Abstract
Type I Interferons (IFNs) are requisite components in antiviral innate immunity. Classically, a Toll-like receptor-dependent pathway induces type I interferons. However, recent recognition of melanoma differentiation associated gene-5 (MDA-5) and retinoic acid inducible gene-I (RIG-I) as primary sensors of RNA viruses for type I interferon induction highlights a potentially unique pathway for innate immunity. Our present investigation tracing the phylogenetic origin of MDA-5 and RIG-I domain arrangement (CARD1-CARD2-helicase-DEAD/DEAH) indicates that these proteins originated specifically in mammals, firmly linking this family of proteins with interferons in a highly derived evolutionary development of innate immunity. MDA-5, but not RIG-I, orthologs are found in fish, indicating that MDA-5 might have evolved before RIG-I. Our analyses also reveal that the MDA-5 and RIG-I domain arrangement evolved independently by domain grafting and not by a simple gene-duplication event of the entire four-domain arrangement, which may have been initiated by differential sensitivity of these proteins to viral infection.
Interferons (IFNs) comprise a family of secreted proteins (cytokines) produced by cells following virus infection and create an antiviral state in neighboring cells (1–6). The relationship between IFN and viral infection is well documented and supported by studies employing neutralizing antibodies and IFN-receptor knockout mice, which document a direct role for IFN in mediating host defense against viruses (7, 8). Moreover, IFNs are now recognized as multifunctional molecules that also provide defense against bacterial infection (especially intracellular parasites), induce antitumor activity (both direct and immune system-mediated), and stimulate or inhibit differentiation depending on cellular context (5, 9). In addition, type I IFNs also induce apoptosis of virus-infected cells and activate natural killer and T cells, thus activating the adaptive immune system as well (3).
The expression of type I IFN is stringently regulated by the activation of pre-existing transcription factors, such as IFN regulatory factor 3 (IRF-3), NF-κB, and ATF-2-c-Jun [supporting information (SI) Fig. S1] (10–12). These transcription factors are activated and, therefore, type I IFN is induced by bacterial components, such as lipopolysaccharide, and CpG DNA in leukocytes, such as macrophages and dendritic cells, as well as by viral infection (13, 14). Targeted-gene-disruption studies indicate that Toll-like receptors (TLRs) recognize pathogen-associated molecular patterns. TLR3, TLR4, mouse TLR7 (human TLR8), and TLR9 function as signaling receptors for extracellular double-stranded RNA (dsRNA), lipopolysaccharide, viral single-stranded RNA, and CpG DNA, respectively (15–18). The interaction of these pathogen-associated molecular patterns with the extracellular leucine-rich repeat of the TLR facilitates the recruitment of adaptor molecules to the cytoplasmic Toll-IL-1 receptor domain of the TLR (19). The adaptors MyD88, IL-1 receptor associated kinase (IRAK), and TRAF6 are recruited by many TLRs and activate signaling cascades. For TLR7 and TLR9, type I IFN is induced by this MyD88-dependent pathway (19). TLR4 and TLR3 activate an additional signaling pathway called the MyD88-independent pathway, which recruits another adaptor molecule, Trif, and activates a second set of genes, including those of type I IFN, through activation of IRF-3 (20, 21). However, for most cell types, it has been postulated that the replication of viruses results in an accumulation of intracellular dsRNA, which triggers host response mechanisms that include expression of type I IFN (22). This signaling pathway is apparently distinct from that mediated by TLR3 and constitutes a major pathway activated by viral infection.
