SUMMARY
DNA damage initiates a series of p53 pulses. Although much is known about the interactions surrounding p53, little is known about which interactions contribute to p53’s dynamical behavior. The simplest explanation is that these pulses are oscillations intrinsic to the p53/Mdm2 negative feedback loop. Here we present evidence that this simple mechanism is insufficient to explain p53 pulses; we show that p53 pulses are externally driven by pulses in the upstream signaling kinases, ATM and Chk2, and that the negative feedback between p53 and ATM, via Wip1, is essential for maintaining the uniform shape of p53 pulses. We propose that p53 pulses result from repeated initiation by ATM which is re-activated by persistent DNA damage. Our study emphasizes the importance of collecting quantitative dynamic information at high temporal resolution for understanding the regulation of signaling pathways and opens new ways to manipulate p53 pulses to ask questions about their function in response to DNA damage.
INTRODUCTION
The tremendous recent progress in identifying the parts that make up cells, and the parallel advances in identifying interactions between the parts has led to an increasing appreciation of the complexity of biological networks. The challenge facing us now is to understand how network structure determines the functional behavior of these systems: in other words, how the concentration, location, specific interactions and dynamical behavior of individual proteins controls the processing of information within the cell. To build this level of understanding we need quantitative information on the dynamics of key proteins under many different conditions; we also need to be able to make a variety of perturbations to the normal network structure and determine how these changes affect the dynamic behavior. The network surrounding the tumor suppressor protein p53 is a natural choice for studies of this kind: p53 is a master regulator of many essential cellular decisions, its interactions have been well studied, and it has been shown to undergo complex, tightly controlled dynamical behavior in response to DNA damage.
The p53 protein is kept at low levels in cells under normal conditions, primarily through a negative feedback loop with Mdm2; p53 positively regulates Mdm2 by activating Mdm2 transcription, and Mdm2 negatively regulates p53 by promoting its ubiquitination and degradation (Barak et al., 1993; Haupt et al., 1997; Kubbutat et al., 1997; Wu et al., 1993). Under various stress conditions, p53 is activated through upstream mediators. Most of these mediators induce post-translational modification of p53 that disrupt the p53-Mdm2 interaction. For example, DNA double-strand breaks (DSBs) induce rapid autophosphorylation and activation of ataxia-telangiectasia mutated protein (ATM) (Bakkenist and Kastan, 2003), which then phosphorylates both p53 (Banin et al., 1998; Canman et al., 1998) and Mdm2 (Khosravi et al., 1999; Maya et al., 2001), as well as other substrates. These events lead to disassociation of the p53-Mdm2 complex, stabilizing p53 and increasing p53 protein levels. ATM also phosphorylates the checkpoint kinase Chk2 (Ahn et al., 2000; Matsuoka et al., 2000), which directly phosphorylates p53, further contributing to p53 stabilization (Chehab et al., 1999). After DNA damage caused by γ-irradiation, p53 and Mdm2 show repeated pulses that were originally characterized as damped oscillations (Lev Bar-Or et al., 2000). We later showed in single-cell experiments that individual cells show varying numbers of p53 pulses of fixed amplitude and duration (Lahav et al., 2004). The appearance of damped oscillations results from the averaging of the pulses across a population of cells. A recent in vivo study showed pulses of p53 activity in a transgenic mouse suggesting that these dynamics are not limited to cultured cancer cells (Hamstra et al., 2006).
The p53/Mdm2 negative feedback loop is composed of interactions on two different timescales: a slow positive transcriptional arm and a fast negative protein-protein interaction arm (Figure 1A). Mathematical models predict that such feedback loops can exhibit oscillatory behavior (Goodwin, 1965; Tyson et al., 2003); thus, the simplest explanation of the repeated pulses we observed was that they were intrinsic oscillations of the p53/Mdm2 loop (Lev Bar-Or et al., 2000; Mihalas et al., 2000; Monk, 2003; Tiana et al., 2002) (Figure 1A). Other models include additional features such as an additional positive feedback on p53 (Ciliberto et al., 2005; Zhang et al., 2007), or assume high constant levels of active upstream DNA damage sensor proteins (such as ATM) to account for the sustained oscillations (Ma et al., 2005; Wagner et al., 2005). An interesting and distinct proposal, from us, Uri Alon and colleagues, suggests that p53 oscillations might depend on pulses of the upstream damage signaling elements (Geva-Zatorsky et al., 2006) (Figure 1B).
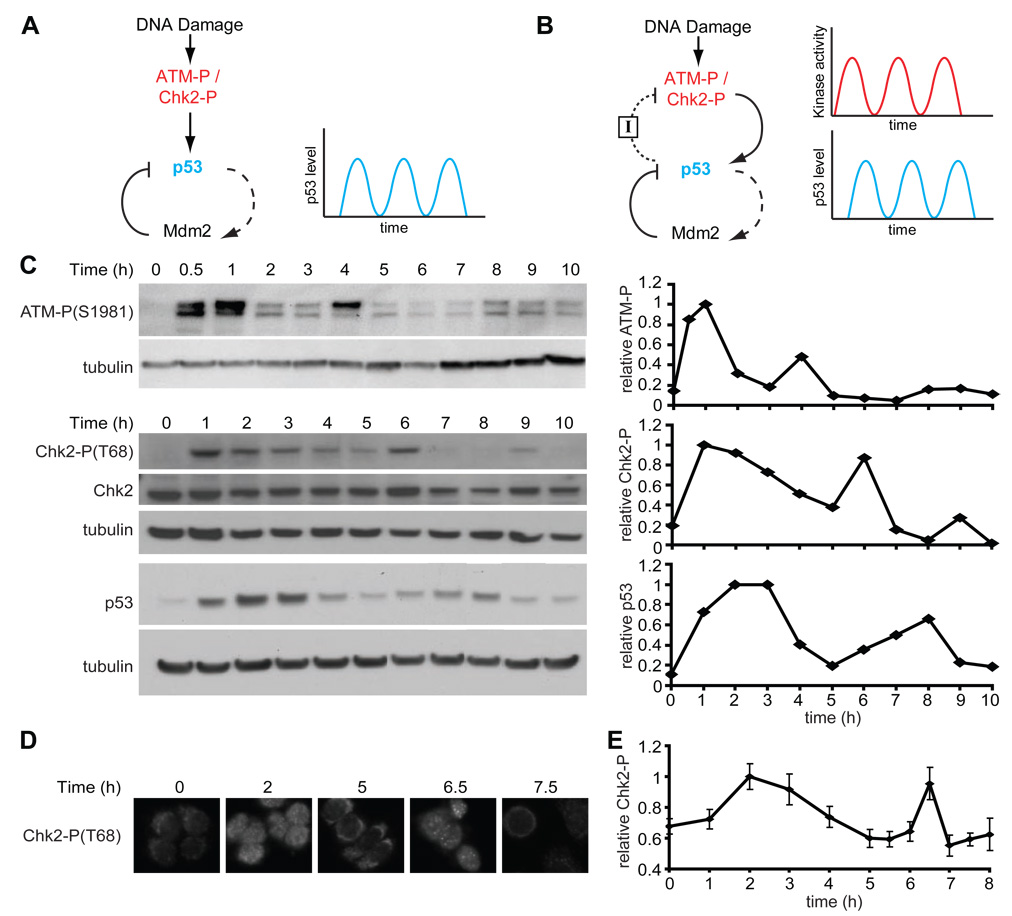
Figure 1. Dynamics of DNA damage kinases.
