Abstract
Kallikreins play an important role in tumour microenvironment and as cancer biomarkers in different cancer entities. Previous studies suggested an upregulation of KLK10 and KLK6 in pancreatic ductal adenocarcinoma (PDAC). Therefore, we evaluated the clinicopathological role of these kallikreins and their value as biomarkers in PDAC.
Differential expression was validated by DNA-microarrays and immunohistochemistry in normal and malignant pancreatic tissues. Sera concentrations of both kallikreins were evaluated using ELISA. In silico analysis of possible protein interactions and gene silencing of KLK10 in vitro using siRNAs gave further insights in the pathomechanisms.
Gene expression analysis and immunohistochemistry demonstrated a strong expression for KLK10 and KLK6 in PDAC. Statistical analysis showed that co-expression of these kallikreins correlated with an R1-resection status (P=0.017) and worse outcome for overall survival (P=0.031). Multivariate analysis proofed that co-expression is an independent prognostic factor for survival (P=0.043). Importantly, KLK10 knockdown in AsPC-1 cells significantly reduced cell migration, whereas computational analysis suggested interaction of KLK6 with angiogenetic factors as an important mechanism.
Co-expression of KLK10 and KLK6 plays an unfavourable role in PDAC. Our results suggest that this effect is likely mediated by an interaction with the factors of the extracellular matrix and enhancement of cancer cell motility.
Keywords: pancreatic cancer, KLK10 , KLK6 , DNA-microarray, microenvironment
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive cancers with an incidence rate of 6.3/100 000 (Lowenfels and Maisonneuve, 2006; Jemal et al, 2008). It is characterised by early metastasising and aggressive, infiltrative growth along the endothelium basement membrane and neurons (Eccles and Welch, 2007). This leaves more than 85% of the patients inoperable at the time of diagnosis. Unfortunately pancreatic carcinoma also shows an unsatisfactory response to oncological treatment (Wolff et al, 2000). This demonstrates the need for new therapeutic approaches and also for biomarkers, which make early diagnosis possible. Recently, we (Grutzmann et al, 2003) and others (Iacobuzio-Donahue et al, 2003; Yousef et al, 2004) showed that human kallikrein 10 and human kallikrein 6 are among the most highly and specifically overexpressed genes in pancreatic cancer compared with normal and benign pancreas tissues.
KLK10 and KLK6 are members of the kallikrein family of 15 known proteases in humans, which play an emerging role in tumour microenvironment, invasion and angiogenesis (Borgono and Diamandis, 2004). Kallikreins exert this function as secreted trypsin and chymotrypsin-like proteases by degradation of the extracellular matrix, which is an important reservoir for cytokines and growth factors such as VEGF, TGF-β and kininogens (Borgono and Diamandis, 2004). Moreover, KLK3, also known as prostate-specific antigen, is of great clinical value as a serological marker in prostate cancer (Borgono and Diamandis, 2004). Other members of the kallikrein family might also have a utility in screening of malignancies, like KLK11 and KLK6 for ovarian cancer (Diamandis et al, 2003; McIntosh et al, 2007). This leaves kallikreins as advantageous candidate genes for diagnosis and therapy in pancreatic cancer. Therefore, the aim of this study was to evaluate the clinicopathological role of these kallikreins and their value as biomarkers in PDAC.
KLK10, also known as the normal epithelial cell-specific 1 is one of the newly identified members of the kallikrein family. Its role in carcinogenesis is unsolved as it is downregulated in some tumours such as breast cancer and acute lymphoblastic leukaemia, whereas overexpressed in ovarian, prostate or renal cancer (Luo et al, 2003; Petraki et al, 2003, 2006; Zhang et al, 2006). Unfortunately, the function of KLK10 protein remains poorly documented, neither the activators nor the substrates for KLK10 are actually known (Zhang et al, 2006). KLK6, or protease M, is highly expressed in several malignancies like ovarian, breast, colon or gastric cancer. It is correlated with lymphatic invasion and poor prognosis in gastric cancer (Yousef et al, 2004; Nagahara et al, 2005). KLK6 might exert this effect by the degradation of matrix proteins and thereby the augmentation of cancer cell motility and proliferation (Ghosh et al, 2004).
In this study, we show that kallikrein 10 and 6 demonstrate a strong protein expression in pancreatic carcinoma and are associated with poor patient prognosis and R1 resection status and thereby might contribute to the aggressive character of this malignancy.
The results indicate that this effect is most likely mediated by the interaction of KLK6 with factors of the extracellular matrix and the enhancement of cancer cell motility by KLK10.
Materials and methods
Patients and demographic data
For immunohistochemical analysis we used samples from 54 patients, operated from July 1996 until December 2003. None of the patients received adjuvant chemotherapy prior to operation. The eligibility criterion was a histologically confirmed PDAC. A positive microscopic resection margin (R1) was operationally defined as at least one cancer cell within 1 mm of any surface of the resected specimen. We included patients with the finding of metastases to the intra-aortocaval lymph nodes. Previous reports have shown that these metastases should not exclude patients in good condition from oncologic resection of the PDAC (Shrikhande et al, 2007). Relevant patient characteristics are summarised in Table 1.