Recent studies identified two proteins, melanoma differentiation associated gene-5 (MDA-5) and retinoic acid inducible gene I (RIG-I), as intracellular sensors of dsRNA responsible for induction of type I IFN (23, 24). Analysis of mda-5 or RIG-I knockout mice demonstrates that this TLR-independent pathway is central for innate immunity against viral infection (25–27). Moreover, both MDA-5 and RIG-I are also IFN-stimulated genes, thereby creating a positive feedback-loop generating a potent anti-viral state (28, 29). MDA-5 and RIG-I contain two N-terminal caspase (cysteine-dependent aspartate-specific proteases) recruitment domains (CARDs), followed by a DEAD/DEAH box helicase domain (Fig. 1A) (24). The DEAD/DEAH box helicase domains are large domains (over 300 aa) with eight conserved short motifs (including two Walker-like boxes) distributed throughout the larger domain (called I, Ia, Ib, and II-VI) (30–33). The DEAD and DEAH box helicase domains are each other's closest relatives with respect to other RNA helicase families and are named after “the single-letter designation of the amino acid sequence of motif II” (31). DEAD/DEAH box domains exhibit ATPase activity while the helicase domain is involved in unwinding of RNA. Functionally, DEAD-box proteins use ATP as a substrate, while DEAH-box proteins are promiscuous in their NTP usage (33). MDA-5 and RIG-I are structurally similar proteins, having 23% and 35% amino acid identity in their N-terminal tandem CARD and C-terminal helicase domains, respectively. The helicase domain binds to dsRNA, which leads to activation of the CARD domains (24). MDA-5 and RIG-I interact with the CARD domain of the mitochondrial protein IFN-β promoter stimulator-1 (IPS-1, also known as MAVS, VISA, and CARDIF), followed by recruitment of TNF-receptor associated factor-3 (TRAF-3) and activation of TRAF family member-associated NF-κB-activator binding kinase-1 (TBK1) and inducible IκB kinase (IKKε) (34–39). These kinases phosphorylate IRF-3 and IRF-7 and activate NF-κB, and these transcription factors translocate to the nucleus to induce type I IFN expression (40). In addition to MDA-5 and RIG-I, LGP2, a third member of this family containing of only the helicase domain but no CARD domains, has been identified, which functions as a dominant negative-inhibitor of MDA-5 and RIG-I antiviral action (41).
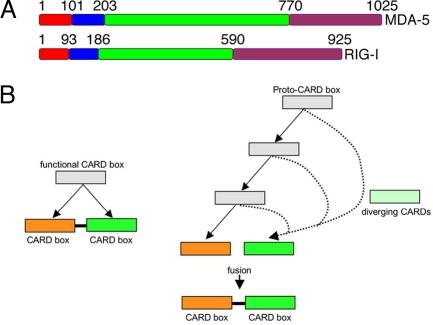
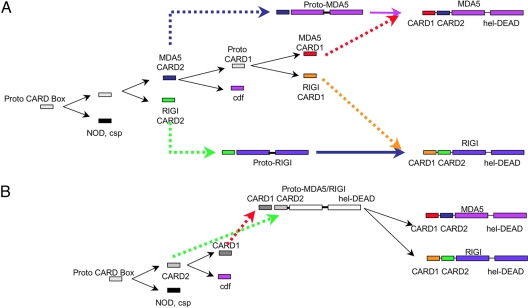
Fig. 1.
Diagrammatic representation of two possible pathways for the creation of linked but divergent CARD domains. (A) Domain structure of MDA-5 and RIG-I. Red box, CARD1; blue box, CARD2; green box, helicase domain; purple box, DEAD/DEAH domain. The numbers represent amino acid positions. (B) (Left) A simple domain duplication event to produce two linked domains. In this scenario, the two domains would be expected to have similar evolutionary histories. (Right) A more complicated pathway where linked domains, produced by independent and divergent evolution (dotted lines) of duplicated domains originally not in linkage, fuse to form linked domains. This second scenario would produce domains with differing evolutionary histories.