(A) In one class of previous models, repeated p53 pulses result from the negative feedback loop between p53 and Mdm2. In this model, DNA damage activates the p53-Mdm2 module through ATM and Chk2, and subsequent oscillation results from the timing delay between p53 upregulation and Mdm2 activation. Dashed lines correspond to slow rate responses (e.g. transcription). Solid lines correspond to fast rate responses (e.g. protein-protein interaction or phosphorylation). (B) In an alternative model, in addition to the p53-Mdm2 negative feedback loop, p53 creates a separate negative feedback loop with the upstream kinases ATM and Chk2 by upregulating an inhibitor, I. In this proposal, the signaling kinase activities may oscillate as well. The nature and rate of feedback between p53 and ATM through I is unknown. (C) Left: Immunoblots of ATM-P(S1981), Chk2-P(T68), and p53 kinetics in MCF7 cells irradiated with 10Gy of γ-irradiation. Right: protein quantification of the presented blots normalized by tubulin, in units relative to peak intensity. (D) Representative Chk2-P(T68) immunofluorescence images of MCF7 cells fixed at the indicated times after irradiation with 10Gy of γ-irradiation. (E) Quantification of the 10% brightest cells from Chk2-P(T68) immunofluorescence analysis. Bars = standard deviations. (n = 127 cells/time point)
To clarify the origin of the p53 pulses, we set out to study the dynamics of the p53 network in more detail, by performing quantitative population and single cell measurements of the dynamics of several proteins in the p53 signaling pathway. We integrated this information into a theoretical framework to generate predictions that we then tested with specific genetic and chemical perturbations. Our results show that the p53/Mdm2 loop does not, by itself, drive sustained p53 oscillations: the p53 pulses depend on pulses of the proteins that sense and transfer the damage signal to p53, and on a second negative feedback loop between p53 and the upstream signaling proteins. We identify the phosphatase Wip1 as the central element mediating this second feedback loop. We suggest that the pulses arise from periodic examinations of the damaged DNA by ATM to determine whether the damage has been repaired; if not, additional pulses of both ATM-P and p53 are triggered.
RESULTS
ATM-P and Chk2-P show pulsatile dynamics in response to DNA damage
To broaden our understanding of the dynamics of the system, we first examined the dynamics of the kinases ATM and Chk2 in response to γ-irradiation. ATM is a sensor protein that transmits information about DNA damage to p53; it binds to DSBs, and is activated by autophosphorylation on Ser1981 in response to DNA damage. Phosphorylated ATM phosphorylates Mdm2 and p53, reducing the strength of their interaction and thus stabilizing p53. In addition, ATM-P has an indirect effect on p53 via Chk2, which it phosphorylates on Thr68. As shown in Figure 1C, clear indications of repeated pulses are observed for phosphorylated ATM (ATM-P) and for phosphorylated Chk2 (Chk2-P) in immunoblots. The timing between the first two pulses was 3.6 ± 1.2 h for ATM-P, 4.5 ± 1.0 h for Chk2-P, and 5.2 ± 0.6 h for p53, based on three independent experiments. In all our experiments, the second pulse of ATM-P precedes the second pulse of Chk2-P, as expected. Both the ATM-P pulse and the Chk2-P pulse precede the second pulse of p53, consistent with the possibility that the second pulse of p53 depends on the second induction of ATM-P and Chk2-P.
In immunoblot studies of p53, the amplitude of the second pulse is lower than that of the first. This phenomenon is also seen for ATM-P and Chk2-P (Figure 1C). This could result from a decrease in the amplitude of the second pulse in all cells, or, as was shown for p53 ((Lahav et al., 2004) and Figure S1), from undamped pulses that are present in only a subset of the population. To distinguish between these possibilities, we measured the level of Chk2-P in single cells following γ-irradiation (Figure 1D) and found that the amplitudes of the first and second pulse were essentially the same when probing the brightest 10% of cells (Figure 1E). We were unable to measure ATM-P dynamics at the single cell level since the available antibodies give a poor signal to noise ratio in fixed MCF7 cells. Since Chk2-P level is thought to be an indicator of ATM-P activity, however, it is reasonable to suggest that both ATM-P and Chk2-P pulses are undamped. In many of the experiments described below, we use Chk2-P as a surrogate for ATM-P levels. The demonstration that both ATM-P and Chk2-P show repeated pulses is consistent with the idea that p53 pulses may be repeatedly triggered by upstream events.
Chk2-P pulses depend on p53
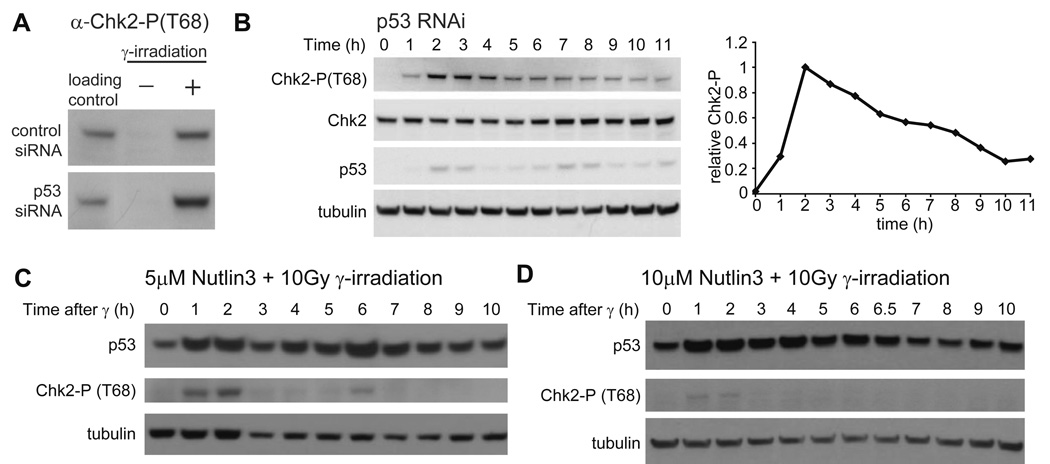
If DNA damage is continuous, and therefore ATM-P is continuously made, what explains the fact that ATM-P and Chk2-P levels are repressed in between pulses? One possibility is that a negative feedback between p53 and the damage signal represses ATM-P/Chk2-P levels (as in the dual negative feedback model; Figure 1B and (Geva-Zatorsky et al., 2006)), while persistent damage causes their repeated reactivation. We therefore looked directly for a negative feedback between p53 and Chk2-P. First, we used RNAi to remove p53 and examined the resulting dynamics of Chk2-P levels following γ-irradiation. MCF7 cells transfected with siRNA against p53 express p53 at a level of ~5% of that seen in cells treated with the RNAi control (data not shown). This residual p53 level was found to be due to a small percentage of cells in the population in which the siRNA was not transfected (Figure S2). The effect on Chk2-P kinetics was two-fold: the overall level of Chk2-P is increased (Figure 2A) and the second pulse is not observed in p53 RNAi cells (Figure 2B) compared with no siRNA (Figure 1C) and with control siRNA cells (data not shown).
Figure 2. p53 affects Chk2-P levels and dynamics.
(A) Immunoblots of Chk2-P(T68) in MCF7 cells transfected with control or p53 siRNA 48h prior to harvesting. Cells were harvested before irradiation or 1h after irradiation with 10 Gy of γ-irradiation. (B) Immunoblots of Chk2-P(T68) and p53 kinetics in MCF7 cells transfected with p53 siRNA 48h prior to irradiation with 10 Gy of γ -irradiation. Cells were harvested at the indicated times after irradiation. The graph shows quantification of Chk2-P levels normalized by tubulin and relative to peak intensity. Blots are representative of triplicate experiments. (C, D) Immunoblots of Chk2-P(T68) and p53 kinetics in MCF7 cells treated with (C) 5 µM Nutlin3 for 2h or (D) 10 µM Nutlin3 for 4h, prior to irradiation with 10 Gy of γ-irradiation. Cells were harvested at the indicated time after irradiation. Blots are representative of duplicate experiments.
Next, we asked whether Chk2-P dynamics could also be changed by artificially increasing p53 levels using the small molecule Nutlin3, which inhibits the interaction between p53 and Mdm2 (Vassilev, 2004). To increase p53 levels and activate the putative negative feedback, we pretreated cells with 5µM Nutlin3 for two hours, and then irradiated the cells to produce DSBs. Compared to untreated cells (Figure 1C), the duration of the first pulse of Chk2-P was reduced from 5 hours (Figure 1C) to 3 hours (Figure 2C and S3). Pre-treatment of cells with a higher concentration (10µM) of Nutlin3 for 4 hours led to even greater inhibition of Chk2-P (Figure 2D and Figure S3). These observations indicate the presence of a negative feedback mechanism connecting p53 with the kinases that signal the presence of DNA damage, such as ATM and Chk2.
