Abstract
The IgA isotype of human antibodies triggers inflammatory responses via the IgA-specific receptor FcαRI (CD89). Structural studies have suggested that IgA1 N-glycans could modulate the interaction with FcαRI. We have carried out detailed biophysical analyses of three IgA1 samples purified from human serum and recombinant IgA1-Fc and compared their binding to FcαRI. Analytical ultracentrifugation revealed wide variation in the distribution of polymeric species between IgA1 samples, and Fourier transform ion cyclotron resonance mass spectrometry showed overlapping but distinct populations of N-glycan species between IgA1 samples. Kinetic and equilibrium data from surface plasmon resonance experiments revealed that variation in the IgA1 CH2 N-glycans had no effect on the kinetics or affinity constants for binding to FcαRI. Indeed, complete enzymatic removal of the IgA1 N-glycans yielded superimposable binding curves. These findings have implications for renal diseases such as IgA nephropathy.
Keywords: FcαRI, IgA1, IgA nephropathy, N-glycosylation, mass spectrometry, analytical ultracentrifugation, surface plasmon resonance
IgA antibodies play an important role in protecting against environmental pathogens and antigens encountered at mucosal sites (1). IgA is the second most prevalent antibody in the serum after IgG and is divided into two subclasses, IgA1 and IgA2. There are several distinct forms of IgA; monomeric, dimeric or higher polymeric forms are found in serum, and secretory IgA is found in mucosal secretions. Monomeric IgA consists of two heavy chains and two light chains, κ or λ. Each IgA heavy chain has an extended C-terminal tailpiece, which can form an intermolecular disulfide bond with joining (J) chain leading to the formation of dimeric or polymeric IgA. In serum, approximately 95% of IgA is monomeric and 5% is polymeric (1, 2). The IgA1 subclass contains an O-glycosylated hinge region connecting the Fc and Fab regions as well as two conserved N-glycosylation sites per heavy chain: one at residue N263 in the CH2 domain and the other at N459 in the tailpiece. IgA2 has a truncated hinge region lacking O-glycans and an additional two or three conserved N-glycans (3, 4).
Inflammatory responses triggered by IgA1 or IgA2 are primarily mediated by the IgA-specific Fc receptor, FcαRI. FcαRI is a type I transmembrane glycoprotein expressed on cells of the myeloid lineage including eosinophils, neutrophils, monocytes and macrophages (5, 6). It consists of two extracellular immunoglobulin-like domains, a transmembrane domain with a positively charged arginine residue that associates with the FcRγ-chain signaling co-receptor, and a short cytoplasmic tail (1). Activation of FcαRI on myeloid cells by IgA immune complexes triggers inflammatory immune responses such as phagocytosis, respiratory burst, antibody-dependent cellular cytotoxicity (ADCC) and cytokine release (1). In addition, cross-linking of cell surface FcαRI by IgA immune complexes causes the shedding of the extracellular domain of FcαRI into circulation (7). This soluble FcαRI still retains its capacity to bind IgA1 and can associate with polymeric IgA1, creating soluble IgA-FcαRI complexes in circulation (8).
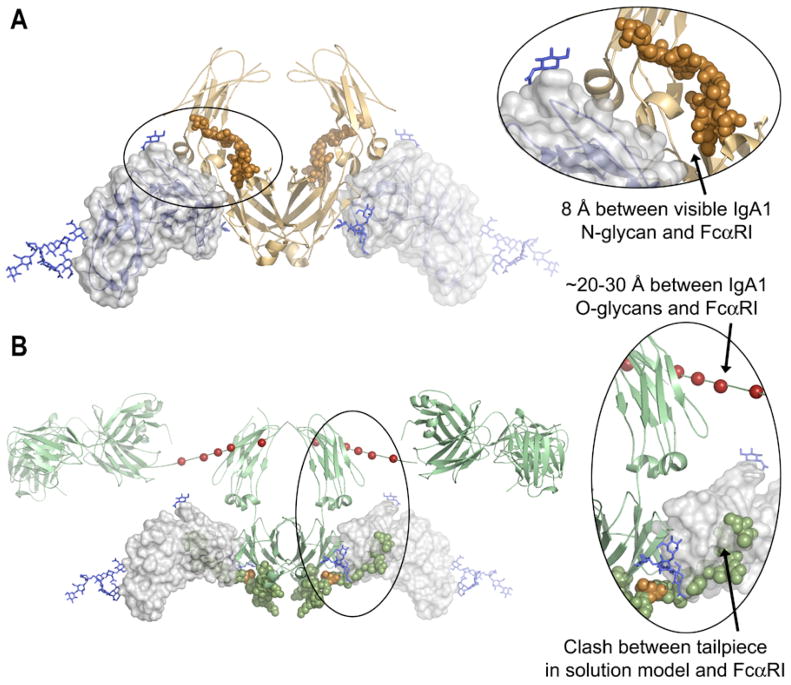
Recently, we solved the crystal structure of soluble FcαRI bound to an IgA1-Fc core fragment (called Fcα) at 3.1 Å (9). The N-terminal domain of FcαRI (D1) binds IgA in the CH2/CH3 junction of Fcα, as predicted by mutational work (9, 10). Two FcαRI molecules bind a single IgA homodimer, which is consistent with prior analytical ultracentrifugation (AUC) and surface plasmon resonance (SPR) studies (11). Although Fcα generally resembles IgG-Fc (Fcγ) and IgE-Fc (Fcε), the location of the CH2 domain N-glycans differs dramatically. The Fcα N-glycans are located on the external surface of the CH2 domains (Figure 1A), unlike Fcγ and Fcε N-glycans, which are found between the upper Fc domains (9, 12, 13). Furthermore, the CH2 N-glycans on Fcα are solvent-accessible and approach within 8 Å of FcαRI. However, the electron density for the Fcα N-glycans in the crystal structure was weak and only one branch of the putative biantennary oligosaccharide could be resolved. Thus, the CH2 N-glycans of IgA1 may directly contact FcαRI and play a role in the FcαRI-IgA1 interaction. In contrast to the IgA1 N-glycans, the O-glycosylated hinge region is located 20–30 Å from FcαRI (based on superposition of the Fcα:FcαRI crystal structure (9) and the solution structure of IgA1 determined by small-angle X-ray and neutron scattering (14), and thus would not be expected to interact with FcαRI at all (Figure 1B).
Figure 1. Structural views of the IgA1 N-glycans and tailpiece.
(A) Crystal structure of the FcαRI:Fcα complex (pdb 1OW0, reference (9), showing that the CH2 N-glycan on IgA1 (copper spheres) approaches FcαRI (blue ribbons and transparent surface) but does not directly contact the receptor. The electron density for the N-glycan was weak and only one branch of the complex carbohydrate was visible.
(B) Docking of FcαRI from the FcαRI:Fcα complex onto the low-resolution solution scattering structure of intact IgA1 (pdb 1IGA, reference (14). The deposited IgA1 model illustrates one possible location for the IgA1 tailpiece, which is shown as green spheres, with N459 (the location of the tailpiece N-glycan) in gold. Note that in this potential orientation the tailpiece clashes with FcαRI, and that the potential hinge O-glycosylation sites (red spheres) are distant from FcαRI.
The physiological role of IgA1 glycosylation is unclear, outside of IgA clearance by hepatocytes via the asialoglycoprotein receptor (15) and glycan-dependent immune exclusion by secretory IgA (2, 16). However, abnormalities in glycosylation have been implicated in several diseases such as Sjögren's syndrome (17), Henoch-Schönlein purpura (18–20) and IgA nephropathy (IgAN) (20, 21). IgAN is the best-understood example of a disease whose pathogenesis has been linked to aberrant IgA1 glycosylation. This disease is characterized by the deposition of IgA1-containing immune complexes in the glomerular mesangium, resulting in mesangial cell proliferation, matrix overproduction, and infiltration of inflammatory immune cells (22, 23). The mechanisms leading to IgA1 deposition in the kidney are still unclear; however, altered glycosylation and polymerization states of IgA1 and immune complex formation have been reported in a large number of IgAN patients. Aberrancies in IgA1 glycans in IgAN include galactose deficiency of O-linked glycans (20, 24–26), sometimes reported to be accompanied by either oversialylation (27) or undersialylation (28, 29). One of these studies (28) also indicated that N-linked glycans of IgA may be affected. These alterations in IgA1 may affect its interaction with receptors; indeed, IgA1 from IgAN patients has been reported to show either increased (30) or decreased (31) binding to FcαRI, and increased binding to transferrin receptor (32). Altered IgA1 affinity for cellular receptors could have important implications for immune complex formation, mesangial deposition of IgA1, and pathogenesis of IgAN. For example, soluble FcαRI complexed with IgA has been implicated in pathogenesis in a mouse model of IgAN (33), and transferrin receptor has been identified as a mesangial cell receptor capable of binding IgA1 from IgAN patients (34). However, there is some level of controversy about the role of these receptors in IgAN. Circulating FcαRI:IgA1 complexes are present in healthy subjects as well as in patients with IgAN, and no correlation between levels of circulating IgA1-FcαRI complexes and IgAN prognosis was found (35). Furthermore, mouse IgA failed to bind FcαRI under conditions favoring binding of human IgA (36), raising questions about the conclusions of the human FcαRI transgenic mouse model of IgAN.
Given the aberrant glycosylation of IgA1 in IgAN, the suggested role of soluble FcαRI in the disease, and the possible interaction between IgA1 N-glycans and FcαRI suggested by the crystal structure, we set out to characterize the contribution that IgA1 N-glycans make to the IgA1-FcαRI interaction. We have analyzed the polymeric state and N-glycosylation profile of different human myeloma IgA1 proteins using analytical ultracentrifugation (AUC) and Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS), respectively. Furthermore, we have compared the FcαRI binding affinities of a recombinant IgA1-Fc fragment and the three purified human IgA1 samples through extensive kinetic and equilibrium binding analyses by surface plasmon resonance (SPR). Finally, we have compared the FcαRI binding affinity of a human IgA1 sample before and after enzymatic removal of the N-glycans to assess their contribution to the interaction with FcαRI.
Experimental Procedures
IgA1 purification
Myeloma patient-derived IgA1κ and λ proteins were obtained from Biodesign International (Cincinnati, OH). IgA1 (Mce), a naturally undergalactosylated myeloma IgA1 protein that resembles IgA1 from IgAN patients, was isolated as described (21). Each protein was purified by size exclusion chromatography using a Hi Load 16/60 Superdex 200 column (Amersham Biosciences) into monomeric, dimeric/trimeric and high-molecular weight fractions. Size exclusion-purified monomeric IgA1 (mIgA1) fractions were used for binding experiments and are referred to as mIgA1κ, mIgA1λ, and mIgA1Mce.
Fcα vector, transfection, expression and purification
The core IgA1-Fc fragment (called Fcα) lacking hinge and tailpiece regions has been described previously (11). This construct was sub-cloned into the pcDNA3 vector using EcoRI and HindIII restriction sites. COS-7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) and supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The COS-7 cells were transfected with 5 μg of Fcα vector using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. 72 h after transfection, stable cells were selected with 500 μg/ml G418. Western blots using anti-IgA1 (Southern Biotech, Birmingham, AL) showed expression of Fcα. Culture medium was harvested from the stably transfected cells and dialyzed against 20 mM Tris, 150 mM NaCl, pH 7.4 (TBS). Fcα was purified by two sequential Ni-affinity chromatography steps, followed by size exclusion chromatography using a Superdex 200 column (Amersham Biosciences). Purity of the protein was determined to be >95% by SDS-PAGE. Protein concentrations were determined using extinction coefficients of 64,940 M−1 cm−1 or Fcα (11) and 208,280 M−1 cm−1 for mIgA1 (14).
