Abstract
Enzymatic activity is dependent on temperature, although some proteins have evolved to retain activity at low temperatures at the expense of stability. Cold adapted enzymes are present in a variety of organisms and there is ample interest in their structure-function relationships. Lysozyme (E.C. 3.2.1.17) is one of the most studied enzymes due to its antibacterial activity against Gram positive bacteria and is also a cold adapted protein. In this work the characterization of lysozyme from the insect Manduca sexta and its activity at low temperatures is presented. Both M. sexta lysozymes natural and recombinant showed a higher content of α-helix secondary structure compared to that of hen egg white lysozyme and a higher specific enzymatic activity in the range of 5−30 °C. These results together with measured thermodynamical activation parameters support the designation of M. sexta lysozyme as a cold adapted enzyme. Therefore, the insect recombinant lysozyme is feasible as a model for structure-function studies for cold-adapted proteins.
Keywords: lysozyme, insect, Manduca sexta, cold adapted, glycohydrolase
INTRODUCTION
Lysozyme (E.C. 3.2.1.17) is probably the best studied enzyme as an antibacterial and as a model for structure-function studies [1]. Antibacterial activity is required at low temperature in many organisms. In general, specific enzymatic activity is decreased at low temperatures, although some proteins have evolved to retain activity at cold temperatures at the expense of stability [2,3]. Innate defense responses in insects are mediated by patterns recognition proteins which signal to expression of effectors which include the antibacterial protein lysozyme [4].
Insect lysozymes have been postulated as cold-adapted enzymes. Structural studies on the lysozyme from silkworm Bombyx mori have reported [5,6] a canonical hen egg white lysozyme (HEWL) structure for this enzyme. However, structural work performed in our laboratory with a recombinant tobacco hornworm Manduca sexta lysozyme (MSlyz) showed an unusual highly α-helical content on compared to HEWL and B. mori lysozyme [7].
Cold adaptation may provide an advantage to poikilothermic insects living at low temperatures. Insects synthesize lysozyme in response to bacterial infections, as part of the innate immune response [8]. Lysozyme, along with other antimicrobial peptides secreted into the hemolymph kill bacteria that have invaded the hemocoel. In the tobacco hornworm, Manduca sexta, lysozyme expression is highly induced after bacterial infection and becomes an abundant protein in the hemolymph [9,10].
Several questions were raised from previous work regarding MSlyz. First, the unusual high α-helical content of recombinant MSlyz may be due to the refolding process. It has been reported that slight differences in secondary structure content occur between natural and recombinant proteins [11]. If the secondary structure content is validated for the recombinant protein using the natural enzyme purified from the tobacco hornworm hemolymph, MSlyz may be an interesting model to study structure-function and folding for a novel lysozyme. Moreover, to demonstrate that it is a cold-adapted protein, thermodynamical parameters needed to be calculated.
This paper focuses on the validation of the secondary structure content of Mslyz using natural enzyme and synchrotron circular dichroism spectroscopy and calculation of thermodynamical parameters.
MATERIALS AND METHODS
Enzymes and chemicals
HEWL was purchased from Sigma Chemical Co. (St. Louis MO). Natural Mslyz was purified from the insect hemolymph using ion exchange and gel filtration chromatography as previously described [12]. Recombinant MSlyz was expressed in Escherichia coli, using the pET system [13], refolded and purified using Micrococcus luteus affinity and ion exchange chromatography as previously described [14]. Natural and recombinant lysozymes were analyzed by electro spray mass spectrometry at the Molecular Structure Facility, University of California, Davis, USA.
Determination of lysozyme activity
The enzymatic activity was measured using the M. luteus lytic assay as described [15]. One unit of lysozyme enzymatic activity (k) is defined as the change of 0.001 units of absorbance at 450 nm per minute of a M. luteus suspension (ΔA450/min). Since the bacterial substrate does not have a defined molecular weight, we used the convention of using k as the rate constant in all calculations and mainly for comparison between lysozymes, namely natural and recombinant MSlyz and HEWL. Thermal stability and optimal temperature was assayed in a Peltier-temperature controlled UV spectrophotometer (Cary 50 Varian Inc.). Arrhenius equation (k= Ae−E/RT) was adjusted to a linear regression and thermodynamic parameters were calculated as previously described [16] for natural and recombinant M. sexta lysozyme and for HEWL.