ATM-P and Chk2-P kinetics affect p53 pulses
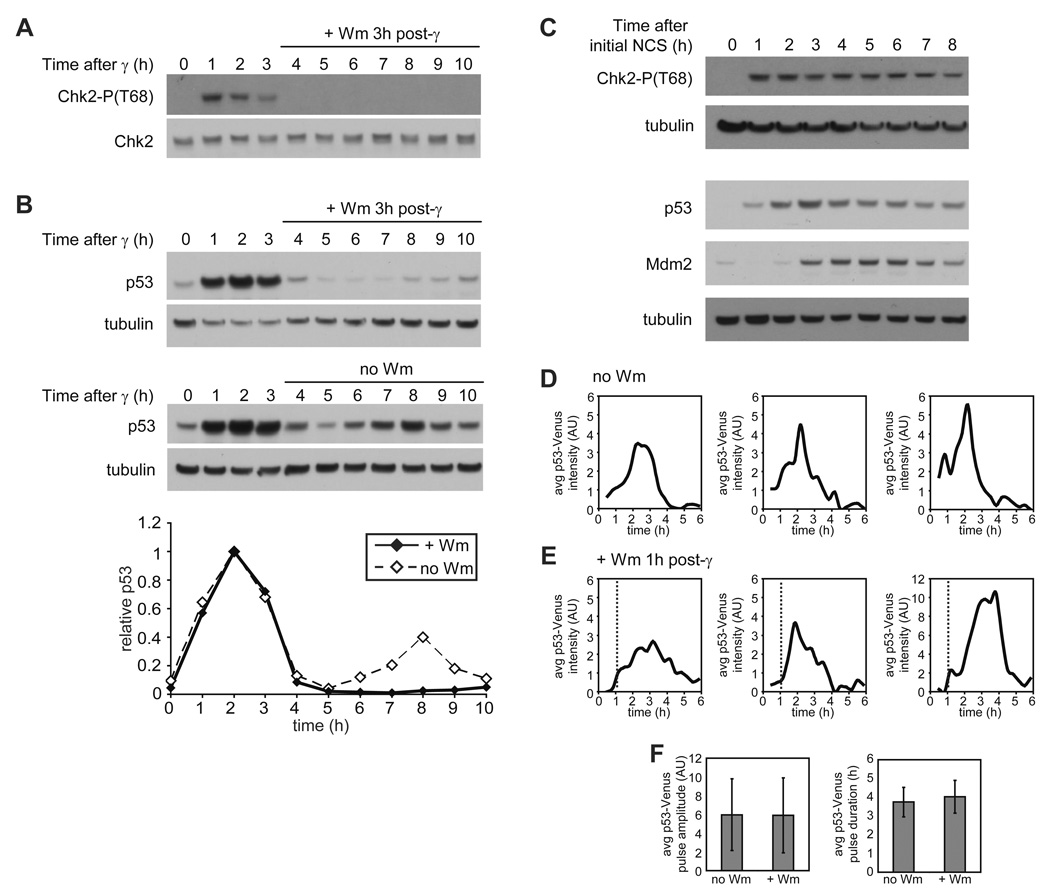
It is well established that activation of ATM is required to up-regulate p53 in response to irradiation (Bakkenist and Kastan, 2003; Banin et al., 1998; Canman et al., 1998; Khosravi et al., 1999; Maya et al., 2001). However, it is not clear whether the repeated pulses of ATM-P/Chk2-P, seen in Figure 1C, are required for the series of p53 pulses. It is possible that, as some models suggest, the p53/Mdm2 feedback loop can independently generate oscillations once triggered. To ask whether the kinase pulses drive the pulses in p53, we took two approaches. First we used wortmannin, an inhibitor of phosphoinositide 3-kinase-related kinases such as ATM (Sarkaria et al., 1998), to block ATM-P and Chk2-P pulses. We added wortmannin to MCF7 cells 3 hours after exposing the cells to γ-irradiation, directly after the first ATM-P pulse is complete. As expected, Chk2-P levels decreased dramatically upon wortmannin addition (Figure 3A). The first pulse of p53 was unaffected, but no subsequent p53 pulses were observed (Figure 3B). Secondly, we examined the effect of constant elevated levels of Chk2-P, caused by re-damaging cells with neocarzinostatin (NCS). This treatment led to high levels of p53 with only a slight decrease at the later time points, but to no pulses (Compare Figure 3C to Figure 1C). We suggest that this decrease in p53 results from increasing levels of Mdm2, which indeed increase in response to constant NCS (Figure 3C). These data support the idea that although Mdm2 is important for regulating p53 levels it is not the only source of p53 pulses; instead, the pulses we observed for ATM-P and Chk2-P (Figure 1C) are required for the p53 pulses, and an initial activation of p53 by ATM and Chk2, is insufficient to drive autonomous, multiple p53 pulses by the p53/Mdm2 module.
Figure 3. p53 pulses depend on ATM dynamics.
(A, B) Immunoblots of Chk2-P(T68) (A), and p53 (B) kinetics in MCF7 cells irradiated with 10 Gy of γ-irradiation. Fresh medium containing 100µM wortmannin (+ Wm) or no wortmannin (no Wm) was added to cells 3h after irradiation. Quantifications are of indicated proteins normalized by tubulin and relative to peak intensity. Blots are representative of triplicate experiments. (C) Immunoblots of Chk2-P(T68), p53, and Mdm2 kinetics in MCF7 cells treated with 400 ng/ml NCS every hour. Blots are representative of triplicate experiments. (D, E) The first pulse of p53 in representative clonal MCF7 cells expressing p53-Venus after 10 Gy of γ-irradiation. Fresh medium containing 100µM wortmannin (D) or no wortmannin (E) was added to cells 1h after irradiation (time of Wm addition is indicated by dashed line). (F) Average p53-Venus pulse amplitude and duration in cells irradiated with 10 Gy of γ-irradiation and left untreated (n = 67) or treated with 100µM wortmannin (n = 32) 1h post irradiation. Bars = standard deviation.
A striking feature of the system’s dynamics is that the initial pulse of Chk2-P is of longer duration than the subsequent pulses (Figure 1C), while this is not true for p53. We therefore wondered whether a shorter initial pulse of ATM-P (and therefore Chk2-P) might be sufficient to produce a full single pulse of p53. We used Wortmannin to inhibit ATM 1 hour post irradiation and followed Chk2-P and p53 dynamics. Shortening the ATM-P pulse did not prevent the initiation of the p53 pulse: p53 levels continued to increase even after Chk2-P levels fell below the level of detection by immunoblot (Figure S4). p53 levels were slightly decreased compared to those in cells not treated with wortmannin, however, and we therefore looked more closely at the dynamics of p53 on the single-cell level. We monitored p53 levels in response to DNA damage using a p53-Venus fusion protein (Figure S1) and time-lapse fluorescence microscopy. We found that the lowered level of p53 is explained by a reduction in the number of cells undergoing a p53 pulse: 76% of control cells show induction of p53 in response to 10Gy of γ-irradiation, but only 65% of the wortmannin-treated cells do so. This result suggests that a threshold level of ATM-P or Chk2-P may be required to initiate the p53 pulse, and that some cells do not reach this threshold within 1 hour. Those cells that did initiate a p53 pulse showed pulses that were indistinguishable in amplitude and duration from those seen in control cells (Figures 3D – 3F). Our results suggest that a short, 1-hour pulse of ATM and Chk2 activity is sufficient to drive a full single pulse of p53.
