptides derived from the full τ sequence yields a minimal hexapeptide interaction motif of 306VQIVYK311 at the beginning of the third internal repeat. This motif coincides with the highest predicted β-structure potential in τ. CD and Fourier transform infrared spectroscopy shows that PHF43 acquires pronounced β structure in conditions of self assembly. Point mutations in the hexapeptide region by proline-scanning mutagenesis prevent the aggregation. The data indicate that PHF assembly is initiated by a short fragment containing the minimal interaction motif forming a local β structure embedded in a largely random-coil protein.
Alzheimer's disease (AD) and other dementias are characterized by abnormal protein deposits in the brain, such as amyloid plaques or neurofibrillary tangles, formed by fibrous assemblies of the Aβ peptide or of τ protein. These aggregates are thought to be toxic to neurons, either by causing some toxic signaling defect (in the case of Aβ) or by obstructing the cell interior (in the case of τ deposits). Therefore, one of the top priorities in Alzheimer research is to understand the reasons for the pathological aggregation and to find methods to prevent it. The principles governing Aβ aggregation are understood in some detail (see review, ref. 1). By contrast, the principles governing τ aggregation have remained elusive (see review, ref. 2). τ can aggregate as an intact protein, 352–441 residues in length (depending on isoform), so that all six τ isoforms are found in Alzheimer paired helical filaments (PHFs) (3). The core of these PHFs is built mainly from the repeat domain (4), and this domain also promotes PHF assembly in vitro (5). τ contains almost no secondary structure but rather appears as a Gaussian random coil, as judged by spectroscopic and x-ray evidence (6, 7). On the other hand, despite its random coil appearance in solution, τ assembles into well-defined fibers, the PHFs. This process can be enhanced by oxidation of SH groups (5) and by polyanions (8–11) and can be described by a nucleation-condensation mechanism (12, 13). A fraction of τ polymers in Alzheimer brains occurs as straight (untwisted) fibers (14), and fibers without axial periodicity have also been described for some in vitro assembly conditions of τ, e.g., with arachidonic acid (15) or other microtubule-associated proteins (MAPs) (16, 17).
Because of their periodicity, one could speculate that PHFs are built from a reproducible secondary structure element. Therefore, we searched for peptides derived from the C-terminal half of τ that would support the aggregation of PHFs. This search led to the identification of the fragment PHF43 containing largely the third repeat R3 plus some adjacent residues. This peptide can self assemble into filaments (not PHFs in the strict sense) but, significantly, nuclei derived from these filaments efficiently promote the assembly of bona fide PHFs from larger τ constructs or intact τ. A search for the motif underlying the interactions between PHF43 molecules yielded the hexapeptide 306VQIVYK311 (termed PHF6), which also shows a high tendency to aggregate into fibrous structures. Assembly of PHF43 or PHF6 is accompanied by a noticeable shift from random-coil to β structure. Moreover, the sequence motif of PHF6 has a high predicted β-structure potential. We therefore conclude that τ filaments can assemble from a small stretch of τ containing the PHF6 sequence, capable of interacting with other τ molecules by a β-sheet-like interaction.
Materials and Methods
Chemicals and Proteins.
Heparin [average molecular weight (MW), 6,000 Da), poly-l-glutamate (average MW, 1,000 Da), tRNA (from bovine liver), and thioflavine S (ThS) were obtained from Sigma. Human τ23 and construct K19 (see Fig. 1) were expressed in Escherichia coli, as described (18). The numbering of the amino acids is that of the isoform hτ40 containing 441 residues (19). The protein was expressed and purified as described elsewhere making use of heat stability and FPLC Mono S (Pharmacia) chromatography (20). The purity of the proteins was analyzed by SDS/PAGE. Protein concentrations were determined by the Bradford assay. Synthetic peptides were obtained from Eurogentec SA (Seraing, Belgium). The mutations of K19, hτ23, and PHF47 were created by site-directed mutagenesis, which was performed by using the Quickchange kit (Stratagene). Cloning of PHF43 was performed after PCR with a hτ23 sequence bearing pNG2-plasmid as template and primers that define both ends of the peptide. The PCR product was cloned by the TA-cloning kit (Invitrogen) and subcloned into expression vector pNG2.
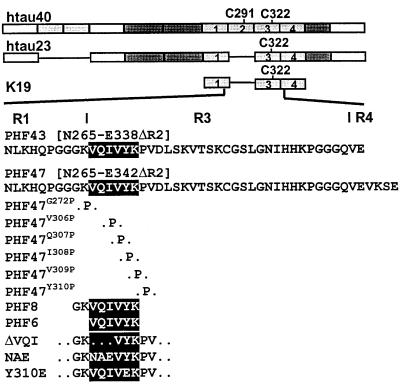
Figure 1.
