Abstract
Zorbamycin (1, ZBM) is a glycopeptide antitumor antibiotic first reported in 1971. The partial structures of 1 were speculated on the basis of its acid hydrolysis products, but the structure of the intact molecule has never been established. The low titer of 1 from the wild-type strains, combined with its acid-instability, has so far hampered its isolation. By random mutagensis of Streptomyces flavoviridis ATCC21892, a wild-type producer of 1, with UV irradiation, two high-producing strains of 1, S. flavoviridis SB9000 and SB9001, were isolated. Under the optimized fermentation conditions, these two strains produced about 10 mg/L of 1, which was about 10-fold higher than the wild-type ATCC21892 strain, as estimated by HPLC analysis. Finally, 1 was isolated both as a 1-Cu complex and Cu-free molecule, and the intact structure of 1 was established on the basis of a combination of mass spectrometry and 1H and 13C NMR spectroscopic analyses.
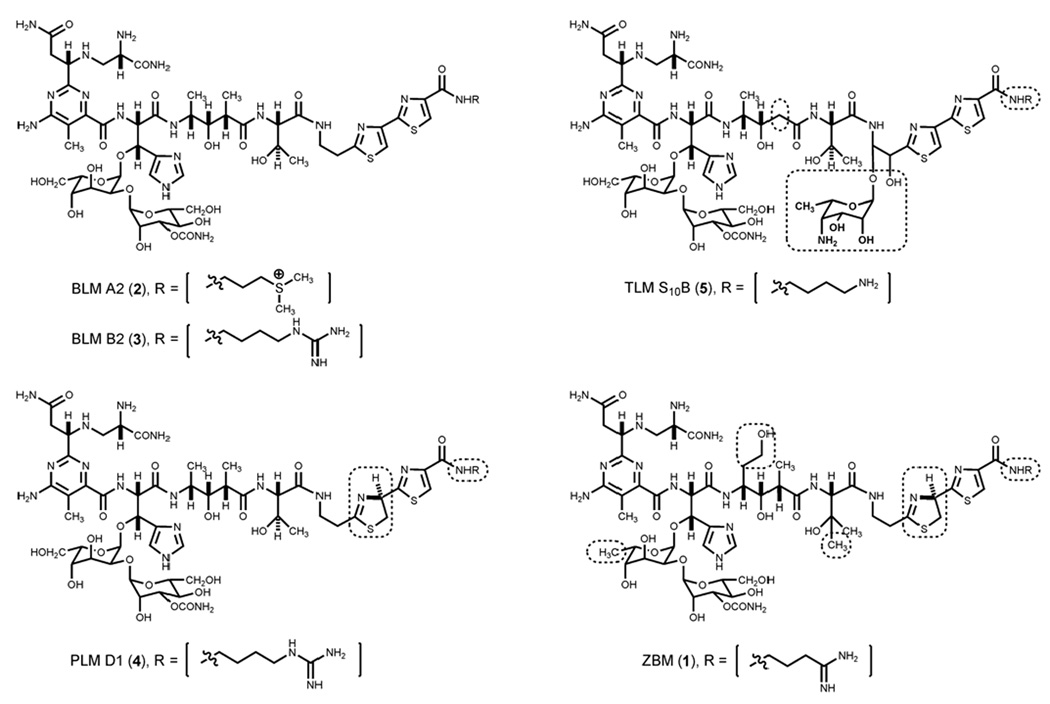
Zorbamycin (1, ZBM) belongs to the bleomycin (BLM) family of glycopeptide antitumor antibiotics, and other members of this family include the BLMs (2 and 3), the phleomycins (4, PLMs), and the tallysomycins (5, TLMs) (Figure 1). The BLMs are currently used clinically under the trade name Blenoxane® in combination with a number of other agents for the treatment of several types of tumors, notably testicular cancer and certain types of lymphoma.1,2 Early development of drug resistance and cumulative pulmonary toxicity are the major limitations of the BLMs in chemotherapy.1–3 Consequently, there have been continuing attempts to synthesize new analogs of the BLM family to search for antitumor drugs with better clinical efficacy and lower toxicity. Although numerous BLM analogs have been synthesized in the past two decades,4–6 total chemical synthesis is inevitably very expensive, thus limiting its practicality, given the structural complexity of the BLM family of natural products. In contrast, recent progress in combinatorial biosynthesis presents an attractive alternative to produce novel BLM analogs by genetic engineering of the biosynthetic machinery of this family of natural products.7,8
Figure 1.
Structures of selected members of the BLM family of antitumor antibiotics: BLM A2 (2), BLM B2 (3), PLM D1 (4), TLM S10B (5), and ZBM (1). Structural differences between BLMs and other members of this family are highlighted by the boxes.