Our observations so far are consistent with the idea of a recurrent initiation mechanism for triggering p53 pulses, in which the DNA damage signaling pathway repeatedly checks for the existence of DSBs. Our overall picture is that ATM-P/Chk2-P levels increase in response to DSBs, and are then shut down by the increase in the level of p53. We theorize that when p53 levels are in turn reduced by p53’s interaction with Mdm2, ATM is released to re-examine the DNA. If the number of breaks remains above a certain threshold, the pathway becomes reactivated, leading to a second pulse of p53, and so on until the number of DNA breaks has dropped below the threshold. In this proposed mechanism, the pulses of ATM-P/Chk2-P are not generated by cellular components acting upstream of the kinases at the site of DNA damage; instead, the pulsatile dynamics are a result of the feedback loops involving ATM, Chk2, p53, and a p53-regulated inhibitor protein(s). To examine this proposal more closely, we formalized our assumptions in a computational model. We used this model to help test candidates for the putative p53-dependent inhibitor of ATM/Chk2 activity.
Predictions connecting the inhibitor levels and ATM-P/Chk2-P dynamics
To develop a picture of the likely dynamics of a putative inhibitor, we constructed a delay differential equation model. Simulation of the model proposed by Geva-Zatorsky et al., 2006, like our observed data, shows pulses in upstream signaling molecules, and we therefore used this model as our starting point. Since the exact kinetic parameters associated with this model are not fully quantified, the output of the model is not expected to offer a perfect recapitulation of the DNA damage response. Instead, by making reasonable assumptions for parameter values based on currently available data and our experimental observations, we aimed to develop a simplified model that would reproduce the overall dynamical behavior of the system and aid in exploring the mechanistic principles responsible for p53’s behavior.
A summary of the model is shown in Box 1. The model of Geva-Zatorsky et al., 2006 used an inhibitor of ATM/Chk2 with parameters similar to those for Mdm2. In our model, we introduced an inhibitor with distinct parameters that has different behavior from Mdm2. We reasoned that since the initial pulse of upstream kinase activity is of relatively long duration, the inhibitor is likely to be present at a low level initially and takes a significant amount of time to build up to the level that begins to inhibit the damage signal. We modeled this dynamic behavior by requiring a low initial concentration of inhibitor and a rate of p53-induced inhibitor production that was slow compared with Mdm2 production. In our model the decay rate of the kinase inhibitor is also relatively slow, allowing the inhibitor concentration to build up and stay relatively high even after p53 is degraded. This would explain the shorter ATM-P/Chk2-P second pulse; after p53 levels are induced for the second time, the inhibitor levels reach the critical inhibition level faster (Figure 4A, 4B, and Figure S5C).
Box 1
We developed a delay differential equation model for ATM-P/Chk2-P dynamics based in part on the model proposed in Geva-Zatorsky et al., 2006. We considered five species: p53active, p53inactive, Mdm2, Signal (representing the active upstream kinases such as ATM-P and Chk2-P), and inhibitor. The equations describing the dynamics of these species are: Our model is similar in essence to that of Geva-Zatorsky et al., 2006, with one key difference. Because the nature of the inhibitor was not known, Geva-Zatorsky et al. chose to model it with identical dynamics to Mdm2. In contrast, ours has explicit parameters for the production rate βi, delay τi, and degradation rate αi of the inhibitor that are distinct from those of Mdm2. We also included a term describing p53-independent Mdm2 production, βmi. In our model, the Signal also negatively regulates Mdm2; this corresponds to the inactivation of Mdm2 by ATM-P (Khosravi et al., 1999; Maya et al., 2001; Stommel and Wahl, 2004). Adding this interaction appears to provide a slightly closer correspondence between the amplitude of the response in simulations and that seen in experiments (Figures S5A and S5B).
For the Signal dynamics, we used a large constant production term based on data showing a rapid induction of ATM following DNA damage. θ(t) is the Heaviside step function, which we used to simulate the presence of DNA damage starting at time t = 0. We modeled the inhibition of the signal by a putative inhibitor as a Hill function with a relatively large saturation rate and a Hill coefficient of ni = 4 due to our observation that later pulses of ATM-P and Chk2-P are relatively sharp and rapid. We selected a half-maximal inhibition threshold value based on our prediction that the inhibitor needs to build up in concentration before efficient inhibition of the signal occurs. We also include a separate degradation term for the signal activity to account for additional inhibitors of the signal, such as constitutive dephosphorylation of the kinases by PP2A (Goodarzi et al., 2004) and degradation of active kinases. All other parameters were selected to best fit the experimentally observed behavior of p53 in our previous single cell studies (Lahav et al., 2004) and the dynamics of the signal kinases as presented in this study. Values for all parameters are given in Table S1.
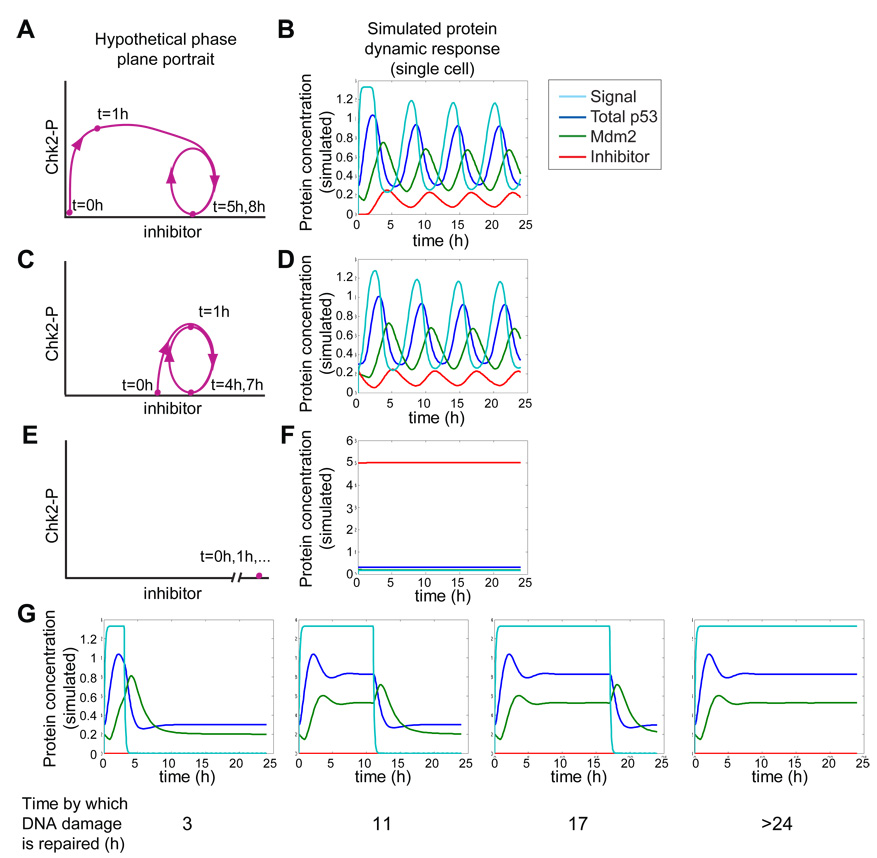
Figure 4. Simulation of the p53 signaling pathway using the recurrent-initiation model.
(A, C, E) Hypothetical phase-plane trajectories of Chk2-P and putative inhibitor concentrations following γ-irradiation based on experimental observations. (B, D, F, G) Numerical simulations of the recurrent initiation model in response to DNA damage. Signal (light blue), total p53 (dark blue), Mdm2 (green), and inhibitor (red) concentrations following γ-irradiation in unperturbed (A, B), elevated-inhibitor (C, D), and over-expressed-inhibitor (E, F) conditions. (G) Simulated dynamics in null inhibitor condition for cells in which DNA damage has been repaired by the indicated times. Total p53 includes both active and inactive p53.
