Abstract
The fatty acid biosynthesis (FAS-II) pathway in Mycobacterium tuberculosis generates long chain fatty acids that serve as the precursors to mycolic acids, essential components of the mycobacterial cell wall. Enzymes in the FAS-II pathway are thought to form one or more noncovalent multi-enzyme complexes within the cell, and a bacterial two-hybrid screen was used to search for missing components of the pathway and to furnish additional data on interactions involving these enzymes in vivo. Using the FAS-II β-ketoacyl synthase, KasA, as bait, an extensive bacterial two-hybrid screen of a M. tuberculosis genome fragment library unexpectedly revealed a novel interaction between KasA and PpsB as well as PpsD, two polyketide modules involved in the biosynthesis of the virulence lipid phthiocerol dimycocerosate (PDIM). Sequence analysis revealed that KasA interacts with PpsB and PpsD in the region of the acyl carrier domain of each protein, raising the possibility that lipids could be transferred between the FAS-II and PDIM biosynthetic pathways. Subsequent studies utilizing purified proteins and radiolabeled lipids revealed that fatty acids loaded onto PpsB were transferred to KasA and also incorporated into long chain fatty acids synthesized using a Mycobacterium smegmatis lysate. These data suggest that in addition to producing PDIMs, the growing phthiocerol product can also be shuttled into the FAS-II pathway via KasA as an entry point for further elongation. Interactions between these biosynthetic pathways may exist as a simple means to increase mycobacterial lipid diversity, enhancing functionality and the overall complexity of the cell wall.
In the early 1990s the World Health Organization acknowledged the reemergence of tuberculosis as a major global health concern (1). The advent of AIDS and the rise in multi-drug-resistant strains of Mycobacterium tuberculosis have been the impetus for a more complete understanding of the complex nature of the bacteria. After the elucidation of the H37Rv genome sequence in 1998, it was concluded that M. tuberculosis contains roughly 250 distinct proteins involved in fatty acid metabolism (2). This large number of genes correlates directly with the important role that lipids play in the pathogenic life-style of the bacillus, allowing the bacteria to overcome the harsh environment of the host, dehydration, and other chemical injury (3, 4). The critical role that lipids play in mycobacterial viability is further substantiated by the current anti-tubercular drugs, isoniazid, ethionamide, and ethambutol, which inhibit cell wall biosynthesis (5). In the post-genomic era there has been an increased focus on drug target validation and in identifying novel protein-protein interactions involving known or suspected drug targets. Both serve as stepping stones for rationally designing new inhibitors and improving the longstanding front line chemotherapeutic agents.
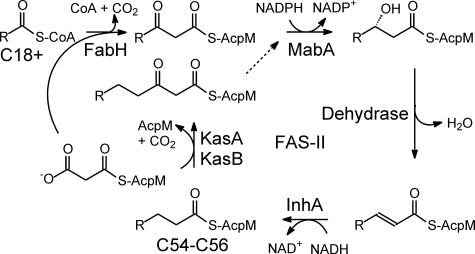
Mycolic acids are long chain (C70–C90) α-alkyl, β-hydroxylated fatty acids containing 70–90 carbons in total, and are the hallmark of mycobacteria, comprising an estimated 60% of the cell wall (3, 6). De novo fatty acid biosynthesis in mycobacteria is performed by the fatty-acid synthase type I complex (FAS-I),3 which is an iterative multi-enzyme system that synthesize short chain fatty acids from acetyl-CoA and malonyl-CoA. These C18:0+ products can be either utilized directly or shuttled off to other pathways, such as the fatty acid biosynthesis type II (FAS-II) cycle or polyketide synthases, for further modification (2, 7). Unlike the large single polypeptide format of the FAS-I synthase, the FAS-II pathway consists of multiple enzymes that each catalyzes a distinct step in the cycle, utilizing substrates presented on an acyl carrier protein, AcpM. The FAS-II pathway generates fatty acids up to C54/56 in length, which are used for mycolic acid biosynthesis (Fig. 1).
FIGURE 1.