To study the biosynthetic pathway of a natural product and to generate new analogs thereof by combinatorial biosynthesis methods, three prerequisites have to be fulfilled: (i) the gene cluster of the target natural product has to be cloned, (ii) the producing strain has to be genetically amenable, and (iii) the natural product has to be produced in sufficient yield to allow for its detection, isolation, and structural elucidation. During our ongoing research on hybrid peptide-polyketide natural product biosynthesis, we have cloned and sequenced the gene clusters for BLM from Streptomyces verticillus ATCC15003,9 TLM from Streptoalloteichus hindustanus E465-94 ATCC31158,10 and 1 from Streptomyces flavoviridis ATCC21892 (unpublished data), respectively. While the yield of BLMs (10–12 mg/L) and TLMs (18–20 mg/L) from the wild-type strains are suitable for laboratory-scale fermentation to isolate and identify the natural products and to search for new analogs that potentially could be produced by engineered recombinant strains, exhaustive efforts to develop an expedient genetic system for the BLM producer S. verticillus or the TLM producer S. hindustanus remain so far with limited success, due to their slow growth and poor sporulation, inefficient introduction of plasmid DNAs into these organisms by protoplast transformation, electroporation, or intergeneric conjugation, and their intrinsic low homologous recombination activity.10 In contrast, S. flavoviridis, the producer of 1, sporulates well, plasmid DNAs can be efficiently introduced by either intergeneric conjugation or protoplast transformation, and homologous recombination occurs with reasonably high frequency in this organism (unpublished data). However, the yield of 1 from the wild-type S. flavoviridis ATCC 21892 strain was very low, the acid-instability of 1 further complicated its isolation, and the structure of the intact molecule of 1 remains to be established.
Also known as YA56-X, 1 was first isolated by two different groups independently and almost simultaneously in 1971 from two different strains, Streptomyces bikiniensis var. zorbonensis and Streptomyces humidus var. antitumoris, respectively.11,12 YA56-X was later found to be identical to 1 by direct comparison of the two antibiotics.13 A series of studies on the hydrolysis products of 1 suggested that it consists of one terminal amine group [3-aminopropionamidine (APA)], six amino acids [β-aminoalanine (β–ALA), β-amino-β-(4-amino-6-carboxy-5-methylpyrimidine-2-yl)propionic acid (PBA), L-erythro-β-hydroxyhistidine (OH-HIS), 2-(2-(2-aminoethyl)-Δ2-thiazoline-4-yl)-thiazole-4-carboxyic acid (ATTC), β-hydroxy-L-valine (OH-VAL), 4-amino-3,6-dihydroxy-2-methylhexanoic acid (ADMH)], and one disaccharide [2-O-(3-O-carbamoyl-α-D-mannosyl)-6-deoxy-L-gulose (CMDG)].14–17 These partial structures, in addition to properties such as water solubility and copper coordination, suggested that 1 is related to the BLM family of antitumor antibiotics. The first two articles concerning this antibiotic were published more than 35 years ago,11,12 but the intact structure of 1 has never been fully established. Here we present strain improvement of S. flavoviridis ATCC21892 and isolation and structure elucidation of the intact molecule of 1. These findings should greatly facilitate our current effort to engineer novel analogs of the BLM family of antitumor antibiotics by combinatorial biosynthesis methods.
Results and Discussion
The BLM family of compounds, in addition to their potent anticancer activity, is known to specifically inhibit growth of Mycobacterium smegmatis.18 A sensitive and specific paper disk agar diffusion bioassay using M. smegmatis as the test organism was therefore adopted to estimate the titer of 1 in the fermentation broth. Since authentic 1 is not commercially available, PLM D1 (4), which was thought to be closely related to 1 structurally (Figure 1), was used instead as an alternative standard to estimate the titer of 1 by the bioassay method. While this bioassay method is convenient, expedient, and high-through-put (see below in section of strain improvement by random mutagenesis), it could suffer from potential pitfalls resulting from (i) the difference in antibacterial activities between 1 and 4 and (ii) the presence of other bioactive secondary metabolites in S. flavoviridis fermentation. Although the production of 1 was apparent from the wild-type S. flavoviridis ATCC21892 strain under the original literature medium and fermentation conditions19 according to the bioassay, all attempts to isolate 1 were unsuccessful, and no 1 could be detected upon HPLC analysis after two steps of chromatographic purification.
To increase the titer of 1, the wild-type strain was subjected to UV irradiation for strain improvement according to literature protocols.20 S. flavoviridis ATCC21892 spores were irradiated for 4 to 5 min, resulting in a survival rate of 0.1% to 1.0% (Table S1, Supporting Information), and 1315 colonies that exhibited different spore colors or colony morphology were then selected. After culturing in 96-well plates for 8 days, they were examined for antimicrobial activity against M. smegmatis by the bioassay method (Figure S2, Supporting Information). Among them, 31 colonies showed higher activities than the wild-type, 764 colonies were comparable to the wild-type, and 520 colonies exhibited lower activities than the wild-type or completely lost activity. Finally, SB9000 and SB9001, two isolates that showed the highest titer of 1 according to bioassay, were selected as the improved strains for medium and fermentation condition optimization.
