Abstract
Cardiac troponin C (cTnC) is the calcium-dependent switch for contraction in heart muscle and a potential target for drugs in the therapy of congestive heart failure. This calmodulin-like protein consists of two lobes connected by a central linker; each lobe contains two EF-hand domains. The regulatory N-terminal lobe of cTnC, unlike that of skeletal troponin C (sTnC), contains only one functional EF-hand and does not open fully upon the binding of Ca2+. We have determined the crystal structure of cTnC, with three bound Ca2+ ions, complexed with the calcium-sensitizer bepridil, to 2.15-Å resolution. In contrast to apo- and 3Ca2+-cTnC, the drug-bound complex displays a fully open N-terminal lobe similar to the N-terminal lobes of 4Ca2+-sTnC and cTnC bound to a C-terminal fragment of cardiac troponin I (residues 147–163). The closing of the lobe is sterically hindered by one of the three bound bepridils. Our results provide a structural basis for the Ca2+-sensitizing effect of bepridil and reveal the details of a distinctive two-stage mechanism for Ca2+ regulation by troponin C in cardiac muscle.
Contraction in skeletal and cardiac muscles is triggered by the binding of Ca2+ to the troponin C (TnC) component of the troponin complex (1, 2). TnC, like other members of the calmodulin (CaM) superfamily, consists of two lobes joined by a flexible central linker (see Fig. 1A legend). The C-terminal or “structural” lobe (C-lobe) comprises two EF-hand motifs (domains III and IV) that bind either Ca2+ or Mg2+. The N-terminal or “regulatory” lobe (N-lobe) also contains two EF-hands (domains I and II) and in skeletal TnC (sTnC), both domains of the N-lobe bind Ca2+ specifically. By contrast, in the N-lobe of cardiac TnC (cTnC), only domain II binds Ca2+. Domain I is nonfunctional because of the mutation of Ca2+ ligands (D29L, D31A) and a V28 insertion. The other two subunits of troponin are troponin I (TnI), which binds to F-actin and inhibits actomyosin ATPase, and troponin T (TnT), which links TnC and TnI to tropomyosin. In the relaxed state of muscle, the interaction between myosin and F-actin is blocked by tropomyosin, whose position on F-actin is determined by its linkage with troponin. The binding of Ca2+ to the N-lobe of TnC produces a structural change so that TnC interacts strongly with TnI. As a result TnI is released from actin, and tropomyosin moves from its blocking position on F-actin. Myosin is then able to interact with F-actin and start its catalytic cycle (3, 4).
Figure 1.
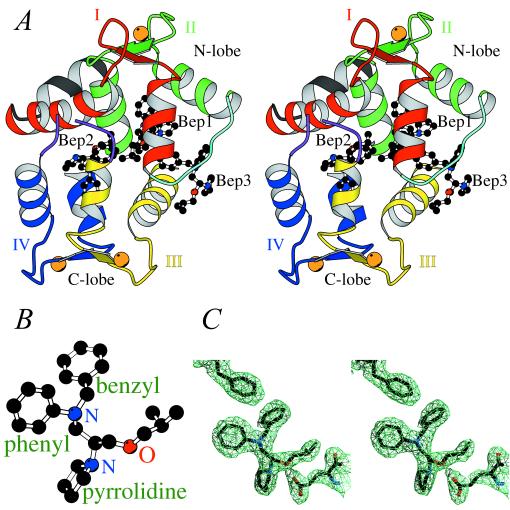
The overall structure of bepridil-3Ca2+-cTnC. (A) Stereo view of bepridil-3Ca2+-cTnC. cTnC comprises an N- and a C-terminal lobe. Each lobe consists of two helix–loop–helix EF-hand domains connected by a linker. The nomenclature is as follows: domain I (red) starts with helix A, followed by loop I, then helix B followed by the N-terminal linker (cyan); domain II (green) starts with helix C, followed by loop II, then helix D followed by the central linker (not included in the current structure because of disorder); domain III (yellow) starts with helix E, followed by loop III, then helix F followed by the C-terminal linker (purple and discontinuous at residues 125–126 because of disorder); domain IV (blue) starts with helix G, followed by loop IV, then helix H. There is also a helix N (black) N-terminal to domain I, which is present in TnC but not in CaM. There are two small antiparallel β-sheets in cTnC: one between loops I and II, and the other between loops III and IV. Three Ca2+ ions, bound to domains II, III, and IV, are shown as orange spheres. Three bepridil molecules (balls and sticks) are bound to cTnC: Bep1 mainly to the N-lobe, Bep2 mainly to the C-lobe, and Bep3 to both lobes. (B) Ball-and-stick representation of the calcium-sensitizing agent bepridil. (C) Electron density around Phe27 (upper left), Bep1 (center), and Glu96 (lower right) in a (2|Fo| − |Fc|) map contoured at 1.0σ. The positively charged pyrrolidine ring of Bep1 forms a salt bridge with the side chain of Glu96.