The simultaneous presence of CARD and RNA helicase domains in a single molecule, as observed in MDA-5 and RIG-I, is unique. RNA helicases play significant roles in diverse processes of RNA metabolism, including splicing, editing, translation initiation, transcription, rRNA processing, export, degradation, and protein displacement from RNP (RNPase) (42). Helicases use the energy provided by hydrolysis of ATP to catalyze the unwinding of nucleic acid duplexes. CARD is a structural domain composed of amphipathic α-helixes, and is predicted to function by protein-protein interactions with both apoptotic and antiapoptotic proteins (43). However, the convergence of these two distinct functional domains into a single molecule for specific antiviral processes represents a very intriguing phenomenon. Considering the uniqueness and importance of MDA-5 and RIG-I in innate immunity, we embarked on studies to trace the phylogenetic origins of these specific domains. Such an approach elucidates the evolutionary pathways involved in the shuffling of domains, which culminated in a unique mechanism for protecting against viral infection. The phylogenetic analyses of these domains also provide insight into the temporal pathways of development of innate immunity.
Results and Discussion
MDA-5 and RIG-I each contain four discrete domains (two CARD domains, a helicase domain, and a DEAD/DEAH domain). Consequently, there are four evolutionary histories to examine in deciphering the origin of these proteins; these involve several steps of investigation. The first step is to examine if any of the domains have ancient linkage. The second step is to identify whether any of the four domains have similar evolutionary histories, thus supporting the hypothesis that fusion events of previously independent domains occurred. The third step is to examine the evolutionary patterns of these singular and/or linked domains.
Step 1: Ancient Linkage and Coevolution of Helicase DEAD Domains.
The linked state of helicase-DEAD/DEAH in Archaea is well known and signifies that the linkage may be presumed ancient for the helicase superfamily of proteins (32, 44). The history of the CARD domain linkage is more complicated. These domains, most often associated with caspases, are found in a wide variety of proteins from a broad array of vertebrates (45, 46).
Most caspases (cysteine-dependent aspartate-specific proteases) have a single CARD domain, with the exceptions being MDA-5, RIG-I, and Nacht proteins. Because the Nacht CARD domains have diverged greatly from the MDA-5 and RIG-I CARD domains (see below), we can rule out an ancient linkage involving Nacht CARD domains. Furthermore, because the linked CARD1/CARD2 state in MDA-5 is found in all vertebrates, we conclude that the linkage of the two CARD domains in this protein occurred in the common ancestor of vertebrates.
Step 2: CARD1/CARD2 Domain Linkage by History.
The linked CARD1/CARD2 arrangement in the common ancestor of vertebrates could have arisen by two very different processes. First, a single CARD domain could have simply duplicated to produce two linked domains. Second, two independently derived CARD domains could have fused to produce the linked state (Fig. 1 B). Consequently, we looked for evidence that any of the four domains have directly coevolved. The domain-by-domain comparisons are shown in Table 1 and indicate that CARD2, helicase and DEAD/DEAH domains are all coevolving, as revealed by the significant incongruence length difference (ILD) statistics (47–49). CARD1, however, shows incongruence with the other domains. This result indicates a different evolutionary history for this domain relative to the other three domains, and suggests that the CARD1 domain was grafted onto an existing CARD2-helicase-DEAD/DEAH structure (as depicted in Fig. 1 B) at a later time in the evolutionary history of these proteins.
Table 1.
ILD statistics for comparison of common evolutionary history of CARD boxes with each other and with the helicase and DEAD/DEAH domains
| CD1 | CD2 | HEL | DED | |
|---|---|---|---|---|
| CD1 | — | 0.68 | 0.45 | 0.91 |
| CD2 | — | 0.01 | 0.01 | |
| HEL | — | 0.05 | ||
| DED | — |
P values given in the cells of the table. Bold values indicate significant ILD scores and hence a common phylogenetic pattern. The CARD1 boxes (CD1) have experienced different evolutionary histories than the helicase domain (HEL), DEAD domain (DED) and CARD2 boxes (CD2).
Step 3: Using Phylogeny to Unravel Domain Fusion.
The coevolution analysis (see Table 1) using the ILD test suggests that there are two independently evolving domain linkages that make up these proteins. Consequently, we constructed phylogenies for the domains in these proteins to examine the evolutionary history of each. We first examined the linked helicase/DEAD-DEAH domains and second, the genealogical relationships of the two CARD domains.