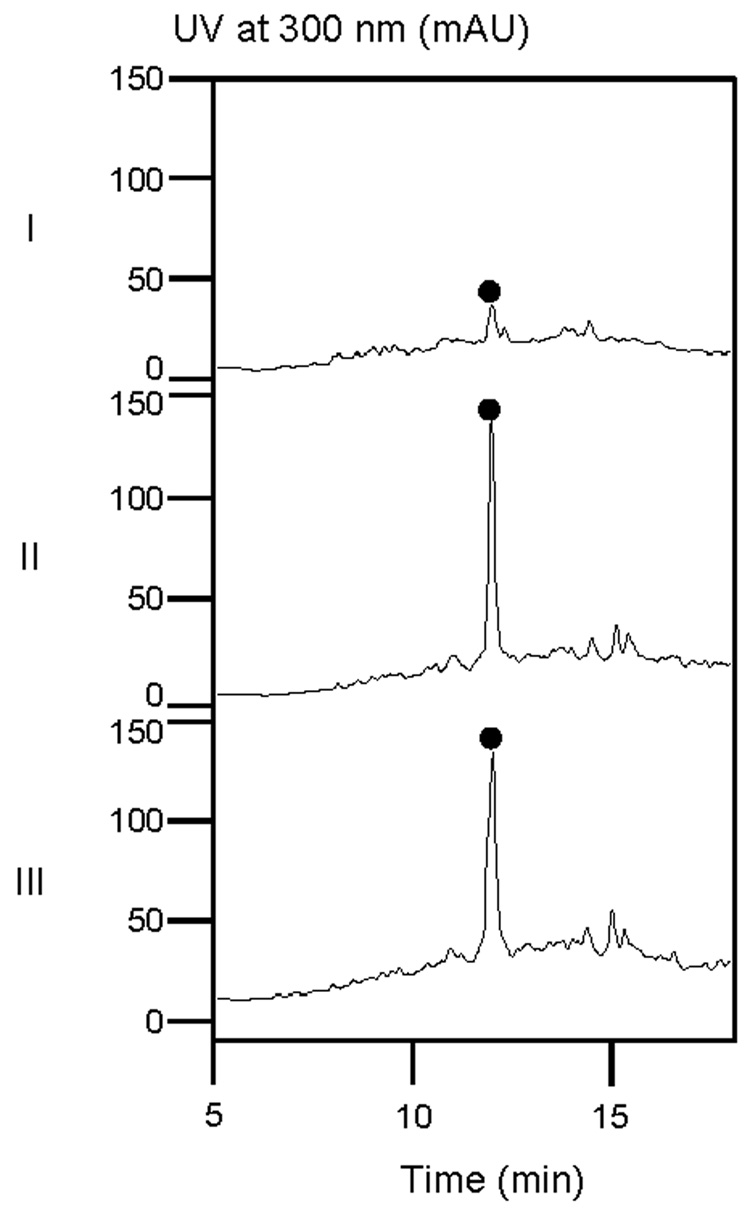
Since the amount of copper and zinc in the fermentation medium has been previously reported to be important to the PLM titer,18 different concentrations of transition metals were next tested in the fermentation medium to optimize the titer of 1 for the SB9001 strain. Those used for PLM fermentation21 turned out to yield the best results. Thus, the concentrations of CuSO4·5H2O and ZnSO4·7H2O were increased from 0.0005% and 0.005% to 0.01% and 0.05%, respectively, and MnCl2·7H2O was eliminated from the original production medium19 for SB9001 fermentation. Under the optimized medium and fermentation conditions, 1 could be reliably and reproducibly isolated from the fermentation broths of both SB9000 and SB9001 and detected by HPLC analysis after two steps of chromatographic purification. The titers of 1 in SB9000 and SB9001 under the optimized medium and fermentation conditions were very similar, estimated to be ~ 10 mg/L according to HPLC analysis, which was about 10-fold higher than that in the wild-type ATCC21892 strain under the identical medium and fermentation conditions (Figure 2).
Figure 2.
HPLC analysis of partially purified 1 (•) from fermentation of the wild-type S. flavoviridis ATCC21892 (I) or the improved S. flavoviridis SB9000 (II) and SB9001 (III) strains under the optimized medium and fermentation conditions.