Circular dichroism (CD)
CD spectra were acquired in a Jasco J-810 CD spectrophotopolarimeter using a 0.1 cm path-length cell in the far-UV range (180−240 nm) and near-UV (240−340) using a 1.0 cm path-length. The CD spectra were recorded at 25°C, in triplicate. Averaged spectra, corrected for the blank (distilled water or buffer) were smoothed using the Savitsky-Golay algorithm. Ellipticities were also recorded at Brookhaven National Laboratory NSLS beam line U9B at 25°C.
Spectra were recorded in a 0.3-ml quartz cuvette with a 0.1-mm optical path at 1-nm intervals and 0.5-s time constant and 50-v sensitivity. A water blank was subtracted from the corresponding sample. Secondary structure content was determined by analysis of ellipticities with the CDSTTR algorithm [17] available via Internet on the DICHROWEB server (http://www.cryst.bbk.ac.uk/cdweb/html/) at Birbeck College, UK [18,19]. Final far-UV CD data was reported as MRE (mean residue ellipticity) degrees cm2 dmol−1 residue−1.
RESULTS
Natural and recombinant MSlyz are similar in mass and structure
To compare mass and secondary structure of the recombinant versus the natural MSlyz, the natural protein was purified from M. sexta hemolymph. The two enzymes were identical when analyzed by gel electrophoresis, while by mass spectrometry there was a difference between natural MSlyz (13,975 Da) and recombinant MSlyz (14,106 Da) that can be explained due to the presence of the extra N-terminal methionine present in the recombinant protein (data not shown). This methionine is required to be present for proper expression in the T7-based pET bacterial expression system [13]. These results are in agreement with a natural MSlyz that is not glycosylated, even though it contains two potential N-linked glycosylation sites in its primary sequence. Moreover, the experimentally determined mass matches that predicted from the cDNA sequence, and the prokaryotic expression system prevents this posttranslational modification. Therefore, E. coli is adequate for expression of MSlyz, although some eukaryotic system might eliminate a need for the refolding process.
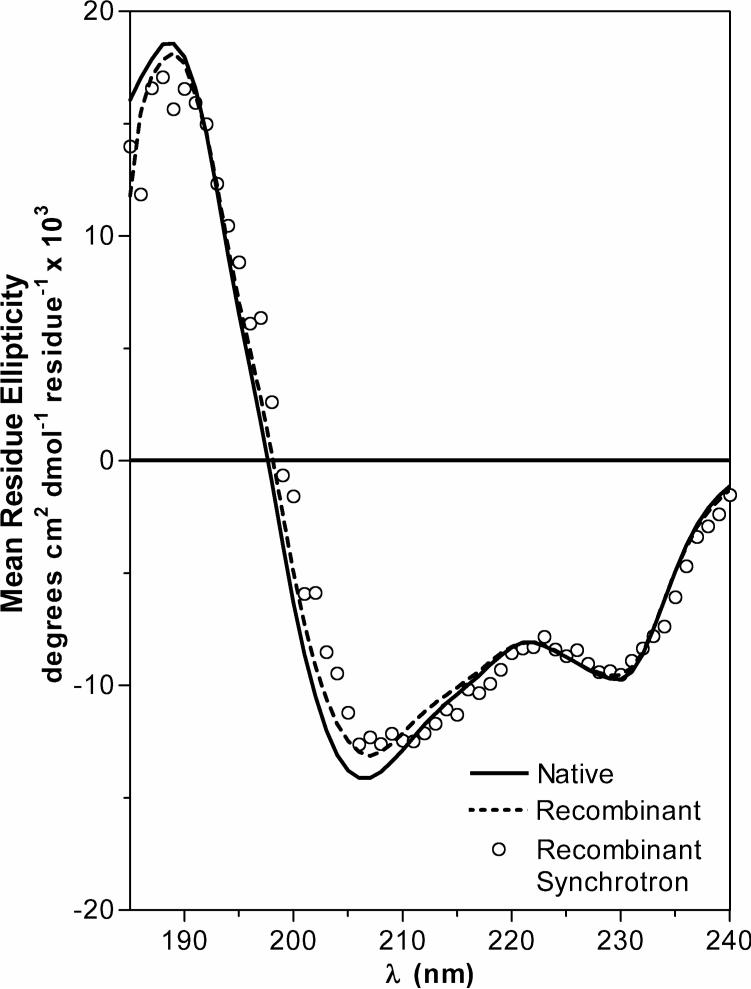
Once the identity between natural and recombinant MSLyz was validated, this is that both polypeptides had the same mass and there were no differences in posttranslational modifications, we proceeded to compare the fold of natural and recombinant MSlyz by circular dichroism (Figure 1).