FcαRI purification
Soluble recombinant FcαRI ectodomain was expressed in baculovirus-infected insect cells by the Caltech Protein Expression Center and purified by Ni-affinity hromatography followed by size exclusion chromatography (11). The resulting FcαRI was monomeric based on size exclusion chromatography and AUC analyses and was estimated to be > 95% pure by SDS-PAGE. FcαRI concentration was calculated using an extinction coefficient of 33,140 M−1 cm−1.
Enzymatic Deglycosylation of IgA1 N-glycans
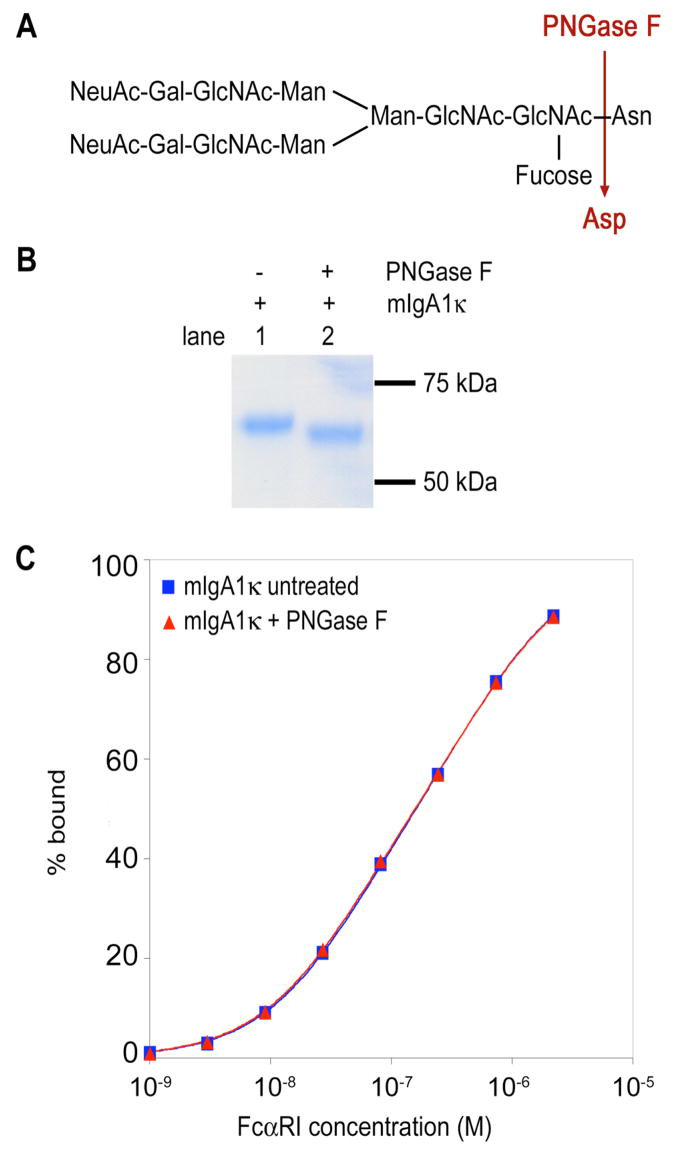
Forty μg of serum-derived mIgA1κ was incubated overnight at 37° C in 100 mM sodium phosphate buffer (pH 7.5) with 0.08 U of PNGase F (N-glycanase; Glyko, San Leandro, CA) in the presence or absence of 0.3 U α-L-fucosidase (Sigma). One-hundred ng of mIgA1κ before and after enzymatic treatment was separated by SDS-PAGE under reducing conditions and stained with Coomassie blue to verify the expected decrease of the apparent molecular mass of the heavy chain after deglycosylation.
Mass spectrometry
For analysis of IgA1 N-glycosylation by reversed phased C18 liquid chromatography (LC) FT-ICR MS, IgA1 glycopeptides were analyzed as tryptic fragments as described (37). The glycopeptide amino acid sequence was confirmed by use of activated ion-electron capture dissociation (AI-ECD) FT-ICR tandem mass spectrometry (MS/MS) as previously described (37), with the exception that the precursor ion populations were photon-irradiated for 100 ms at 8% (1.6 W) laser power. Individual N-glycans were identified from the known sequence of the isolated N-glycopeptide and monoisotopic mass assignment of glycopeptides by use of the GlycoMod tool (http://www.expasy.org). Methods for analysis of PNGase F-released IgA1 N-glycans by use of FT-ICR MS and LTQ tandem mass spectrometry (MS/MS) are described in supplemental material.
Carbohydrate analyses
The monosaccharide composition of glycans of mIgA1κ, mIgA1λ, and mIgA1Mce proteins were determined as trifluoroacetates of methylglycosides by gas-liquid chromatography (38, 39). All monosaccharides were quantified relative to the total protein and their ratios were expressed relative to mannose.
Analytical Ultracentrifugation
To yield the size distribution of all components of myeloma patient-derived IgA1 (κ, λ and Mce), proteins were analyzed by sedimentation velocity experiments in a Beckman XL-I ProteomeLab analytical ultracentrifuge as described (11). 400 μl of IgA1κ, λ and Mce were spun at 36,000 rpm at 20° C and scanned at 230 nm. Data were fitted using the c(s) and c(M) analysis routines in the program SEDFIT, which calculates the differential distribution of sedimentation coefficients or apparent molecular masses after accounting for sample diffusion (40). Each initial IgA1 sample, as well as each monomeric, dimeric and polymeric sub-fraction obtained by size-exclusion chromatography, was subjected to sedimentation velocity analysis to determine the sedimentation coefficients of the sedimenting species.
Biosensor Analyses
Surface plasmon resonance (SPR) biosensor assays were carried out on a Biacore 3000 instrument. For all experiments, 10 μg/ml Fcα, mIgA1κ, mIgA1λ, or mIgA1Mce were diluted in sodium acetate buffer (pH 4.5) and immobilized on CM-5 chips (~300 RU for Fcα and ~600 RU for mIgA1) using random amine chemistry. Previous studies showed that binding parameters derived from oriented and random amine coupling were similar (11). The first flow cell was mock-coupled and its response was subtracted from all other flow cells, followed by subtraction of a buffer blank injection. Soluble FcαRI ectodomain was injected at 25 °C at concentrations ranging from 1 nM to 2.2 μM with a flow rate of 30 μl/minute.
The association and dissociation phases of the kinetic sensogram data were fitted using the bivalent ligand model in the program ClampXP version 3.50 (Tom Morton and David Myszka, University of Utah, 2002), which describes sequential binding of two receptor molecules to a homodimeric ligand (i.e., Fcα or mIgA1). Each binding site is distinguishable (i.e., not constrained to have identical affinity) and is defined as having equal occupancy. Initial binding experiments were carried out with the standard buffer TBS-P (TBS with 0.005% P20 detergent). Data from SPR chips coupled at three different densities (27 binding curves) were fitted globally using the program ClampXP (41) to yield binding parameters for mIgA1κ. Control experiments at different flow rates indicated that the binding was not mass transport limited (11). Salt dependence experiments used 50 mM Tris-HCl (pH 7.4) with 10 mM, 30 mM, 100 mM, 300 mM, or 1 M NaCl and 0.005% P-20. For pH dependence experiments a triple buffer was utilized containing 25 mM sodium acetate, 25 mM MES, and 25 mM bis-Tris propane, adjusted to several pH values ranging from 5.5 to 9.5, with 150 mM NaCl and 0.005% P20. The apparent kinetic rate constants reported by ClampXP were corrected by appropriate statistical factors (i.e., the corrected value k1on = 0.5*k1on, app as derived from ClampXP; corrected k2off = 0.5*k2off, app), as described (11).
Although the kinetic fits do not perfectly overlay the observed data, they are quite reasonable given the fast kinetics and the complexity of the system. We have globally fitted two sets each of association and dissociation rate constants per dataset, and in appropriate cases we globally fitted multiple datasets from different surfaces. If a single-site model was used instead to fit the same data, the residuals were much worse unless we allowed numerous bulk refractive index (RI) shifts, which was not warranted based on the experimental design. Furthermore, adding in the bulk RI shifts increased the % error of the rate constants by 2-3-fold or more, and in some cases prevented the determination of a single best-fit set of parameters.
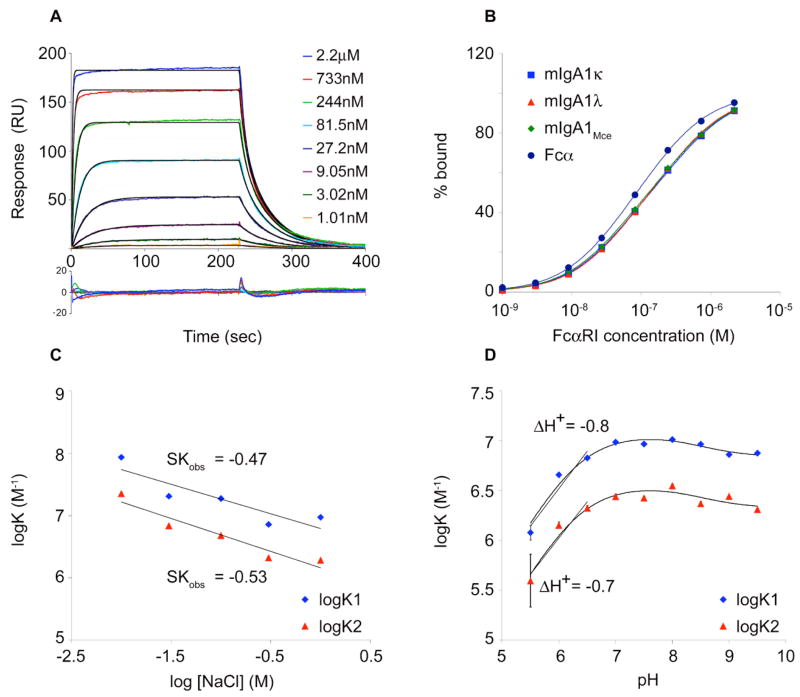
For equilibrium analyses, the equilibrium response (Req) at each injected analyte concentration was averaged over 60 seconds using the program Scrubber version 2 (BioLogic Software, 2005), and the binding isotherms were fitted to equations describing a two-site model in the program Scientist (Micromath Scientific Software, Salt Lake City, Utah), as described (11). Briefly, this model assumes two distinguishable sites with equal occupancy and yields the microscopic affinities for each binding event. Data from three independent experiments were fit globally to yield equilibrium-binding affinities for mIgA1κ. The binding isotherms were shallow (i.e., broader than a single-site isotherm), as expected for two non-identical sites without positive cooperativity. To illustrate this, a 1:1 binding isotherm should go from 10% occupancy to 90% occupancy over an 81-fold range in ligand concentration (as dictated by the binding equation: % bound = Kx / (1+Kx)). In the binding isotherm shown (Figure 3B), the curve increases from 10% to 90% bound over a ~200-fold range of FcαRI concentration (9 nM to 2 μM).
Figure 3. Surface plasmon resonance analysis of the FcαRI:mIgA1 or Fcα interaction.
(A) Representative sensograms derived from injection of different concentrations of FcαRI over mIgA1κ. Kinetic data were fit globally using the bivalent ligand model (black lines) in the program ClampXP. A residual plot is shown beneath the data. (B) Equilibrium binding isotherms for FcαRI binding to Fcα and mIgA1 κ, λ and Mce. Data points were determined by averaging the equilibrium response at the plateau of each kinetic curve and fitted to a two-site model (solid lines) in the program Scientist. (C) Analysis of salt dependence of mIgA1κ binding to FcαRI. Association equilibrium constants K1 (blue diamonds) and K2 (red triangles) for each of the two binding events were determined as a function of [NaCl] from kinetic parameters. The slope of this plot (SKobs) was determined for each binding event by linear regression. (D) Analysis of pH dependence of mIgA1κ binding to FcαRI. Binding constants K1 (blue diamonds) and K2 (red triangles) for each binding event were determined as a function of pH from kinetic parameters. Data were fitted to a model of linked protonation and binding equilibria in the program Scientist. In addition, the slope of each curve yields ΔH+, the net number of protons released or taken up on binding. ΔH+ of −0.8 and −0.7 for the first and second binding event, respectively, suggest, a single ionizable residue is present at each FcαRI:mIgA1 binding interface.