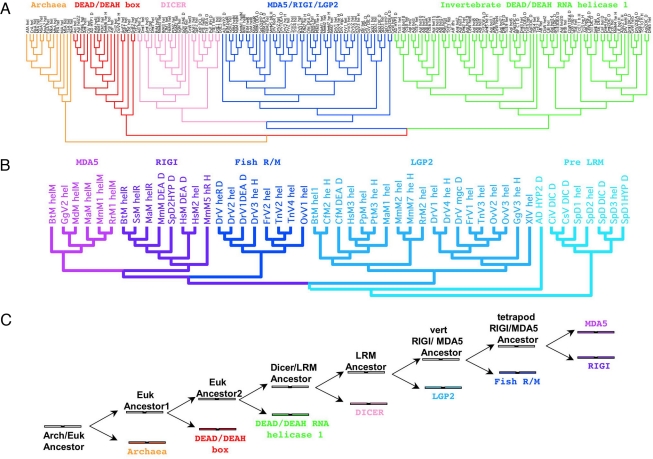
MDA-5 and RIG-I proteins are each other's closest relatives, so the MDA-5 and RIG-I helicase/DEAD domain linkage most likely arose as a result of a duplication event (Fig. 2). These proteins are equally related to the LGP2 helicase family and the best explanation for this arrangement is also a duplication event. The next closest helicase family is the DICER group. Because LGP2, MDA-5, and RIG-I are found only in vertebrates and DICER proteins are broadly distributed phylogenetically, we propose that the duplication event of MDA-5 and RIG-I occurred in the common ancestor of vertebrates. Fig. 2 C shows a cartoon demonstrating the best-supported route for the origin of the helicase/DEAD structure of MDA-5 and RIG-I deduced from the phylogenetic analyses in Fig. 2.
Fig. 2.
Relationships of the helicase-DEAD/DEAH domains. (A) Phylogenetic tree showing the relationships of helicases. See text for details. Accession numbers for all sequences used in this tree are in Table S1. See Fig. S2 for a magnification of this tree to observe the gene names. (B) A magnification of the blue clade from (A). This is the clade circumscribing the MDA-5 and RIG-I helicase domain families. Accession numbers for all sequences used in this tree are in Table S1. (C) Diagram showing the best-supported pathway for generating the helicase/DEAD domain structure in MDA-5 and RIG-I. The colors refer to colors of clades in A. LRM stands for LGP2/RIG-I/MDA-5. Eukaryotic ancestor1 indicates an ancestor where a duplication produced the helicase/DEAD/DEAH structure and a common ancestor of all other helicases. Eukaryotic ancestor2 indicates an ancestor where a duplication occurred that produced the DEAD RNA helicases and all other helicases. Solid black lines indicate duplication events. Colors of major hel-DEAD families correspond to color labels of genes in the trees in A.
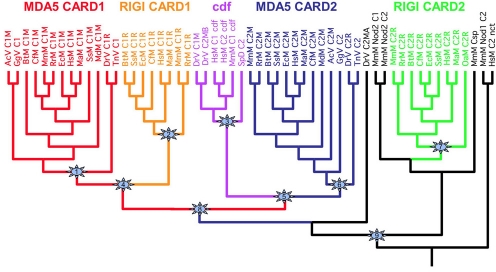
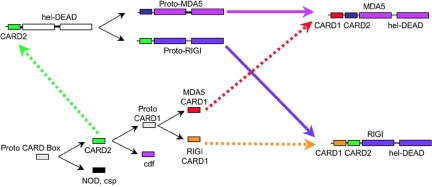
Phylogenetic analysis of the CARD domains is shown in Fig. 3 and Table 2, and is summarized in Fig. 4. The phylogenetic results indicate that the CARD2 boxes were the first N-terminal elements to be grafted to the helicase/DEAD domains. In addition, it appears that the RIG-I CARD2 domain was grafted first, and the MDA-5 CARD2 domain was produced by a duplication event of the grafted RIG-I CARD2 domain. Next, the MDA-5 CARD1 domain was grafted to the CARD2-helicase-DEAD domains. Because all of these domains exist only in vertebrates, we propose that these CARD duplications all occurred in the common ancestor of the vertebrates. Note that MDA-5 proteins are found in fish, but RIG-I proteins are not. We searched the existing fish databases very aggressively and did not find RIG-I genes in any of the available fish genomes. This result implies that the common ancestor of fish either lost the RIG-I gene or the CARD1 domain, for RIG-I did not fuse with the CARD2-helicase-DEAD domain structure. Based on our analysis, the timing of this latter event would be in the common ancestor of tetrapods.