Table 1. Clinicopathological variables and KLK10 and KLK6 co-expression of the tumour cohort (n=54).
| All cases |
Co-expression
|
|||
|---|---|---|---|---|
| Characteristics | n (%) | None/low | Strong | P-value |
| Age at diagnosis (years) 60.5 (range, 31–76) | ||||
| Gender | ||||
| Male | 24 (44.4) | 12 | 12 | 0.808 |
| Female | 30 (55.6) | 16 | 14 | |
| Localisation | ||||
| Head of pancreas | 47 (87.0) | 26 | 21 | 0.186 |
| Tail of pancreas | 7 (13.0) | 2 | 5 | |
| Tumour stage | ||||
| pT1 | 6 (11.1) | 5 | 1 | 0.391 |
| pT2 | 13 (24.1) | 6 | 7 | |
| pT3 | 32 (59.3) | 16 | 16 | |
| pT4 | 3 (5.5) | 1 | 2 | |
| Nodal status | ||||
| N0 | 22 (40.7) | 12 | 10 | 0.565 |
| N1 | 31 (57.4) | 15 | 16 | |
| N2 | 1 (1.9) | 1 | 0 | |
| Interaortocaval metastasis | ||||
| Positive | 7 (13.0) | 4 | 3 | 0.764 |
| Negative | 47 (87.0) | 24 | 23 | |
| Grade | ||||
| G1 | 4 (7.4) | 4 | 0 | 0.096 |
| G2 | 38 (70.4) | 17 | 22 | |
| G3 | 12 (22.2) | 7 | 5 | |
| Residual tumour status | ||||
| R0 | 41 (75.9) | 25 | 16 | 0.017 |
| R1 | 13 (24.1) | 3 | 10 | |
| Perineural invasion | ||||
| Yes | 19 (35.2) | 11 | 8 | 0.513 |
| No | 35 (64.8) | 17 | 18 | |
| Perilymphatic invasion | ||||
| Yes | 24 (44.4) | 11 | 13 | 0.429 |
| No | 30 (55.6) | 17 | 13 | |
Patients with strong immunohistochemical co-expression of KLK6 and KLK10 showed a significant correlation with R1-Resection status (P=0.017).
For serum-ELISA several panels of sera were selected from 130 healthy donors, eight patients with benign tumours, 34 with inflammatory diseases and 28 patients with malignant diseases (Table 2). All sera were from patients treated at the University Hospital Dresden, Germany and University Hospital Schleswig–Holstein, Germany. All patients had given informed consent.
Table 2. Results of the serum ELISA showed no statistical significance between diseases nor between different localisations.
|
Median
|
Mean
|
||||
|---|---|---|---|---|---|
| Diagnosis (Localisation) | KLK6 | KLK10 | KLK6 | KLK10 | P (log-rank) |
| Benign Tumours (n=4) | 12.4 | 2.1 | 10.3±7.8 | 1.4±1.8 | n.s. |
| Inflammatory diseases | |||||
| Bile duct (n=1) | 12.9 | 1.3 | |||
| Gallbladder (n=4) | 13.1 | 2.1 | 14±2.2 | 2.3±0.67 | |
| Pancreas (n=12) | 10 | 2.0 | 7.7±5.7 | 2.4±0.79 | |
| Malignant tumours | |||||
| Bile duct (n=3) | 8.1 | 1.3 | 7.2±4.7 | 1.2±0.31 | |
| Gallbladder (n=3) | 5.7 | 1.0 | 4.5±2.0 | 1.2±0.42 | |
| Pancreas (n=8) | 9.7 | 1.6 | 9.3±4.6 | 1.5±0.95 | |
| Normal (n=65) | 9.5 | 1.8 | 9.0±3.6 | 1.7±0.85 | |
Construction of a virtual subarray and bioinformatic analysis
For the construction of the virtual subarray we used data obtained from the U133 A/B Affymetrix GeneChip using extracted RNA from microdissected tissue as described earlier (Pilarsky et al, 2008).
The Cel Files obtained from the Affymetrix MAS 5.0 software were used for further analysis. The files were loaded into dCHIP 1.3 (www.dchip.org) then normalised, and expression values as well as absolute calls were calculated using the PM/MM model (Grutzmann et al, 2003). We scored genes as differentially expressed if they displayed a fold change >2. To identify signature genes we used the method described elsewhere (Grutzmann et al, 2004). Datasets are accessible (Arrayexpress E-MEXP-1121 and E-MEXP-950).
Immunohistochemistry study and evaluation
Immunostaining was carried out on paraffin-embedded tissue sections by streptavidin—biotin–peroxidase complex method using polyclonal antibodies for KLK6 and KLK10 (1 : 150). The ductal epithelium and the Langerhans' islets served as positive controls for both kallikreins (Petraki et al, 2001, 2002a, 2003). Negative controls were performed for all studied tissues by omitting the primary antibody (KLK6 and KLK10) or by replacing it by non-immune serum (dilution 1 : 500) (see Supplementary Data 1).
All sections were examined by one observer (CDP) blinded to both clinical and pathological data. Protein expression in PDACs was quantified using a visual grading system with a range between 0 and 2, based on the intensity and the proportion of positive tumour cells on the studied section: 0=no immunoexpression or weak staining in any proportion of the cancerous tissue, or moderate expression in </=5% of the cancerous tissue, 1=moderate staining in 5–50% of the cancerous tissue, 2=moderate staining in >50% of the cancerous tissue or strong staining in any proportion of the cancerous tissue. According to staining intensity, cancers were classified as low, medium or high expressing tumours. Coexpression of both KLKs was defined as strong if the sample showed strong expression for both KLKs (2/2) and/or at least showed moderate expression for one KLK (2/1). KLK6 and 10 immunoexpression were also screened in the normal pancreatic parenchyma (acinar, ductal and endocrine cells) and in the ampulla of Vater region of the small intestine.