The binding of Ca2+ has strikingly different structural consequences in the regulatory N-lobes of cTnC and sTnC. In sTnC, the N-lobe switches from a “closed” (5, 6) to an “open” (7–9) conformation upon the binding of 2 Ca2+ ions. Consequently, a hydrophobic pocket is exposed as the binding site for skeletal TnI (sTnI). This Ca2+-dependent structural transition was proposed by Herzberg, Moult, and James (ref. 10, the so-called “HMJ model”) and later confirmed by the crystal and NMR structures of sTnC (7–9). By contrast, NMR studies have shown that the N-lobe of cTnC opens only partially upon the binding of a Ca2+ ion to the single regulatory site (domain II) (refs. 11 and 12; see Fig. 2 A and B). (Although the conformation of the N-lobe of 3Ca2+-cTnC has been described as “closed” in ref. 11, we prefer the term “partially open,” to distinguish this structure from the fully closed conformation of the N-lobe of 2Ca2+-cTnC). Moreover, Li et al. (13) have recently shown that a C-terminal fragment of cardiac TnI (cTnI, residues 147–163) induces the fully open conformation in the N-lobe in which a single Ca2+ ion is bound (see Fig. 2 B and C). Thus TnC-based Ca2+ regulation appears to have distinctive mechanisms in skeletal and cardiac muscles. Although the structures of sTnC, in both open and closed states, have been determined to high resolution by x-ray crystallography, no crystal structure of cTnC has previously been reported.
Figure 2.
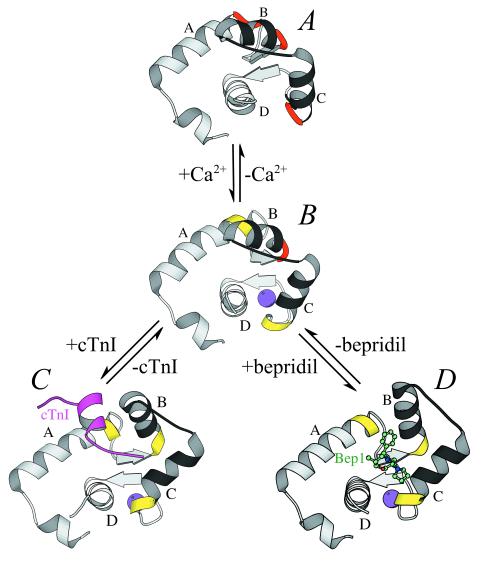
An overview of the stepwise mechanism for the opening of the N-lobe of cTnC. The N-lobes of Apo- or 2Ca2+-cTnC (12) (A), 3Ca2+-cTnC (11) (B), 3Ca2+-cTnC-cTnI147–163 (13) (C), and bepridil-3Ca2+-cTnC (D) are shown as ribbons. The A–D and B–C units are colored in gray and in black, respectively. Ca2+, bepridil, and cTnI147–163 are shown as spheres, balls-and-sticks, and ribbons, respectively. The “hinge” regions (residues 27–28, 37–40, and 64–65) are colored in red when in a nonhelical conformation and in yellow when in a helical conformation.
The essential role of cTnC in the regulation of contraction suggests that this protein is a logical target for drugs in the therapy of congestive heart failure. “Calcium-sensitizers” are a newly described class of inotropic drugs that enhance the responsiveness of the cardiac myofibril to calcium (14). The effect of these agents on short- or long-term treatment of heart failure is uncertain. In fact, no agent has yet been developed that has high specificity as a calcium sensitizer of the thin filament. Several CaM antagonists bind directly to cTnC and are thus good model compounds for the design of calcium sensitizers (15). For example, although bepridil (see Fig. 1B) is not used clinically for congestive heart failure, this drug has been shown to increase both the Ca2+ affinity of cTnC and the activity of actomyosin ATPase in vitro (16, 17).
We have determined the crystal structure of 3Ca2+-cTnC complexed with bepridil to 2.15-Å resolution. Our results indicate that the calcium-sensitizing effect of bepridil arises from the stabilization of the fully open N-lobe by this drug. The current structure also reveals how the special properties of cTnC lead to a distinctive, stepwise mechanism for the opening of its N-lobe.
Materials and Methods
Crystallization and Data Collection.