Bar diagram of τ constructs and peptides. (A) hτ40, containing four repeats of ≈31 residues (labeled 1–4) and two inserts near the N terminus. Residue numbering follows that of hτ40 throughout. (B) hτ23, the smallest and fetal isoform. (C) K19 represents only the repeat domain of hτ23, i.e., R1, R3, R4. (D) PHF43, containing the end of R1, all of R3, and the beginning of R4. PHF47 was used for proline-scanning mutagenesis. (E) PHF8. (F) PHF6, highlighted also in the sequences of PHF43 and PHF8. (G) K19 and hτ23 mutants in the region 306-311. ΔVQI mutants lack 306VQI308, NAE mutants contain NAE instead of 306VQI308, and Y310E, Tyr-310 is exchanged to Glu.
Preparation of Dimeric τ.
Dimers of τ isoforms or τ constructs were allowed to form by incubation at 10 mg/ml after removal of DTT. The dimers were separated from monomers by gel filtration on a Superdex 75 column (Pharmacia) equilibrated with 200 mM NH4Ac, pH 7.0. Fractions were collected and assayed for dimeric K19 by 15% SDS/PAGE under nonreducing conditions.
Generation of Proteolytic Fragments of K19.
Different fragments of K19 were generated by proteolysis with several proteases. Extensive proteolysis by trypsin (ratio wt/wt 1:100, in 50 mM NH4CO3, pH 8.4, 37°C for 2 h), limited proteolysis by chymotrypsin [ratio wt/wt 1:200, in 50 mM NH4CO3, pH 8.4, room temperature (RT)], for 30 min), and extensive proteolysis by GluC (wt/wt 1:100, in 50 mM NH4Ac, pH 4.0, or 50 mM NaHPO4, pH 7.8, RT, over night).
Purification and Analysis of Proteolytic Peptides.
Peptides were purified by reverse-phase HPLC on a C18 column (Vydac, Hesperia, CA) on a SMART HPLC system (Pharmacia) with a standard gradient from 0–50% buffer B (80% acetonitrile vol/vol in 10 mM NH4Ac, pH 7.0), buffer A (10 mM NH4Ac, pH 7.0) in 8 ml and 50–100% B in 2 ml. The peptide PHF43 was purified by gel filtration on a Peptide Gelfiltration column (Pharmacia) with 100 mM NH4Ac, pH 7.0, at a flow rate of 40 μl/min. Peptides were lyophilized after HPLC and analyzed by matrix-assisted laser desorption ionization mass spectrometry (Maldi II, Shimadzu) and N-terminal sequencing (Applied Biosystems 476). Peptide concentrations were determined by fluorescamine assay (21).
Spot Membrane-Binding Assay.
Iodo beads (Pierce) were preincubated with Na125I (0.5 mCi in 100 mM Tris, pH 7.0) for 5 min at RT. The purified peptide PHF43 (50 μg) was iodinated at Tyr-310 for 5 min at RT. The reaction was stopped by removing the solution, and the mixture was applied to a 1-ml C18 cartridge (Waters), equilibrated with 0.1% trifluoroacetic acid (TFA). The unbound radioactivity was washed out with 4 ml TFA (0.1%), and the radioactive peptide was eluted from the column with 66% acetonitrile, 0.1% TFA. All fractions were analyzed by scintillation counting. The eluate of the labeled peptide was lyophilized and diluted in 50 mM ammonium acetate. The membrane containing the library of overlapping peptides was synthesized according to Frank (22). Before use, the membrane was incubated in 100% ethanol, then washed three times with Tris-buffered saline (TBS) (5 min, RT). The labeled peptide solution was diluted 10-fold in TBS (final concentration 0.1 mg/ml). The membrane was incubated in the peptide solution for 2 h at RT, then washed three times with TBS. Autoradiography on Kodak X-omat film was done over night.
PHF Assembly.
Aggregation was induced by incubating varying concentrations of τ isoforms or τ constructs and fragments (typically in the range of 1–100 μM) in volumes of 20–500 μl at 37°C in 50 mM NH4Ac, pH 7.0, or 20 mM Mops–NaOH, pH 7.0, containing anionic cofactors (polyglutamate, RNA, or heparin), as described (10). Incubation times varied from minutes to several days. The formation of aggregates was ascertained by ThS fluorescence, electron microscopy, or sedimentation (see below).
Fluorescence Spectroscopy.
Fluorescence was measured with a Fluoroskan Ascent (Labsytems, Chicago) with an excitation filter of 480 ± 5 nm and an emission filter of 510 nm ± 5 nm in a 384-well plate. Measurements were carried out at RT in 50 mM ammonium acetate, pH 7.0, or 20 mM Na–Mops, pH 7.0, with 5 μM ThS unless otherwise stated. Background fluorescence and light scattering of the sample without thioflavine were subtracted when needed.
Electron Microscopy.
Protein solutions diluted to 1–10 μM protein were placed on 600-mesh carbon-coated copper grids for 1 min and negatively stained with 2% uranyl acetate for 45 sec. The specimens were examined in a Philips (Eindhoven, the Netherlands) CM12 electron microscope at 100 kV.
Sedimentation Assay.
τ constructs were analyzed for their ability to form aggregates by ultracentrifugation of the reaction mixture for 2 h by 40,000 rpm in a TLA45 rotor in a TL-100 ultracentrifuge (Beckman Coulter). The supernatants were collected and the pellets washed once with buffer (50 mM NH4Ac, pH 7.0) and repelleted. The pellets were dissolved in ×2 SDS/PAGE sample buffer (5% SDS/20% glycerol/0.1% bromophenol blue/20 mM Tris, pH 6.8/4% β-mercaptoethanol) by incubation for 5 min at 65°C.