Patient serum selection and ELISA for kallikrein measurement in serum
ELISA-type immunofluorometric procedures developed in-house were used to measure KLK6 and 10 levels in these sera. Assays used in this study were of the ‘sandwich’ type with the primary antibody used for capture and the secondary one for detection. Monoclonal–monoclonal combinations were used in this study. All ELISAs were tested negative for cross-reactivity against other kallikreins. Assay precision within the dynamic range was <10%. These assays were standardised with recombinant proteins produced in yeast or mammalian expression systems. More details about the kallikrein ELISA have recently been published (Shaw and Diamandis, 2007).
Protein interaction prediction
To evaluate the interactions we queried databases with known protein–protein interactions such as NetPro (www.molecularconnections.com), SCOPPI (www.scoppi.org) and HPRD (www.hprd.org) and compared them to our data. To find possible novel interactions we used structure- and sequence-based prediction of protein interactions as described earlier (Altschul et al, 1997; Mishra et al, 2006; Dawelbait et al, 2007).
Cell culture and transfection conditions
The AsPC-1 cell line (ATCC Number CRL-1682), established from malignant ascites of a 62-year-old female caucasian, was used for this study. Cells were grown in RPMI-1640 (Invitrogen, Karlsruhe, Germany) with 2 mM L-glutamine, 1 mM sodium pyruvate, 4.5 g l−1 glucose and 10% FCS in a humidified atmosphere containing 5% CO2 at 37°C. AsPC-1 cells (50 000) in media with 1% FCS were transfected with 600 ng of siRNA using oligofectamine (Invitrogen GmbH, Karlsruhe, Germany). Target sense sequences that effectively mediated silencing were as follows: KLK10.1 (UACAUGUCCUGGAUCAAUA) and KLK10.2 (UGACGUGCCUACCUCUUAG) (all from MWG Biotech, Ebersberg, Germany). Knockdown was confirmed by RT–PCR and western blot. siRNA against the green fluorescent protein (GGCUACGUCCAGGAGCGCACC) served as a negative control. For RT–PCR the following cell lines were used: Capan-1 (ATCC No. HTB-79), Capan-2 (ATCC No. HTB-80), MiaPaCa-2 (CRL-1420), Panc1 (ECACC No. 7092802), BXPC3 (CRL-1687) and Panc 89, Colo357, PancTUI, PT45P1 (all from Professor H Kalthoff, Kiel, Germany, (Sipos et al, 2003)).
Reverse transcription–polymerase chain reaction
Using RNeasy Mini Kit (Qiagen, Hilden, Germany) we isolated total RNA and subjected 500 ng to cDNA synthesis using random primer and SuperScript II (Invitrogen GmbH, Karlsruhe, Germany). Of the synthesised cDNA 2% were used for quantitative RT–PCR. Analysis was performed using an ABI PRISM 5700 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). The genes were amplified with the Power SybrGreen PCR Master Mix according to the manufacturer's instructions. Gene expression was quantified by the comparative Ct-Method, normalising Ct-values to a housekeeping gene (β-actin) and calculating the relative expression values. Each experiment was repeated two times in duplicate. The following primers were used: β-Actin: Actb 1498 f (AAGCCACCC-CACTTCTCTCTAA) and actb-1570R (AATGCTATCACCTCCCCTGTGT), KLK10: KLK10_f1 ex (GTCCTGGTGGACCAGAGTTG) and KLK10_r1_ex (GAGCTG-CTCTCCCTGAAGAA), KLK6: KLK6_f1 (GTG TGC TGG GGA TGA GAA GT) and KLK6_r1 (GGG ATG TTA CCC CAT GAC AC) (all from MWG Biotech, Ebersberg, Germany).
Western blotting
Cells were washed and lysed with LDS sample buffer (Invitrogen, Karlsruhe, Germany). Proteins were electrophoresed under reducing conditions on 4–12% acrylamide gels (Invitrogen, Karlsruhe, Germany) and then transferred to a nitrocellulose membrane (Hybond ECL, GE Healthcare, Munich, Germany). To block nonspecific binding, the membrane was incubated overnight in PBS with 0.1% Tween 20 (T-PBS) containing 5% BSA at 4°C. Subsequently, the membrane was incubated for 1 h with the antibody against KLK10 (1 : 1000, see (Petraki et al, 2001, 2002a, 2003)) or β-actin loading control (1 : 5000, no. ab6276, Abcam, Cambridge, UK) in T-PBS and 5% BSA. After washing in T-PBS, protein on the membrane was visualised using the ECL detection kit (GE Healthcare, Munich, Germany) with a peroxidase-labelled anti-mouse antibody for β-Actin (1 : 25000, no. NIF 825, Amersham Pharmacia, Amersham, United Kingdom) and peroxidase-labelled anti-rabbit antibody for KLK10 (1 : 5000, no. NIF 824, Amersham Pharmacia, Amersham, United Kingdom) as per manufacturer's instructions. Protein expression was measured by AIDA evaluation software (Raytest, Straubenhardt, Germany) as the ratio of KLK10-staining intensity to actin-staining intensity.
Boyden chamber assay
Invasion in vitro was measured in Boyden chamber assay (no. 353097, BD Falcon, Heidelberg Germany). The PET membrane had a pore size of 8 μm with a pore density of 1.0 × 105 cm−2. Cells were transfected using the above-mentioned protocol and incubated for 48 h. Cells were then trypsinised, counted and cell suspensions of the two groups (5 × 105 cells per 250 μl) were transferred in 1% FCS medium onto the membrane. Then the chambers were put in 24-well plates containing 10% FCS medium and cultured for 72 h. Cells infiltrated through the reconstituted basement membrane and appeared on the outer surfaces. By HE staining the number of the cells was counted microscopically. Migration assays were repeated three times.