The A-Cys (C35S, C84S) mutant (18) of chicken cTnC studied here has the same functional properties as the wild-type protein. Orthorhombic (P212121) crystals with unit cell dimensions a = 36.27 Å, b = 62.04 Å, c = 62.04 Å were grown by vapor diffusion at 4°C from a mixture of 4 μl of protein solution (1.2 mM cTnC/6 mM CaCl2/2.5 mM bepridil⋅HCl/10 mM Hepes, pH 7.2) and 4 μl of reservoir buffer (30% PEG 3350/20 mM MgCl2/50 mM Hepes, pH 7.2). The partial specific volume is 1.9 Å3/Da, with one molecule per asymmetric unit. Data were collected at 95 K to 2.15-Å resolution by using synchrotron radiation at the Cornell High Energy Synchrotron Source (CHESS; beamline A1, λ = 0.935 Å). Data were reduced with the programs denzo and scalepack (19). (See Table 1 for data collection statistics.)
Table 1.
Data collection and refinement statistics
| Measurement | Value |
|---|---|
| Data collection | |
| Resolution, Å | 2.15 |
| Measured reflections | 54,641 |
| Unique reflections | 7,998 |
| Completeness, % | 99.7 (99.2)* |
| Rmerge† | 0.066 (0.126)* |
| Refinement | |
| Resolution range, Å | 30.0–2.15 |
| No. of reflections | 7,829 |
| Sigma cutoff | None |
| Rcryst‡ | 0.213 |
| Rfree§ | 0.276 |
| No. of protein atoms | 1,185 |
| No. of bepridil atoms | 81 |
| No. of Ca2+ atoms | 3 |
| No. of water molecules | 136 |
| rms deviation bond lengths, Å | 0.017 |
| rms deviation angles, ° | 1.48 |
Values for the highest resolution bin (2.23–2.15 Å) are shown in parentheses.
†Rmerge = Σhkl|I − 〈I〉|/ΣIhkl.
‡ Rcryst = Σhkl||Fobs| − |Fcalc||/Σhkl|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
§ Rfree is the same as Rcryst except that the summation is over 5% of the reflections, which were randomly selected and excluded from the refinement (21).
Structure Determination.
A molecular replacement solution was obtained with the program AMoRe (20), using the C-lobe of rabbit sTnC (8) and the 2Ca2+-bound N-lobe of avian sTnC (9) as independent search models, with data from 8.0- to 3.5-Å resolution. The final correlation coefficient was 38.8 (R factor 0.473). Positional refinement using the program xplor (21) (R factor 0.377, with data to 2.15 Å) yielded electron density maps (2|Fobs| − |Fcalc| and |Fobs| − |Fcalc|) which allowed manual rebuilding of the protein model with the program o (22). Two bepridil molecules were also modeled into the electron density. Subsequent maximum likelihood refinement and simulated annealing with the program cns_solve (23) aided in the modeling of ordered waters and a third bepridil. Residues 1–4, 87–91, 125–126, and 161 are disordered and were not included in the final structure. Main-chain dihedral angles of all non-glycine residues lie in allowed regions of the Ramachandran plot, with 92.4% in most-favored regions.
Results and Discussion
Overall Description of the Structure.
Bepridil-3Ca2+-cTnC displays a globular (46 × 38 × 35 Å) shape resembling that of CaM–trifluoperazine (TFP) and CaM–peptide complexes (Fig. 1A) (24–26). The central helical linker is unwound at residues 87–91, producing a 108° bend that brings the lobes close together. The two lobes are related by pseudo-twofold symmetry so that their hydrophobic pockets merge to form a channel largely occupied by two bepridil molecules (Bep1 and Bep2). A third bepridil (Bep3) is located at one end of the hydrophobic channel, near the N-terminal linker and helices B, C, and E, and partially blocks solvent access to the interior of the molecule. The structure contains 3 Ca2+ ions, bound to domains II, III, and IV. Despite the absence of Ca2+ in domain I, the N-lobe of the current structure reveals a strong similarity to the 2 Ca2+-bound, fully open N-lobe of chicken sTnC (9), with an rms deviation of 0.621 Å for backbone overlay. The C-terminal lobe of bepridil-3Ca2+-cTnC is similar to that of rabbit 4Ca2+-sTnC (ref. 8, Protein Data Bank entry 1tn4) with an rms deviation of 0.608 Å.
Bepridil-Induced Changes in the N-Lobe.