CD Spectroscopy.
All measurements were made with a Jasco J-710 CD-Dichrograph (Jasco, Tokyo) in a cuvette with 0.05-cm path length. For each experiment, 10 spectra were summed up, and the molar ellipticity was determined after normalizing for the protein concentrations.
Infrared Spectroscopy.
Fourier transform infrared spectroscopy (FTIR) experiments were performed on a Bruker IFS 66v spectrometer. Atmospheric water vapor was removed by evacuation of the spectrometer [P < 8 millibar (1 millibar = 100 Pa)] except for the sample chamber, which was purged with dry air (for technical details, see ref. 23). Two hundred interferograms at a spectral resolution of 4 cm−1 were averaged, apodized with a Blackman–Harris three-term function, zero filled with a level of two, and Fourier transformed. Spectra were acquired with the attenuated total reflection (ATR) technique. A thermostated (20°C) microATR unit was used with a diamond disk as internal reflection element (24). Protein solution 5 μl (18 μg/μl in D2O) was applied, and the absorbance spectrum of the sample was acquired. Besides scaling of minor concentration differences and subtraction of the D2O and the heparin spectrum, further data treatment was avoided.
Results
(i) Proteolytic Degradation Products of the Repeat Domain Cause Efficient Assembly of PHFs.
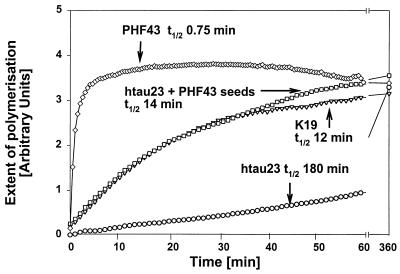
Earlier efforts to reconstitute PHFs in vitro had shown that τ constructs containing roughly the repeat domain polymerized much more readily than the full-length protein (5). This domain is also present in the core of PHFs from Alzheimer brains (4). We therefore decided to study this issue systematically with proteolytic peptides of τ. We focused on construct K19, a derivative of the fetal τ isoform (hτ23; Fig. 1). Construct K19 was digested with chymotrypsin, trypsin, and GluC (Table 1). One of the GluC products was a fragment termed PHF43, comprising only 43 residues (Fig. 1). This fragment assembled much more rapidly than other fragments or intact τ, as judged by electron microscopy and the ThS fluorescence assay (half time 0.75 min, compared with 12 min for K19 and 180 min for hτ23; Fig. 2).
Table 1.
Peptides derived from τ by proteolysis and their ability for PHF assembly
Figure 2.
Kinetics of PHF assembly. The assembly of τ or τ fragments was measured by the fluorescence of ThS (10). Protein concentrations were 20 μM in the presence of 5 μM heparin and 20 mM NH4Ac, at RT. PHF43 assembles rapidly and spontaneously (diamonds, top curve, t1/2 = 0.75 min), but hτ23 is very slow by comparison (circles, bottom curve, t1/2 = 180 min). K19 shows intermediate kinetics (triangles, t1/2 = 12 min). Hτ23 can be speeded up to a similar rate as K19 in the presence of seeds from sonicated fibers obtained after PHF43 assembly (squares, t1/2 = 14 min).
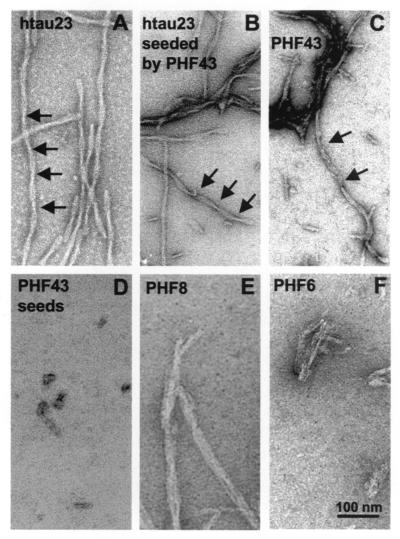
(ii) PHF43 Alone Forms Straight Filaments but Rapidly Nucleates Bona Fide PHFs from hτ23.
By electron microscopy, most fibers obtained after self assembly of the peptide PHF43 appeared as straight thin filaments, lacking the periodic ≈80-nm supertwist that is characteristic of Alzheimer PHFs, in contrast to fibers assembled from construct K19 or hτ23. The absence of a twist could have been taken as a sign of a different assembly form than that of PHFs. Using sonicated PHF43 fibers, we observed that the elongation of these seeds by hτ23 led to bona fide PHFs (Fig. 3B). This aggregation means that the interaction between PHF43 molecules must be very similar to that of PHFs, in both a kinetic sense (requiring dimerization and polyanions) and in a structural sense (nucleation of twisted fibers). The smaller diameter of PHF43 fibers could be accounted for by the smaller size of the peptide, compared with the larger τ constructs studied previously (5, 11).
Figure 3.