Statistical analysis
P-values were assigned using χ2 (Pearson) for the cross tables, the log-rank test (Mantel–Cox) for the survival univariate analysis by the Kaplan–Meier test, and the Cox regression analysis for multivariate survival analysis. For statistical analysis ‘SPSS 13.0’ for Windows was used.
Results
Virtual subarray
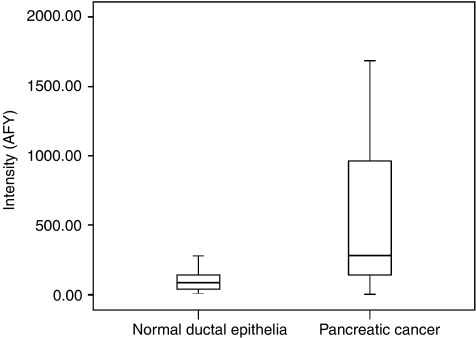
Gene expression profiles of 19 patients with PDAC and normal ductal cells from 13 individuals were generated (Pilarsky et al, 2008). We then constructed a virtual subarray to identify gene expression changes of the 15 kallikreins. The nine probe sets identified with the virtual subarray analysis represented six differentially expressed genes (Table 3). Of these, KLK10 was overexpressed whereas KLK3, 12, 13 and 15 were downregulated in PDAC compared with microdissected normal ductal cells. Upregulation of KLK10 was strong (Figure 1) compared with normal individuals (P=0.009). We also decided to consider KLK6 for further evaluation because upregulation was found in PDAC by other groups (Iacobuzio-Donahue et al, 2003; Yousef et al, 2004).
Table 3. (A): Results of the GeneChip analysis. The upregulated genes (fold-change >2, P<5%) are listed in the upper part, the downregulated genes in the lower part of the table; (B): Sequence-based and structure-based protein interaction prediction showed four possible interaction partners for KLK6.
| Probe set | HGNC symbol | Fold change | P-value | |
|---|---|---|---|---|
| A | ||||
| Upregulated genes | ||||
| KLK10@209792_s_at | KLK10 | 5.2 | < 0.01 | |
| KLK10@215808_at | KLK10 | 1.6 | 0.01 | |
| Downregulated genes | ||||
| KLK13@216670_at | KLK13 | −1.7 | 0.01 | |
| KLK3@204583_x_at | KLK3 | −6.6 | 0.03 | |
| KLK12@233586_s_at | KLK12 | −9.4 | < 0.01 | |
| KLK12@234316_x_at | KLK12 | −10.1 | < 0.01 | |
| KLK1@216699_s_at | KLK1 | −10.2 | < 0.01 | |
| KLK12@220782_x_at | KLK12 | −10.5 | < 0.01 | |
| KLK15@221462_x_at | KLK15 | −15.3 | < 0.01 | |
| B | ||||
| Protein 1 | Predicted partner | Also known as | Function | Compartment |
| KLK6 | SERPINA3 | α-1 antiproteinase | DNA-binding, endopeptidase inhibitor activity, inflammatory response | Extracellular, intracellular, nucleus |
| SERPINC1 | AT III | Blood coagulation | Extracellular region | |
| SERPINF2 | Pigment epithelium-derived factor | Endopeptidase inhibitor activity, protein-binding, acute phase response | Extracellular region | |
| SNCA | Synuclein | Anti-apoptosis, central nervous system, development, protein binding | Cytoplasm | |
| KLK10 | No interaction partner found | |||
Figure 1.
Results of the GeneChip analysis. KLK10 showed a marked upregulation in pancreatic cancer samples compared with normal individuals (P=0.009).
Immunohistochemical expression of KLK6 and KLK10
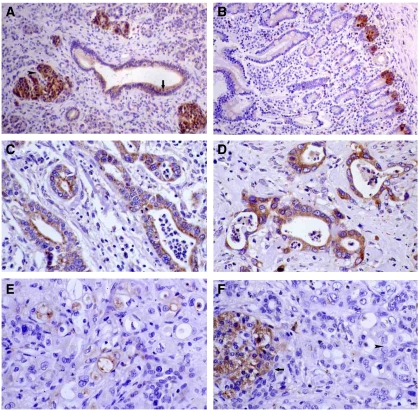
In the endocrine pancreas, the immunohistochemistry for KLK6 and KLK10 showed strong staining in the endocrine cells of the Langerhans' islets and in scattered endocrine cells in connection with pancreatic ducts and acinar cells.
The exocrine part of the pancreas displayed a cytoplasmic expression in the small intercalated pancreatic ducts, the intra- and inter-lobular pancreatic ducts, the main pancreatic duct and the common bile duct. Staining was absent in the acinar cells (Figure 2A and E). In the region of the ampulla of Vater in the small intestine, a strong cytoplasmic, mostly supranuclear immunoexpression was observed in the epithelium of the intestinal crypts. The absorptive cells in the surface villous epithelium showed a moderate cytoplasmic and brush border expression, whereas goblet cells were mostly negative (Figure 2B).
Figure 2.
Immunohistochemical staining of PDAC samples. Moderate KLK6 immunoexpression in pancreatic ducts (arrow) and strong expression in Langerhans' islets (arrowhead) no staining in acini ( × 100) (A). Strong KLK10 immunoexpression in the crypts of the intestinal epithelium of the ampulla of Vater ( × 100) (B). Strong KLK6 immunoexpression in pancreatic adenocarcinomas ( × 200) (C and D). Moderate KLK10 immunoexpression in pancreatic adenocarcinomas ( × 200) (E). Strong KLK10 immunoexpression in Langerhans' islets (arrow), absence of expression in pancreatic adenocarcinoma (arrowhead) ( × 200) (F).