The most important effect of bepridil on 3Ca2+-cTnC is the full opening of the N-lobe. No crystal structure is presently available for 3Ca2+-cTnC, so that our analysis relies on the comparison between the current structure and the NMR structure of the N-lobe of chicken 3Ca2+-cTnC (11). In that study, the N-lobe hydrophobic pocket of 3Ca2+-cTnC (without bepridil) is largely unexposed, in contrast to that of 4Ca2+-sTnC. In the current study, upon the binding of bepridil, the A–B and C–D interhelical angles decrease from 138° and 108° to 92° and 89°, respectively. The N-lobe opens as helices B and C move as a pair away from helices A and D (Fig. 2 B and D) so that this bepridil-induced transition in cTnC resembles the Ca2+-induced transition in the N-lobe of sTnC (5–9).
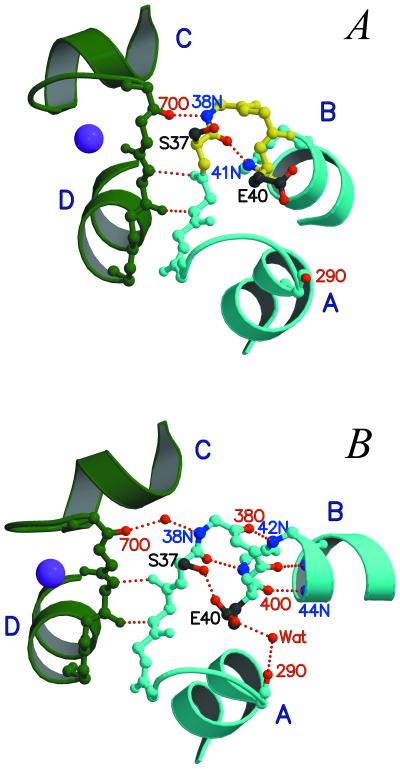
As in sTnC (7–9), the opening of the N-lobe in cTnC is accompanied by a backbone rearrangement in helix B, although domain I of cTnC is nonfunctional (Fig. 3). In the solution study of the partially open N-lobe of 3Ca2+-cTnC (11), helix B displays a kink in its N-terminal region (residues 37–40) (Fig. 3A). By contrast, in the current structure, residues 37–40 adopt a helical conformation, and the C terminus of helix B is swung away from helix A (Fig. 3B). Several changes lead to the straightening of helix B upon the binding of bepridil. In the small β-sheet between domains I and II, the Thr38N-Gly70O backbone hydrogen bond is broken. The carbonyl oxygens of Thr38, Lys39, and Glu40 then form three hydrogen bonds with the amino groups of Gly42, Lys43, and Val44, respectively, and a carboxylate oxygen of Glu40 also forms a short hydrogen bond with the hydroxyl group of Ser37 (Oγ–Oɛ distance = 2.35 Å), providing stabilization to the newly formed helical region. As shown in the NMR study by Gagné et al. (27), in sTnC helix B is straightened only when the side chain of a glutamate (Glu41) in loop I moves toward a Ca2+ ion as a bidentate ligand. Thus it appears that in cTnC, the side-chain interaction between Glu40 and Ser37 plays a role related to that of Ca2+ ligation by Glu41 in sTnC in the opening of the N-lobe.
Figure 3.
A detailed view of the bepridil-induced opening of the N-lobe. (A) In 3Ca2+-cTnC (11), helix B displays a kink due to the nonhelical torsion angles of residues 37–40 (yellow). (B) The binding of bepridil results in a rearrangement of the hydrogen bonding pattern in the small β-sheet between loops I and II and in the N-terminal region of helix B. Consequently, helix B is extended at the N terminus to include residues 37–40 (cyan), and its C terminus is swung away from helix A. Note that the breaking of the 38N⋅⋅⋅⋅ 70O hydrogen bond also leads to a small decrease in the C–D interhelical angle. Bep1 is not shown.
The two N-lobe domains of cTnC, like those of sTnC (8, 9), are structurally coupled through the small β-sheet. The breaking of the 38N⋅⋅⋅⋅ 70O hydrogen bond in the β-sheet not only contributes to the straightening of helix B but also releases the conformational constraints on Gly70 and Thr71 in the middle of Ca2+-binding loop II. Changes in the φ–ψ angles of these two residues then allow helix C to move in a concerted fashion with helix B. As a result, the C–D interhelical angle in bepridil-3Ca2+-cTnC is smaller than that in 3Ca2+-cTnC.
Binding Sites for Bepridil.
Bep1 plays the key role in the stabilization of the open N-lobe. Its phenyl group is located at the bottom of the N-lobe pocket in a small hydrophobic “cavity” (see Table 2) that seems to be the primary anchoring site for this drug molecule. Similar cavities have been found in CaM and accommodate certain bulky groups of several drugs (e.g., the trifluoromethyl group of TFP) (24, 25, 28). The benzyl group of Bep1 protrudes toward the N-terminal linker and has extensive van der Waals contacts with helices B and C. The two aromatic rings of Bep1 are inserted as a “wedge” between the A-D and B-C units, sterically hindering the closing of the N-lobe (Fig. 2D). Overall, Bep1 interacts with 13 hydrophobic residues in the N-lobe (Table 2) and shields 96 Å2 of N-lobe apolar surface from the solvent (calculated with Insight '95, Molecular Simulations, Waltham, MA).