Fig. 3.
Phylogenetic tree showing the relationships of the CARD boxes examined in this study. Accession numbers of genes used to generate this phylogeny are available in Table S2. The numbered stars refer to nodes in the tree where jackknife and Bayes probabilities were calculated and are summarized in Table 2.
Table 2.
Support values for Fig. 3
| Node | Group | Gonn | GI | Bayes1 | Bayes2 |
|---|---|---|---|---|---|
| 1 | MC1 | 100 | 100 | 100 | 100 |
| 2 | RC1 | 100 | 100 | 100 | 100 |
| 3 | Cdf | 100 | 99 | 100 | 100 |
| 4 | RC1/MC1 | 100 | 89 | 69 | 100 |
| 5 | RC1/MC1/cdf | 99 | 85 | —* | —* |
| 6 | MC2 | 99 | 100 | 100 | 100 |
| 7 | RC2 | 100 | 100 | 100 | 100 |
| 8 | RC1/MC1/cdf/MC2 | 95 | 96 | 95 | 100 |
| 9 | Exclude node | 93 | 51 | 95 | 100 |
Numbers in the first column refer to the nodes as numbered in Fig. 3. Accession numbers for all sequences used in this tree are in Table S1. Abbreviations: cdf, CARDif box; GI, Genetic Identity transformation weighting; Gonn, Gonnett transformation weighting; MC1, MDA-5 CARD1 box; MC2, MDA-5 CARD2 box; RC1, RIG-I CARD1 box; RC2, RIG-I CARD2 box.
*CARDif boxes associated with MDA-5-CARD2 boxes
Fig. 4.
Diagram showing the best-supported pathway for generating the CARD1 and CARD2 domains and ultimately the MDA-5 and RIG-I proteins. Abbreviations: csp, caspase; cdf, CARDif. Solid black lines indicate duplication events. Dotted lines indicate fusion events that we infer from the CARD phylogenetic analysis in Fig. 3. Colors correspond to domain family labels in Fig. 3. Alternative scenarios with fewer fusion events are less likely to represent the evolutionary history of MDA-5 and RIG-I, as explained in Fig. 5.
The Evolutionary Path to Innate Immunity: A Continuing Process of Domain Duplication and Grafting.
Immunity against prokaryotes and virus infections evolved very early in animal evolution. Toll, Toll-like receptors, and Toll-IL-1 receptor proteins that recognize pathogen-associated molecular patterns and provide protection from them have been identified in organisms in the phyla Porifera and Cnidaria (50). In addition, the prototypic complement-effector pathway, consisting of C3 and membrane-attack complex-perforin proteins, has also been identified in Cnidarians. The evolution of type I IFNs occurred at a much later time and type I IFNs are identified only in vertebrates (51). Knocking out type I IFN signaling by targeted deletion of type I IFN-receptor genes augments sensitivity of mice to even minute levels of most viral infections, indicating that type I IFN confers primary innate antiviral immunity (8). The observation that type I IFNs evolved only in vertebrates, as well as our current findings that MDA-5 and RIG-I homologs and orthologs are also detected only in vertebrates, indicate that vertebrates may have acquired additional weapons to combat invading pathogens. Because RNA viruses are responsible for the majority of severe and lethal diseases, there may have been intense selection pressure for an additional pathway, consisting of MDA-5 and RIG-I, which would sense invading RNA viruses, trigger type I IFN production, and provide immunity. This argument is supported by our observation that MDA-5 and RIG-I CARD domain orthologs are found mainly in mammals and marsupials, indicating a much later time-scale of evolutionary origin. Studies with RIG-I and mda-5 knock-out mice demonstrate that RIG-I recognizes paramyxoviruses, influenza virus, vesicular stomatitis virus, and Japanese encephalitis virus, whereas MDA-5 recognizes picornaviruses (25–27, 52, 53). MDA-5 but not RIG-I orthologs are identified in fish, the first chordates to evolve, indicating that MDA-5 might precede RIG-I in evolutionary history. The sequential evolution of MDA-5 and RIG-I might reflect temporal exposure of organisms to different viruses, as exemplified by differential RNA virus recognition properties of MDA-5 and RIG-I.