This model captures the basic features of the dynamics of the upstream kinases (Signal), p53, and Mdm2 in terms of pulse ordering and relative pulse duration, and allowed us to generate several testable predictions for the dynamics of the inhibitor and how it affects the dynamics of ATM-P/Chk2-P and p53, including the following: (1) The inhibitor should show pulses, and its dynamics should lag those of p53 (Figure 4B). (2) The inhibitor should start at a low initial concentration, increase to a certain threshold, and then show several pulses of a higher concentration. As long as p53 continues to pulse, the inhibitor will not return to the original low concentration (Figure 4B). (3) If the initial level of the inhibitor is increased to the threshold level, the duration of the first pulse of ATM-P/Chk2-P will be shortened and its initiation will be delayed (Figure 4C, 4D, and Figure S5D). (4) If the inhibitor is expressed at levels significantly above the threshold, up-regulation of ATM and Chk2 (and consequently p53) will be suppressed (Figure 4E, 4F, and Figure S5E). (5) If the inhibitor is eliminated, ATM-P/Chk2-P and p53 levels will remain high following induction as long as DNA damage persists (Figure 4G). We next performed a set of experiments to test these predictions and to identify the putative inhibitor.
Identifying the putative inhibitor
Based on our experimental observations and theoretical predictions we next searched for the mechanism that links p53 activation and ATM/Chk2 inhibition. There are at least 6 known negative feedbacks involving p53 (Harris and Levine, 2005), two of which may regulate ATM/Chk2 function. One is cyclin G, a transcriptional target gene of p53, which has been shown to recruit protein phosphatase 2A (PP2A) (Okamoto et al., 1996) to dephosphorylate Mdm2 and ATM (Goodarzi et al., 2004; Ohtsuka et al., 2004). However, cyclin G does not fit our profile for the putative inhibitor responsible for ATM-P/ Chk2-P shut-down. Breast cancer cells, such as MCF7, have been shown to have high constant levels of cyclin G mRNA and protein in unstressed conditions (Reimer et al., 1999). We confirmed the presence of high levels of cyclin G protein in MCF7 cells in the absence of DNA damage using Western blots, and also found that these levels did not increase in response to γ-irradiation (data not shown). At least in MCF7 cells, then, cyclin G does not appear to play an essential role in the generation of p53 pulses.
The second candidate inhibitor is the phosphatase Wip1 (PPM1D). Wip1 is a downstream target of p53 (Fiscella et al., 1997); it dephosphorylates p53 (Lu et al., 2005), Chk2 (Fujimoto et al., 2005; Oliva-Trastoy et al., 2006), and ATM (Shreeram et al., 2006). Consistent with these observations, RNAi against Wip1 leads to high constant levels of ATM-P and Chk2-P after DNA damage (Fujimoto et al., 2005; Shreeram et al., 2006). In addition, a recent study showed that Wip1 also stabilizes Mdm2 by dephosphorylating it on Ser 395 (Lu et al., 2007). Since the exact kinetic parameters of the interactions within the system are not known, a more complex model including the direct interactions between Wip1 and p53 (Lu et al., 2005), and Wip1 and Mdm2 (Lu et al., 2007), can give rise to the same dynamic behavior that results from our simplified model (data not shown).
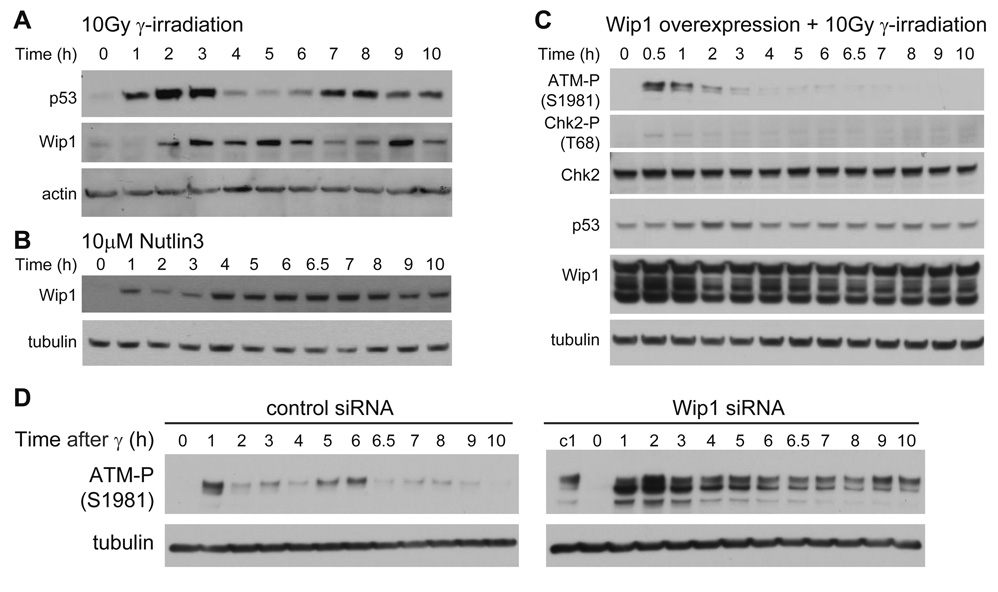
To explore the hypothesis that Wip1 is the inhibitor assumed in our model we first followed Wip1 dynamics in response to γ-irradiation. In our simulations, the putative inhibitor shows repeated pulses that lag behind the p53 pulses (Figure 4B). Experimentally, we see an increase in Wip1 levels 2 hours after γ-irradiation (Figure 5A), one hour after p53 levels begin to rise. Wip1 levels decrease in the 7th hour post-irradiation, several hours after the decrease in p53 levels, and as predicted they do not appear to fall back to the initial low level. Wip1 levels then increase again shortly after the second increase in p53 levels (Figure 5A). These results are consistent with behavior predicted for the putative inhibitor.
Figure 5. The Wip1 phosphatase shows behavior similar to the simulated behavior of the putative inhibitor.
(A) Immunoblots of p53 and Wip1 kinetics in MCF7 cells irradiated with 10 Gy of γ-irradiation. (B) Immunoblot of Wip1 kinetics in MCF7 cells in response to 10 µM Nutlin3 (C) Immunoblots of ATM-P(S1981), Chk2-P(T68), and p53 kinetics in MCF7 cells transfected with pCMV-Wip1 48h prior to irradiation with 10 Gy of γ-irradiation. Blots are representative of duplicate experiments. (D) Immunoblots of ATM-P kinetics in MCF7 cells transfected with control or Wip1 siRNA and irradiated with 40 Gy of γ-irradiation. Lane “c1” in the Wip1 RNAi blot is the 1h control RNAi sample for comparison. Blots are representative of duplicate experiments.
Our model predicts that increasing the level of the inhibitor should shorten the duration of the first ATM-P/Chk2-P pulse (Figures 4C and 4D). Indeed, we did observe a shortening in the Chk2-P pulse when we used Nutlin3 to increase p53 levels prior to irradiation (Figure 2C, D). Figure 5B shows that Wip1 levels are also elevated in response to Nutlin3 treatment, consistent with the possibility that it might be responsible for the inhibition of Chk2-P (and by implication ATM-P). To test this possibility directly, we overexpressed Wip1 in MCF7 cells by transfecting a plasmid containing Wip1 cDNA under the constitutive CMV promoter. After exposure to γ-irradiation, these Wip1-overexpressing cells showed partial inhibition of ATM phosphorylation and significant inhibition of Chk2 phosphorylation (Figure 5C and Figure S6A), behavior that is consistent with the possibility that Wip1 is the inhibitor assumed in our model (Figures 4E and 4F). As expected, the level of p53 induction was also reduced (Figure 5C). On the single-cell level, ~74% of the population showed increased Wip1 expression (Figure S6B); cells with induced p53 levels showed low expression of Wip1, suggesting that they may not have been successfully transfected with the Wip1-expressing plasmid (Figure S6C). Lastly, we have shown that removal of Wip1 leads to high levels of ATM-P with no pulses (Figure 5D), a behavior consistent with that predicted for the putative inhibitor (Figure 4G). Taken together, our results indicate that Wip1 functions to link p53 with Chk2 and ATM and therefore might play a role in controlling p53 pulses in response to γ-irradiation.