Table 2.
Residues involved in contacts (<4.5 Å) with bepridil
| Molecule | Region | Residues |
|---|---|---|
| Bep1 | N-lobe | F27, I36, L41, V44, M45, L48, M60, I61, V64, V72, F77, M80, M81, S84 |
| C-lobe | E96, L97, L100 | |
| Bep2 | N-lobe | F20, A23, I26, F27, F77, M81 |
| C-lobe | L100, F104, I112, L117, M120, L136, F153, F156, M157 | |
| Bep3 | N-lobe | M45, M50, P52, E56, M60 |
| C-lobe | E96, D99, L100, R102, M103 |
Residues whose side chains form the hydrophobic “cavity” at the bottom of each lobe are underlined. Residues that form salt bridges with the pyrrolidine rings of the bepridils are in boldface.
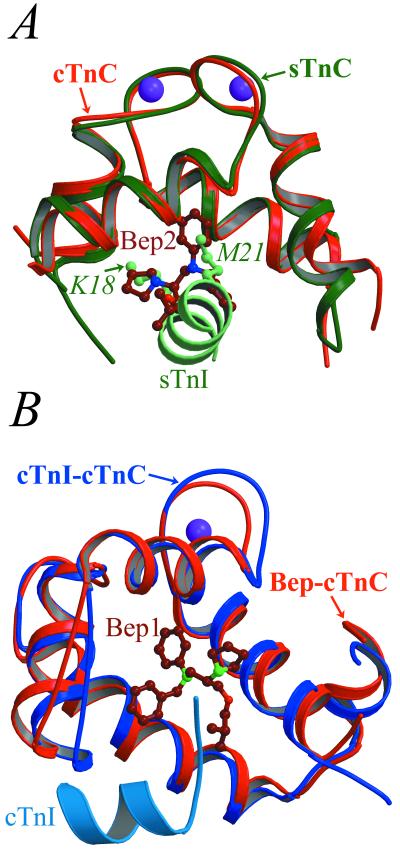
Bep2 and Bep3 appear to mimic the binding of TnI to the C-lobe. An overlay of the current structure and that of rabbit sTnC bound to the sTnI1–47 fragment (29) shows that the phenyl ring of Bep2 takes the place of the side chain of Met21 of sTnI (equivalent to Met53 of cTnI) in the small hydrophobic cavity at the bottom of the C-lobe (see Table 2 and Fig. 4A). Correspondingly, the binding of Bep2 results in a bend in helix F at Ile119, which is not observed in 3Ca2+-cTnC. Consequently, Met120 in the current structure (equivalent to Ile121 of rabbit sTnC; see ref. 29 for numbering scheme) occupies the position of Phe122 of sTnC and has contacts with both the phenyl and benzyl groups of Bep2. Bep3 makes contacts with several residues on helix E (Table 2), which have also been shown to interact with TnI (29). In fact, both bepridil and cTnI induce backbone conformational changes in the C terminus of helix F and the F-G linker (30). Because Bep2 and Bep3 occupy the same region of the C-lobe as TnI, it appears that these two molecules probably would not bind to cTnC in the troponin complex. By contrast, Bep1 is not likely to be displaced by TnI (see below).
Figure 4.
Comparison of the binding of bepridil and TnI to cTnC. (A) Backbone overlay of the C-lobes of bepridil-3Ca2+-cTnC (red) and 2Ca2+-sTnC-sTnI1–47 (green) (29) shows that Bep2 (dark red) mimics the binding of TnI (light green) to the C-lobe. Most notably, the pyrrolidine and phenyl groups of Bep2 occupy the same sites as the side chains of Lys18 and Met21 of sTnI (light green balls and sticks), respectively. (B) By contrast, Bep1 (dark red) and cTnI (sky blue) bind to different regions of the N-lobe pocket, as shown in the overlay of the N-lobes of bepridil-3Ca2+-cTnC (red) and 3Ca2+-cTnC-cTnI147–163 (blue) (13).