Thermodynamic analyses
Determination of salt and pH dependence of binding was carried out at 25 °C as described (11). The electrostatic contribution to the overall free energy of binding was determined by comparing the ΔG at 10 mM NaCl to that at 1 M NaCl, conditions under which electrostatic interactions are abolished (42). For pH studies, log K versus pH was plotted. The slope of this plot (between pH 5.5 and pH 7.5) yields –ΔH+, where ΔH+ is the net number of protons bound or released when FcαRI binds to Fcα. The pH dependence of binding can be described as linked equilibria by the following equation (11):
where Kb, obs is the observed binding constant as a function of pH. Fitting our binding data to this equation, with two independent ionizable residues at each receptor binding site on Fcα or mIgA1 yields the following parameters; Kb, binding constant of FcαRI to Fcα or mIgA1 in the unprotonated state; (pKa1)f and (pKa2)f, the pKa’s of the two ionizable residues in the free state; and (pKa1)b and (pKa2)b the pKa’s of the two ionizable residues in the bound state.
Results
Sedimentation velocity analysis of myeloma patient-derived IgA1κ, λ and Mce
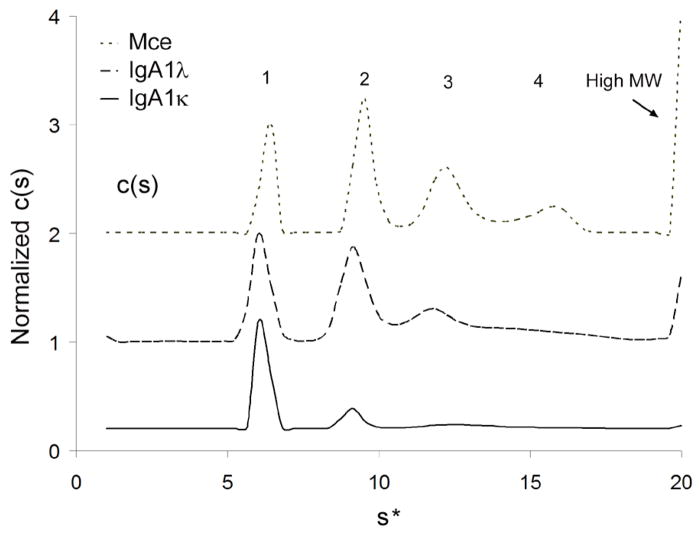
To determine the oligomeric species present in each serum-derived IgA1 sample, IgA1κ, IgA1λ and IgA1Mce were subjected to sedimentation velocity experiments in an analytical ultracentrifuge. The data were analyzed using the c(s) size distribution analysis method in the program SEDFIT (40). Distinct peaks corresponding to monomeric, dimeric, and trimeric IgA1, as well as high-molecular-weight forms (presumably aggregates) could be resolved in all the three samples, although the relative levels varied considerably (Figure 2). The apparent sedimentation coefficients of the species present in each IgA1 sample are listed in Supplementary Table S1. Perhaps due to its undergalactosylated state, the IgA1Mce distribution was skewed towards dimeric and trimeric species and also showed high levels of very large-molecular-weight species. For binding analyses, each IgA1 sample was further purified by size exclusion chromatography and the monomeric fraction was used.
Figure 2. Sedimentation Velocity Analysis of IgA1 κ, λ and Mce.
IgA1 κ, λ and Mce samples were spun at 36,000 rpm in a Beckman XL-I analytical ultracentrifuge. Data were fitted using the c(s) and c(M) sedimentation analysis routines in the program SEDFIT, which yielded the differential distribution coefficients of the sedimenting species for each sample. Distinct peaks corresponding to monomeric (labeled “1”), dimeric (“2”), trimeric (“3”) and polymeric (“4”) IgA1 species could be resolved from high molecular weight aggregates in all three samples analyzed.
SPR analysis of FcαRI binding to Fcα and mIgA1κ
Kinetic and equilibrium SPR experiments were carried out in order to compare the binding of soluble FcαRI to either recombinant Fcα or serum-purified intact mIgA1κ (Figure 3A). For kinetic analyses, a bivalent ligand model was used to derive the rate constants and a statistical correction was applied, as described (11) (Table 1). The KDs for Fcα binding to FcαRI are 60.4 ± 0.3 nM and 121 ± 1 nM, while those for mIgA1κ binding to FcαRI are 74.9 ± 0.4 nM and 244 ± 4 nM for the first and second binding events, respectively. For equilibrium binding analyses, the steady-state response at each injected FcαRI concentration was measured, and the resulting binding isotherms were fitted to a two-site binding model (Figure 3B and Table 2). Fcα binds FcαRI with KDs of 36.1 ± 1.6 nM and 204 ± 15 nM, whereas serum-purified intact mIgA1κ binds FcαRI with KDs of 40.2 ± 1.9 nM and 452 ± 58 nM. Both kinetic and equilibrium analyses indicate that mild negative cooperativity exists between the two binding sites, as seen previously in SPR analyses of FcαRI binding to Fcα (11). Based on both the kinetic and equilibrium analyses, Fcα and mIgAκ bind FcαRI with similar, although not identical affinities. The differences between the affinities may be due to the presence of the tailpiece in mIgA1κ, which is lacking in the Fcα fragment used here. Alternatively, differences between the N-glycans between recombinant Fcα (expressed in COS-7 cells) and human mIgA1κ could play a role.
TABLE 1. Kinetic parameters for FcαRI interactions with Fcα, mIgA1 κ, λ and Mce at 25° C.
Kinetic rate constants were determined using the bivalent ligand model in the program ClampXP.
| Ligand | Analyte | k1on (M s)−1 a | k1off (s−1) | k2on (M s)−1 | k2off (s−1) a | KD1 (nM) | KD2 (nM) |
|---|---|---|---|---|---|---|---|
| Fcα | FcαRI | 4.61 × 105 | 0.0278 | 4.47 × 105 | 0.054 | 60.4 ± 0.3 | 121 ± 1 |
| mIgA1κ b | FcαRI | 3.55 × 105 | 0.0266 | 3.04 × 105 | 0.074 | 74.9 ± 0.4 | 244 ± 4 |
| mIgA1λ | FcαRI | 3.12 × 105 | 0.0254 | 5.25 × 105 | 0.102 | 81.5 ± 0.7 | 195 ± 6 |
| mIgA1Mce | FcαRI | 3.65 × 105 | 0.0264 | 4.56 × 105 | 0.101 | 72.3 ± 0.8 | 222 ± 8 |
|
| |||||||
| mIgA1κ c | FcαRI | 3.17 × 105 | 0.0267 | 3.15 × 105 | 0.096 | 84.1 ± 0.6 | 303 ± 8 |
| Deglycosylated mIgA1κ c | FcαRI | 3.29 × 105 | 0.0258 | 1.87 × 105 | 0.060 | 78.4 ± 0.6 | 320 ± 7 |
Rate constants k1on and k2off have been corrected by statistical factors, as described in Experimental Procedures.
Kinetic parameters for mIgA1κ listed here were derived from a global analysis of three independent experiments, each with eight different concentrations of injected analyte.
Kinetic parameters for mIgA1κ before and after deglycosylation were determined from a single batch of mIgA1κ in order to more precisely determine the specific effects of glycosylation.
TABLE 2. Equilibrium parameters for FcαRI binding to Fcα, mIgA1 κ, λ and Mce.
KD values were determined using a two-site model in the program Scientist.
| KD1 (nM) | KD2 (nM) | |
|---|---|---|
| Fcα | 36.1 ± 1.6 | 204 ± 15 |
| mIgA1κ a | 40.2 ± 1.9 | 452 ± 58 |
| mIgA1λ | 47.6 ± 1.2 | 371 ± 21 |
| mIgA1Mce | 40.8 ± 1.7 | 403 ± 40 |
|
| ||
| mIgA1κ | 45.0 ± 0.9 | 571 ± 37 |
| Deglycosylated mIgA1κ | 42.6 ± 1.0 | 582 ± 45 |
Equilibrium constants for mIgA1κ were derived from global analysis of three independent experiments each with eight different concentrations of injected analyte.
Effect of electrostatics on Fcα:FcαRI and mIgAκ:FcαRI interactions
To look for subtle differences between Fcα and mIgA1κ binding to FcαRI, the electrostatic contribution to the binding interaction was examined. The solution scattering study of IgA1 by Boehm et al. suggested that the tailpiece of IgA1 is likely to fold back close to the receptor binding site (14). Therefore, as the tailpiece contains a histidine, two acidic residues, and potential negatively-charged sialylated N-glycans, the presence of the tailpiece within the IgA1-FcαRI interface would be expected to confer a significant electrostatic component of binding. To determine the electrostatic contribution to the binding free energy, we carried out kinetics-based biosensor binding experiments with different concentrations of NaCl in the running buffer ranging from 10 mM NaCl to 1 M NaCl; 1 M NaCl is widely accepted to effectively screen charge-charge interactions (42). The logarithms of the equilibrium association constants for Fcα or mIgA1κ binding to FcαRI were plotted as a function of log [NaCl]. The slope (SKobs) of each of these plots gives the net number of Na+ and Cl− ions taken up or released upon binding of FcαRI to Fcα or mIgA1κ (42) (Figure 3C). The salt dependence of mIgA1κ:FcαRI interaction yielded slopes of −0.47 and −0.53 for the sequential binding events while the Fcα:FcαRI interaction had slopes of −0.32 and −0.36, respectively. These shallow slopes for both mIgA1κ and Fcα binding to FcαRI suggest a minor role for counterion release upon complex formation (Table 3). The overall electrostatic contribution to the free energy of binding was calculated by comparing the Gibbs free energy (ΔG) for binding of FcαRI to Fcα or mIgA1κ at 10mM and 1 M NaCl. Less than 15% of the ΔG of binding for each interaction was contributed by electrostatics, and no significant difference was seen between the Fcα:FcαRI and mIgA1κ:FcαRI interactions.
TABLE 3. NaCl dependence of Fcα and mIgA1 κ, λ and Mce binding to FcαRI.