The staining for KLK6 in primary PDAC showed a moderate to strong expression in 91.5% of the cases, whereas it was only 64.4% for KLK10.
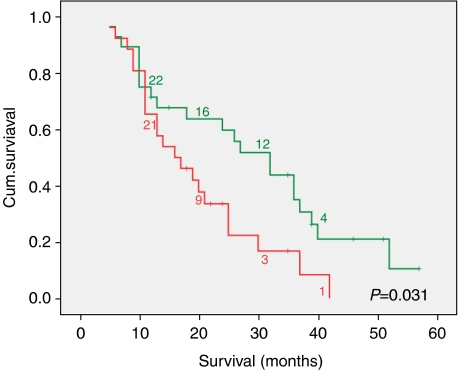
KLK6 showed a diffuse cytoplasmic immunostaining in the cancerous epithelium, whereas KLK10 mostly showed a patchy expression, often with luminal pattern (Figure 2). Analysis of immunohistochemistry revealed that patients with strong KLK6 and KLK10 co-expression had significant lower medium survival time of 20 months (15.0–24.0) compared with patients without/weak expression of these kallikreins (29 months (22.8–35.8)) (P=0.031) (Figure 3). Neither KLK10 (P=0.259) nor KLK6 (P=0.452) expression could be correlated with survival alone. We could further associate high KLK6 and KLK10 immunoreactivity with R1-resection status (P=0.017) (Table 1).
Figure 3.
The survival curve shows a lower medium survival time of 20 months (15.0–24.0) in the subgroup of patients with strong KLK6 and KLK10 co-expression compared with patients without/weak expression of these kallikreins (29 months (22.8–35.8)) (P=0.031).
Cox regression analysis identified KLK10 and KLK6 co-expression as an independent prognostic factor with a statistically significant relationship to survival in PDAC (P=0.043, RR 2.002) (Table 4).
Table 4. Cox regression model, including conventional variables and co-expression of KLK6 and KLK10 in all cases (n=54).
| Relative risk | 95% CI | P-value | |
|---|---|---|---|
| Co-expression KLK6/KLK10 | 2.022 | 1.021–4.006 | 0.043 |
| Resection status | 0.571 | 0.247–1.321 | 0.190 |
| Grading | 1.964 | 1.030–3.746 | 0.040 |
| pT stage | 0.498 | 0.310–0.801 | 0.004 |
| Nodal status | 1.814 | 0.937–3.513 | 0.077 |
| Metastases | 0.545 | 0.177–1.679 | 0.290 |
Co-expression is a strong independent prognostic factor for survival in patients with PDAC (P=0.043).
KLK10 and KLK6 serum concentration
Using ELISA immunoassays developed in-house, we tested KLK6 and KLK10 serum levels in patients with malignant, inflammatory and benign diseases; additionally there was a panel of healthy patients.
There was no significant correlation of KLK6 and KLK10 serum levels with survival of the patients. Also, there was no statistical significance between healthy donors and patients with PDAC nor between different localisations of inflammatory, benign and malignant diseases of the pancreatico-biliary tract (Table 2).
Protein interaction prediction
To find potential interaction partners for KLK6 and 10, which might explain poor survival, we used two different computational methods: the structure- and the sequence-based protein interaction prediction. By means of these methods we could identify four potential interaction partners for KLK6: α-1 antiproteinase, AT III, pigment epithelium-derived factor and synuclein (Table 3). However, no additional interaction partner for KLK10 other than the already described could be identified (Dawelbait et al, 2007).
Expression of KLK10 and KLK6 in pancreatic cancer cell lines
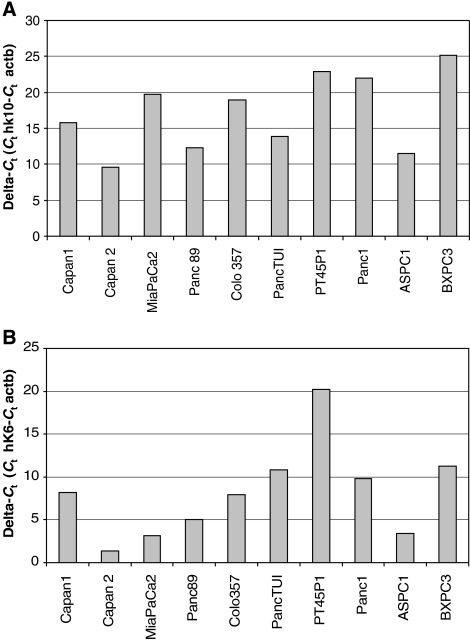
To evaluate expression of KLK10 and KLK6 in established cell lines we conducted a qRT–PCR. Capan-2, Panc89 and AsPC-1 cell lines displayed the highest KLK10 expression. For KLK6 the cell lines Capan-2, Mia PaCa-2 and AsPC-1 showed a good expression. Like in immunohistochemistry of native tumour, KLK6 expression in cell lines was more distinct than that of KLK10 (Figure 4).
Figure 4.
A qRT–PCR of different established pancreatic cancer cells lines showed Capan-2 and AsPC-1 with relevant KLK10-expression (A). KLK6-expression was high in nearly all measured cell lines (B).
Gene silencing of KLK10 in AsPC-1 cells
To elucidate the relevance of dysregulations of KLK10 for carcinogenesis, we established a siRNA assay. For this purpose, we used the human pancreatic cancer cell line AsPC-1. This cell line showed detectable KLK10-expression as well as KLK6-expression (Figure 4).