The compact shape of the current structure seems to be a direct result of drug binding. The positively charged pyrrolidine groups of the bepridils bring the N- and C-lobes close together by neutralizing the negative charges at their interface. Moreover, bepridil bridges the lobes through two ionic pairs: one between Bep1 pyrrolidine and Glu96 (Fig. 1C) and the other between Bep3 pyrrolidine and Asp99. A similar effect on CaM has been observed for the inhibitor TFP, whose positively charged piperazine ring stabilizes a compact conformation in CaM (24, 25). It has also been suggested that CaM cannot bind to its target proteins in this compact form (24). By contrast, cTnC, when bound to the N-terminal region of cTnI (residues 33–80), adopts a more extended conformation than that of bepridil-3Ca2+-cTnC and CaM-TFP (31). Correspondingly, as noted above, bepridil appears to activate rather than inhibit cTnC.
Stepwise Mechanism for the Opening of the N-Lobe of cTnC.
Previous NMR studies have shown that the 2Ca2+, 3Ca2+, and cTnI147–163-bound states of cTnC assume three distinct conformations (11–13). A comparison of these results with the current structure and with the crystal structures of sTnC (5, 6, 8, 9) reveals the details of a stepwise mechanism for the opening of the N-lobe of cTnC. In sTnC, the interhelical angles of domains I and II, which define the “openness” of the N-lobe, depend on the conformation of three groups of “hinge residues”: Phe29-Asp30 at the C terminus of helix A, Ser38–Glu41 at the N terminus of helix B, and Val65-Asp66 at the C terminus of helix C (8). In the 2Ca2+ or closed state of sTnC, the hinge residues display nonhelical φ-ψ angles. Two critical changes lead to the opening of the N-lobe upon the binding of Ca2+ (see refs. 8 and 9 for the detailed mechanism in sTnC): in the small β-sheet between domains I and II, the first and the last of the four backbone hydrogen bonds are broken; the three hinge regions then adopt a helical conformation, leading to the lengthening of helices A, B, and C. Similar changes also occur in cTnC, but in two steps. As shown by NMR, in apo- or 2Ca2+-cTnC, residues Phe27-Val28, Ser37–Glu40, and Val64-Asp65 all display nonhelical torsion angles (Fig. 2A) (12), and the small β-sheet is stabilized by four backbone hydrogen bonds (74N⋅⋅⋅34O, 36N⋅⋅⋅72O, 72N⋅⋅⋅36O, and 38N⋅⋅⋅70O) (32). A comparison of the NMR structures of apo- and 3Ca2+-cTnC (11, 12) indicates that Asp65 and Glu76 in loop II move toward the incoming Ca2+ ion as carboxylate ligands during the first step of the conformational change. Consequently, helix C is extended at the C terminus to include Val64-Asp65 (Fig. 2B), and the first hydrogen bond (74N⋅⋅⋅34O) is broken. The size of the N-lobe hydrophobic pocket increases slightly as the C–D interhelical angle decreases by about 10°. It should be noted that although the binding of Ca2+ to domain II does not alter the A–B interhelical angle, it does seem to “prime” domain I for further changes: helix A is lengthened by two residues (Phe27 and Val28, see Fig. 2B)—either because the side chain of Phe27 moves farther into the slightly enlarged N-lobe pocket or because the constraints imposed by the 74N⋅⋅⋅34O hydrogen bond are released. When cTnI (13) or bepridil binds during the second step, the last hydrogen bond (38N⋅⋅⋅70O) in the β-sheet is broken, and helix B is extended at the N terminus to include Ser37-Glu40 (Figs. 2 C and D and 3). The N-lobe then opens fully.
In summary, the lengthening of helices A, B, and C is required for the full opening of the N-lobe in both sTnC (7–9, 27) and cTnC (11–13). In both cases, however, the lengthening of helix B (which has the longest hinge region) produces the largest increase in the hydrophobic surface area of the lobe. As mentioned earlier, in sTnC the change in helix B requires the movement of a glutamate side chain toward the Ca2+ ion in domain I (27). By contrast, helix B of cTnC cannot be extended in such a fashion because domain I of this isoform does not bind Ca2+. Moreover, in cTnC the binding of a single Ca2+ ion to domain II introduces relatively minor changes (11, 12), which, as described above, are nonetheless essential for “priming” the opening of the N-lobe. As shown in the NMR study by Li et al. (13) and in the current work, the lengthening of helix B and the subsequent opening of the N-lobe are achieved by an agent such as cTnI or bepridil, which shields the N-lobe hydrophobic pocket from the solvent. This two-stage mechanism provides an extension to the HMJ model for sTnC (10), in which the N-lobe opens upon the binding of two Ca2+ ions.
Structural Basis for the Calcium-Sensitizing Effect of Bepridil.