The FAS-II pathway. The FAS-II pathway in M. tuberculosis extends the C18+-CoA products of the FAS-I pathway to C54–C56 fatty acids. The β-ketoacyl synthase FabH condenses the C18+ acyl group with malonyl-AcpM to form a β-ketoacyl-AcpM. This is subsequently reduced, dehydrated, and reduced by the actions of the NADPH-dependent β-ketoacyl reductase MabA, a dehydrase, and the NADH-dependent enoyl reductase InhA, respectively. The β-ketoacyl synthases KasA and KasB initiate additional cycles of elongation by catalyzing the condensation of the growing acyl-AcpM and malonyl-AcpM.
Complexities in the inhibition of the FAS-II enoyl reductase InhA by isoniazid led us to speculate that enzymes in the FAS-II pathway interact with each other inside the cell (8). This hypothesis was supported by the observation that a large non-covalently associated complex of proteins showing FAS-II activity could be isolated from Mycobacterium smegmatis (9). Although some of the interactions between the members of the FAS-II pathway have now been elucidated (10–12), we now have evidence of additional interactions that connect the FAS-II pathway with a polyketide synthase pathway involved in the production of the virulence lipid, phthiocerol dimycocerosate (PDIM). Present only in pathogenic mycobacteria, this 50-kb gene cluster contains 15 open reading frames (7, 13–17). The locus includes the ppsA-ppsE genes, which all encode large multidomain proteins containing active sites for multiple enzymatic functions, as well as an acyl carrier protein (ACP) domain to which the growing acyl chain is tethered (16, 17). In the current studies, we have demonstrated substrate transfer from the ACP domain of PpsB to the FAS-II β-ketoacyl synthase, KasA, which initiates each cycle of fatty acid elongation in the FAS-II pathway (Fig. 2). The subsequent incorporation and elongation of this substrate into long chain fatty acids demonstrates the functional nature of this interaction in vitro, and suggests a novel mechanism by which modifications such as hydroxylation could be incorporated into long chain fatty acids and mycolic acids within the cell. PDIM biosynthesis is up-regulated during infection, which may be responsible for causing a subset of Pps side-products to be shuttled into the FAS-II pathway (15). The studies presented here show the intricate interplay that can exist between previously unrelated pathways and open the door for defining alternate lipid modification routes.
FIGURE 2.
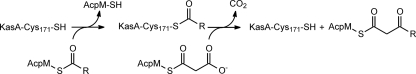
The reaction catalyzed by KasA. The β-ketoacyl synthase KasA catalyzes the Claisen condensation of the growing acyl chain carried by AcpM with malonyl-AcpM. In the first half of this ping-pong bi-bi reaction, the acyl chain is transferred from AcpM to the active site Cys-171 in KasA. After the AcpM has dissociated, malonyl-AcpM binds, and the enzyme generates a resonance-stabilized carbanion, which then attacks the acyl-enzyme thioester to form the β-ketoacyl-AcpM product.
EXPERIMENTAL PROCEDURES
Bacterial Strains—M. smegmatis mc2155 (ATCC 700084) was obtained from ATCC (Manassas, VA). Escherichia coli XL1-Blue, XL1-Blue MRF′ Kan library-competent cells and BacterioMatch II validation reporter-competent cells were purchased from Stratagene.
Bacteria Two-hybrid Library Construction and Cloning—M. tuberculosis strain H37Rv genomic DNA was partially digested with DpnII utilizing a dilution gradient of 0.001 to 10 u of enzyme in triplicate for 15, 30, and 60 min. DNA fragments ranging in size from 0.5 to 5 kb were ligated into the bacterial two-hybrid target vector, pTRG (Stratagene), to produce C-terminal fusions with the N-terminal domain of the RNA polymerase α-subunit (designated pTRG-MTB). Approximately 100,000 clones were harvested and isolated for use in library screening efforts. The pBT-kasA bait vector, in which KasA is expressed as a C-terminal fusion with the bacterial phage λcl protein, was constructed as described previously (12).
Library Screening—The pTRG-MTB library was screened with the pBT-kasA bait vector using a standard co-transformation protocol and using nonselective, selective, or dual selective media as described previously (12). Interaction candidates were checked for nonspecific interaction by transformation with control bait proteins. Plasmids that produced a hybrid protein capable of specifically interacting with the λcl-KasA fusion were sequenced using standard dideoxynucleotide methodology.