Electron micrographs of fibers obtained from the self assembly of τ or τ peptides. Assembly conditions were the same as in Fig. 2 (20 μM protein, 5 μM heparin), except E and F (660 μM peptide, 660 μM heparin). A–C show mostly twisted filaments (width ≈10–20 nm) resembling Alzheimer PHFs polymerized from, (A) hτ23, (B) hτ23 plus seeds made from PHF43, and (C) PHF43. D shows the seeds obtained by sonication of PHF43 fibers. E and F show filamentous aggregates with variable diameters obtained from the short peptides PHF8 and PHF6.
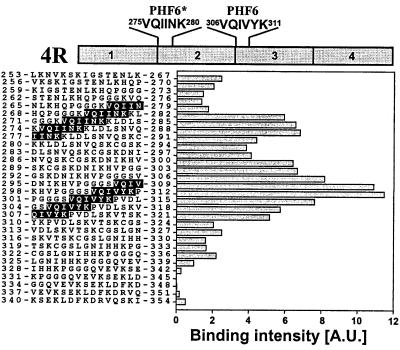
(iii) The Hexapeptide PHF6 (306VQIVYK311) Represents a Minimal Interaction Motif with a Predicted β Conformation.
To check the τ–τ interaction under nonpolymerizing conditions, we used a spot membrane of immobilized 15-mer peptides, covering the sequence of the repeats with overlaps of three residues between successive peptides (22). The peptide spot membrane was probed against the PHF43 peptide, radioactively labeled by iodination with 125I at Tyr-310. The most prominent interaction between PHF43 and the spot membrane was found at peptides surrounding Tyr-310 (Fig. 4). A motif overlapping with the interacting peptides was the hexapeptide 306VQIVYK311 (termed PHF6, highlighted in Fig. 4). A second maximum of interaction was centered around Ile-278, covering the sequence 275VQIINK280 (PHF6*).
Figure 4.
Peptide spot membrane interactions between PHF43 and peptides derived from the repeat region of τ. The region of τ from L253 in repeat 1 to D348 in repeat 4 (encompassing the sequence of PHF43) was subdivided into consecutive 15-mer peptides staggered by three residues, synthesized and covalently attached to a cellulose membrane (22), and incubated with iodinated PHF43. Bars represent the bound radioactivity determined by autoradiography. The strongest interaction occurs around the spots that contain the sequence of PHF6 (306VQIVYK311, highlighted) at the beginning of R3. Another cluster of strong interaction occurs around the spots that harbor the analogous sequence in R2 (275VQIINK280, highlighted).
(iv) CD Spectroscopy of τ Fragments Suggests a β Structure in PHF Assembly Conditions.
To study the conformation of τ and peptides, we used CD (Fig. 5) and infrared spectroscopy (Fig. 6) under several conditions:
Figure 5.
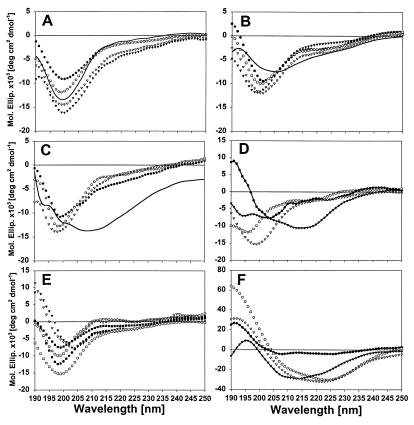
CD spectroscopy of τ constructs and τ polymers. CD spectra were obtained at 50 μM protein concentrations in 10 mM NH4 acetate at RT, in the absence or presence of 50 μM heparin and/or 50% TFE. (A) hτ23 monomers (circles), dimers (triangles), and assembled PHFs (solid line) show similar CD curves with or without heparin (filled or open symbols). (B) K19 monomers (without heparin, open circles; with heparin, filled circles) and dimers (without heparin, open triangles; with heparin, filled triangles; after aggregation, solid line). (C) PHF43 monomers (without heparin, open circles; with heparin, filled circles) and dimers without heparin (open triangles) and after aggregation (solid line). (D) PHF8 (without heparin, open circles; with heparin, filled circles) and PHF6 (without heparin, open triangles; with heparin, filled triangles). (E) Three peptides derived from PHF43, but not including 306-311 (313VDLSKVTSK321; without heparin, open circles; with heparin, filled circles; 318VTSKCGSLGNIHHKPGGG335 without heparin, open triangles; with heparin, filled triangles; 335GQVEVKSE342, without heparin, open squares; with heparin, filled squares). (F) K19 (open circles) and PHF43 dimers (open triangles) and PHFs (K19, filled circles; PHF43, filled triangles) in the presence of 50% TFE.
Figure 6.
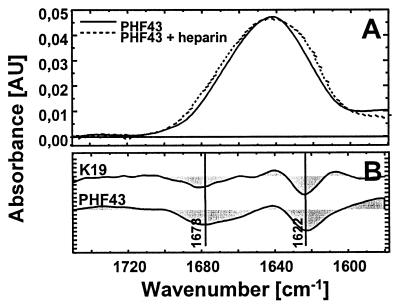
FTIR of τ constructs and τ-derived peptides. (A) Infrared absorbance spectrum of the peptide PHF43 in the absence (solid line) and presence (dotted line) of heparin. Spectral contributions of heparin and the solvent D2O have been subtracted. (B) Aggregation-induced difference spectra of K19 and PHF43.