To purify 1, SB9001 was fermented under the optimized conditions, the fermentation broth (12.5 L) was centrifuged (3000 rpm for 30 min), and the supernatant was collected and adjusted to pH 7.0. After sequential column chromatography on Amberlite® IRC50 and Diaion HP-20 followed by HPLC on a C18 column, pure 1 (8 mg) was obtained as a 1-Cu complex. Significant amount (> 90%) of 1 was lost during its isolation mainly as a result from the acid-instability of the thiazoline moiety of 1, which is known to be labile for hydrolysis.22 To isolate Cu-free 1, the 1·Cu complex was further treated with 8-hydroxyquinoline and EDTA to remove Cu and subsequently subjected to HPLC on a C18 column to afford Cu-free 1 (3.8 mg) as a pale white powder.10
Structure elucidation of the intact molecule of 1 was carried out by a combination of mass spectrometry (MS) and 1H and 13C NMR spectroscopic analyses, as well as by comparison of the resultant NMR data with those reported in the literature, in particular to those of 3 and the hydroperoxide of Co (III)-4 complex.23,24 Upon electrospray ionization (ESI)-MS analysis, the 1-Cu complex yielded a molecular ion peak (m/z) at 737.3, consistent with the M2+ ion for the 1-Cu2+ complex (calcd 1474). Upon MALDI-FTMS analysis, Cu-free 1 showed a molecular ion peak (m/z for [M + H]+) at 1412.5664, which agreed well with the Cu-free 1 molecular formula of C55H85N19O21S2 (calcd 1411.5602).
Table 1 summarizes the 1H and 13C NMR data of Cu-free 1 in D2O. The 1H NMR spectrum of 1 is very similar to that of 3 except that (i) only one signal (H-47 at δ 8.16, s) from the thiazole group was detected, and (ii) additional signals attributed to the thiazoline group appeared at δ 5.89 (dd, J = 9.0 and 7.0 Hz) for the methine proton of C-45 and at δ 3.58 (m) and δ 3.90 (m) for the two methylene protons of C-44. These data suggested that 1 has the same thiazolinylthiazole moiety as PLMs instead of the bithiazole moiety as BLMs and TLMs.23,24
Table 1.
13C NMR and 1H NMR Data of Zorbamycin (1) [D2O, TSP, δ (ppm) (J = Hz)]a
| positionb | δC | δH |
|---|---|---|
| 1 | 173.8 | |
| 2 | 55.2 | 4.07 mc |
| 3 | 49.8 | 2.88–3.06 m |
| 4 | 179.0 | |
| 5 | 43.1 | 2.78 mc, 2.69 dd (15.0, 9.0) |
| 6 | 62.5 | 4.05 mc |
| 7 | 168.2 | |
| 8 | 167.6 | |
| 9 | 115.5 | |
| 10 | 154.8 | |
| 11 | 13.8 | 2.16 s |
| 12 | 170.8 | |
| 13 | 60.4 | 5.09 d (8.0) |
| 14 | 75.6 | 5.28 d (8.0) |
| 15 | 100.8 | 5.25 d (3.5) |
| 16 | 66.0 | 4.20 mc |
| 17 | 74.4 | 3.64 mc |
| 18 | 70.9 | 4.12 mc |
| 19 | 73.4 | 3.99 mc |
| 20 | 101.3 | 5.02 brs |
| 21 | 71.2 | 4.06 mc |
| 22 | 77.1 | 4.65 mc |
| 23 | 67.4 | 3.82 mc |
| 24 | 76.5c | 3.84 mc |
| 25 | 63.8 | 3.80 mc, 3.94 mc |
| 26 | 160.8 | |
| 27 | 137.6 | |
| 28 | 120.0 | 7.30 s |
| 29 | 139.6 | 7.91 s |
| 30 | 172.2 | |
| 31 | 51.8 | 3.82 mc |
| 32 | 34.5 | 1.70 m |
| 33 | 76.5c | 3.74 mc |
| 34 | 45.2 | 2.36 m |
| 35 | 14.3 | 1.07 d (7.0) |
| 36 | 179.6 | |
| 37 | 63.6 | 4.29 s |
| 38 | 74.3 | |
| 39 | 28.3d | 1.19 s |
| 40 | 174.2 | |
| 41 | 40.1 | 3.58 mc |
| 42 | 42.1 | 2.92 m |
| 43 | 180.3 | |
| 44 | 41.4 | 3.58 mc, 3.90 mc |
| 45 | 78.4 | 5.89 dd (9.0, 7.0) |
| 46 | 175.0 | |
| 47 | 128.3 | 8.16 s |
| 48 | 151.0 | |
| 49 | 165.9 | |
| 50 | 17.3 | 0.92 d (8.0) |
| 51 | 61.4 | 3.50 mc |
| 52 | 28.6d | 1.22 s |
| 53 | 39.4 | 3.81 mc |
| 54 | 35.5 | 2.80 mc |
| 55 | 171.8 |
Assignment confirmed by the combination of DEPT, 1H-1H COSY, TOCSY, HSQC, and HMBC, spectra obtained at 500 MHz for 1H and 125 MHz for 13C, respectively.