Slopes (SKobs) for each binding event were determined by fitting log KA (derived from kinetic rate constants) to a linear regression model. The electrostatic component of the Gibbs free energy of binding (ΔGelec/ΔG) was determined by comparing the ΔG at 10mM with that at 1M NaCl. ΔG was determined using the formula ΔG= − RTlnKA where KA is the association constant, R is the gas constant in cal (K−1 mol−1), and T is the temperature in Kelvin.
| High affinity site | Low affinity site | |||
|---|---|---|---|---|
| SKobs | ΔGelec/ΔG | SKobs | ΔGelec/ΔG | |
| Fcα | −0.32 | 8.2% | −0.36 | 9.3% |
| mIgA1κ | −0.47 | 12% | −0.53 | 15% |
| Deglycosylated mIgA1κ | −0.45 | 11% | −0.51 | 14% |
| mIgA1λ | −0.51 | 12% | −0.51 | 14% |
| mIgA1Mce | −0.60 | 15% | −0.42 | 11% |
Effect of pH on Fcα:FcαRI and mIgA1κ:FcαRI interactions
To compare the number of ionizable residues in the Fcα:FcαRI and mIgA1κ:FcαRI binding interfaces, pH studies were carried out as described (11). Kinetics-based biosensor experiments were performed over a pH range from 5.5 to 9.5. The logarithm of the association constants (KA) were plotted as a function of pH, yielding sigmoidal plots (Figure 3D). The affinities of both mIgA1κ and Fcα for FcαRI decreased as the pH was lowered from 7.0 to 5.5 (at pH 5.5, KD1 = 803 nM, KD2 = 3.43 μM for Fcα; and KD1 = 837 nM, KD2 = 2.53 μM for mIgA1κ binding to FcαRI). The negative slope of these plots (ΔH+) describes the net number of protons (H+ ions) released or taken up upon binding of FcαRI to Fcα or mIgA1κ. The values of ΔH+ for FcαRI binding to Fcα and mIgA1k are approximately -1 for both sequential binding events, suggesting the presence of a single ionizable residue below pH 7.0 in the binding interface as previously reported (11), indicating the tailpiece does not contribute additional ionizable residues to the interface. The data were also fitted to a model describing linked protonation and binding equilibria (43) as described (11). Given the slight decrease in affinity at high pH as well as the large decrease at low pH, we used a model with two protonation events. This model yields Kb1 and Kb2, the dissociation constants for the first and second binding events between FcαRI and Fcα or mIgA1κ in the unprotonated state. The low-pH transition is described by (pKa1)f and (pKa1)b, the pKa values for the ionizable residue when FcαRI and Fcα or mIgA1κ are free or bound, respectively. Likewise, (pKa2)f and (pKa2)b describe the high-pH transition in the free and bound states, respectively. The fitted values for FcαRI binding to Fcα and FcαRI binding to mIgA1κ are very similar (Table 4). The pH dependence of binding for both complexes are consistent with the presence of an ionizable residue in each interface with a free pKa value of 6.3–6.5 that becomes buried in the complex; this agrees with the Fcα:FcαRI crystal structure that revealed the burial of His85 of FcαRI in a primarily hydrophobic pocket on Fcα (9). The high-pH protonation event is likely due to the FcαRI N-terminus and has very little effect on binding affinity of FcαRI for Fcα or mIgA1κ.
TABLE 4. Linked binding/protonation equilibrium analysis of Fcα and mIgA1κ binding to FcαRI.
Parameters were determined using a linked binding/protonation model with two ionizable residues per binding site in the program Scientist, as described in Experimental Procedures.
| Ligand | High affinity site Kb (nM) | Low affinity site Kb (nM) | (pKa1)f | (pKa1)b | (pKa2)f | (pKa2)b |
|---|---|---|---|---|---|---|
| Fcα | 79.9 ± 1.95 | 187 ± 45.7 | 6.5 ± 0.25 | ∼4.0 | 8.1 ± 0.93 | 8.4 ± 0.92 |
| mIgA1κ | 88.0 ± 1.15 | 287 ± 37.6 | 6.3 ± 0.17 | ∼4.0 | 8.4 ± 0.60 | 8.6 ± 0.61 |
FT-ICR mass spectrometry analysis of N-glycan populations of mIgA1κ and mIgA1λ
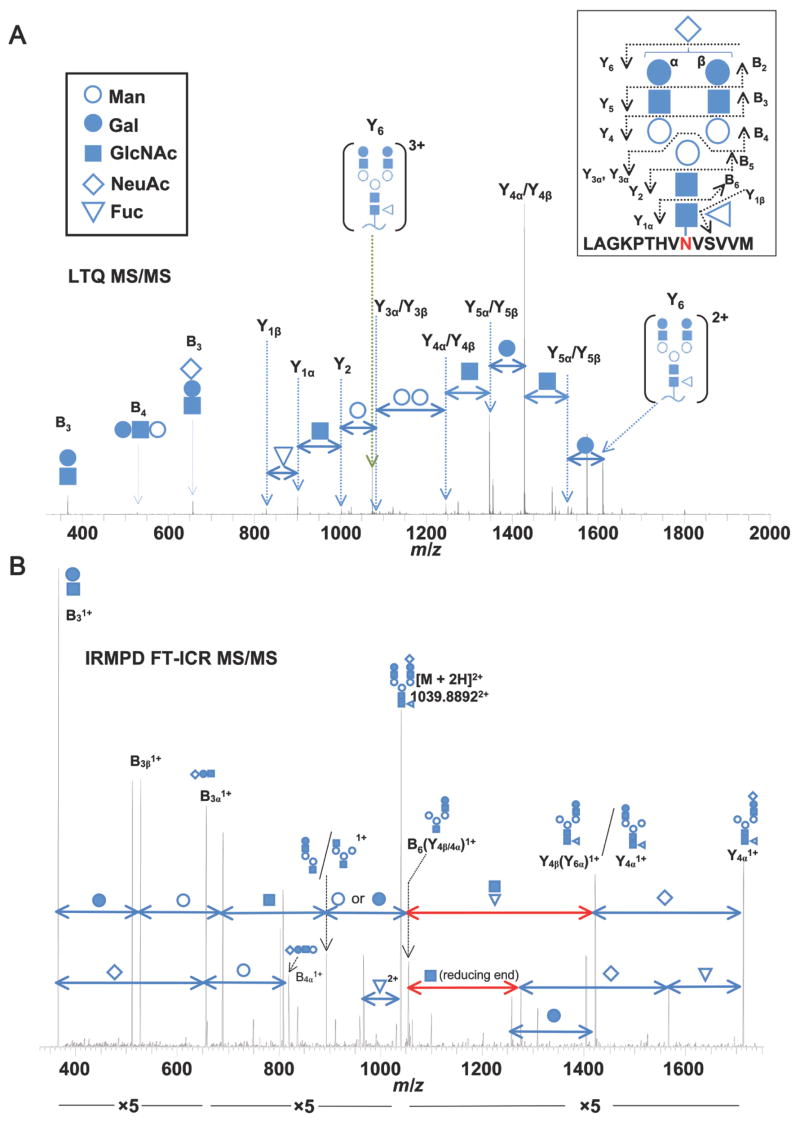
In order to analyze the importance of IgA1 N-glycosylation in the interaction with FcαRI, we first carried out a detailed analysis of the populations of N-glycans present on mIgA1 samples by monosaccharide compositional analysis and LC FT-ICR MS. Gas-liquid chromatographic analysis identified fucose, mannose, galactose, N-acetylgalactosamine, N-acetylglucosamine, and sialic acid in the preparations of mIgA1, indicating the presence of both N-linked and O-linked glycans. mIgA1Mce had more galactose and N-acetylgalactosamine and less sialic acid compared to mIgA1κ Relative proportion of mannose to other monosaccharides suggested absence of high-mannose glycans. N-glycans of mIgA1 κ and λ were then analyzed as glycopeptide tryptic fragments. IgA1 N-glycopeptides were identified by accurate mass and simultaneous data-dependent MS/MS of candidate glycopeptides, as previously described for IgA1 O-glycopeptides (37). This approach allowed confirmation of specific ion species as N-glycosylated peptides and provided N-glycan structural information as well (Figure 4). The dominant series of fragments originated from the terminal end of the N-glycan and allowed a stepwise assignment of the N-glycan structure. While the LTQ MS/MS spectra do not distinguish between galactose and mannose residues, our monosaccharide compositional analysis detected the presence of both sugars, in ratios consistent with the presence of the well-established GlcNAc2Man3 core of predominantly complex glycans. IgA1 N-glycans were also removed by PNGase F treatment and analyzed by direct infusion FT-ICR MS, LTQ MS/MS and infra-red multiphoton dissociation (IRMPD) FT-ICR MS/MS (see supplemental Figures S1 and S2). The same N-glycan covalently linked to the IgA1 tryptic fragment in Figure 4 was analyzed by IRMPD MS/MS as a released N-glycan (Figure 4B). A similar fragment ion series indicating the same N-glycan structure as Figure 4A was observed. Additionally, fragments from the reducing end of the chain were observed.
Figure 4. Representative LTQ tandem mass spectrum of an IgA1 N-glycopeptide.
(A) The collision-induced dissociation LTQ MS/MS spectrum confirms the structure of the IgA1 N-glycopeptide. Fragmentation of the IgA1 N-glycopeptide proceeds in at stepwise fashion from the terminal end of the N-glycan. The precursor mass in the FT-ICR MS spectrum (not shown) corresponded to the IgA1 tryptic peptide [L451-M464 + (NeuAc)1(Gal)2(GlcNAc)2(Fuc)1(Man)3(GlcNAc)2]3+. Fragment ions (3+ and 2+) corresponding to the loss of a single NeuAc are observed (Fragment ion Y6). From the Y62+ ion, the loss of subsequent monosaccharide residues can be observed down to the GlcNAc attached to the N459 confirming the expected N-glycan structure. Alternating fragments of the branched chains are observed (Y5 and Y4) followed by the loss of the core Man residues (Y3 and Y2). Additional fragments from the terminal end (B3, B4) further confirm the N-glycan structure. Symbols for the individual monosaccharide residues are defined in the key. Distinction between Gal and Man monosaccharides is assumed based on our monosaccharide compositional analysis and previously reported IgA1 N-glycan analysis (4, 52–54). Some individual N-glycan fragments are assigned to two possible bonds due to their isobaric composition. B) IRMPD FT-ICR MS/MS spectrum of the same IgA1 N-glycan shown in panel A, analyzed as a PNGase F-released glycan. In addition to the pattern of N-glycan fragments similar to those in A, monosaccharide losses which can only be attributed to the reducing end are observed (highlighted in red). The NeuAc residue is arbitrarily assigned to the β branch to simplify the labeled spectra.
In the course of cataloguing the individual IgA1 N-glycopeptide species, several of them could not be identified based on the theoretical mass of expected IgA1 tryptic fragments with attached N-glycans. AI-ECD fragmentation of these candidate glycopeptides was performed to confirm the amino acid sequence (Figure 5). All together, the combination of N-glycopeptide, N-glycan, and base peptide analyses provided high confidence in all of the assigned IgA1 N-glycans, as summarized in Table 5. Analysis of the glycopeptides revealed that mIgA1κ and mIgA1λ had overlapping but distinct patterns of N-glycosylation, including biantennary, triantennary, and tetraantennary complex carbohydrates (Figure 5 and Table 5).
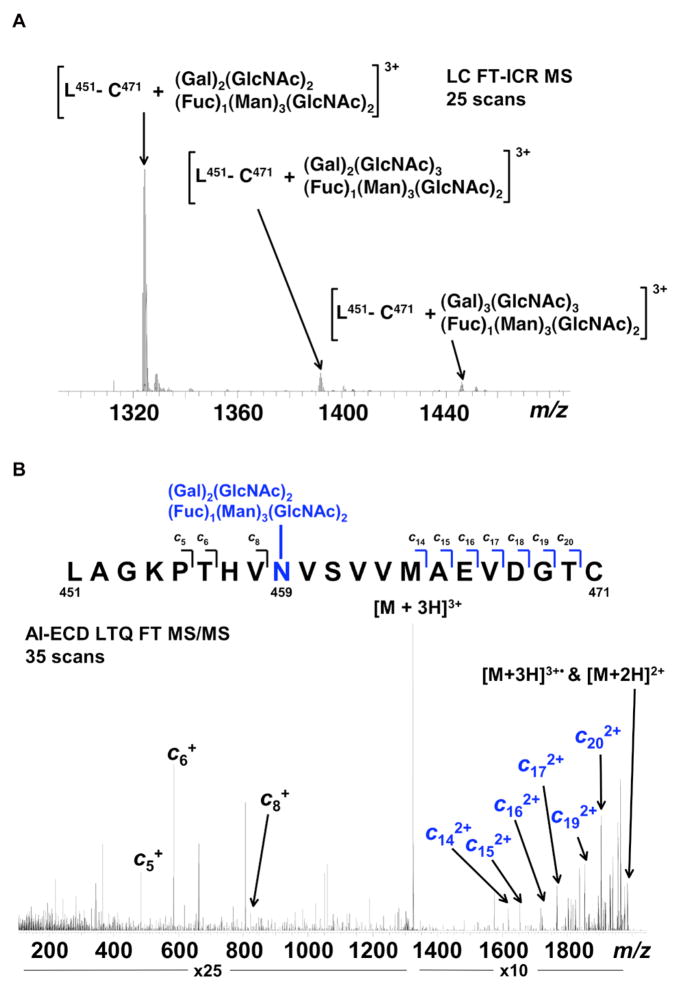
Figure 5. Representative FT-ICR data for mIgA1κ N-glycans.