• The monomeric state is obtained in the presence of DTT, because this agent keeps Cys-322 reduced and prevents dimer formation.
• The dimeric state is obtained by allowing air oxidation of Cys-322, resulting in almost complete conversion from monomers to dimers (see ref. 25).
• Monomers with added polyanions, e.g., heparin.
• Dimers with added polyanions, which induces polymerization; spectra were taken after polymerization was complete.
• Monomers, dimers, or aggregates in the presence of trifluoroethanol.
The quantitation of CD spectra in terms of secondary structure components depends on the algorithm used and is often not reliable; however, more significant are the general shapes of the spectra that reveal gross conformational states. All curves obtained for hτ23 show a minimum of around 200 nm, characteristic of largely random coil structures, which confirms our earlier observations (25). Neither dimerization nor heparin changes the spectra substantially, arguing that the random coil structure dominates in all cases, even after PHF assembly (solid curve in Fig. 5A). Even if a local β structure were formed during PHF assembly, this change does not become noticeable with full-length τ. The situation changes as the τ constructs become smaller (Fig. 5B): K19 also displays a mostly random coil structure in the monomeric or dimeric state, even with heparin, but polymerization induces a noticeable change in the spectra so that the minimum becomes wider and is shifted toward higher wavelengths, indicating a substantial change from random coil to β structure. This behavior is reiterated in the case of PHF43 (Fig. 5C), showing mostly random coil structures except after filament assembly, where the content of β structure is increased. Fig. 5D compares the curves of PHF6 before and after assembly, as well as PHF8; again, there is a substantial change from random coil to β structure after assembly. Other peptides spanning the PHF43 sequence do not show a conformational change in the presence of heparin [313V-K321, 318V-G335, kindly provided by R. Hoffmann and L. Otvos (University of Pennsylvania, Philadelphia) (26), and 335G-E342; Fig. 5E].
The helix inducer TFE (50%) was used to analyze the fraction of the molecules not locked in the interactions within the PHFs (Fig. 5F): In all cases tested, TFE induced a strong shift toward a more α-helical spectrum, as seen by the positive molar ellipticities below 200 nm and the broad minimum between 210 and 230 nm. The fraction convertible from random coil to α-helix increased with the size of the protein (i.e., smallest for PHF43, largest for hτ23; data not shown).
(v) Infrared Spectroscopy.
The detection of β sheet by CD spectroscopy was confirmed by FTIR. As an example, Fig. 6A (straight line) depicts the infrared spectrum of PHF43 in the amide I region. The band maximum around 1,640 cm−1 indicates that the peptide is mostly random in structure. After the aggregation of PHF43, the amide I band exhibits an apparent broadening (dashed spectrum in Fig. 6A). The difference spectrum (dotted line) is characterized by two distinct maxima. The frequency at 1,622 cm−1 is typical for β structure. The lower frequency as compared with a classical β sheet (1,625 cm−1) argues for an aggregated state (27). The additional appearance of a difference maximum around 1,680 cm−1 is an indication for antiparallel β strands (28). This band (at 1,678 cm−1) is observed for the peptides PHF43 and K19 and is consistent with the antiparallel arrangement of τ dimers observed by electron microscopy (16).
(vi) Proline-Scanning Mutations in the Minimal Interaction Motif Prevent PHF Aggregation.
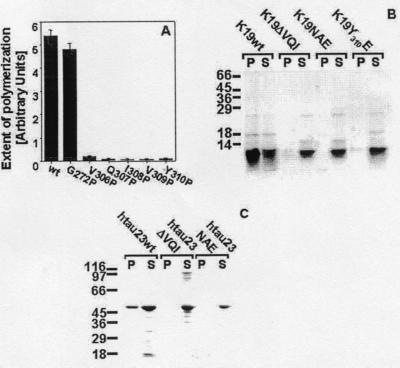
To verify the importance of the hexapeptide PHF6 for the conformational change during τ aggregation, we performed a proline-scanning mutagenesis in the region of PHF6, following a similar approach used for the Aβ peptide (29). For these studies, we used PHF47, a construct that was slightly longer than PHF43 (Fig. 1) but that otherwise showed the same assembly behavior. The rationale behind the proline-scanning approach is that Pro disrupts a potential β strand and should therefore inhibit the aggregation. This inhibition is indeed observed for all residues in the PHF6 sequence mutated into Pro (Fig. 7A). As a control, we synthesized the mutant G272P (construct PHF47G272P). In this case, there was no significant change in aggregation.
Figure 7.