See Figure 2 for hydrogen and carbon numbering of ZBM.
Overlapped signals.
Interchangeable signals.
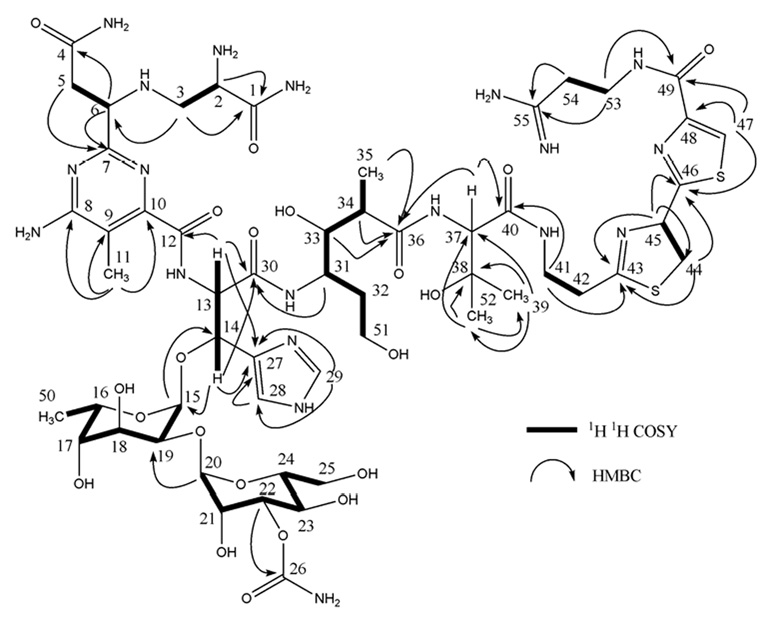
The 13C NMR spectrum combined with the HSQC spectra of 1 showed a total of 55 signals accounting for 5 methyl, 10 methylene, 22 methine, and 18 quaternary carbons. The protonated carbons and their bonded protons were assigned unambiguously by HSQC experiments. Comparison of these NMR data with those of 3 combined with the detailed analysis of 2D 1H-1H COSY, TOCSY, HSQC, and HMBC data for 1 finally resulted in the full 1H and 13C NMR spectroscopic assignments of the structural units of one amine (APA), six constitutional amino acids (β–ALA, PBA, OH-HIS, ATTC, OH-VAL, ADMH), and one disaccharide (CMDG) to 1 (Figure 3), which corresponded well with the partial structures of 1 reported earlier.17
Figure 3.
1H-1H COSY and selected key HMBC correlations of 1.
The connections between the various structural units of 1 were confirmed finally by HMBC experiments, including two-bond and three-bond correlations between H-3 (δ 2.88 and 3.06) and C-6 (δ 62.5), between H-13 (δ 5.09) and C-12 (δ 170.8) and C-30 (δ 172.2), between H-14 (δ 5.28) and C-15 (δ 100.8), between H-31 (δ 3.82) and C-30 (δ 172.2), between Me-35 (δ 1.07), H-34 (δ 2.36), H-33 (δ 3.74), H-37 (δ 4.29) and C-36 (δ 179.6), between H-41 (δ 3.58) and C-40 (δ 174.2), and between H-47 (δ 8.16), H-53 (δ 3.81) and C-49 (δ 165.9), as summarized in Figure 3. The small coupling constants of the anomeric protons (H-15 at δ 5.25, d, J = 3.5 Hz and H-20 at δ 5.02, brs) indicated α glycosidic linkages for the disaccharide moiety.
Comparison of 1 to other members of the BLM family of antitumor antibiotics revealed that, while they shared β–ALA, PBA, and OH-HIS as common constituents, 1 and PLMs are characterized with the ATTC unit, the thiazolinylthiazole chromophore, while the BLMs and TLMs are characterized with the bithiazole chromophore. In addition, 1 differs from the other members of this family by the presence of the OH-VAL and ADMH constituents as well as the CMDG disaccharide and the APA terminal amine (Figure 1).
In spite of the recent progress in cloning and sequencing the biosynthetic gene clusters for three members (i.e., BLM, TLM, and 1) of the BLM family of antitumor antibiotics, application of combinatorial biosynthetic methods to this family of metabolites to produce novel analogs remains limited due to either the lack of an expedient genetic system for the producing organism (i.e., S. verticillus for BLM or S. hindustanus for TLM)9,10 or the structural ambiguity and low titer of the target natural product (i.e., 1 in S. flavoviridis). The strain improvement, isolation, and structural elucidation of 1 from S. flavoviridis reported here finally fulfills all three prerequisites (i.e., the gene cluster cloned, an expedient genetic system established, and the target molecule structure elucidated and titer improved) for combinatorial biosynthesis. Comparing and contrasting the biosynthetic gene clusters for BLM, TLM, and 1 has already been extremely rewarding in unveiling new molecular insight into their biosynthesis.9,10 The availability of the three gene clusters and now the improvement for the production of 1, the structural elucidation of the intact molecule of 1, as well as the demonstrated feasibility to engineer the biosynthetic machinery of 1 in S. flavoviridis set the stage to produce novel structural analogs of the BLM family of antitumor antibiotics by the judicial application of combinatorial biosynthesis strategies.