(A) FT-ICR MS spectrum from the average of 25 LC FT-ICR MS scans revealing the presence of three distinct mIgA1κ N-glycopeptides. Each N-glycopeptide corresponds to the mass of the IgA1 peptide L451-C471 with three related but distinct glycans. (B) AI-ECD FT-ICR tandem mass spectrum of the [L451-C471 + (Gal)2(GlcNAc)2(Fuc)1(Man)3(GlcNAc)2]3+ triply charged ion species confirms the peptide sequence as the tailpiece (N459) site of mIgA1κ. A total of ten c type fragments were observed, originating from the N-terminus. Seven fragments C-terminal to N459 correspond to the mass of the IgA1 tryptic peptide plus the mass of the attached glycan, in contrast to the three fragments N-terminal to N459. The mass accuracy of the three IgA1 tailpiece N -glycans shown in (A) are as follows: L451-C471 + (Gal)2(GlcNAc)2(Fuc)1(Man)3(GlcNAc)2—theoretical mass 3968.714, measured mass 3968.724, error 2.52 ppm; L451-C471 + (Gal)2(GlcNAc)3(Fuc)1(Man)3(GlcNAc)2—theoretical mass 4171.793, measured mass 4171.806, error 3.24 ppm; L451-C471 + (Gal)3(GlcNAc)3(Fuc)1(Man)3(GlcNAc)2—theoretical mass 4333.846, measured mass 4333.848, error 0.67 ppm. Table 5 lists the identified IgA1 N-glycans identified for both mIgA1κ and mIgA1λ.
TABLE 5. FT-ICR Mass spectrometry analysis of N-glycans on mIgA1κ and mIgA1λ.
Glycopeptides from tryptic digests were analyzed to determine site-specific N-glycoforms. In parallel, N-glycans released by PNGase F treatment were analyzed for comparison. Both sets of samples were tested with and without neuraminidase treatment. The summary lists the assigned location(s) of each observed glycoform: either N263 (CH2) or N459 in the tailpiece (TP).
| mIgA1κ | mIgA1λ | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypsin: | ✓ | ✓ | Summary | ✓ | ✓ | Summary | ||||||
| PNGase F: | ✓ | ✓ | ✓ | ✓ | ||||||||
| Neuraminidase: | ✓ | ✓ | CH2 | TP | ✓ | ✓ | CH2 | TP | ||||
|
| ||||||||||||
| Glycoforms | ||||||||||||
| Biantennary | ||||||||||||
| (Gal)2(GlcNAc)2 + (Man)3(GlcNAc)2 | ◆ | ◆ | ■ | □ | ■ | |||||||
| (NeuAc)1(Gal)1(GlcNAc)2 + (Man)3(GlcNAc)2 | ◆ | ■ | ||||||||||
| (NeuAc)1(Gal)2(GlcNAc)2 + (Man)3(GlcNAc)2 | □ | ◆ | ◆ | ■ | ◆ | ■ | ||||||
| (NeuAc)2(Gal)2(GlcNAc)2 + (Man)3(GlcNAc)2 | ✤ | ■ | ||||||||||
| (Gal)2(GlcNAc)2 + (Fuc)1(Man)3(GlcNAc)2 | □ | ◆ | ◆ | ■ | □ | ◆ | ■ | |||||
| (NeuAc)1(Gal)2(GlcNAc)2 + (Fuc)1(Man)3(GlcNAc)2 | □ | □ | ◆ | ◆ | ■ | □ | ■ | |||||
| (NeuAc)2(Gal)2(GlcNAc)2 + (Fuc)1(Man)3(GlcNAc)2 | □ | ■ | □ | ■ | ||||||||
| Triantennary | ||||||||||||
| (Gal)1(GlcNAc)3 + (Man)3(GlcNAc)2 | ◆ | ◆ | ■ | |||||||||
| (Gal)2(GlcNAc)3 + (Man)3(GlcNAc)2 | ◆ | ■ | ||||||||||
| (NeuAc)1(Gal)2(GlcNAc)3 + (Man)3(GlcNAc)2 | ◆ | ■ | ||||||||||
| (Gal)2(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | □ | ◆ | ■ | □ | ◆ | ■ | ||||||
| (NeuAc)1(Gal)2(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | □ | ◆ | ■ | □ | ■ | |||||||
| (NeuAc)2(Gal)2(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | □ | ■ | □ | ■ | ||||||||
| (Gal)3(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | □ | ◆ | ■ | □ | ◆ | ■ | ||||||
| (NeuAc)1(Gal)3(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | ◆ | ■ | ||||||||||
| (NeuAc)2(Gal)3(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | □ | ■ | ||||||||||
| (NeuAc)3(Gal)3(GlcNAc)3 + (Fuc)1(Man)3(GlcNAc)2 | □ | ■ | ||||||||||
| Tetraantennary | ||||||||||||
| (Gal)3(GlcNAc)4 + (Fuc)1(Man)3(GlcNAc)2 | □ | ◆ | ■ | |||||||||
| (NeuAc)2(Gal)3(GlcNAc)4 + (Fuc)1(Man)3(GlcNAc)2 | □ | ◆ | ■ | |||||||||
| (NeuAc)3(Gal)3(GlcNAc)4 + (Fuc)1(Man)3(GlcNAc)2 | □ | ■ | ||||||||||
denotes verified tailpiece N-glycans from tryptic glycopeptide analysis
denotes verified CH2 N-glycans from tryptic glycopeptide analysis
denotes N-glycans released by PNGase F treatment
denotes the assignment of the N-glycans to the CH2 or tailpiece sites.
For simplicity, all glycans containing (Gal)2(GlcNAc)3 are listed as triantennary rather than bisecting biantennary; likewise, all glycans containing (Gal)3(GlcNAc)4 are listed as tetraantennary.
Analysis of tryptic glycopeptides revealed a wide range of N459 (tailpiece) N-glycoforms that were similar between mIgA1κ and mIgA1λ, although only mIgA1λ exhibited tetraantennary N-glycans. In contrast, only a single highly abundant N-glycopeptide was observed for the CH2 (N263) site among the tryptic glycopeptides. The amino acid sequence surrounding this site has a predicted negative charge. As a result, there could be several N263 glycopeptide species that would go undetected in positive ion MS. Analysis of the released N-glycans did reveal several additional N-glycans not observed as glycopeptides for both IgA1 proteins (Table 5). These glycans include three non-fucosylated biantennary N-glycans and one fucosylated triantennary N-glycan from mIgA1κ that were not observed for the tailpiece N-glycans. For mIgA1λ, one biantennary and three triantennary non-fucosylated N-glycans were identified that were not observed among its tailpiece glycans. Of significant note is that only one N-glycan with two sialic acid residues was observed in the released glycans, in contrast to the seven N-glycans observed as N-glycopeptides with two and three sialic acid residues. The presence of the peptide attached to the glycan provides more sites to be protonated in the ionization process. For the N-glycans indirectly assigned to the CH2 site, this would indicate that there are more non-fucosylated N-glycans with two and three sialic acid residues present, but we were not able to detect them in positive ion MS. In general, more than 90% of the tailpiece glycans of both mIgA1κ and mIgA1λ were fucosylated, consistent with previous data (44), whereas only one fucosylated N-glycan was observed among all the CH2 glycans,
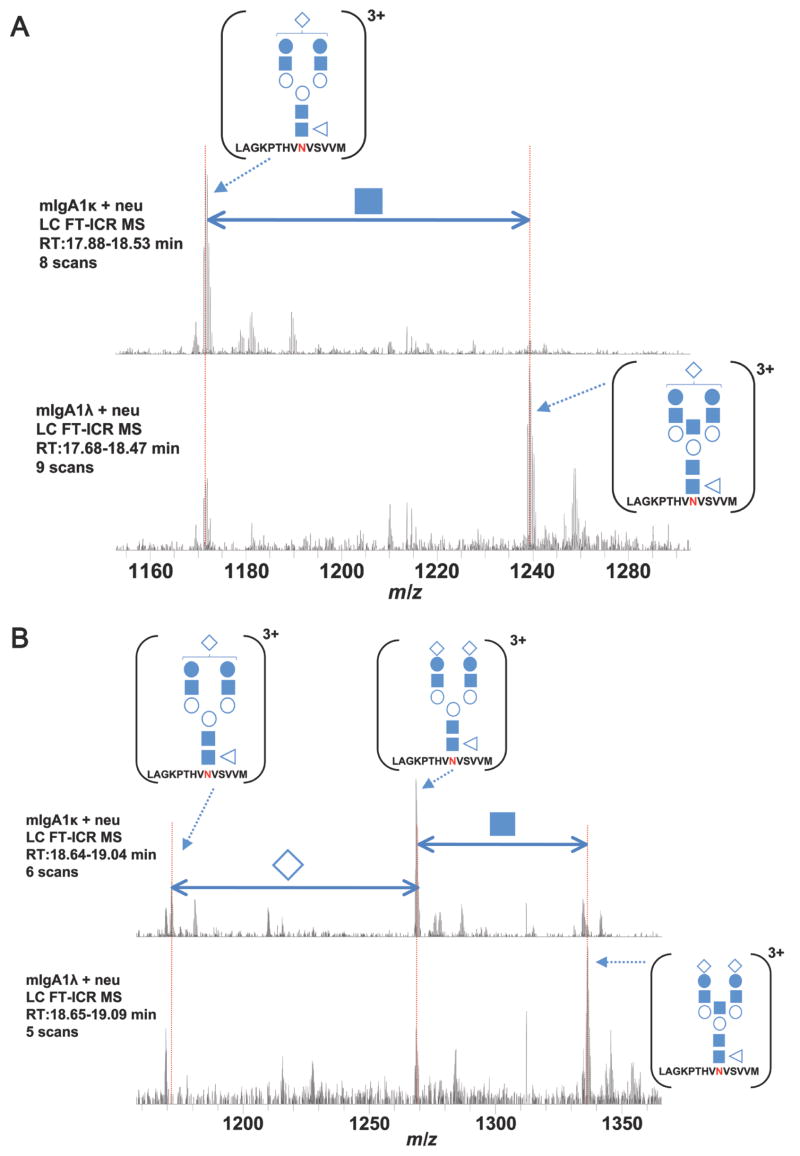
In addition to the qualitative assessment of the N-glycan chains at each site, the MS analysis can provide relative quantitative information on the population of N-glycoforms present within a single sample and compared to each other. Figure 6 is a comparison of two independent LC-MS analyses of mIgA1κ and mIgA1λ. Biantennary N-glycopeptides were the most abundant N-glycans observed on mIgA1κat the tailpiece site. For mIgA1λ, the triantennary N-glycopeptides dominate the population of IgA1 N-glycans at the tailpiece. Figures 6A and B demonstrate these observations in the FT-ICR MS spectra at two different points in the aligned chromatograms. In Figure 6A, the mIgA1κ spectrum shows the N-glycopeptide L112-M123 with N-glycans that differ only by a single GlcNAc residue (i.e., biantennary vs. triantennary). For mIgA1κ, only the biantennary N-glycopeptide is present with no triantennary glycoform observed. For mIgA1λ, both N-glycoforms are observed but the triantennary form dominates. In Figure 6B, the same trend holds for the same IgA1 tryptic glycopeptide with an additional sialic acid residue that elutes later in the LC-MS analysis. This shift towards a higher branched population in mIgA1λ is further supported by detection of tetraantennary forms. The same trend of a higher branched population in mIgA1λ compared to mIgA1κ is observed in the released N-glycans (data not shown). Thus, although the populations of IgA1 N-glycoforms at the tailpiece are qualitatively similar, there are distinct differences in their relative abundances within each sample. This is also true for the N-glycans indirectly assigned to the CH2 site.