It is now clear how bepridil might increase the contractile force of the cardiac myofilament (17). Biochemical studies have indicated that bepridil increases the Ca2+ affinity of cTnC (16, 17). From our results, it is apparent that bepridil hinders the closing of the N-lobe, a step that accompanies the release of Ca2+ from domain II. The resultant increased Ca2+ affinity, in turn, would stimulate actomyosin ATPase at intermediate Ca2+ levels. Bepridil would have a similar effect on the Ca2+ affinity of sTnC, in which the closing of the N-lobe is also coupled to the release of Ca2+. Moreover, by maintaining the open conformation of the N-lobe, bepridil may enhance its affinity for cTnI and thus increase the activity of actomyosin ATPase. In such a scheme, bepridil and cTnI would bind to the N-lobe simultaneously. Support for this mechanism comes from a study by Lin et al. (33), which showed that a synthetic peptide covalently linked to Met45 (in the Bep1 binding site) had little effect on regulation. Moreover, Kleerekoper et al. (15) have recently identified three bepridil binding sites in cTnC and have proposed that a bepridil molecule bound to the site that includes Met45, Met60, and Met80 would not be displaced by cTnI. This prediction appears to be correct. In the current structure, Bep1 is in close contact with these three methionines (Table 2). Moreover, an overlay of bepridil-3Ca2+-cTnC and 3Ca2+-cTnC-cTnI147–163 (13) indicates that Bep1 and cTnI147–163 bind to different regions of the hydrophobic core of the N-lobe: the former to the bottom of the N-lobe pocket, and the latter mainly to the apolar surfaces of helices A and B (Fig. 4B). Due to the predicted displacement of Bep2 and Bep3 by TnI, it appears that the Ca2+-sensitizing effect of the drug resides in Bep1 alone.
Implications for the Future Design of Calcium-Sensitizing Agents for cTnC.
Bepridil belongs to a group of pharmacological agents that also inhibit CaM from binding to its target proteins (34). Most of these drugs have a large apolar region and a positively charged group at least 3 atoms away (25, 35). The apolar region seems to be essential for the binding of these agents to the hydrophobic pockets of both cTnC and CaM. By contrast, the positively charged group is probably not required for cTnC-specific drugs, since the pyrrolidine ring of bepridil does not contribute to the opening of the N-lobe in the current structure. In fact, such a group, when bound to the N-lobe, might hinder the binding of the positively charged cTnI to cTnC. Moreover, because of the structural homology between cTnC and CaM, several drugs that bind to cTnC, including bepridil and TFP, have been shown to bind to CaM as well (13). As noted above, such drugs with positively charged groups may trap CaM in an inactive form. To avoid such an inhibitory effect on CaM, neutral agents resembling bepridil could be designed to modulate the activity of cTnC. Although bepridil itself, as described earlier, is not a good pharmacological agent for heart failure, the current study provides insights into the features that may be important for the design of cTnC-specific calcium-sensitizers.
Acknowledgments
We are grateful to M. N. G. James, Z. Grabarek, L. S. Tobacman, and J. H. Brown for helpful comments on the manuscript, and to W. Liu for expression and purification of cTnC. This study was supported by grants to C.C. from the National Institutes of Health and the Muscular Dystrophy Association and to J.A.P. from the National Institutes of Health and the Robert Welch Foundation.
Abbreviations
- TnC
troponin C
- TnI
troponin I
- TnT
troponin T
- cTnC
cardiac TnC
- cTnI
cardiac TnI
- sTnC
skeletal TnC
- sTnI
skeletal TnI
- CaM
calmodulin
- TFP
trifluoperazine
Footnotes
Data Deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1dtl).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090098997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090098997
References
- 1.Farah C S, Reinach F C. FASEB J. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- 2.Tobacman L S. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 3.Phillips G N, Jr, Fillers J P, Cohen C. J Mol Biol. 1986;192:111–131. doi: 10.1016/0022-2836(86)90468-7. [DOI] [PubMed] [Google Scholar]