Experimental Section
General Experimental Procedures
The optical rotation was measured on a Perkin-Elmer 241 polarimeter, and UV data were recorded on a Beckman Coulter DU® 800 spectrophotometer. IR data were recorded using a Bruker EQUINOX55/S FT-IR/NIR spectrophotometer. 1H and 13C NMR data were acquired on a VARIAN Inova-500 (500 MHz) spectrometer. Cu-free 1 was dissolved in D2O (pH unadjusted) with sodium 3-trimethyl silyl [2,2,3,3,-2H4] propionate (TSP) as an internal standard. ESI-MS analysis of the 1-Cu complex was performed on an Agilent 1100 HPLC-MSD SL quadrupole mass spectrometer or an Agilent 1100 HPLC-MSD SL ion trap mass spectrometer. MALDI-FTMS analysis of Cu-free 1 was performed on an IonSpec HiResMALDI FT-mass spectrometer.
Strains
Mycobacterium smegmatis ATCC607 and S. flavoviridis ATCC21892 were both obtained from the American Type Culture Collection.
Paper Disk Bioassay
PLM D1 (4), as a Cu-complex, was purchased from CAYLA (Toulouse, France) and used as a standard for the bioassay. To generate the standard curve, 4 was dissolved in H2O, and a series of this solution was applied to filter paper disks (6-mm diameter). The solvent was allowed to evaporate, and the disks were placed on agar plates (1% glycerol, 1% peptone, 1% desiccated beef extract, 3% NaCl, and 1.7% agar) pre-seeded with M. smegmatis. Disks with H2O were included as negative controls. After culturing at 37 °C for three to five days, the diameters of the growth inhibition zones were determined. While the disks impregnated with 4 showed increasing inhibition zones as the concentrations of 4 increased, the control disk did not elicit any inhibition. The calibration curve for 4 was constructed by plotting log applied doses (µg) versus diameters (mm) of the inhibition zones, which showed a good linearity between 0.1 and 100 µg of 4 (Figure S2, Supporting Information).
To screen for production of 1 in S. flavoviridis, the fermentation culture was centrifuged, and the supernatant was collected. The resultant supernatant (30 µL) was then applied to the paper disks, and the bioassay was performed as described above. The titer of 1 was estimated on the basis of the diameters of the inhibition zone in comparison with the 4 standard curve.
Mutagenesis of the Wild-type S. flavoviridis ATCC21892 Strain by UV Irradiation
The survival rate for S. flavoviridis spores upon UV irradiation was determined by exposure of spores (103) to UV radiation (254 nm) at a distance of 12 cm for different time periods following the standard protocol.20 The spores were then plated on ISP4 medium and incubated at 30 °C for 10 days. The recommended survival rate of 0.1% to 1% was achieved by 4 to 5 min UV exposure (Table S1, Supporting Information). Colonies (obtained after 4 to 5 min UV irradiation) showing different spore colors or morphology were picked and inoculated into 96-well plates on agar medium (1.5% glucose, 1.5% starch, 2.0% soybean meal, 0.5% yeast extract, 0.25% NaCl, 0.32% CaCO3, 0.0005% CuSO4·5H2O, 0.0005% MnCl2·4H2O, 0.005% ZnSO4·7H2O, pH 7.4, and 0.7% agar). After incubation for 8 days at 30 °C, each of the agar blocks was placed on a bioassay plate pre-seeded with M. smegmatis. After three to five days of cultivation at 37 °C, colonies showing a clear inhibition zone were selected as producers of 1, whose relative titers were estimated by comparison with the wild-type S. flavoviridis ATCC21892 strain as a control (Figure S1, Supporting Information). Among the 1315 colonies examined, two of them, which showed the highest titer of 1 in the bioassays, were selected and named as S. flavoviridis SB9000 and SB9001 strains. Medium and fermentation optimization was subsequently carried out with the S. flavoviridis SB9001 strain only.
Fermentation
The wild-type S. flavoviridis ATCC21892 strain was cultured initially under the original literature medium and fermentation conditions19 and subsequently under the same optimized medium and fermentation conditions as those used for the improved strains. The improved S. flavoviridis SB9001 strain was cultured in 250-mL baffled flasks containing 50 mL of the seed medium (1.5% glycerin, 1.5% pharmamedia, 0.3% NaCl, 0.2% asparagine, with the pH adjusted to 7.2 with 1 N HCl). After growth at 30 °C and 250 rpm for 2 days, 50 mL of the seed cultures were inoculated into 2-L baffled flasks containing 500 mL of the production medium (1.5% glucose, 1.5% starch, 2.0% soy flour, 0.5% yeast extract, 0.25% NaCl, 0.32% CaCO3, 0.01% CuSO4·5H2O, 0.05% ZnSO4·7H2O, pH 7.4). The resulting cultures were fermented at 30 °C and 250 rpm for 12 days.