KasA and ppsB PCR and Cloning—The mycobacterial expression vector pFPCA1 was a gift from Scott Franzblau, University of Illinois at Chicago. To generate plasmids for expressing the full-length KasA and PpsB proteins, the M. tuberculosis kasA gene and ppsB gene were PCR-amplified from M. tuberculosis H37Rv genomic DNA. The following primer sets were used for directional cloning: kasA1, 5′-GAAGATCTATGAGTCAGCCTTCCACCGC-3′ (forward); kasA2, 5′-CGGAATTCCCATGGGTAACGCCCGAAGGCAAG-3′ (reverse); ppsB1, 5′-GGGAATTCCATATGATGCGAACGGCTTTCAGCCGG-3′ (forward); and ppsB2, 5′-GTGCTCGAGTTGTGTTCCTCTTAGTCG-3′ (reverse). PCR products were digested with the appropriate restriction enzymes, and kasA was ligated into the mycobacterial expression vector pFPCA1 (18); ppsB was ligated into the E. coli expression vector pET23b.
Recombinant KasA Protein Expression and Purification—The pFPCA1-kasA plasmid was electroporated into electrocompetent M. smegmatis mc2155 cells and plated onto Middlebrook 7H10 agar containing kanamycin (30 μg/ml) (19). Single colony transformants were grown at 35.5 °C overnight in 10 ml of Middlebrook 7H9 medium supplemented with albumin-dextrose saline (19). The starter culture was then used to inoculate 500 ml of 7H9 medium containing kanamycin (30 μg/ml), 0.5% glycerol, and 0.05% Tween 80. The cells were induced at an A600 of 0.450 with 1 ml of 44% acetamide, and expression was carried out at 35.5 °C overnight. Cells were harvested by centrifugation, the pellet was resuspended in 40 ml of standard His-Bind binding buffer, and cells were lysed by passing the cell suspension three times through a French press (1000 psi). Soluble mycobacterial proteins were then isolated by ultracentrifugation at 4 °C for 1 h at 33,000 rpm. KasA was purified from the supernatant by Ni2+ affinity chromatography on His-Bind resin (Novagen). Fractions containing KasA were identified by SDS-PAGE, pooled, and exchanged into 50 mm Tris-HCl, 0.3 m NaCl, pH 8.5 (20). SDS-PAGE demonstrated that the purity of the recombinant KasA was >95%.
Recombinant PpsB Protein Expression, Purification, and Modification—The pET29b-Sfp phosphopantetheinyl transferase expression plasmid was a kind gift from Mike Burkart, University of California, San Diego. The pET23b-ppsB plasmid was transformed into competent BL21(DE3) pLysS E. coli cells and plated on LB agar containing 200 μg/ml ampicillin (LB-amp). Single colony transformants were cultured in 10 ml of LB-amp medium at 37 °C for 16 h (overnight). This starter culture was subsequently used to inoculate 500 ml of LB-amp medium, which was shaken at 37 °C until an A600 of 1.5 was reached. Protein expression was induced with isopropyl 1-thio-β-d-galactopyranoside (360 μg/ml), and the culture was incubated at 25 °C overnight. Cells were harvested and lysed as described above, and PpsB was purified using His-Bind resin, also as described above. SDS-PAGE demonstrated that recombinant PpsB was >95% pure, and the protein was subsequently exchanged into 100 mm phosphate buffer, pH 7.0, for storage.
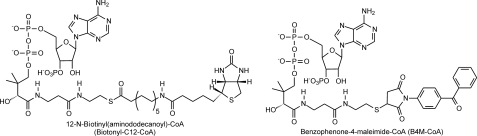
Modification of apoPpsB Protein—PpsB was obtained in the predominantly apoform in which the phosphopantetheine was missing from the carrier domain. The apoPpsB was subsequently converted into malonyl-PpsB, biotinyl C-12-PpsB, or [1-14C]palmitoyl-PpsB using the Sfp phosphopantetheinyl transferase. Modification reactions were performed for 1 h at 37 °C in 10 mm MgCl2, 5 mm dithiothreitol, 100 mm sodium phosphate buffer, pH 6.8. The phosphopantetheinyl donor was 100 μm malonyl-CoA, 12-N-biotinyl(aminododecanoyl)-CoA (Fig. 3, biotinyl C-12-CoA; Avanti Polar Lipids, Inc.), or 1 μCi (53 mCi/mmol) of [1-14C]palmitoyl-CoA (Sigma), respectively (21, 22).