Analysis of the ability of τ mutants to form filaments. (A) A series of substitution mutations to proline was analyzed by ThS fluorescence in comparison to wild-type τ after 24-h incubation under PHF-assembly conditions. (B) PHF assembly of K19 wild type and the mutants K19ΔVQI, K19NAE, and K19Y310E was performed for 24 h, and the supernatants and pellets after ultracentrifugation of the reaction mixtures were applied to a 15% SDS/PAGE and stained by Coomassie blue. (C) PHF assembly of hτ23 wild type and the mutants hτ23ΔVQI and hτ23NAE was performed for 3 days, and the supernatants and pellets after ultracentrifugation of the reaction mixtures were applied to a 10% SDS/PAGE and stained by Coomassie blue.
Further confirmation was provided by several mutants derived from the construct K19, in which residues with high β propensity in the PHF6 region were replaced or deleted (Fig. 7B). Thus, residues 306VQI308 were deleted (construct K19ΔVQI) or exchanged for NAE (construct K19NAE), or Tyr-310 was replaced by E (construct K19Y310E). In all three cases, the aggregation of the K19 mutants was strongly suppressed compared with the K19 control. Thus, even after 24 h when the aggregation of K19 was complete, the aggregation of the K19 mutants was still at background level (Fig. 7B), and no fibers were detected by electron microscopy (not shown). Even in the full-length isoform hτ23, the deletion of 306VQI308 and the substitution of these amino acids to 306NAE308 led to a complete loss of PHF formation (Fig.7C).
Discussion
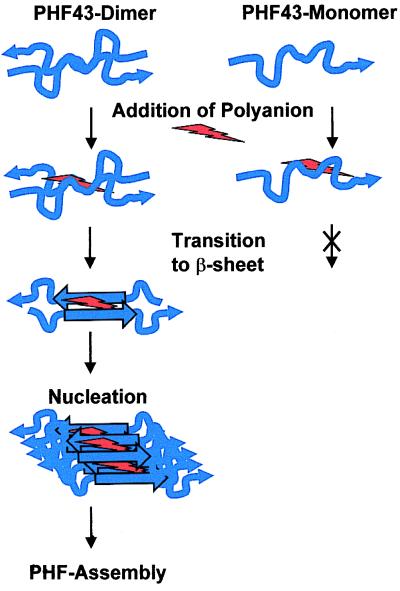
The aim of this work was to define the smallest possible sequence of τ that is capable of initiating the assembly of τ into the pathological PHFs. The results can be summarized as follows (Fig. 8): (i) A contiguous sequence motif supporting PHF assembly indeed exists. (ii) PHF43 assembles within seconds into thin fibers under the same conditions as full-length τ (favored by dimerization and anionic cofactors such as heparin; Fig. 2). Seeds derived by sonication from such fibers are capable of greatly accelerating the assembly of full-length τ into bona fide PHFs (Fig. 3). Thus the nucleation capacity appears to reside within PHF43, even though the morphology of the assembled fibers may be influenced by sequences that lie outside of PHF43. (iii) The interaction between τ or τ-derived peptides under nonpolymerizing conditions is dominated by the hexapeptide PHF6. An equivalent sequence is that of (PHF6*). (iv) PHF assembly is accompanied by a substantial increase in β-sheet formation (Figs. 5 and 6). This is not observed on dimerization or addition of anionic cofactors alone, indicating that β-sheet formation is an essential part of PHF assembly. (v) The hexapeptide PHF6 in R3 and its counterpart PHF6* in R2 are the only two spots in the τ sequence where a β conformation is strongly predicted. The pronounced tendency for β-structure is explained by the local clustering of strong β-sheet-inducing residues such as V, I, Y, and Q, whereas the corresponding hexapeptides in R1 (244QTAPVPM250) and R4 (337VEVKSE342) contain β-sheet breakers such as P and E (30). When the propensity for β conformation is lowered by mutations in the PHF6 motif, PHF aggregation is inhibited as well (Fig. 7).
Figure 8.
Model of structural changes during PHF assembly. PHF43 dimer and PHF43 monomer (in blue) are able to bind polyanions (red), but only the dimer shows a transition from random coil (wiggle) to β sheet (straight) during aggregation.
Several conclusions can be drawn:
(i) The sequence of PHF6 at the beginning of the third repeat (exon 11) is present in all τ isoforms and provides an explanation why all τ isoforms can form PHFs. The counterpart PHF6* lies at the beginning of R2 (exon 10) and is therefore present only in τ isoforms containing four repeats. If the two hotspots for β-sheet formation operated in a cooperative fashion, they could possibly enhance PHF assembly, and this might explain why PHF formation is enhanced in dementias where the four-repeat isoforms are overrepresented (e.g., FTDP-17; see refs. 31–33).
(ii) Dimerization of τ via oxidation of Cys-322 into an intermolecular disulfide bridge strongly promotes PHF assembly (5). The results shown here argue that the peptide containing Cys-322 (peptide 318–335; Fig. 5) does not promote PHF assembly by itself. However, dimerization at Cys-322 could bring two PHF6 motifs into close vicinity, which would facilitate β-sheet interactions. In this view, dimerization would act as an effective enhancer of local τ concentration.
(iii) The role of heparin or other anionic cofactors in AD remains unknown in detail but could effectively help reduce the repulsion between the cationic τ molecules in a general way, as discussed by several authors previously (8–12, 34). This ability would also be equivalent to enhancing the effective local concentration of τ. Heparin does not appear to change the conformation of τ by itself, as judged by the CD spectra.