Figure 1.
Structural comparison between natural and recombinant MSlyz. Data for natural and recombinant enzyme collected on a Jasco 810 spectropolarimeter are drawn with solid and dotted lines. Recombinant MSlyz ellipticities were collected at U9B NSLS Brookhaven synchrotron are represented with circles. Inset graph represents the near-UV circular dichroism spectra.
We used a Jasco-810 spectropolarimeter and the set-up at Line U9B of the National Synchrotron Light Source at the Brookhaven National Laboratory for circular CD measurements. We had previously reported the biophysical characterization of recombinant MSlyz, with an increased content of α-helical structure compared with HEWL [7].
After far-UV CD determination, we found that natural MSlyz secondary structure content is comprised of 43% helices1 (24% α- and 19% 310) while for HEWL was 30% α-helix. The α-helix content of HEWL is similar to the values deduced from an NMR structural determination [20]. In the same way we calculated β-sheet content of 15% for MSlyz and 13% for HEWL; 17% of turns for MSlyz and 27% for HEWL and 25% of unordered structure for MSlyz and 30% for HEWL. This result is a more robust determination than our previous analysis of recombinant MSlyz since the natural enzyme was obtained from the organisms and is not likely to have conformational heterogeneity such as in the case of the refolded recombinant lysozyme, and the RMSD values are much lower compared to our previous work.
To confirm the structural determination by an independent experiment and to a higher resolution, a synchrotron CD confirming the data obtained with the laboratory spectropolarimeter (Figure 1). We also recorded the near-UV spectra, and in the absence of quantitative methods for comparison, it appears that aromatic side chains are in similar environments (inferred from near-UV CD spectra) and therefore have a similar tertiary structure (Figure 1, insert graph).