Uniform p53 pulses depend on Wip1
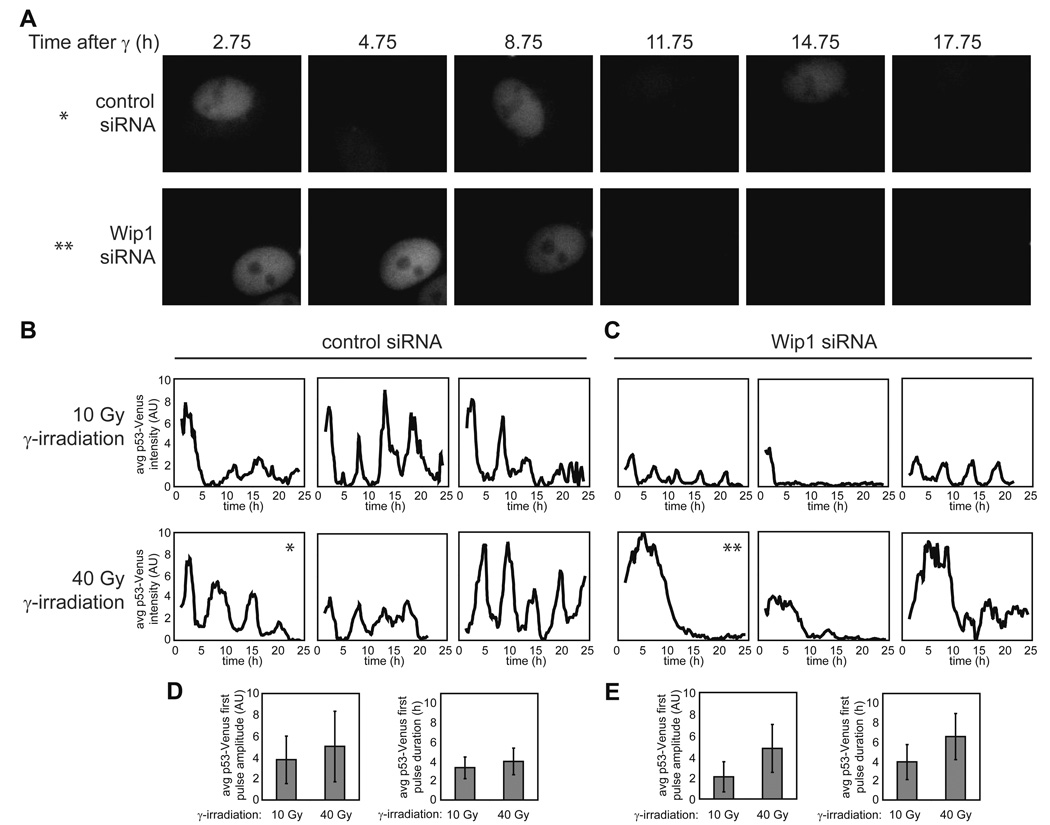
The fact that RNAi against Wip1 leads to high levels of ATM-P and Chk2-P (Fujimoto et al., 2005; Shreeram et al., 2006) and eliminates ATM-P pulses after DNA damage (Figure 5D) suggests that removal of Wip1 might also cause changes in p53 dynamics (Figure 4G). We therefore treated MCF7 cells expressing a p53-Venus fusion protein (Figure S1) with Wip1 siRNA. The p53 response is significantly distorted in these cells. Figure 6 shows p53 expression in cells transfected with control siRNA (Figure 6A, upper panel, and Figure 6B) or with Wip1 siRNA (Figure 6A, lower panel, and Figure 6C) following treatment with 10Gy and 40Gy of γ-irradiation. To ensure that we tracked cells that were transfected with siRNA, we co-transfected cells with a red fluorescently-labeled siRNA and analyzed only the cells that show red signal. In control cells, the p53 pulses show their characteristic fixed amplitude and duration at both radiation doses (Figures 6B and 6D). The Wip1 RNAi-treated cells, however, show deviations from the stereotypical pulse (Figures 6C and 6E); as predicted by our model, a high irradiation dose led to an increase in the average duration of the p53 pulse (4.0 ± 1.4 h vs 6.6 ± 2.4 h). In addition, pitch analysis of p53 dynamics, as describe in (Geva-Zatorsky et al., 2006), showed that Wip1 RNAi reduced the percentage of cells with detectable pitch (Figure S7).
Figure 6. Wip1 RNAi treatment alters p53 dynamics.
(A) Images of clonal MCF7 cells expressing p53-Venus following 40 Gy of γ-irradiation. The upper panel shows an example of a cell treated with control siRNA. The lower panel shows an example of a cell treated with Wip1 siRNA. (B, C) Average nuclear p53 levels in representative clonal MCF7 cells expressing p53-Venus transfected with control siRNA (B) or Wip1 siRNA (C) irradiated with 10 Gy or 40 Gy of γ-irradiation. Asterisks indicate the correspondence between these plots and the images presented in (A). (D, E) Average amplitude and duration of the first pulse of p53-Venus for control cells (D) (10 Gy n = 84; 40 Gy n = 54) and Wip1 RNAi-treated cells (E) (10 Gy n = 62; 40 Gy n = 63). Bars = standard deviation.
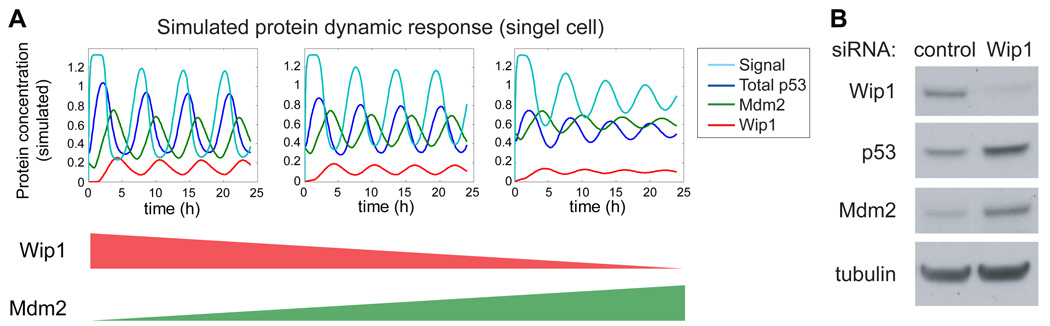
We also observed a surprising second effect of Wip1 RNAi: our model predicts that the amplitude of the p53 pulse should be unaffected in treated cells (Figure 4G), but we found that a lower irradiation dose led to a 45% reduction in the average amplitude of the p53 pulse (Figures 6B–E). To explore this discrepancy, we returned to our model. In simulation studies we found that an increase in Mdm2 level leads to a decrease in the amplitude of the p53 pulse (Figure 7A). One possible explanation for the amplitude decrease, therefore, is that Mdm2 levels are changed during the 72h knock-down of Wip1 prior to irradiation. This is plausible, since removing the Wip1 negative feedback would be expected to increase levels of ATM-P, in turn increasing p53 levels. To compensate for the loss of Wip1, the p53 network might have reached a new steady state in which Mdm2 expression was elevated to counteract the elevated p53 levels. Using Western blot analysis, we were able to show that both Mdm2 and p53 levels are increased in cells treated with Wip1 RNAi compared with control cells (Figure 7B). Taken together these observations suggest that Wip1 has an important role in controlling p53 basal level in unstressed cells, and in defining the shape and maintaining the uniformity of p53 pulses in response to various levels of DNA damage.
Figure 7. Wip1 RNAi treatment sets the p53 network to a new steady state.
(A) Simulation of the p53 response to γ-irradiation in cells treated with: (i) control RNAi, (ii) lower efficiency Wip1 RNAi and (iii) higher efficiency Wip1 RNAi, with concomitant increase in Mdm2 production. Values for the Wip1 production rate βi (Cs h−1), the p53-independent Mdm2 production rate βmi (Cs h−1), the initial active p53 concentration p53active0 (Cs), and the initial Mdm2 concentration Mdm20 (Cs)] were (i) [0.25, 0.2, 0, 0.2], (ii) [0.2125,0.35, 0.1, 0.35], and (iii) [0.175, 0.5, 0.2, 0.5]. All other parameter values were as indicated in Table S1. (B) Immunoblots of Wip1, p53, Mdm2, and tubulin in MCF7 cells transfected with control or Wip1 siRNA 72h prior to harvesting. Blots are representative of triplicate experiments.