Figure 6. The distribution of N-glycans differs between mIgA1κ and mIgA1λ.
Extracted FT-ICR MS spectra from two independent LC MS analyses of mIgA1κ and mIgA1λ. The two LC MS runs are compared at different points in the chromatogram. At the first aligned retention time (A) the spectra reveal a mono-sialylated IgA1 tryptic peptide [L451-M464]3+ that is observed predominantly as a biantennary N-glycopeptide in mIgA1κ (biantennary vs. triantennary abundance > 10:1). In contrast, the same two IgA1 N-glycopeptides observed in mIgA1λ have a different relative abundance (biantennary vs. triantennary abundance ~1:3), with the triantennary N-glycoform dominating. This same pattern holds later in the analysis of a disialylated version of the same IgA1 tryptic peptide (B) Similar relative abundances were observed for each sample in the released N-glycan analysis (data not shown). The position of the bisecting biantennary IgA1 N-glycan is assumed based on previous work (4, 53). Monosaccharide nomenclature is the same as in Figure 4.
SPR analyses of FcαRI binding to three serum-derived mIgA1 species (κ, λ and Mce)
Given the heterogeneity of N-glycosylation at the CH2 site, SPR kinetic and equilibrium experiments were performed to compare the binding affinities of FcαRI for mIgA1κ, mIgA1λ, and mIgA1Mce. Each of these IgA1 samples are of the same isotype and thus will have the same amino acid sequence in their Fc region, so the only relevant differences between samples would be due to N-glycan variability. Our data from kinetic experiments showed very similar association and dissociation constants for FcαRI binding to all the three IgA1 monomer samples (Table 1). Furthermore, our equilibrium analyses showed very similar affinities for FcαRI binding to IgA1κ, λ and Mce monomer; indeed, the equilibrium binding isotherms almost perfectly overlaid (Figure 3B; Table 2). Analysis of the salt dependence of binding showed very similar slopes for each IgA1 sample (Table 3), indicating similar electrostatic interactions.
Enzymatic deglycosylation of IgA1 N-glycans does not affect its interaction with FcαRI
To establish more conclusively whether the N-glycans of IgA1 modulate the interaction with FcαRI, we enzymatically removed the IgA1 N-glycans (Figure 7A). Gas chromatographic analysis of the final deglycosylated sample confirmed that about 85-87% of the N-glycans were removed (data not shown). Kinetic SPR analyses revealed similar on- and off-rates for the binding of FcαRI to untreated or deglycosylated IgA1, and the calculated dissociation constants were nearly identical, indicating that loss of the N-glycans had essentially no effect on the binding affinity (Table 1). Furthermore, equilibrium SPR binding analyses of the native and deglycosylated samples gave superimposable binding curves (Figure 7C; Table 2). Finally, the salt dependence of the native and deglycosylated IgA1 samples were nearly identical, with SKobs values of −0.47 and−0.45, respectively, for the first binding event and SKobs values of −0.53 and −0.51 for the second event (Table 3). These results indicate that removal of the IgA1 N-glycans causes no detectable alteration in the binding affinity or electrostatic component of binding.
Figure 7. Enzymatic deglycosylation of mIgA1κ N-glycans.
(A) Structure of a typical biantennary complex N-glycan and the site of enzymatic deglycosylation. (B) Reducing SDS-PAGE gel stained with Coomassie blue to show the shift in migration of heavy chain after deglycosylation. (C) Equilibrium binding isotherm for FcαRI binding to mIgA1κ before and after enzymatic deglycosylation. Data points were determined by averaging the equilibrium response at the plateau of each kinetic curve and fitted to a two-site model (solid lines) in the program Scientist. FcαRI bound similarly to both native and deglycosylated mIgA1κ samples.
Discussion
Human IgA shares the same basic architecture with other human antibody isotypes, although it has several unusual properties. For example, similar to IgM, IgA is capable of forming higher-order polymeric forms in serum that are formed via interactions between the tailpiece and the J-chain polypeptide. Furthermore, the IgA1 subclass features both N- and O-glycosylation, with the O-glycans restricted to the hinge region whereas the N-glycans are present at N263 in the CH2 domain and N459 in the tailpiece of each heavy chain. The CH2 N-glycans are themselves atypical, in that they are found on the solvent-exposed surface of the Fc region rather than being sequestered between the upper Ig domains of the Fc region, as in IgG and IgE. This exposed nature of the IgA1 N-glycans suggests that they may play a role in modulating interactions with IgA-specific receptors such as FcαRI and Fcα/μR, or with lectin-like receptors such as mannose-binding lectin and the asialoglycoprotein receptor (45, 46). Although the crystal structure of the FcαRI:Fcα complex yielded a wealth of information about the interface formed between the two proteins, several important questions remained unresolved. In particular, what are the contributions of the CH2 N-glycan and the IgA tailpiece to the IgA1-FcαRI interface? The structure of the complex showed that the CH2 N-glycan approaches at least within 8 Å of the receptor. However, due to poorly resolved electron density, only one branch of the complex N-glycan was visible. Furthermore, the Fcα fragment crystallized was produced in CHO cells rather than human B cells and thus may not represent the physiological glycoform. Secondly, the Fcα fragment crystallized did not contain the hinge or tailpiece regions. Superimposing our FcαRI:Fcα complex structure on a model of intact IgA1 determined by small-angle X-ray and neutron scattering suggests that the likely folded-back tailpiece conformations predicted by solution scattering could interfere with or modulate FcαRI binding (Figure 1B) (9, 14). Thus, we have carried out detailed studies of the interaction between FcαRI and full-length monomeric IgA1 purified from human serum and compared the binding parameters to those for the Fcα:FcαRI interaction. In humans, circulatory IgA1 is present mainly in its monomeric form with very little polymeric IgA1 (1). However, in diseases such as IgAN, the ratio of polymeric IgA1 to monomeric IgA1 is increased (1, 2). Our sedimentation velocity experiments on serum-derived intact myeloma IgA1 proteins κ, λ and Mce showed well-resolved peaks corresponding to IgA1 monomer, dimer/trimer, higher polymer as well as very large-molecular-weight species that could be protein aggregates. This analytical ultracentrifugation approach yields higher-resolution discrimination of the components of serum-derived IgA1 than can be achieved by size exclusion chromatography. Thus, AUC analysis could be useful in studying the polymeric distribution of IgA1 in patients with IgAN or other diseases associated with increased polymeric IgA levels. Interestingly, the mIgA1Mce myeloma protein that exhibits undergalactosylation of the hinge region O-glycans (similar to IgAN antibodies) showed a size distribution that was skewed toward larger polymeric species compared to the IgA1κ and IgA1λ samples, suggesting a possible link between undergalactosylation of the hinge region and altered polymeric distribution, as previously proposed (47–51).
Our kinetic and equilibrium biosensor studies comparing the binding of FcαRI to either Fcα or mIgA1 showed small but significant differences in affinity, suggesting that the hinge or tailpiece regions may modulate the interaction. An overlay of the Fcα-FcαRI crystal structure and the intact IgA1 solution structure suggested that the hinge region is 20–30 Å away from the receptor-binding site (Figure 1A). Therefore, neither the hinge region nor its O-glycans are likely to approach FcαRI in the complex and would at most be expected to contribute only minor long-range electrostatic effects. In contrast, the predicted location of the IgA1 tailpiece clashes with FcαRI in the modeled complex (Figure 1B). We therefore carried out detailed analyses of the pH- and NaCl-dependence of binding in order to compare the chemical nature of the binding interfaces when Fcα or mIgA1 bind FcαRI. The binding affinity was reduced as the pH was lowered from 7.0 to 5.5, consistent with protonation of a single histidine residue between neutral and acidic pH, as previously reported for Fcα (11). The NaCl dependence of binding showed a similar slope for both Fcα and mIgA1 binding to FcαRI, indicating that less than 15% of the free energy of binding was contributed by electrostatics. Thus, the number and nature of ionizable groups in the interface as well as the electrostatic component of binding are similar in the presence or absence of the tailpiece, which contains a histidine, two acidic residues, and potential sialylated N-glycans. This suggests that the tailpiece may not play an important role in the IgA1:FcαRI interface, although these results could depend on the experimental setup used for SPR.
We analyzed the N-glycan population on mIgA1κ and mIgA1λ samples by use of high-resolution FT-ICR mass spectrometry. Our work expands on prior studies analyzing IgA1 N-glycosylation (16, 52–54), given that our comprehensive analyses of N-glycopeptides and released N-glycans allows discrimination of individual glycoforms present within a particular IgA1 sample at each glycosylation site. The high mass accuracy and resolution of FT-ICR MS allows the unambiguous assignment of individual glycoforms. This is especially true in the analysis of N-glycosylation where the varied combination of sugars and base peptides can result in a series of N-glycan assignments with mass differences less than one atomic mass unit. MS/MS analysis by collision-induced dissociation and IRMPD confirmed the N-glycan structure. In some cases, individual N-glycans were observed attached to two or three unique tryptic IgA1 fragments due to unexpected trypsin cleavage sites and variably modified C471 and M461. Taken together, the complete analysis of N-glycans as N-glycopeptides and PNGase F-released N-glycans both with and without neuraminidase treatment provided us with a high level of confidence in the assigned glycans at the two IgA1 sites (Table 5). Furthermore, the combined analyses give us high confidence that we completely characterized the tailpiece N-glycans for mIgA1κ and mIgA1λ and have identified those N-glycans that are unique to the CH2 site. We observed that many of the N-glycan structures were found in both samples, although their relative abundances were distinctly different and there were individual glycoforms that differed between samples, such as tetraantennary N-glycans being observed only in the tailpiece of mIgA1λ. Additionally, mostly biantennary N-glycans were observed at the CH2 site in mIgA1κ, whereas mostly triantennary N-glycans were assigned to the CH2 in mIgA1λ. In fact, a greater abundance of triantennary N-glycans was observed at both sites in mIgA1λ.
A comparison of our results with previous analyses of IgA N-glycosylation (4, 52–54) shows generally similar conclusions, in that the most commonly observed N-glycans were complex biantennary and triantennary (or bisecting biantennary) N-glycans showing high levels of sialylation and fucosylation. Baenziger and Kornfeld identified three complex N-glycans from a myeloma IgA1 sample; all were sialylated biantennary complex N-glycans, one of which had a bisecting GlcNAc and another glycan with core fucosylation (52). We observed all the glycans reported by Baenziger, except that we found a monosialylated variant of their disialylated species (IIB). Endo and colleagues analyzed several myeloma IgA1 and IgA2 samples and reported a high degree of variability in the N-glycan species between samples (4), similar to our findings here. The majority of the glycans reported by Endo et al. are biantennary complex N-glycans, about half of which contain bisecting GlcNAc and half of which are fucosylated. They also reported low levels of degalactosylated (i.e., Gal0) complex N-glycans and high-mannose N-glycans, neither of which we observed. Field et al. analyzed serum-derived (not myeloma-derived) IgA and reported primarily biantennary complex N-glycans with variable levels of sialylation and fucosylation, as well as a few triantennary glycans (54). Like Endo et al., they also found low levels of degalactosylated and high-mannose N-glycans. Finally, Mattu et al. analyzed serum-derived and recombinant IgA and reported a range of biantennary and triantennary species with variable sialylation and fucosylation (53). We observed all the glycans they reported in their serum-derived IgA sample (although our approach cannot resolve the sialic acid linkages or core vs, outer-arm fucose attachment). They also observed low levels of degalactosylated and high-mannose N-glycans, but only in their recombinant IgA sample. Our data differs from the previous studies in that we observed tetraantennary N-glycans (or possibly triantennary glycans with bisecting GlcNAc) in the mIgA1λ sample that had not previously been reported from serum-derived IgA samples, although Mattu et al. did report a single tetraantennary species from recombinant IgA (53). Our results also reveal the degree of variability between individual N-glycosylation sites; in particular, only 1 of 8 observed CH2 glycans for both mIgA1κ and mIgA1λ were fucosylated, compared to 19 of 21 tailpiece glycans. These results suggest that differences in accessibility of the N-glycans attached to the CH2 and tailpiece sites likely modulate the activity of the fucosyltransferases involved.