A Circuitous Pathway to Innate Immunity.
The evolution of MDA-5 and RIG-I demonstrate an intriguing and circuitous pathway that is consistent with intense selection pressure for the existence and maintenance of these genes. The easiest way to construct these proteins would have been to make one and then duplicate the entire assemblage (Fig. 5A). However, according to the phylogenetic analyses, there was independent fusion of CARD2 to the RIG-I or MDA-5 helicase/DEAD domains and then another independent fusion of CARD1 to the CARD2/helicase/DEAD domain, ruling out simple duplications as the route to the eventual structure of the RIG-I and MDA-5 proteins (see Fig. 5 A). Another scenario that could potentially explain the observed structure of these two proteins is explored in Fig. 5 B. While the scenario presented in this figure is not an exhaustive presentation of all alternatives, it is the shortest alternative with respect to number of fusions and duplications we can propose, given the phylogenetic evidence. This alternative is less preferred because it hypothesizes four fusion events and three duplication events and is less parsimonious than the scenario we present in Fig. 5 (three fusions and two duplications). We suggest, from these data, that the MDA-5 and RIG-I domain structures are a case where the most parsimonious evolutionary path has not been taken, because the scenario the trees support have nearly twice as many steps in them than required to perform simple duplication events.
Fig. 5.
Cartoon showing two alternative pathways for generating the CARD1 and CARD2 domains and ultimately the MDA-5 and RIG-I proteins. (A) Dotted lines indicate fusion events that we infer from the CARD phylogenetic analysis in Fig. 3. Colors correspond to domain family labels in Fig. 4. This scenario suggests four fusion events, one each for the MDA-5 CARD1 and CARD2 boxes and the RIG-I CARD1 and CARD2 boxes. This scenario is less preferred because it infers one extra fusion event that cannot be supported by the phylogeny in Fig. 3. (B) This scenario suggests that MDA-5 and RIG-I CARD2 boxes have coevolved tightly after their fusion and before the duplication of MDA-5 and RIG-I whole genes. This scenario is less preferred because it suggests tight coevolution of CARD1 and CARD2 domains, which are clearly refuted by Table 1. Abbreviations: cdf, CARDif; csp, caspase. Solid black lines indicate duplication events.
LGP2, the dominant negative inhibitor of MDA-5 and RIG-I, contains no CARD domain, so it preceded both MDA-5 and RIG-I in evolution. In this regard, it is useful to examine the genetic ramifications of deleting LGP2. LGP2−/− mice demonstrate highly elevated type I IFN induction upon poly (I:C) stimulation, and LGP2−/− mouse embryonic fibroblasts are more resistant to vesicular stomatitis virus infection, supporting the hypothesis that the function of LGP2 is to inhibit IFN induction upon viral infection (41). RNA interference (RNAi) technology, which produces dsRNAs, works efficiently in nonchordates but not in vertebrates, owing to nonspecific activation of the IFN pathway (54). The absence of MDA-5 and RIG-I and the presence of LGP2 and DICER in nonchordates might explain why these organisms can employ RNAi to combat viral infection.
Methods
Phylogenetic Matrix.