Analysis and Isolation of 1
To analyze the production of 1 in the S. flavoviridis wild-type ATCC21892 or the improved SB9000 and SB9001 strains, a typical fermentation broth (200 mL) was adjusted to pH 7.0 with 1.0 N HCl and loaded to an Amberlite® IRC50 column (NH4+ type, 24 mL). After washing the column with ten bed volumes of H2O, 1 was eluted with 50 mL of 20% NH4OAc. The resulting Amberlite® IRC50 eluate was mixed with 1/8 volume of Diaion® HP-20 resin and incubated at room temperature under gentle agitation for 45 min. The Diaion® HP-20 resin was then packed into a column, washed with ten bed volumes of H2O, and drained of excess water. The column was then eluted with eight bed volumes of 80% methanol, and the fractions containing 1 were combined and concentrated in vacuo to 1 mL. The isolated 1 at this step was about 90% pure, and the yield of 1 was about 10 mg/L as estimated by HPLC analysis. Analytical HPLC was carried out on an Apollo C18 column (5 µm, 250 × 4.6 mm, Alltech Associates, Inc., Deerfield, IL). The column was equilibrated with 80% solvent A (1% NH4OAc) and 20% solvent B (100% methanol) and developed with a linear gradient (0 to 15 min, from 100% A to 40% A/60% B; 15 to 20 min, from 40% A/60% B to 100% B) at a flow rate of 0.7 mL/min and with UV detection at 300 nm using a Varian Prostar 330 PDA detector (Varian, Palo Alto, CA). Under these conditions, the 1-Cu complex was eluted with a retention time of 12.0 min.
Large-Scale Isolation of 1
The fermentation culture (12.5 L) was centrifuged at 3000 rpm for 30 min, and the supernatant (10 L) was collected, adjusted to pH 7.0 with 1.0 N HCl, and loaded to an Amberlite® IRC50 column (NH4+ type, 1 L). After washing the column with ten bed volumes of H2O, 1 was eluted with 2 L of 20% NH4OAc. The resulting Amberlite® IRC50 eluate was mixed with 600 mL of Diaion® HP-20 resin and incubated at room temperature under gentle agitation for 45 min. The Diaion® HP-20 resin was then packed into a column, washed with ten bed volumes of H2O, and drained of excess water. The column was then eluted with eight bed volumes of 80% methanol, and the fractions containing 1 were combined and concentrated in vacuo to 10 mL. Final purification of 1 was achieved by semipreparative HPLC on an Altima C18 column (5 µm, 250 × 10 mm, Alltech Associates, Inc., Deerfield, IL). HPLC isolation was carried out under the following conditions. Instrument and detector were the same as stated above. The column was equilibrated with 100% solvent A (0.1% NH4OAc, 0.01% trifluoroacetic acid) and 0% solvent B (methanol) and developed with a linear gradient (0 to 5 min, from 100% A/0% B to 80% A/20% B; 5 to 15 min, from 80% A/20% B to 40 % A/60% B; and 15 to 20 min, from 40% A/60% B to 0% A/100% B) at a flow rate of 3 mL/min and with UV detection at 300 nm. Under these conditions, 1 was eluted with a retention time of 13.5 min. The methanol was removed from the eluate by evaporation at 40 °C, and the final pure 1·Cu complex (8.0 mg) was obtained by freeze drying. Cu-free 1 was obtained by treating the 1·Cu complex with 20 mg/mL 8-hydroxyquinoline followed by 0.5 M EDTA-Na (pH 7.3) solution. After the final HPLC purification, Cu-free 1 was obtained as a pale white power (3.8 mg).
Zorbamycin (1)
1-Cu complex: light blue powder; [α]D + 50 (c 0.1, H2O); UV (H2O) λmax 243 (ε 6269), 299 (ε 2081) nm; IR (neat) υmax 3400–3200, 1720 (shoulder), 1643, 1556, 1466, 1379 cm−1; ESI-MS m/z 737.3 [1-Cu]2+, calcd for 1-Cu, 1474. Cu-free 1: pale white power; 1H and 13C NMR data, see Table 1; 1H-1H COSY and selected key HMBC data, see Figure 3; HRMALDI-FTMS m/z 1412.5664 [M + H]+, calcd for C55H85N19O21S2, 1411.5602.