MSlyz thermodynamical parameters
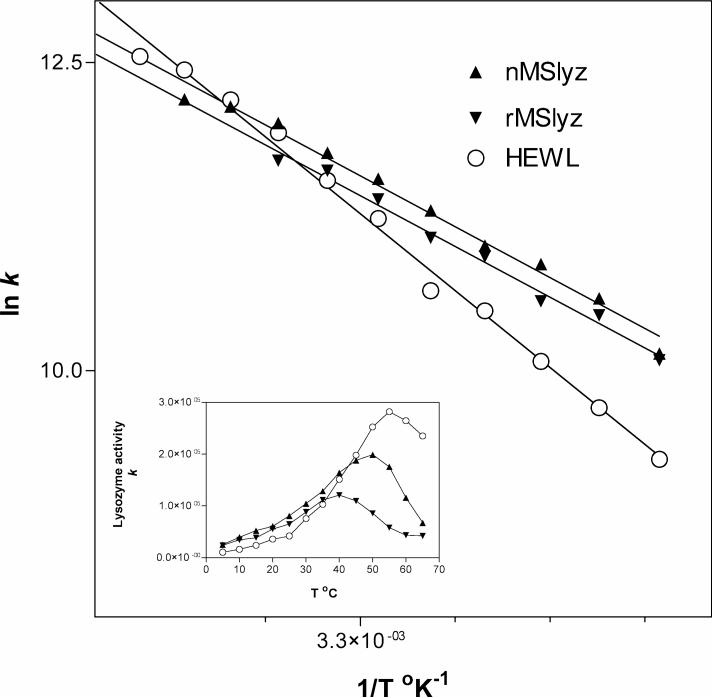
Considering that recombinant and natural MSlyz have practically the same structure and conformation, we aimed to determine whether this insect lysozyme is a bona fide cold adapted enzyme. Analysis of the thermodynamic activation parameters showed that the absolute thermodynamic values are not as relevant as the comparison with corresponding mesophilic or thermophilic homologs. Therefore, comparison of the parameters between MSlyz and HEWL provides the most useful information [16]. In Figure 2 we present the Arrhenius plots of the natural, recombinant and HEWL derived from measuring lysozyme enzymatic activity at different temperatures as shown in the figure insert. From the graph several thermodynamic parameters were calculated which are presented in Table 1.
Figure 2.
Arrhenius plot for temperature dependence of lysozyme activity. Natural (nMSlyz) and recombinant (rMSlyz) data are compared with hen egg white lysozyme (HEWL) as a reference. Inset graph represents raw experimental data on arbitrary units. Lysozyme activity k is described in Materials and Methods.
Table 1.
Thermodynamic activation parameters for glycohydrolases
| Enzyme | Source |
ΔG# (kJ/mol) |
ΔH# (kJ/mol) |
TΔS# (kJ/mol) |
Δ(ΔG#)p-m (kJ/mol) |
Δ(ΔH#)p-m (kJ/mol) |
TΔ(ΔS#)p-m (kJ/mol) |
Ref. |
|---|---|---|---|---|---|---|---|---|
| Lysozyme (E.C. 3.2.1.17) | Recombinant Manduca sextaa,p | 45.5 | 31.8 | −13.7 | −0.7 | −17.6 | −16.9 | This work |
| Natural Manduca sextaa,p | 45.1 | 31.9 | −13.2 | −1.1 | −17.5 | −16.4 | This work | |
| HEWL Gallus gallusa,m | 46.2 | 49.4 | 3.2 | - | - | - | This work | |
| Chitinase (E.C. 3.2.1.14) | Artrobacter sp.b,p | 69.2 | 60.2 | −9.0 | 2.0 | −14.1 | −16.1 | Lonhienne et al. (2000) |
| Serratia marcesensb,m | 67.2 | 74.3 | 7.1 | - | - | - | ibid | |
| Chitobiase (E.C. 3.2.1.52) | Artrobacter sp.b,p | 59.5 | 44.7 | −14.8 | −4.0 | −26.8 | −22.8 | ibid |
| Serratia marcesensb,m | 63.5 | 71.5 | 8.0 | - | - | - | ibid |
Temperature 25°C
Temperature 15°C
Psychrophile
Mesophile
The differential Δ(ΔG#)p-m reflects the difference in rate constants between psychrophilic and mesophilic enzymes. The enthalpic differential parameter Δ(ΔH#)p-m reflects the differences in activation energy (Ea). Lower Ea is characteristic of cold-adapted enzymes and is the most useful parameter for identifying this adaptation. The negative value of this parameter reflects the fact that the insect lysozyme has a lower activation energy compared to HEWL, and consequently maintains more catalytic activity at lower temperatures.
The entropic parameter TΔ(ΔS#)p-m reflects the conformational flexibility of the psychrophilic active sites, which is less restricted and makes this parameter always negative [16]. Interpretation of this entropic parameter is more related to folding stability, which in the case of a disulfide-bonded protein may be less critical for its activity at lower temperatures.