MDA-5 and RIG-I orthologs and paralogs were obtained by BLAST searches of the following fully sequenced genomes: from mammals, Homo sapiens, Pan troglodytes, Macaca macaque (all three primates), Mus musculus, Canis familiaris, and Bos Taurus; from Aves, Gallus gallus; from Osteichthyes, Tetraodon nigrovidis, Fugu rubripes, and Danio rerio; from Urochordata, Ciona intestinallis; and from Echinodermata, Strongylocentrotus puprturatus (Tables S1 and S2). Searches were also made in several insect databases and fungal databases, with no successful hits produced. Table S1 shows all of the MDA-5 and RIG-I genes we obtained from the database. In addition, we searched the unfinished mammalian genome databases and obtained several more orthologues for these two genes. However, with the exception of the marsupial Monodelphus, we did not obtain both gene categories. Therefore, we included only sequences from species with finished genomes and from Monodelphus. We were only able to find orthologs of MDA-5 and RIG-I in vertebrates, and specifically in mammals and marsupials, and a matrix was constructed with these domains (see Table S1). Helicase and DEAD/DEAH box domain orthologues and paralogues were obtained in a similar manner from all of the fully sequenced genomes. The helicase/DEAD/DEAH data set was trimmed to exclude multiple orthologues from several species. In addition, we searched the unfinished mammalian genomes in the database and obtained several more orthologues for these two genes. Table S2 shows all of the helicase and DEAD/DEAH proteins we found in the database.
Alignment and Phylogenetic Analysis.
To explore the alignment space for these genes we constructed matrices using the elision method with gap scores of 0.1, 0.5, 1.0, 2.0, 5.0, and 10.0 (55). Alignments were performed using MAFFT (multiple alignment using fast Fourier transform) (56, 57) for each of these gap scores and then concatenated into a single matrix. Phylogenetic trees were constructed using parsimony (58) and Bayesian (59) approaches. We used jackknife support measures to assess robustness in our parsimony analyses, and in addition we performed analyses with two character weighting schemes: Gonnett and genetic identity. For the Bayesian approach we performed two analyses: Bayes1, where we set the model parameter to parsimony and used 1.5 million generations, and Bayes2, where we set the model to JTT+Gamma using alpha (gamma-shape parameter) of 3.341776 with 1.5 million generations. For the CARD origin phylogeny we used NOD domains to root the phylogeny, and for the helicase/DEAD origin phylogeny we used the archaeal helicase domains as roots. The data matrices used in the phylogenetic analyses are provided as NEXUS files in Table S3 (hel-DEAD analyses) and Table S4 (CARD analyses).
Incongruence Tests.
To assess whether the four protein domains relevant to this study (CARD1, CARD2, helicase, and DEAD/DEAH) are evolving in concert, we used the ILD test (60). The ILD test was developed to test the congruence of two partitions of data relevant to the same taxa. The ILD index measures the amount of disagreement between trees generated from two different partitions (61). The ILD index can be used in a test that compares an observed ILD index with a null distribution of ILD indices generated by random permutation. Significant departure from the null distribution indicates strong congruence and, hence, correlated evolutionary histories.
Supplementary Material
Acknowledgments.
R.D. thanks the Lewis and Dorothy Cullman Program in Systematic Biology and the Sackler Institute for Comparative Genomics, both at the American Museum of Natural History. The present study was supported by National Institutes of Health Grant GM068448. D.S. is the Harrison Endowed Scholar in Cancer Research at the Massey Cancer Center. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research at the Massey Cancer Center and is a Samuel Waxman Cancer Research Foundation investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804956105/DCSupplemental.