(iv) The increase of β-sheet structure encompasses only a small fraction of the whole sequence and therefore is not detected reliably by spectroscopic or scattering techniques in the full-length protein. The presence of a β-sheet interaction also removes a previous dilemma: How could a disordered protein give rise to an ordered filament? The answer is that only a kernel of the structure needs to be ordered. The observations also explain why only the repeat domain of τ is important for PHF assembly, whereas the N- or C-terminal tails protrude as “fuzzy coat” (4).
(v) In AD and related “τ-opathies,” τ is both abnormally phosphorylated and aggregated, and it was thought that phosphorylation primes τ for aggregation. However, we have shown recently that particularly the phosphorylation at the three or four KXGS motifs in the repeats (Ser-262, -293, -324, and -356; see ref. 35) is antagonistic to PHF assembly. It is not known whether these sites interact with the PHF6 motif, but at least Ser-324 is very close to the dimer-forming residue Cys-322. Consistent with this proximity, phosphorylation at KXGS motifs inhibits dimerization and thus the subsequent assembly via the PHF6 motif, in contrast to other phosphorylation sites that allow dimerization but inhibit PHF assembly at a later stage (e.g., Ser-214).
(vi) τ mutations have recently been discovered for several familial frontotemporal dementias (32, 33, 36, 37). Some of these mutations are intronic mutations that affect the splicing of τ; the exonic mutations cause changes or loss of residues in the repeat domain of τ. It is notable that several of these mutations lie close to the motifs of PHF6 [e.g., G272V in 3R-τ, P301L in 4R-τ, both of which have been reported to form PHFs more readily (38)]. Other mutations lie near the motif of PHF6*, e.g., G272V in 4R-τ, N279K, ΔK280, or in an analogous position at the beginning of R4, e.g., V337 M, for which also an increased polymerization rate was reported (39).
Finally, we note that these results offer new approaches to testing the assembly properties of τ into PHFs and to searching for inhibitors of PHF formation. Because PHF43 aggregates particularly rapidly and has the appropriate substructure for nucleating PHFs, this fragment or related ones can be used in screening assays to investigate the assembly properties of PHFs, their dependence on cofactors, mutations, etc. Likewise, inhibitors of PHF could be found through the use of PHF43 assembly tests. Because the assembly appears to be mediated by β-sheet-like interactions of the PHF6 motif, drugs interacting with that motif should be inhibitory for the τ–τ association leading to PHFs.
Acknowledgments
We are grateful to B. Krüger, A. Konopatzki, K. Blume, and N. Habbe for expert technical assistance. The peptide spot membrane was generously provided by R. Frank and J. Wehland (Gesellschaft für Biotechnologische Forschung, Braunschweig, Germany), and the peptide 318–335 by R. Hoffmann and L. Otvos (University of Pennsylvania, Philadelphia). We thank L. Li for help with the proline-scanning mutagenesis. This project was supported by a grant from the Deutsche Forschungsgemeinschaft.
Abbreviations
- AD
Alzheimer's disease
- PHF
paired helical filaments
- ThS
thioflavine S
- FTIR
Fourier transform infrared spectroscopy
- RT
room temperature
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Koo E H, Lansbury P T, Jr, Kelly J W. Proc Natl Acad Sci USA. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedhoff P, Mandelkow E. In: Guidebook to the Cytoskeletal and Motor Proteins. Kreis T, Vale R, editors. Oxford: Oxford Univ. Press; 1998. pp. 230–236. [Google Scholar]
- 3.Goedert M, Spillantini G, Cairns N J, Crowther R A. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 4.Wischik C, Novak M, Thogersen H, Edwards P, Runswick M, Jakes R, Walker J, Milstein C, Roth M, Klug A. Proc Natl Acad Sci USA. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wille H, Drewes G, Biernat J, Mandelkow E M, Mandelkow E. J Cell Biol. 1992;118:573–584. doi: 10.1083/jcb.118.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland D W, Hwo S Y, Kirschner M W. J Mol Biol. 1977;116:227–247. doi: 10.1016/0022-2836(77)90214-5. [DOI] [PubMed] [Google Scholar]
- 7.Schweers O, Schönbrunn-Hanebeck E, Marx A, Mandelkow E. J Biol Chem. 1994;269:24290–24297. [PubMed] [Google Scholar]
- 8.Perez M, Valpuesta J M, Medina M, Degarcini E M, Avila J. J Neurochem. 1996;67:1183–1190. doi: 10.1046/j.1471-4159.1996.67031183.x. [DOI] [PubMed] [Google Scholar]