Cells were transfected with two KLK10 sequences, KLK10.1 and KLK10.2. Untreated cells and cells transfected with unspecific siRNA served as a negative control. As shown in Figure 5 the expression of KLK10 was strongly inhibited compared with controls in RT–PCR (P<0.001). The same holds true for protein synthesis in western blot (P=0.025). β-actin levels showing equal quantities of protein were loaded (Figure 5); all experiments were repeated four times. Because transfection with KLK10.1 showed the best inhibition of protein synthesis we chose this siRNA for our further studies. To examine off-target gene-silencing in our cells, we used the BLAST database (www.ncbi.nlm.nih.gov/BLAST) to search the human genome for any complementary sequences with at least 11 contiguous nucleotides matching the RNAi sites we used. No gene appeared to have sequence similarity. Transfection of AsPC-1 cells with KLK10.2 siRNA did not change the KLK6 expression levels as measured qRT–PCR (see Supplementary Data 1).
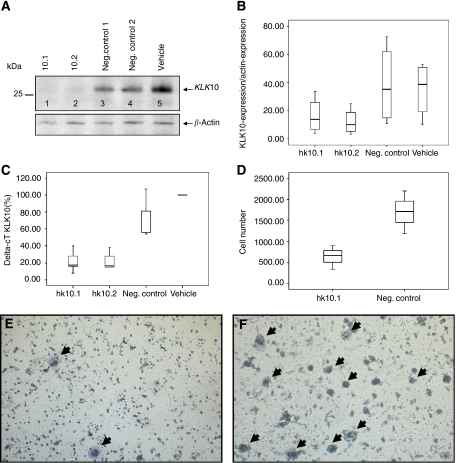
Figure 5.
AsPC-1 cells were transfected with two KLK10-sequences, KLK10.1 and KLK10.2. Transfection resulted in a strong downregulation of KLK10 protein synthesis in the western blot. Cell lysates of KLK10.1- and KLK10.2-transfected cells showed nearly no staining (lanes 1 and 2) compared with the controls (lanes 3–5) (A). Statistical analysis showed a statistical downregulation of the transfected group compared with the control group (P=0.025) (B). The same holds true for gene expression in RT–PCR (P<0.001) (C). The Boyden chamber migration assay: Invasion in vitro was measured as described in ‘Materials and methods’. Statistical analysis showed that KLK10.1-transfected cells had a significant decrease in cell motility compared with the controls (P=0.05) ( × 200) (D). Only few KLK10.1-transfected AsPC-1 cells migrated through the membrane (arrow) (E). The control group displayed normal cell migration (arrows) (F).
Migration assay
AsPC-1 cells transfected with KLK10.1 siRNA showed a significant decrease in cell motility compared with the GFP-transfected cells. Although only 636 cells (±285) could be isolated from the membrane in KLK10 knockout cells, it was 1189 (±508) in the control group (P=0.05) (Figure 5).
Discussion
KLK10 and KLK6 are among the most highly and specifically overexpressed genes in pancreatic cancer compared with normal and benign pancreas tissues (Grutzmann et al, 2003; Iacobuzio-Donahue et al, 2003; Yousef et al, 2004).
Our study confirmed a marked overexpression of KLK10 in PDAC by means of a virtual subarray. Immunohistochemistry in native tumour tissue could prove not only an intense expression for KLK10 in 64.4% of the malignant cells, but also for KLK6 in 91.5%. Both proteins were located in the cytoplasm, from where they are likely to be secreted (Borgono et al, 2004).
Co-expression of different kallikreins, similar to the situation found in our study, was already reported in skin and different glands. In these tissues the kallikreins can act independently, but also together as part of proteolytic cascades (Petraki et al, 2002b; Borgono and Diamandis, 2004). The latter seems to be an important mechanism in pancreatic cancer, because expression of KLK10 itself could not be associated with poor survival in PDAC, whereas the co-expression of both kallikreins was significantly associated with poor survival and an R1-resection status, which is an indirect sign for infiltrative and aggressive growth. In multivariate analysis, the co-expression of KLK10 and KLK6 was also an independent risk factor for survival.
It is most interesting, in which ways kallikreins affect cellular signalling and thereby contribute to cancer progression. It was already reported that kallikreins influence communication between malignant cells and their environment by degradation of extracellular matrix and thereby facilitate tumour invasion and metastasis (Borgono and Diamandis, 2004). In case of KLK6, degradation of fibrinogen, laminin, fibronectin and collagen types I and IV are documented (Bernett et al, 2002; Magklara et al, 2003). This cleavage of fractions of the ECM might be of specific importance in pancreatic carcinoma, which is a tumour type with a very high content of stromal tissue (Pilarsky et al, 2008). In contrast, functional data on KLK10 are very limited. Although Zhang et al (2006) suggested, that KLK10 was not even an active protease, it was stated in the same report that neither the protein relevant for conversion of KLK10 into its active form nor the physiological substrates for KLK10 are known. So, the importance of KLK10 in tumour progression remains unclear.