Supplementary Material
Data summarizing UV-irradiation time and S. flavoviridis ATCC21892 spore viability, screening for high-producing strains of 1 upon UV irradiation of the wild-type S. flavoviridis ATCC21892 strain by bioassays against M. smegmatis, and the standard curve for 4 against M. smegmatis by the paper disk bioassay. This material is available free of charge via the Internet at hhtp://pubs.acs.org.
Acknowledgment
We thank the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison, for support in obtaining MS and NMR data. This work was supported in part by the NIH grant CA94426. N. P. G is supported in part by NIH grant T32 GM08505, U.G is a Postdoctoral Fellow of the Deutsche Forschungsgemeinschaft (DFG), and B. S. is the recipient of an NIH Independent Scientist Award (AI51687).
References and Notes
- 1.Hecht SM. J. Nat. Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Stubbe J. Nature Rev. Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- 3.Ute G, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Chem. Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 4.Borger DL, Cai H. Angew. Chem. Int. Ed. 1999;38:448–476. doi: 10.1002/(SICI)1521-3773(19990215)38:4<448::AID-ANIE448>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 5.Leitheiser CJ, Smith KL, Rishel MJ, Hashimoto S, Konishi K, Thomas CJ, Li C, McCormick MM, Hecht SM. J. Am. Chem. Soc. 2003;125:8208–8227. doi: 10.1021/ja021388w. [DOI] [PubMed] [Google Scholar]
- 6.Leitheiser CJ, Rishel MJ, Wu X, Hecht SM. Org. Lett. 2000;2:3397–3399. doi: 10.1021/ol0002469. [DOI] [PubMed] [Google Scholar]
- 7.Van Lanen SG, Shen B. Curr. Opin. Microbiol. 2006;9:252–260. doi: 10.1016/j.mib.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Van Lanen SG, Shen B. Drug Disc. Today Technol. 2006;3:285–292. doi: 10.1016/j.ddtec.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Du L, Sánchez C, Chen M, Edwards DJ, Shen B. Chem. Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 10.Tao MF, Wang L, Wendt-Pienkowski E, George NP, Ute G, Zhang GD, Coughlin JM, Shen B. Mol. BioSyst. 2007;2:60–74. doi: 10.1039/b615284h. [DOI] [PubMed] [Google Scholar]
- 11.Argoudelis D, Bergy E, Pyke R. J. Antibiot. 1971;24:543–557. doi: 10.7164/antibiotics.24.543. [DOI] [PubMed] [Google Scholar]
- 12.Ito Y, Oashi Y, Egawa Y, Yamaguchi T, Furmai T. J. Antibiot. 1971;24:727–731. doi: 10.7164/antibiotics.24.727. [DOI] [PubMed] [Google Scholar]
- 13.Ito Y, Oashi Y, Kawabe S, Sakurazawa M, Ogawa T. J. Antibiot. 1973;26:77–83. doi: 10.7164/antibiotics.26.77. [DOI] [PubMed] [Google Scholar]
- 14.Ohashi Y, Abe H, Ito Y. Agri. Biol. Chem. 1973;37:2277–2282. [Google Scholar]
- 15.Ohashi Y, Abe H, Ito Y. Agri. Biol. Chem. 1973;37:2283–2287. [Google Scholar]
- 16.Ohashi Y, Kawabe S, Kono T, Ito Y. Agri. Biol. Chem. 1973;37:2379–2385. [Google Scholar]
- 17.Ohashi Y, Abe H, Kawabe S, Ito Y. Agri. Biol. Chem. 1973;37:2387–2391. [Google Scholar]
- 18.Takita T. J. Antibiot. 1959;12:285–289. [PubMed] [Google Scholar]
- 19.Okuda T, Awataguchi S. Jpn. Tokkyo Koho, JP 47002557. 1972 [Google Scholar]
- 20.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. pp. 102–103. [Google Scholar]
- 21.Umezawa H, Takita T, Muraoka Y, Fujii A. 3953594. U.S. Patent. 1976
- 22.Doi T, Yoshida M, Shin-ya K, Takahashi T. Org. Lett. 2006;8:4165–4167. doi: 10.1021/ol061793i. [DOI] [PubMed] [Google Scholar]
- 23.Calafat AM, Won H, Marzilli LG. J. Am. Chem. Soc. 1997;119:3656–3664. [Google Scholar]
- 24.Wu W, Vanderwall DE, Turner CJ, Hoehn S, Chen JY, Kozarich JW, Stubbe J. Nucleic Acids Res. 2002;30:4881–4891. doi: 10.1093/nar/gkf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data summarizing UV-irradiation time and S. flavoviridis ATCC21892 spore viability, screening for high-producing strains of 1 upon UV irradiation of the wild-type S. flavoviridis ATCC21892 strain by bioassays against M. smegmatis, and the standard curve for 4 against M. smegmatis by the paper disk bioassay. This material is available free of charge via the Internet at hhtp://pubs.acs.org.