Despite the observed variation in N-glycans between mIgA1 samples, kinetic and equilibrium SPR analyses revealed that mIgA1κ, mIgA1λ, and mIgA1Mce bound FcαRI with very similar kinetic parameters and with nearly identical affinities (Figure 3, Tables 1 and 2). Moreover, binding studies with different salt concentrations indicated that all three IgA1 monomers showed similar behavior on binding to FcαRI (Table 3), indicating that variation in the CH2 N-glycans did not affect FcαRI affinity. Indeed, even after near-complete enzymatic removal of the IgA1 N-glycans, the binding curves for deglycosylated and control mIgA1κ superimposed precisely, with essentially identical kinetic rate constants, equilibrium dissociation constants, and salt dependence parameters (Figure 7, Tables 1, 2, and 3). Taken together, these results establish that despite physical proximity, the CH2 N-glycans of IgA1 do not play a significant role in the interaction with FcαRI.
Contradictory results have been published regarding the importance of IgA1 CH2 N-glycans in binding to FcαRI. Other groups have shown that removal of the IgA1 CH2 N-glycan by mutagenesis or enzymatic deglycosylation results in either: i) no effect on FcαRI binding (53), ii) a moderate increase in initial binding (55), or iii) complete loss of binding to FcαRI (56). Many previous binding analyses have tended toward either quantitative biophysical binding analyses using recombinant IgA1 or IgA1-Fc (11, 57) or qualitative binding experiments using intact human IgA1 (53, 55, 56). We have used three different intact IgA1 proteins with native glycosylation purified from human serum in careful quantitative SPR analyses in an attempt to resolve the controversy.
A second ongoing question relates to the role that the IgA tailpiece plays in FcαRI binding. Our kinetic and equilibrium SPR results are in agreement with the studies suggesting that neither the tailpiece peptide nor the N-glycans play a significant role in FcαRI binding (53, 56, 58). In these studies, IgA1 mutants, including N263A that lacks the CH2 N-glycan and a tailpiece mutant lacking the tailpiece and its N-glycan, were able to bind neutrophil FcαRI and mediate rosette formation to the same extent as wild type IgA1. However, our results are in contrast to qualitative SPR results from Oortwijn et al. (55), who reported that removal of the N-glycans from serum IgA resulted in apparent enhanced association and rapid dissociation of mIgA1 with FcαRI by SPR. The two SPR studies were set up differently, however; Oortwijn et al. coupled an Fc-FcαRI fusion to the SPR chip and injected IgA1, whereas we coupled IgA1 to the chip and injected soluble FcαRI. Coupling FcαRI to the chip mimics the physiological setting, although we prefer coupling IgA1 in order to improve precision of KD determinations (11). Finally, our results differ from those of Carayannopoulos et al, who reported that an IgA1 mutant (N263Q using our numbering scheme) lacking the CH2 N-glycan expressed in insect cells did not interact with neutrophil FcαRI (56). This discrepancy might be due to a specific effect of the glutamine residue introduced or the expression of IgA1 in insect cells rather than mammalian cells.
Our results are particularly relevant to studies of pathogenesis of IgA nephropathy, as IgAN patients have been widely reported to show aberrant glycosylation of IgA1, along with higher prevalence of polymeric IgA1 forms in serum. Moreover, soluble FcαRI has been implicated in the pathogenesis of IgAN using an FcαRI-transgenic mouse model of IgAN (33). IgA1 glycan abnormalities reported in IgAN patients include oversialylation (27, 51), undersialylation (29), and galactose deficiency of hinge-region O-glycans (20, 21, 25, 26, 29, 51, 59, 60), as well as oversialylation (24) or truncation (28) of the N-glycans. Our results indicate that N-glycan truncation is not responsible for modulating the interaction with FcαRI. It is, however, possible that altered IgA1 polymerization may play a role in modulating the functional interaction with FcαRI, and that aberrantly glycosylated IgA1 could play an indirect role as well. Undergalactosylation and undersialylation of IgA1 O-glycans have been reported to cause the IgA1 to aggregate or form immune complexes (21, 25, 47–50), which is consistent with our sedimentation analysis of IgA1Mce. Higher-order IgA1 polymeric forms would be expected to show increased avidity of binding to cell-surface FcαRI and would likely be more effective at activating inflammatory pathways and inducing shedding of FcαRI. Future studies will be needed to carefully compare the intrinsic affinity of monomeric and polymeric forms of IgA1 for FcαRI, as well as their functional properties.
Supplementary Material
Supplementary experimental procedures and figures providing information about the analysis of PNGase F-released N-glycans, and a supplementary table listing sedimentation coefficients for observed species of IgA1κ, IgA1λ, and IgA1Mce. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank George Ibrahim for making the Fcα construct, Dr. Jost Vielmetter and Inderjit Nangiana of the Caltech Protein Expression Center for expression of FcαRI, Mrs. Rhubell Brown for technical assistance, and Dr. Sohaib Khan for use of the Biacore 3000 instrument.
Abbreviations
- IgA1
Immunoglobulin alpha 1
- Fc
crystallizable fragment
- GlcNAc
N-acetyl glucosamine
- ADCC
antibody-dependent cell-mediated cytotoxicity
- D1
domain 1
- AUC
analytical ultracentrifugation
- SPR
surface plasmon resonance
- IgG
Immunoglobulin gamma
- IgE
Immunoglobulin epsilon
- IgAN
IgA nephropathy
- FT-ICR MS
Fourier transform ion cyclotron resonance mass spectrometry
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
fetal bovine serum
- TBS
Tris-buffered saline
- SDS-PAGE
sodium dodecylsulfate-polyacrylamide gel electrophoresis
- PBS
phosphate-buffered saline
- RU
response units
- MES
morpholino-ethane sulfonic acid
- LTQ
two-dimensional linear quadrupole ion trap
- IRMPD
infra-red multiphoton dissociation
Footnotes
This work was supported by NIH grants DK071802, DK078244, DK080301, DK061525, DE013694, DK047322, and DK064400 and funds from the State of Ohio Eminent Scholar Program.
References
- 1.Monteiro RC, Van De Winkel JG. IgA Fc receptors. Annu Rev Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 2.Mestecky J, Moro I, Kerr MA, Woof JM. In: Mucosal Immunology. Mestecky J, Bienenstock J, Lamm ME, Mayer L, McGhee JR, Strober W, editors. Vol. 3. Elsevier Academic Press; Amsterdam: 2005. pp. 153–181. [Google Scholar]
- 3.Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the α1 heavy chain. J Biol Chem. 1979;254:2865–2874. [PubMed] [Google Scholar]
- 4.Endo T, Mestecky J, Kulhavy R, Kobata A. Carbohydrate heterogeneity of human myeloma proteins of the IgA1 and IgA2 subclasses. Mol Immunol. 1994;31:1415–1422. doi: 10.1016/0161-5890(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 5.Maliszewski CR, March CJ, Schoenborn MA, Gimpel S, Shen L. Expression cloning of a human Fc receptor for IgA. J Exp Med. 1990;172:1665–1672. doi: 10.1084/jem.172.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monteiro RC, Kubagawa H, Cooper MD. Cellular distribution, regulation, and biochemical nature of an Fcα receptor in humans. J Exp Med. 1990;171:597–613. doi: 10.1084/jem.171.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Zandbergen G, Westerhuis R, Mohamad NK, van De Winkel JG, Daha MR, van Kooten C. Crosslinking of the human Fc receptor for IgA (FcαRI/CD89) triggers FcR gamma-chain-dependent shedding of soluble CD89. J Immunol. 1999;163:5806–5812. [PubMed] [Google Scholar]
- 8.van der Boog PJ, van Zandbergen G, de Fijter JW, Klar-Mohamad N, van Seggelen A, Brandtzaeg P, Daha MR, van Kooten C. FcαRI/CD89 circulates in human serum covalently linked to IgA in a polymeric state. J Immunol. 2002;168:1252–1258. doi: 10.4049/jimmunol.168.3.1252. [DOI] [PubMed] [Google Scholar]
- 9.Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcαRI and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- 10.Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 11.Herr AB, White CL, Milburn C, Wu C, Bjorkman PJ. Bivalent Binding of IgA1 to FcαRI Suggests a Mechanism for Cytokine Activation of IgA Phagocytosis. J Mol Biol. 2003;327:645–657. doi: 10.1016/s0022-2836(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 12.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor FcεRIα. Nature. 2000;406:259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 13.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-FcγRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 14.Boehm MK, Woof JM, Kerr MA, Perkins SJ. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J Mol Biol. 1999;286:1421–1447. doi: 10.1006/jmbi.1998.2556. [DOI] [PubMed] [Google Scholar]
- 15.Rifai A, Fadden K, Morrison SL, Chintalacharuvu KR. The N-glycans determine the differential blood clearance and hepatic uptake of human immunoglobulin (Ig)A1 and IgA2 isotypes. J Exp Med. 2000;191:2171–2182. doi: 10.1084/jem.191.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Royle L, Roos A, Harvey DJ, Wormald MR, van Gijlswijk-Janssen D, Redwan el RM, Wilson IA, Daha MR, Dwek RA, Rudd PM. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J Biol Chem. 2003;278:20140–20153. doi: 10.1074/jbc.M301436200. [DOI] [PubMed] [Google Scholar]
- 17.Basset C, Durand V, Jamin C, Clement J, Pennec Y, Youinou P, Dueymes M, Roitt IM. Increased N-linked glycosylation leading to oversialylation of monomeric immunoglobulin A1 from patients with Sjögren's syndrome. Scand J Immunol. 2000;51:300–306. doi: 10.1046/j.1365-3083.2000.00685.x. [DOI] [PubMed] [Google Scholar]
- 18.Saulsbury FT. Alterations in the O-linked glycosylation of IgA1 in children with Henoch-Schönlein purpura. J Rheumatol. 1997;24:2246–2249. [PubMed] [Google Scholar]
- 19.Lau KK, Wyatt RJ, Moldoveanu Z, Tomana M, Julian BA, Hogg RJ, Lee JY, Huang WQ, Mestecky J, Novak J. Serum levels of galactose-deficient IgA in children with IgA nephropathy and Henoch-Schönlein purpura. Pediatr Nephrol. 2007;22:2067–2072. doi: 10.1007/s00467-007-0623-y. [DOI] [PubMed] [Google Scholar]
- 20.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang WQ, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 2007;71:1148–1154. doi: 10.1038/sj.ki.5002185. [DOI] [PubMed] [Google Scholar]
- 21.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J. Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest. 1999;104:73–81. doi: 10.1172/JCI5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monteiro RC, Moura IC, Launay P, Tsuge T, Haddad E, Benhamou M, Cooper MD, Arcos-Fajardo M. Pathogenic significance of IgA receptor interactions in IgA nephropathy. Trends Mol Med. 2002;8:464–468. doi: 10.1016/s1471-4914(02)02405-x. [DOI] [PubMed] [Google Scholar]