References
- 1.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang SY, et al. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol Rev. 2007;220:225–236. doi: 10.1111/j.1600-065X.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 4.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202(1):8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 5.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 6.Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 7.Lydon NB, et al. Immunochemical mapping of alpha-2 interferon. Biochemistry. 1985;24:4131–4141. doi: 10.1021/bi00336a048. [DOI] [PubMed] [Google Scholar]
- 8.Hwang SY, et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher PB, Grant S. Effects of interferon on differentiation of normal and tumor cells. Pharmacol Ther. 1985;27:143–166. doi: 10.1016/0163-7258(85)90067-1. [DOI] [PubMed] [Google Scholar]
- 10.Wathelet MG, et al. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 11.Lenardo MJ, Fan CM, Maniatis T, Baltimore D. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 12.Du W, Maniatis T. An ATF/CREB binding site is required for virus induction of the human interferon beta gene. Proc Natl Acad Sci USA. 1992;89:2150–2154. doi: 10.1073/pnas.89.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Ann Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 14.Sing A, et al. Bacterial induction of beta interferon in mice is a function of the lipopolysaccharide component. Infect Immun. 2000;68:1600–1607. doi: 10.1128/iai.68.3.1600-1607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 16.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 17.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 18.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 19.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 20.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 22.Diebold SS, et al. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 24.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 25.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23(1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 27.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang DC, et al. Mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su ZZ, Sarkar D, Emdad L, Barral PM, Fisher PB. Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J Cell Physiol. 2007;213:502–510. doi: 10.1002/jcp.21128. [DOI] [PubMed] [Google Scholar]
- 30.Lasko P. The Drosophila melanogaster genome: translation factors and RNA binding proteins. J Cell Biol. 2000;150:F51–F56. doi: 10.1083/jcb.150.2.f51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdelhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 32.Heung LJ, Del Poeta M. Unlocking the DEAD-box: a key to cryptococcal virulence? J Clin Invest. 2005;115:593–595. doi: 10.1172/JCI200524508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 35.Matsui K, et al. Cutting edge: Role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J Immunol. 2006;177:5785–5789. doi: 10.4049/jimmunol.177.9.5785. [DOI] [PubMed] [Google Scholar]
- 36.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 37.Oganesyan G, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 38.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 41.Venkataraman T, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 42.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Damiano JS, Reed JC. CARD proteins as therapeutic targets in cancer. Curr Drug Targets. 2004;5:367–374. doi: 10.2174/1389450043345470. [DOI] [PubMed] [Google Scholar]
- 44.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc Natl Acad Sci USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kersse K, Vanden Berghe T, Lamkanfi M, Vandenabeele P. A phylogenetic and functional overview of inflammatory caspases and caspase-1-related CARD-only proteins. Biochem Soc Trans. 2007;35:1508–1511. doi: 10.1042/BST0351508. [DOI] [PubMed] [Google Scholar]
- 46.Inohara N, Chamaillard C, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 47.Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- 48.Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G. Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol. 1999;16:826–838. doi: 10.1093/oxfordjournals.molbev.a026167. [DOI] [PubMed] [Google Scholar]
- 49.Leszczyniecka M, DeSalle R, Kang DC, Fisher PB. The origin of polynucleotide phosphorylase domains. Mol Phylogenet Evol. 2004;31(1):123–130. doi: 10.1016/j.ympev.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Hemmrich G, Miller DJ, Bosch TC. The evolution of immunity: a low-life perspective. Trends Immunol. 2007;28:449–454. doi: 10.1016/j.it.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Krause CD, Pestka S. Evolution of the Class 2 cytokines and receptors, and discovery of new friends and relatives. Pharmacol Ther. 2005;106:299–346. doi: 10.1016/j.pharmthera.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Barral PM, et al. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81:3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 55.Wheeler WC, Gatesy J, DeSalle R. Elision: a method for accommodating multiple molecular sequence alignments with alignment-ambiguous sites. Mol Phylogenet Evol. 1995;4(1):1–9. doi: 10.1006/mpev.1995.1001. [DOI] [PubMed] [Google Scholar]
- 56.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 58.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, Massachusetts: Sinauer Associates; 2000. [Google Scholar]
- 59.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 60.Farris JS, Kallersjo M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44:570–572. [Google Scholar]
- 61.Mickevich MF, Johnson MS. Congruence between morphological and allozyme data in evolutionary inference and character evolution. Syst Zool. 1976;25:260–270. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.