It therefore seems crucial to further pinpoint some of the components, which might be responsible for the pathophysiological effect of KLK10. To find possible interaction partners for both kallikreins we used the in silico method of protein interaction prediction. By means of this method we could identify four potential interaction partners for KLK6. Although α-1 antiproteinase seems to be an inhibitor for KLK6 action, the interaction with AT III shows a branching between kallikreins and blood coagulation cascade, as already reported earlier (Borgono et al, 2004). Another interaction partner was pigment epithelium-derived factor (PEDF), which is the major circulating inhibitor of plasmin. With this interaction, PEDF is linked to the plasminogen activator/plasmin system, which is one of the main protease systems involved in tumour cell invasion and metastasis (Hayashido et al, 2007). Only recently PEDF was also identified as a key inhibitor of stromal vasculature in the mural pancreas. In vivo androgen ablation increased PEDF in human cancer biopsies, which might also be an indirect sign for the interaction of the androgen-responsive kallikrein family and PEDF (Doll et al, 2003). The interaction between KLK6 and PEDF seems highly significant and studies are under way which will further evaluate this topic. Another interaction partner we found is synuclein, which integrates presynaptic signalling and membrane trafficking in neurons. The high expression of KLK6 might thereby play an important role in various pathologic processes of pancreatic cancer.
Although a specific interaction partner for KLK10 could not be found, our study implies that it might have a role in the pathophysiology of PDAC. To ascertain the contribution of KLK10 to pancreatic cancer microenvironment, we used siRNA-mediated gene-silencing (Hammond et al, 2000). AsPC-1 cells, which inherently express high levels of KLK10 mRNA, were transfected with specific siRNA. We could not observe an effect on proliferation or apoptosis in KLK10-silenced cells (data not shown). But KLK10-suppressed clones had markedly reduced cell motility in the Boyden chamber assay. The number of cells migrating through the membrane along an FCS-gradient dropped more than 50%. This is highly significant, as KLK6 was also shown to reduce cell motility (Ghosh et al, 2004).
Although high expression in pancreatic carcinoma indicates that KLK6 and 10 could be promising tumour markers, we could not assess the use of KLK6 and KLK10 as serum biomarkers in PDAC. The serum levels of both proteins showed no significant differences between patients with PDAC and healthy donors. In addition, the serum concentrations were not able to predict the localisation of malignant lesions in the pancreatico-biliary tract. This circumstance can be because of the mainly local action of the kallikreins or fast degradation. Probably future studies including more patients can prove a use for KLK6 or KLK10 as tumour biomarkers in PDAC.
In conclusion, this study shows that KLK10 and KLK6 co-expression has an unfavourable influence on the survival in patients with PDAC and was significantly associated with R1 resection status. This effect might be mediated by direct or indirect interaction of the two kallikreins. The pathophysiological mechanisms are most likely degradation of the extracellular matrix and interaction with angiogenic factors by KLK6, whereas KLK10 augments cell motility. However, our findings suggest a high complexity of interactions between the kallikreins, which leaves it difficult to generally make statements about properties of single kallikreins.
It seems very promising to find out more about the physiological role of KLK10. Consequently, it might be possible to use inhibitors of kallikreins to disrupt interactions between the tumour and its environment and thereby reduce disease progression in patients with pancreatic cancer.
Acknowledgments
We thank Ms Beatrix Jahnke and Mrs Anne Lehner for technical support.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernett MJ, Blaber SI, Scarisbrick IA, Dhanarajan P, Thompson SM, Blaber M (2002) Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system. J Biol Chem 277: 24562–24570 [DOI] [PubMed] [Google Scholar]
- Borgono CA, Diamandis EP (2004) The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer 4: 876–890 [DOI] [PubMed] [Google Scholar]
- Borgono CA, Michael IP, Diamandis EP (2004) Human tissue kallikreins: physiologic roles and applications in cancer. Mol Cancer Res 2: 257–280 [PubMed] [Google Scholar]
- Dawelbait G, Winter C, Zhang Y, Pilarsky C, Grutzmann R, Heinrich JC, Schroeder M (2007) Structural templates predict novel protein interactions and targets from pancreas tumour gene expression data. Bioinformatics 23: i115–i124 [DOI] [PubMed] [Google Scholar]
- Diamandis EP, Scorilas A, Fracchioli S, Van Gramberen M, De Bruijn H, Henrik A, Soosaipillai A, Grass L, Yousef GM, Stenman UH, Massobrio M, Van Der Zee AG, Vergote I, Katsaros D (2003) Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J Clin Oncol 21: 1035–1043 [DOI] [PubMed] [Google Scholar]
- Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE (2003) Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med 9: 774–780 [DOI] [PubMed] [Google Scholar]
- Eccles SA, Welch DR (2007) Metastasis: recent discoveries and novel treatment strategies. Lancet 369: 1742–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh MC, Grass L, Soosaipillai A, Sotiropoulou G, Diamandis EP (2004) Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells. Tumour Biol 25: 193–199 [DOI] [PubMed] [Google Scholar]