DISCUSSION
The tumor suppressor protein p53 is the protein most frequently inactivated in human cancer, and perhaps one of the most intensely investigated proteins. Only recently have technological developments allowed detailed analysis of p53 kinetics in individual cells. Our discovery that single cells show a series of p53 pulses in response to DNA damage opened several new questions in the field; although several mathematical models have been suggested to explain p53’s dynamical behavior it is still unclear how p53 pulses arise and which feedbacks control their shape and timing. To obtain a clearer picture of the dynamics of the p53 system we studied the temporal changes in levels and activity of a number of proteins in the p53 pathway. The individual interactions between the proteins of this system were previously identified, but their relative contributions to the behavior of the system were unclear. Here we show that two upstream regulators of p53, ATM-P and Chk2-P, undergo repeated pulses in response to DNA damage, which are also controlled by p53. Even after the first pulse of p53 has been initiated, subsequent pulses of p53 depend on continued activity of ATM. Additionally, a short, 1-hour induction of ATM is sufficient to drive a full first pulse of p53. The behavior of the system controlling p53 dynamics thus in some ways resembles excitable behavior described in other biological systems (for example (Hodgkin and Huxley, 1952; Suel et al., 2006; Tyson, 1991)). Our observations suggest that the ATM-P pulses drive the p53 pulses, and that p53 then feeds back on ATM-P to shut down its activity. The fact that ATM-P levels indeed decline opens the possibility that ATM may need to be repeatedly activated by the presence of DNA double-strand breaks in order to give rise to p53 pulses. This recurrent initiation model of p53 dynamics not only identifies the feedback from p53 to the upstream kinases as important for shaping p53 dynamics, but also suggests that the functional form of p53 dynamics is successive “pulses” rather than autonomous “oscillations”.
We used a mathematical simulation approach to make predictions about the behavior of the putative mediator of the feedback between p53 and ATM-P. Using criteria determined from the simulation, we proposed and tested the hypothesis that Wip1 is responsible for some aspects of p53 dynamics. The Wip1 gene was already known to be a transcriptional target for p53, and Wip1 was known to dephosphorylate ATM-P and Chk2-P. We show that the dynamics of Wip1 are consistent with those predicted for the p53/ATM-P feedback mediator; that increasing the level of Wip1 inhibits the induction of p53 pulses; and that reducing Wip1 levels breaks the control mechanism that leads to the precise timing and amplitude of p53 pulses independently of the irradiation dose.
The stereotyped nature of the p53 pulses was one of the most striking features about them when they were discovered (Lahav et al., 2004), and the mechanism for achieving the precision in timing and amplitude was completely unknown. Our study suggests that although the negative feedback between p53 and Mdm2 down-regulates p53 levels after initial activation, it is not sufficient to trigger the repeated uniform pulses of p53 in response to various levels of DNA damage; if Wip1 is removed, larger radiation doses result in pulses with longer durations. In addition, silencing Wip1 leads to higher levels of Mdm2 pre-irradiation, which results in a decrease in the amplitude of p53 pulses in lower radiation doses. The exact role of Wip1 in controlling the shape of the p53 pulses, and the downstream consequences of distortions in pulse amplitude and duration, are now open areas for further study.
Our mathematical simulation recapitulates many of the important features of the dynamics we have observed, but gaps that could be important still remain. For example, our model shows that the pulses of p53 and the Signal have similar frequencies, while experimentally the frequency of the first few ATM-P pulses is lower than that for p53 pulses (Figure 1C). It is also not clear why the timing of p53 pulses is relatively robust between different experiments while the timing of Chk2-P and ATM-P is more variable. It is possible that the information from several pathways that respond to DSBs is integrated to ensure precise timing of p53 dynamics. For example, recent studies show that the ATR/Chk1 pathway also responds to DSB formation (Adams et al., 2006; Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006). Wip1 could also feed back on the ATR/Chk1 pathway since it was shown to dephosphorylate Chk1-P (Lu et al., 2005). Interestingly, the levels of active Chk1 (phosphorylated at Ser317) do not show pulses in response to irradiation (Figure S8A) and its induction is subtle under these conditions in comparison to the UV response (Figure S8B). Further studies are required to test the relative role of the ATR pathway in controlling p53 dynamics and to fill the gaps that still exist in our understanding of the p53 pathway.
Ultimately, we wish to understand what function, if any, p53 pulses have. One possibility is that p53 pulses translate into pulses in its target genes, which are then important for enacting downstream effects. Our initial studies show that p21, one of p53’s target genes and a mediator of cell cycle arrest, also undergoes pulsatile dynamics in response to DSBs (Figure S9), raising the possibility that pulses in p21, or other target genes may be important for regulating cell cycle arrest. A second possibility is that the pulses may be required for an upstream function: perhaps the interval between pulses is used to re-evaluate cell state by checking whether DNA damage remains. In this scenario ATM might leave the breaks to allow repair, and the repair proteins might then leave the breaks to allow reevaluation of the damage. This idea is supported by a recent study showing that ATM and the repair protein XRCC attach and detach from DSBs with an anti-phase correlation (Berkovich et al., 2007). We suggest that if the damage has not been repaired after one pulse of p53, a new p53 pulse is initiated. This proposal raises a further question, however: why would p53 need to be degraded to allow the re-evaluation, instead of sequestered or otherwise deactivated? A possible explanation for this lies in the fact that p53 activation is not simply a question of increased levels: stresses of many kinds also cause post-translational modifications that are thought to be important in p53’s transcriptional activity, and may selectively increase the transcription of one set of downstream target genes relative to another. To erase the information encoded in these modifications, which may no longer be relevant after the pulse is complete, it may be simplest to degrade the modified protein and make unmodified protein in the next pulse (Lahav, 2004). Such a mechanism might, for example, allow the cell to activate DNA repair and cell cycle arrest for the first few pulses, and trigger apoptosis only after several attempts to repair the damage (Lahav, 2004; Zhang et al., 2007).
If p53 pulses are important for stress responses, one might expect that pre-cancerous cells would sometimes use modulation of p53 dynamics as a step towards immortalization. A single nucleotide polymorphism in the Mdm2 promoter, SNP309G, has been shown to cause defects in the activation of p53-dependent transcriptional programs, and has also been suggested to be associated with accelerated tumor formation in humans (Bond et al., 2006; Bond et al., 2004). Cell lines carrying SNP309G allow p53 up-regulation but do not show pulses (Hu et al., 2007). Although there may be many downstream effects of the SNP309G polymorphism, the correlation between lack of pulsing and changes in the activation of p53-dependent transcription is suggestive. These cells also offer a new system in which to explore how the activation of downstream programs is affected by the form of p53 pulses, and what the eventual outcome is for the damaged cell. In this context the ability that we now have to manipulate the duration or amplitude of the pulses, by removing Wip1 and/or altering other parts of the network, should prove invaluable.
EXPERIMENTAL PROCEDURES
Plasmids and cells
We maintained human breast cancer epithelial MCF-7 cells at 37° in RPMI supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 µg/ml streptomycin, and 250 ng/ml fungizone (Gemini Bio-Products). Nutlin3 (Cayman Chemical) was added to the medium at 5µM or 10µM. For wortmannin treatment, we supplemented the medium with 100 µM wortmannin (Tocris Cookson). Due to the instability of this compound in solution, we replaced medium with fresh wortmannin-containing medium every hour during experiments. For Wip1 overexpression, we transiently transfected MCF7 cells with TrueClone plasmid DNA containing the CMV promoter driving expression of PPM1D (TC117895, OriGene) (referred to in the text as pCMV-Wip1) using FuGene6 (Roche). We performed all subsequent analysis 48h after transfection. Additional plasmid and cell line construction are described in Supplemental Experimental Procedures.