DISCUSSION
Cold adapted proteins reduce the activation energy barrier that permits that more molecules cross the reaction pathway. Two factors contribute to this effect, ΔH# and ΔS# by a decrease of the first parameter or an increase of the second. The enthalpy term is more significant at lower temperatures since it reflects the decrease in activation energy. It appears that structural changes have evolved to decrease ΔH# or Ea to reduce the dependence of kcat on temperature [16]. Features such as the lower optimal temperature for the psychrophilic enzyme (MSlyz) compared to the mesophilic (HEWL) were found. There is no consensus on whether the optimal temperature has to be lower of a certain cut-off value [2]. However, cold-adapted proteins have lower optimal temperatures compared with their mesophilic or thermophylic counterparts. Therefore, the thermodynamical activation parameters obtained in this work support the denomination of M. sexta lysozyme as a cold-adapted protein.
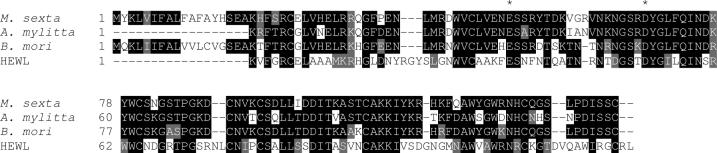
To establish a structure-function relationship between secondary structure and cold adaptation, the effect of temperature on structure is required. Recent work using NMR has addressed this question using human lysozyme as a model [21]. Comparison of lysozyme solution structure at 4 vs. 35°C indicates that low temperature leads to a conformational change where α-helix is reduced from 37.7 to 33.1% and 310 helices increase to a final value of 3.1% of. In MSlyz, there is a significant content of 310 helix (19%) compared to α-helix (24%), which might be a determinant in cold adaptation based on the NMR study. Comparison of the amino acid sequence of insect lysozymes does not show any apparent reason for these differences in secondary structure content (Figure 3). X-ray crystallography or NMR would be the way forward to test and determine the three-dimensional structure of MSlyz and advance on the structural understanding of the determinants of its cold-adaptation.
Figure 3.
Sequence alignment of lysozymes. The catalytic residues E35 and D52 are labeled with an asterisk. Manduca sexta (Genbank accession number 7327646), Antherea mylitta (accession number 17943393), Bombix mori (accession number 567099), HEWL (hen egg white lysozyme, accession number 31616035).
From the biological point of view, it appears that lysozyme has other roles besides degrading bacterial cell wall. It has been shown that lysozyme inhibits the enzymatic activity of mosquito phenoloxidase, therefore modulates melanization [22] and its cold adaptation may have effects in such novel regulatory functions. Growth and development are also dependent on temperature [23], and the role of lysozyme during ontogeny as part of the innate defense will require further investigation. From many points of views, MSlyz is an interesting model to understand the structural basis of enzyme cold adaptation.
ACKNOWLEDGMENTS
A. A. Lopez-Zavala and A.A. Arvizu-Flores received graduate scholarships and RR Sotelo-Mundo acknowledges grant (36928-B) from National Science and Technology Research Council (CONACYT) Mexico. EF Velazquez acknowledges support from Subsecretaría de Educación Superior/SEP grant P/PIFI 2001−26-F0−08 for equipment instrumentation. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. MK acknowledges support from NIH grant GM41247. We thank Dr. William Jewell from the Molecular Structure Facility at University of California-Davis for help with mass spectrometry analysis. We are also grateful to Drs. John Sutherland, John Trunk and Denisse Monteleone from NSLS-Brookhaven Nat. Lab., Dr. Bonnie Wallace from Birbeck College-University of London and Dr. Robert W. Janes from School of Biological Sciences, Queen Mary University of London for training at the BioCD 2005 course and beamline time at U9B. We thank Dr. Maria Islas-Osuna for critical reading and suggestions.
Footnotes
This paper is dedicated to the memory of Prof. Michael A. Wells, who was a mentor for some of the authors, made fundamental contributions to insect biochemistry and molecular biology, and was a promoter of Manduca sexta as a model organism and as a teaching resource.