- 23.Emancipator SN. In: Heptinstall's Pathology of the Kidney. Jennette JC, Olson JL, Schwartz MM, Silva FG, editors. Lippincott-Raven Publishers; Philadelphia: 1998. pp. 479–539. [Google Scholar]
- 24.Linossier MT, Palle S, Berthoux F. Different glycosylation profile of serum IgA1 in IgA nephropathy according to the glomerular basement membrane thickness: normal versus thin. Am J Kidney Dis. 2003;41:558–564. doi: 10.1053/ajkd.2003.50117. [DOI] [PubMed] [Google Scholar]
- 25.Mestecky J, Tomana M, Crowley-Nowick PA, Moldoveanu Z, Julian BA, Jackson S. Defective galactosylation and clearance of IgA1 molecules as a possible etiopathogenic factor in IgA nephropathy. Contrib Nephrol. 1993;104:172–182. doi: 10.1159/000422410. [DOI] [PubMed] [Google Scholar]
- 26.Hiki Y, Odani H, Takahashi M, Yasuda Y, Nishimoto A, Iwase H, Shinzato T, Kobayashi Y, Maeda K. Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int. 2001;59:1077–1085. doi: 10.1046/j.1523-1755.2001.0590031077.x. [DOI] [PubMed] [Google Scholar]
- 27.Leung JC, Tang SC, Chan DT, Lui SL, Lai KN. Increased sialylation of polymeric lambda-IgA1 in patients with IgA nephropathy. J Clin Lab Anal. 2002;16:11–19. doi: 10.1002/jcla.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amore A, Cirina P, Conti G, Brusa P, Peruzzi L, Coppo R. Glycosylation of circulating IgA in patients with IgA nephropathy modulates proliferation and apoptosis of mesangial cells. J Am Soc Nephrol. 2001;12:1862–1871. doi: 10.1681/ASN.V1291862. [DOI] [PubMed] [Google Scholar]
- 29.Odani H, Hiki Y, Takahashi M, Nishimoto A, Yasuda Y, Iwase H, Shinzato T, Maeda K. Direct evidence for decreased sialylation and galactosylation of human serum IgA1 Fc O-glycosylated hinge peptides in IgA nephropathy by mass spectrometry. Biochem Biophys Res Commun. 2000;271:268–274. doi: 10.1006/bbrc.2000.2613. [DOI] [PubMed] [Google Scholar]
- 30.Grossetete B, Launay P, Lehuen A, Jungers P, Bach JF, Monteiro RC. Down-regulation of Fcα receptors on blood cells of IgA nephropathy patients: evidence for a negative regulatory role of serum IgA. Kidney Int. 1998;53:1321–1335. doi: 10.1046/j.1523-1755.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 31.van Zandbergen G, van Kooten C, Mohamad NK, Reterink TJ, de Fijter JW, van de Winkel JG, Daha MR. Reduced binding of immunoglobulin A (IgA) from patients with primary IgA nephropathy to the myeloid IgA Fc-receptor, CD89. Nephrol Dial Transplant. 1998;13:3058–3064. doi: 10.1093/ndt/13.12.3058. [DOI] [PubMed] [Google Scholar]
- 32.Moura IC, Arcos-Fajardo M, Sadaka C, Leroy V, Benhamou M, Novak J, Vrtovsnik F, Haddad E, Chintalacharuvu KR, Monteiro RC. Glycosylation and size of IgA1 are essential for interaction with mesangial transferrin receptor in IgA nephropathy. J Am Soc Nephrol. 2004;15:622–634. doi: 10.1097/01.asn.0000115401.07980.0c. [DOI] [PubMed] [Google Scholar]
- 33.Launay P, Grossetete B, Arcos-Fajardo M, Gaudin E, Torres SP, Beaudoin L, Patey-Mariaud de Serre N, Lehuen A, Monteiro RC. Fcα receptor (CD89) mediates the development of immunoglobulin A (IgA) nephropathy (Berger's disease). Evidence for pathogenic soluble receptor-IgA complexes in patients and CD89 transgenic mice. J Exp Med. 2000;191:1999–2009. doi: 10.1084/jem.191.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moura IC, Centelles MN, Arcos-Fajardo M, Malheiros DM, Collawn JF, Cooper MD, Monteiro RC. Identification of the transferrin receptor as a novel immunoglobulin (Ig)A1 receptor and its enhanced expression on mesangial cells in IgA nephropathy. J Exp Med. 2001;194:417–425. doi: 10.1084/jem.194.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Boog PJ, De Fijter JW, Van Kooten C, Van Der Holst R, Van Seggelen A, Van Es LA, Daha MR. Complexes of IgA with FcαRI/CD89 are not specific for primary IgA nephropathy. Kidney Int. 2003;63:514–521. doi: 10.1046/j.1523-1755.2003.00756.x. [DOI] [PubMed] [Google Scholar]
- 36.van der Boog PJ, van Kooten C, van Zandbergen G, Klar-Mohamad N, Oortwijn B, Bos NA, van Remoortere A, Hokke CH, de Fijter JW, Daha MR. Injection of recombinant FcαRI/CD89 in mice does not induce mesangial IgA deposition. Nephrol Dial Transplant. 2004;19:2729–2736. doi: 10.1093/ndt/gfh459. [DOI] [PubMed] [Google Scholar]
- 37.Renfrow MB, Mackay CL, Chalmers MJ, Julian BA, Mestecky J, Kilian M, Poulsen K, Emmett MR, Marshall AG, Novak J. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal Bioanal Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 38.Tomana M, Prchal JT, Garner LC, Skalka HW, Barker SA. Gas chromatographic analysis of lens monosaccharides. J Lab Clin Med. 1984;103:137–142. [PubMed] [Google Scholar]
- 39.Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, Hiki Y, Toma K, Emmett MR, Mestecky J, Marshall AG, Novak J. Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation fourier transform-ion cyclotron resonance mass spectrometry. J Biol Chem. 2005;280:19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 40.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myszka DG, Morton TA. CLAMP: a biosensor kinetic data analysis program. TIBS. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 42.Record MT, Jr, Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 43.Bradshaw JM, Waksman G. Calorimetric investigation of proton linkage by monitoring both the enthalpy and association constant of binding: application to the interaction of the Src SH2 domain with a high-affinity tyrosyl phosphopeptide. Biochemistry. 1998;37:15400–15407. doi: 10.1021/bi9814991. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka A, Iwase H, Hiki Y, Kokubo T, Ishii-Karakasa I, Toma K, Kobayashi Y, Hotta K. Evidence for a site-specific fucosylation of N-linked oligosaccharide of immunoglobulin A1 from normal human serum. Glycoconj J. 1998;15:995–1000. doi: 10.1023/a:1006989910120. [DOI] [PubMed] [Google Scholar]
- 45.Gomes MM, Herr AB. IgA and IgA-specific receptors in human disease: structural and functional insights into pathogenesis and therapeutic potential. Springer Semin Immunopathol. 2006;28:383–395. doi: 10.1007/s00281-006-0048-x. [DOI] [PubMed] [Google Scholar]
- 46.Hamburger AE, Bjorkman PJ, Herr AB. Structural insights into antibody-mediated mucosal immunity. Curr Top Microbiol Immunol. 2006;308:173–204. doi: 10.1007/3-540-30657-9_8. [DOI] [PubMed] [Google Scholar]
- 47.Iwase H, Ohkawa S, Ishii-Karakasa I, Hiki Y, Kokubo T, Sano T, Tanaka A, Toma K, Kobayashi Y, Hotta K. Study of the relationship between sticky human serum IgA1 and its O-glycan glycoform. Biochem Biophys Res Commun. 1999;261:472–477. doi: 10.1006/bbrc.1999.1006. [DOI] [PubMed] [Google Scholar]
- 48.Iwase H, Tanaka A, Hiki Y, Kokubo T, Sano T, Ishii-Karakasa I, Toma K, Kobayashi Y, Hotta K. Aggregated human serum immunoglobulin A1 induced by neuraminidase treatment had a lower number of O-linked sugar chains on the hinge portion. J Chromatogr B Biomed Sci Appl. 1999;724:1–7. doi: 10.1016/s0378-4347(98)00552-0. [DOI] [PubMed] [Google Scholar]
- 49.Kokubo T, Hiki Y, Iwase H, Tanaka A, Toma K, Hotta K, Kobayashi Y. Protective role of IgA1 glycans against IgA1 self-aggregation and adhesion to extracellular matrix proteins. J Am Soc Nephrol. 1998;9:2048–2054. doi: 10.1681/ASN.V9112048. [DOI] [PubMed] [Google Scholar]
- 50.Kokubo T, Hiki Y, Iwase H, Horii A, Tanaka A, Nishikido J, Hotta K, Kobayashi Y. Evidence for involvement of IgA1 hinge glycopeptide in the IgA1-IgA1 interaction in IgA nephropathy. J Am Soc Nephrol. 1997;8:915–919. doi: 10.1681/ASN.V86915. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki H, Moldoveanu Z, Hall S, Brown R, Vu HL, Novak L, Julian BA, Tomana M, Wyatt RJ, Edberg JC, Alarcon GS, Kimberly RP, Tomino Y, Mestecky J, Novak J. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baenziger J, Kornfeld S. Structure of the carbohydrate units of IgA1 immunoglobulin. I Composition, glycopeptide isolation, and structure of the asparagine-linked oligosaccharide units. J Biol Chem. 1974;249:7260–7269. [PubMed] [Google Scholar]
- 53.Mattu TS, Pleass RJ, Willis AC, Kilian M, Wormald MR, Lellouch AC, Rudd PM, Woof JM, Dwek RA. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J Biol Chem. 1998;273:2260–2272. doi: 10.1074/jbc.273.4.2260. [DOI] [PubMed] [Google Scholar]
- 54.Field MC, Amatayakul-Chantler S, Rademacher TW, Rudd PM, Dwek RA. Structural analysis of the N-glycans from human immunoglobulin A1: comparison of normal human serum immunoglobulin A1 with that isolated from patients with rheumatoid arthritis. Biochem J. 1994;299(Pt 1):261–275. doi: 10.1042/bj2990261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oortwijn BD, Roos A, van der Boog PJ, Klar-Mohamad N, van Remoortere A, Deelder AM, Daha MR, van Kooten C. Monomeric and polymeric IgA show a similar association with the myeloid FcαRI/CD89. Mol Immunol. 2007;44:966–973. doi: 10.1016/j.molimm.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 56.Carayannopoulos L, Max EE, Capra JD. Recombinant human IgA expressed in insect cells. Proc Natl Acad Sci U S A. 1994;91:8348–8352. doi: 10.1073/pnas.91.18.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wines BD, Hulett MD, Jamieson GP, Trist HM, Spratt JM, Hogarth PM. Identification of residues in the first domain of human Fcα receptor essential for interaction with IgA. J Immunol. 1999;162:2146–2153. [PubMed] [Google Scholar]
- 58.Pleass RJ, Dunlop JI, Anderson CM, Woof JM. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcα receptor (FcαR) CD89. J Biol Chem. 1999;274:23508–23514. doi: 10.1074/jbc.274.33.23508. [DOI] [PubMed] [Google Scholar]
- 59.Lai KN, Chan LY, Tang SC, Tsang AW, Guo H, Tse KC, Yip T, Leung JC. Characteristics of polymeric lambda-IgA binding to leukocytes in IgA nephropathy. J Am Soc Nephrol. 2002;13:2309–2319. doi: 10.1097/01.asn.0000026497.82930.73. [DOI] [PubMed] [Google Scholar]
- 60.Moore JS, Kulhavy R, Tomana M, Moldoveanu Z, Suzuki H, Brown R, Hall S, Kilian M, Poulsen K, Mestecky J, Julian BA, Novak J. Reactivities of N-acetylgalactosamine-specific lectins with human IgA1 proteins. Mol Immunol. 2007;44:2598–2604. doi: 10.1016/j.molimm.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary experimental procedures and figures providing information about the analysis of PNGase F-released N-glycans, and a supplementary table listing sedimentation coefficients for observed species of IgA1κ, IgA1λ, and IgA1Mce. This material is available free of charge via the Internet at http://pubs.acs.org.