Immunoblots
We harvested cells and obtained protein samples by lysis, and quantified the total protein concentration by Bradford assay. We separated equal amounts of total protein by electrophoresis on 3–8% Tris-actetate gradient gels (Invitrogen) for analysis of ATM, or 4–12% Bis-Tris gradient gels (Invitrogen) for analysis of all other proteins. We detected p53 using DO-1 monoclonal antibody (mAb; Santa Cruz Biotechnoology) or FL-393 rabbit polyclonal antibody (pAb; Santa Cruz Biotechnology), phospho-ATM (Ser1981) using rabbit pAb (Rockland) or mouse mAb (Rockland), total ATM using 5C2 mouse mAb (Santa Cruz Biotechnology), phospho-Chk2 (Thr68) using rabbit pAb (Cell Signaling), total Chk2 using rabbit pAb (Cell Signaling), Wip1 using WC10 mouse mAb (Trevigen) or H-300 rabbit pAb (Santa Cruz Biotechnology), Mdm2 using SMP14 mouse mAb (Santa Cruz Biotechnology), β-tubulin using E7 mouse mAb (Developmental Studies Hybridoma Bank), and actin using JLA20 mouse mAb (Developmental Studies Hybridoma Bank). We quantified blot images using ImageJ (NIH).
Immunofluorescence
We grew cells on coverslips coated with poly-L-lysine (Sigma-Aldrich). We fixed cells with 4% paraformaldehyde and permeabilized cells with −20° methanol. To quench cellular autofluorescence, we treated cells with 0.1% sodium borohydride. We blocked cells in goat serum supplemented with 1% bovine serum albumin. For secondary antibodies, we used antibodies conjugated with AlexaFluor 488 (Molecular Probes) or Cy3 (Jackson ImmunoResearch Laboratories, Inc.). The Alexa488 filter set was 480 nm/ 30 nm excitation, 505 nm dichroic beam splitter, and 535 nm / 40 nm emission (Chroma). The Cy3 filter set was 540 nm/ 25 nm excitation, 565 nm dichroic beam splitter, and 620 nm / 60 nm emission (Chroma).
RNAi
To target p53, we used a siGENOME SMARTpool of siRNA (Dharmacon). To target Wip1, we used a custom siRNA with the sequence 5’-UUGGCCUUGUGCCUACUAAUU-3’ as was previously described (Fujimoto et al., 2005). For all controls, we used siCONTROL non-targeting siRNA #2 (Dharmacon). To identify cells that received siRNA in time lapse fluorescence microscopy experiments, we co-transfected cells with siGLO Red (Dharmacon). We performed all RNA transfections using DharmaFECT 1 transfection reagent following the manufacturer’s protocol (Dharmacon). We assayed knock-down of p53 48h after transfection, and knock-down of Wip1 72h after transfection.
Time-Lapse microscopy
Two days prior to microscopy, we grew cells in RPMI lacking riboflavin and phenol red to reduce medium autofluorescence in poly-D-lysine-coated glass bottom plates (MatTek Corporation). The medium was supplemented with 2% fetal calf serum, 100 U/ml penicillin, 100 µg/ml streptomycin, and 250 ng/ml fungizone (Gemini Bio-Products). We viewed cells with a Nikon Eclipse TE2000-E inverted fluorescence microscope in which the stage was surrounded by a custom enclosure to maintain constant temperature, CO2 concentration, and humidity. The Venus filter set was 500 nm/ 20 nm excitation, 515 nm dichroic beam splitter, and 535 nm/ 30 nm emission (Chroma). The siGLO Red filter set was 545 nm/ 20–25 nm excitation, 570 nm dichroic beam splitter, and 600 nm / 50-25 nm emission (Chroma). We acquired and analyzed images using MetaMorph software (Molecular Devices).
Computational Modeling
We performed numerical integration and optimization using Matlab software (MathWorks).
Supplementary Material
ACKNOWLEDGMENTS
We thank Uri Alon for encouragement and discussion, Tim Mitchison, Roy Kishony, Ron Milo, Michael Springer, Shalev Itzkovitz, Nitzan Rosenfeld, Bodo Stern, Rebecca Ward and all members of our laboratory for comments and discussions. We thank the Nikon Imaging Center at Harvard Medical School for assistance with immunofluorescence microscopy. This research was supported by the Smith Family New Investigator Awards Program (The Medical Foundation), the Giovanni Armenise-Harvard Foundation and by the NIH grant GM083303. E.B was partially supported by the American Cancer Society. I.B. was supported by fellowships from Aid for Cancer Research and from the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. Embo J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto A, Novak B, Tyson JJ. Steady states and oscillations in the p53/Mdm2 network. Cell Cycle. 2005;4:488–493. doi: 10.4161/cc.4.3.1548. [DOI] [PubMed] [Google Scholar]
- Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, Minami Y. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2005 doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Rosenfeld N, Itzkovitz S, Milo R, Sigal A, Dekel E, Yarnitzky T, Liron Y, Polak P, Lahav G, Alon U. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:E1–E13. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. Embo J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin BC. Oscillatory behavior in enzymatic control processes. Adv Enzyme Regul. 1965;3:425–438. doi: 10.1016/0065-2571(65)90067-1. [DOI] [PubMed] [Google Scholar]
- Hamstra DA, Bhojani MS, Griffin LB, Laxman B, Ross BD, Rehemtulla A. Real-time evaluation of p53 oscillatory behavior in vivo using bioluminescent imaging. Cancer Res. 2006;66:7482–7489. doi: 10.1158/0008-5472.CAN-06-1405. [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Feng Z, Ma L, Wagner J, Rice JJ, Stolovitzky G, Levine AJ. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res. 2007;67:2757–2765. doi: 10.1158/0008-5472.CAN-06-2656. [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lahav G. The strength of indecisiveness: oscillatory behavior for better cell fate determination. Sci STKE. 2004;2004:pe55. doi: 10.1126/stke.2642004pe55. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wagner J, Rice JJ, Hu W, Levine AJ, Stolovitzky GA. A plausible model for the digital response of p53 to DNA damage. Proc Natl Acad Sci U S A. 2005;102:14266–14271. doi: 10.1073/pnas.0501352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas GI, Simon Z, Balea G, Popa E. Possible oscillatory behavior in p53-Mdm2 interaction computer simulation. Journal of Biological Systems. 2000;8:21–29. [Google Scholar]
- Monk NA. Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Jensen MR, Kim HG, Kim KT, Lee SW. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene. 2004;23:5405–5408. doi: 10.1038/sj.onc.1207693. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kamibayashi C, Serrano M, Prives C, Mumby MC, Beach D. p53-dependent association between cyclin G and the B' subunit of protein phosphatase 2A. Mol Cell Biol. 1996;16:6593–6602. doi: 10.1128/mcb.16.11.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene. 2006 doi: 10.1038/sj.onc.1209927. [DOI] [PubMed] [Google Scholar]
- Reimer CL, Borras AM, Kurdistani SK, Garreau JR, Chung M, Aaronson SA, Lee SW. Altered regulation of cyclin G in human breast cancer and its specific localization at replication foci in response to DNA damage in p53+/+ cells. J Biol Chem. 1999;274:11022–11029. doi: 10.1074/jbc.274.16.11022. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. Embo J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Tiana G, Jensen MH, Sneppen K. Time delay is a key to apoptosis induction in the p53 network. European Physical Journal B. 2002;29:135–140. [Google Scholar]
- Tyson JJ. Modeling the cell division cycle: cdc2 and cyclin interactions. Proc Natl Acad Sci U S A. 1991;88:7328–7332. doi: 10.1073/pnas.88.16.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- Vassilev LT. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3:419–421. [PubMed] [Google Scholar]
- Wagner J, Ma L, Rice JJ, Hu W, Levine AJ, Stolovitzky GA. p53- Mdm2 loop controlled by a balance of its feedback strength and effective dampening using ATM and delayed feedback. Syst Biol (Stevenage) 2005;152:109–118. doi: 10.1049/ip-syb:20050025. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Zhang T, Brazhnik P, Tyson JJ. Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle. 2007;6:85–94. doi: 10.4161/cc.6.1.3705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.