Root mean standard deviation of MSlys secondary structure calculation was 0.021
REFERENCES
- 1.Jolles P, Jolles J. Mol Cell Biochem. 1984;63:165–89. doi: 10.1007/BF00285225. [DOI] [PubMed] [Google Scholar]
- 2.Siddiqui KS, Cavicchioli R. Annual Review of Biochemistry. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 3.Georlette D, Blaise V, Collins T, D'Amico S, Gratia E, Hoyoux A, Marx JC, Sonan G, Feller G, Gerday C. FEMS Microbiol Rev. 2004;28:25–42. doi: 10.1016/j.femsre.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Kanost MR, Jiang H, Yu XQ. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 5.Masaki K, Aizawa T, Koganesawa N, Nimori T, Bando H, Kawano K, Nitta K. J Protein Chem. 2001;20:107–13. doi: 10.1023/a:1011073206353. [DOI] [PubMed] [Google Scholar]
- 6.Matsuura A, Yao M, Aizawa T, Koganesawa N, Masaki K, Miyazawa M, Demura M, Tanaka I, Kawano K, Nitta K. Biochemistry. 2002;41:12086–92. doi: 10.1021/bi016099j. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Zavala AA, de-la-Re-Vega E, Calderon-Arredondo SA, Garcia-Orozco KD, Velazquez EF, Islas-Osuna MA, Valdez MA, Sotelo-Mundo RR. Protein Pept Lett. 2004;11:85–92. doi: 10.2174/0929866043478374. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie JP, Kanost MR, Trenczek T. Annual Review of Entomology. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Kanost MR, Dai W, Dunn PE. Archives of Insect Biochemistry and Physiology. 1988;8:147–164. [Google Scholar]
- 10.Mulnix AB, Dunn PE. Insect Biochemistry and Molecular Biology. 1994;24:271–281. doi: 10.1016/0965-1748(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 11.Soulages JL, Pennington J, Bendavid O, Wells MA. Biochem Biophys Res Commun. 1998;243:372–6. doi: 10.1006/bbrc.1998.8099. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal GA, Dahlman DL. J Biol Chem. 1991;266:15684–7. [PubMed] [Google Scholar]
- 13.Studier FW, Moffatt BA. J Mol Biol. 1986;189:113–30. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Orozco KD, Lopez-Zavala AA, Puentes-Camacho D, Calderon-de-la-Barca AM, Sotelo-Mundo RR. Biotechnol Lett. 2005;27:1075–80. doi: 10.1007/s10529-005-8452-1. [DOI] [PubMed] [Google Scholar]
- 15.Shugar D. Biochim Biophys Acta. 1952;8:302–9. doi: 10.1016/0006-3002(52)90045-0. [DOI] [PubMed] [Google Scholar]
- 16.Lonhienne T, Gerday C, Feller G. Biochim Biophys Acta. 2000;1543:1–10. doi: 10.1016/s0167-4838(00)00210-7. [DOI] [PubMed] [Google Scholar]
- 17.Sreerama N, Woody RW. Anal Biochem. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 18.Lobley A, Whitmore L, Wallace BA. Bioinformatics. 2002;18:211–2. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 19.Whitmore L, Wallace BA. Nucleic Acids Res. 2004;32:W668–73. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knubovets T, Osterhout JJ, Connolly PJ, Klibanov AM. Proc Natl Acad Sci U S A. 1999;96:1262–7. doi: 10.1073/pnas.96.4.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumeta H, Miura A, Kobashigawa Y, Miura K, Oka C, Nemoto N, Nitta K, Tsuda S. Biochemistry. 2003;42:1209–16. doi: 10.1021/bi026730w. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Paskewitz SM. Journal of Insect Physiology. 2006;52:936–942. doi: 10.1016/j.jinsphys.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert N, Raworth DA. Canadian Entomologist. 1996;128:1–13. [Google Scholar]