- 4.Squire J M, Morris E P. FASEB J. 1998;12:761–771. doi: 10.1096/fasebj.12.10.761. [DOI] [PubMed] [Google Scholar]
- 5.Herzberg O, James M N G. J Mol Biol. 1988;203:761–779. doi: 10.1016/0022-2836(88)90208-2. [DOI] [PubMed] [Google Scholar]
- 6.Satyshur K A, Rao S T, Pyzalska D, Drendel W, Greaser M, Sundaralingam M. J Biol Chem. 1988;263:1628–1647. [PubMed] [Google Scholar]
- 7.Gagné S M, Tsuda S, Li M X, Smillie L B, Sykes B D. Nat Struct Biol. 1995;2:784–789. doi: 10.1038/nsb0995-784. [DOI] [PubMed] [Google Scholar]
- 8.Houdusse A, Love M L, Dominguez R, Grabarek Z, Cohen C. Structure. 1997;5:1695–1711. doi: 10.1016/s0969-2126(97)00315-8. [DOI] [PubMed] [Google Scholar]
- 9.Strynadka N C, Cherney M, Sielecki A, Li M X, Smillie L B, James M N G. J Mol Biol. 1997;273:238–255. doi: 10.1006/jmbi.1997.1257. [DOI] [PubMed] [Google Scholar]
- 10.Herzberg O, Moult J, James M N G. J Biol Chem. 1986;261:2638–2644. [PubMed] [Google Scholar]
- 11.Sia S K, Li M X, Spyracopoulos L, Gagné S M, Liu W, Putkey J A, Sykes B D. J Biol Chem. 1997;272:18216–18221. doi: 10.1074/jbc.272.29.18216. [DOI] [PubMed] [Google Scholar]
- 12.Spyracopoulos L, Li M X, Sia S K, Gagné S M, Chandra M, Solaro R J, Sykes B D. Biochemistry. 1997;36:12138–12146. doi: 10.1021/bi971223d. [DOI] [PubMed] [Google Scholar]
- 13.Li M X, Spyracopoulos L, Sykes B D. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 14.The Advisory Council to Improve Outcomes Nationwide in Heart Failure. Am J Cardiol. 1999;83:1A–38A. [PubMed] [Google Scholar]
- 15.Kleerekoper Q, Liu W, Choi D, Putkey J A. J Biol Chem. 1998;273:8153–8160. doi: 10.1074/jbc.273.14.8153. [DOI] [PubMed] [Google Scholar]
- 16.MacLachlan L K, Reid D G, Mitchell R C, Salter C J, Smith S J. J Biol Chem. 1990;265:9764–9770. [PubMed] [Google Scholar]
- 17.Solaro R J, Bousquet P, Johnson J D. J Pharmacol Exp Ther. 1986;238:502–507. [PubMed] [Google Scholar]
- 18.Putkey J A, Dotson D G, Mouawad P. J Biol Chem. 1993;268:6827–6830. [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Navaza J, Saludjian P. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T. x-plor, A System for X-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1993. , Version 3.1. [Google Scholar]
- 22.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewksi J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Vandonselaar M, Hickie R A, Quail J W, Delbaere L T. Nat Struct Biol. 1994;1:795–801. doi: 10.1038/nsb1194-795. [DOI] [PubMed] [Google Scholar]
- 25.Cook W J, Walter L J, Walter M R. Biochemistry. 1994;33:15259–15265. doi: 10.1021/bi00255a006. [DOI] [PubMed] [Google Scholar]
- 26.Meador W E, Means A R, Quiocho F A. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 27.Gagné S M, Li M X, Sykes B D. Biochemistry. 1997;36:4386–4392. doi: 10.1021/bi963076+. [DOI] [PubMed] [Google Scholar]
- 28.Osawa M, Swindells M B, Tanikawa J, Tanaka T, Mase T, Furuya T. J Mol Biol. 1998;276:165–176. doi: 10.1006/jmbi.1997.1524. [DOI] [PubMed] [Google Scholar]
- 29.Vassylyev D G, Takeda S, Wakatsuki S, Maeda K, Maeda Y. Proc Natl Acad Sci USA. 1998;95:4847–4852. doi: 10.1073/pnas.95.9.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasmi-Seabrook G M C, Howarth J, W, Finley N, Abusamhadneh E, Gaponenko V, Brito R M M, Solaro R J, Rosevear P R. Biochemistry. 1999;38:8313–8322. doi: 10.1021/bi9902642. [DOI] [PubMed] [Google Scholar]
- 31.Kleerekoper Q, Howarth J W, Guo X, Solaro R J, Rosevear P R. Biochemistry. 1995;34:13343–13352. doi: 10.1021/bi00041a010. [DOI] [PubMed] [Google Scholar]
- 32.Spyracopoulos L, Gagné S M, Li M X, Sykes B D. Biochemistry. 1998;37:18032–18044. doi: 10.1021/bi9816960. [DOI] [PubMed] [Google Scholar]
- 33.Lin X, Dotson D G, Putkey J A. J Biol Chem. 1996;271:244–249. doi: 10.1074/jbc.271.1.244. [DOI] [PubMed] [Google Scholar]
- 34.Itoh H, Ishikawa T, Hidaka H. J Pharmacol Exp Ther. 1984;230:737–741. [PubMed] [Google Scholar]
- 35.Prozialeck W C, Weiss B. J Pharmacol Exp Ther. 1982;222:509–516. [PubMed] [Google Scholar]