- 9.Goedert M, Jakes R, Spillantini M G, Hasegawa M, Smith M J, Crowther R A. Nature (London) 1996;383:550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 10.Friedhoff P, Schneider A, Mandelkow E M, Mandelkow E. Biochemistry. 1998;37:10223–10230. doi: 10.1021/bi980537d. [DOI] [PubMed] [Google Scholar]
- 11.Kampers T, Friedhoff P, Biernat J, Mandelkow E M. FEBS Lett. 1996;399:344–349. doi: 10.1016/s0014-5793(96)01386-5. [DOI] [PubMed] [Google Scholar]
- 12.Friedhoff P, von Bergen M, Mandelkow E M, Davies P, Mandelkow E. Proc Natl Acad Sci USA. 1998;95:15712–15717. doi: 10.1073/pnas.95.26.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King M E, Ahuja V, Binder L I, Kuret J. Biochemistry. 1999;38:14851–14859. doi: 10.1021/bi9911839. [DOI] [PubMed] [Google Scholar]
- 14.Crowther R A. Proc Natl Acad Sci USA. 1991;88:2288–2292. doi: 10.1073/pnas.88.6.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson D M, Binder L I. Am J Pathol. 1997;150:2181–2195. [PMC free article] [PubMed] [Google Scholar]
- 16.Wille H, Mandelkow E M, Mandelkow E. J Biol Chem. 1992;267:10737–10742. [PubMed] [Google Scholar]
- 17.DeTure M A, Zhang E Y, Bubb M R, Purich D L. J Biol Chem. 1996;271:32702–32706. doi: 10.1074/jbc.271.51.32702. [DOI] [PubMed] [Google Scholar]
- 18.Biernat J, Mandelkow E M, Schröter C, Lichtenberg-Kraag B, Steiner B, Berling B, Meyer H E, Mercken M, Vandermeeren A, Goedert M, et al. EMBO J. 1992;11:1593–1597. doi: 10.1002/j.1460-2075.1992.tb05204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goedert M, Wischik C, Crowther R, Walker J, Klug A. Proc Natl Acad Sci USA. 1988;85:4051–4055. doi: 10.1073/pnas.85.11.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustke N, Trinczek B, Biernat J, Mandelkow E M, Mandelkow E. Biochemistry. 1994;33:9511–9522. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- 21.Udenfriend S, Stein S, Bohlen P, Dairman W, Leimgruber W, Weigele M. Science. 1972;178:871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- 22.Frank R. J Biotechnol. 1995;41:259–272. doi: 10.1016/0168-1656(95)00062-u. [DOI] [PubMed] [Google Scholar]
- 23.Zscherp C, Heberle J. J Phys Chem B. 1997;101:10542–10547. [Google Scholar]
- 24.Heberle J. Rec Res Dev Appl Spectrosc. 1999;2:147–159. [Google Scholar]
- 25.Schweers O, Mandelkow E M, Biernat J, Mandelkow E. Proc Natl Acad Sci USA. 1995;92:8463–8467. doi: 10.1073/pnas.92.18.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann R, Dawson N F, Wade J D, Otvos L., Jr J Pep Res. 1997;50:132–142. doi: 10.1111/j.1399-3011.1997.tb01178.x. [DOI] [PubMed] [Google Scholar]
- 27.Fabian H, Szendrei G I, Mantsch H H, Otvos L., Jr Biochem Biophys Res Commun. 1993;191:232–239. doi: 10.1006/bbrc.1993.1207. [DOI] [PubMed] [Google Scholar]
- 28.Schladitz C, Vieira E P, Hermel H, Mohwald H. Biophys J. 1999;77:3305–3310. doi: 10.1016/S0006-3495(99)77161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood S J, Wetzel R, Martin J D, Hurle M R. Biochemistry. 1995;34:724–730. doi: 10.1021/bi00003a003. [DOI] [PubMed] [Google Scholar]
- 30.Street A G, Mayo S L. Proc Natl Acad Sci USA. 1999;96:9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spillantini M G, Murrell J R, Goedert M, Farlow M R, Klug A, Ghetti B. Neuron. 1998;21:955–958. [Google Scholar]
- 32.Hutton M, Lendon C L, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Nature (London) 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 33.Clark L N, Poorkaj P, Wszolek Z, Geschwind D H, Nasreddine Z S, Miller B, Li D, Payami H, Awert F, Markopoulou K, Andreadis A, et al. Am J Pathol. 1998;153:1359–1363. doi: 10.1073/pnas.95.22.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasegawa M, Crowther R A, Jakes R, Goedert M. J Biol Chem. 1997;272:33118–33124. doi: 10.1074/jbc.272.52.33118. [DOI] [PubMed] [Google Scholar]
- 35.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow E M. Biochemistry. 1999;38:3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 36.Spillantini M G, Crowther R A, Kamphorst W, Heutink P, van Swieten J C. Am J Pathol. 1998;153:1359–1363. doi: 10.1016/S0002-9440(10)65721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poorkaj P, Bird T D, Wijsman E, Nemens E, Garruto R M, Anderson L, Andreadis A, Wiederholt W C, Raskind M, Schellenberg G D. Ann Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 38.Goedert M, Jakes R, Crowther R A. FEBS Lett. 1999;450:306–311. doi: 10.1016/s0014-5793(99)00508-6. [DOI] [PubMed] [Google Scholar]
- 39.Nacharaju P, Lewis J, Easson C, Yen S, Hackett J, Hutton M, Yen S H. FEBS Lett. 1999;447:195–199. doi: 10.1016/s0014-5793(99)00294-x. [DOI] [PubMed] [Google Scholar]