- Grutzmann R, Foerder M, Alldinger I, Staub E, Brummendorf T, Ropcke S, Li X, Kristiansen G, Jesnowski R, Sipos B, Lohr M, Luttges J, Ockert D, Kloppel G, Saeger HD, Pilarsky C (2003) Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch 443: 508–517 [DOI] [PubMed] [Google Scholar]
- Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, Sipos B, Foerder M, Alldinger I, Jahnke B, Schackert HK, Kalthoff H, Kremer B, Kloppel G, Saeger HD (2004) Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 6: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hayashido Y, Hamana T, Ishida Y, Shintani T, Koizumi K, Okamoto T (2007) Induction of alpha2-antiplasmin inhibits E-cadherin processing mediated by the plasminogen activator/plasmin system, leading to suppression of progression of oral squamous cell carcinoma via upregulation of cell-cell adhesion. Oncol Rep 17: 417–423 [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH (2003) Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res 63: 8614–8622 [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ (2008) Cancer statistics, 2008. CA Cancer J Clin 58: 71–96 [DOI] [PubMed] [Google Scholar]
- Lowenfels AB, Maisonneuve P (2006) Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol 20: 197–209 [DOI] [PubMed] [Google Scholar]
- Luo LY, Katsaros D, Scorilas A, Fracchioli S, Bellino R, van Gramberen M, de Bruijn H, Henrik A, Stenman UH, Massobrio M, van der Zee AG, Vergote I, Diamandis EP (2003) The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res 63: 807–811 [PubMed] [Google Scholar]
- Magklara A, Mellati AA, Wasney GA, Little SP, Sotiropoulou G, Becker GW, Diamandis EP (2003) Characterization of the enzymatic activity of human kallikrein 6: autoactivation, substrate specificity, and regulation by inhibitors. Biochem Biophys Res Commun 307: 948–955 [DOI] [PubMed] [Google Scholar]
- McIntosh MW, Liu Y, Drescher C, Urban N, Diamandis EP (2007) Validation and characterization of human kallikrein 11 as a serum marker for diagnosis of ovarian carcinoma. Clin Cancer Res 13: 4422–4428 [DOI] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun KS, Sharma S, Chandrika KN, Deshpande N, Palvankar K, Raghavnath R, Krishnakanth R, Karathia H, Rekha B, Nayak R, Vishnupriya G, Kumar HG, Nagini M, Kumar GS, Jose R, Deepthi P, Mohan SS, Gandhi TK, Harsha HC, Deshpande KS, Sarker M, Prasad TS, Pandey A (2006) Human protein reference database--2006 update. Nucleic Acids Res 34: D411–D414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara H, Mimori K, Utsunomiya T, Barnard GF, Ohira M, Hirakawa K, Mori M (2005) Clinicopathologic and biological significance of kallikrein 6 overexpression in human gastric cancer. Clin Cancer Res 11: 6800–6806 [DOI] [PubMed] [Google Scholar]
- Petraki CD, Gregorakis AK, Papanastasiou PA, Karavana VN, Luo LY, Diamandis EP (2003) Immunohistochemical localization of human kallikreins 6, 10 and 13 in benign and malignant prostatic tissues. Prostate Cancer Prostatic Dis 6: 223–227 [DOI] [PubMed] [Google Scholar]
- Petraki CD, Gregorakis AK, Vaslamatzis MM, Papanastasiou PA, Yousef GM, Levesque MA, Diamandis EP (2006) Prognostic implications of the immunohistochemical expression of human kallikreins 5, 6, 10 and 11 in renal cell carcinoma. Tumour Biol 27: 1–7 [DOI] [PubMed] [Google Scholar]
- Petraki CD, Karavana VN, Luo LY, Diamandis EP (2002a) Human kallikrein 10 expression in normal tissues by immunohistochemistry. J Histochem Cytochem 50: 1247–1261 [DOI] [PubMed] [Google Scholar]
- Petraki CD, Karavana VN, Revelos KI, Luo LY, Diamandis EP (2002b) Immunohistochemical localization of human kallikreins 6 and 10 in pancreatic islets. Histochem J 34: 313–322 [DOI] [PubMed] [Google Scholar]
- Petraki CD, Karavana VN, Skoufogiannis PT, Little SP, Howarth DJ, Yousef GM, Diamandis EP (2001) The spectrum of human kallikrein 6 (zyme/protease M/neurosin) expression in human tissues as assessed by immunohistochemistry. J Histochem Cytochem 49: 1431–1441 [DOI] [PubMed] [Google Scholar]
- Pilarsky C, Ammerpohl O, Sipos B, Dahl E, Hartmann A, Wellmann A, Braunschweig T, Lohr M, Jesnowski R, Friess H, Wente MN, Kristiansen G, Jahnke B, Denz A, Ruckert F, Schackert HK, Kloppel G, Kalthoff H, Saeger HD, Grutzmann R (2008) Activation of Wnt signalling in stroma from pancreatic cancer identified by gene expression profiling. J Cell Mol Med [e-pub ahead of print] [DOI] [PMC free article] [PubMed]
- Shaw JL, Diamandis EP (2007) Distribution of 15 human kallikreins in tissues and biological fluids. Clin Chem 53: 1423–1432 [DOI] [PubMed] [Google Scholar]
- Shrikhande SV, Kleeff J, Reiser C, Weitz J, Hinz U, Esposito I, Schmidt J, Friess H, Buchler MW (2007) Pancreatic resection for M1 pancreatic ductal adenocarcinoma. Ann Surg Oncol 14: 118–127 [DOI] [PubMed] [Google Scholar]
- Sipos B, Moser S, Kalthoff H, Torok V, Lohr M, Kloppel G (2003) A comprehensive characterization of pancreatic ductal carcinoma cell lines: towards the establishment of an in vitro research platform. Virchows Arch 442: 444–452 [DOI] [PubMed] [Google Scholar]
- Wolff RA, Chiao P, Lenzi R, Pisters PW, Lee JE, Janjan NA, Crane CH, Evans DB, Abbruzzese JL (2000) Current approaches and future strategies for pancreatic carcinoma. Invest New Drugs 18: 43–56 [DOI] [PubMed] [Google Scholar]
- Yousef GM, Borgono CA, Popalis C, Yacoub GM, Polymeris ME, Soosaipillai A, Diamandis EP (2004) In-silico analysis of kallikrein gene expression in pancreatic and colon cancers. Anticancer Res 24: 43–51 [PubMed] [Google Scholar]
- Zhang Y, Bhat I, Zeng M, Jayal G, Wazer DE, Band H, Band V (2006) Human kallikrein 10, a predictive marker for breast cancer. Biol Chem 387: 715–721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.