FIGURE 3.
Structures of biotinyl C-12-CoA and B4M-CoA. In the reaction catalyzed by the Sfp phosphopantetheinyl transferase, the phosphopantetheine group, together with the attached biotin or benzophenone, is transferred to a conserved serine residue in PpsB or AcpM. AMP is the other product of the transferase reaction.
Acylation of KasA—Recombinant KasA protein was acylated in a 250-μl reaction mixture containing 25 mm MgCl2, 5 mm dithiothreitol, and 100 mm sodium phosphate, pH 6.8, at 37 °C for 1–2 h with either 100 μm biotinyl C-12-CoA or [1-14C]palmitoyl-CoA.
In Vitro Acyl Chain Transfer of the Biotinyl C-12 Group from PpsB to KasA—The purified and concentrated biotinyl C-12-PpsB was combined with an equimolar amount of purified KasA protein and incubated at 37 °C.
Transfer of the Biotinyl C-12 Group from KasA to PpsB or AcpM—Biotinyl C-12-KasA was generated as described above, and an equimolar amount of malonyl-PpsB or malonyl-AcpM was added. Malonyl-AcpM was synthesized as described previously (23). All transfer reactions were performed for 1 h at 37 °C. Biotinyl C-12 transfer was monitored via Western blot utilizing an anti-biotin-alkaline phosphatase antibody and developed with BCIP/NBT.
Preparation of Benzophenone-labeled CoA—A 5 mg/ml solution of benzophenone-4-maleimide (B4M) was prepared in N,N-dimethylformamide and combined at 1:1 with a solution of CoA (9 mg/ml) in 50 mm Tris-HCl, pH 7.5, under light-restrictive conditions. The mixture was incubated at 25 °C until titration with 5,5′-dithiobis(2-nitrobenzoic acid) demonstrated that no free thiol remained. Subsequently, the B4M-CoA (Fig. 3) was purified using an analytical HPLC Phenomenex Luna C18 5 μm (4.6 × 250 mm) column at a flow rate of 1 ml/min. The fractions containing the desired product were concentrated in vacuo, redissolved in water, and analyzed using mass spectrometry (supplemental Fig. S1).
Photoreactive Protein Preparation—The B4M-phosphopantetheinyl group was transferred from B4M-CoA to either apoAcpM or apoPpsB using the Sfp phosphopantetheinyl transferase. Briefly, 60 μm apoAcpM or apoPpsB was incubated with 200 nm Sfp and 250 μm B4M-CoA for 90 min at 37 °C (24).
Mass Spectrometry of B4M-AcpM—The modified AcpM was analyzed by LC/MS with an Agilent 1100 LC/MSD VL instrument using a Phenomenex Jupiter C4 HPLC column (5 μm, 300 Å, 4.6 × 250 mm). Chromatography was performed at 1 ml/min with a column temperature of 35 °C. Solvent A was 0.2% acetic acid, 0.02% trifluoroacetic acid, v/v in H2O, and solvent B was 0.2% acetic acid and 0.02% trifluoroacetic acid in a 1:1 mixture of acetonitrile:2-propanol. Samples were eluted with 25% B for 0 to 2 min followed by a gradient of 25–85% B over 2 to 32 min. The column eluant was analyzed in positive electrospray mode with the following spray chamber parameters: capillary voltage, 4.5 kV; gas temperature, 350 °C; drying gas flow, 13 liters/min; nebulizer pressure, 50 psig. The fragmentor voltage was 125 V, and mass spectra were recorded over the m/z range of 300 to 1500 atomic mass units. The average molecular weight of the proteins was calculated by deconvolution of the averaged mass spectra over the full width at half maximum of the peaks using Agilent ChemStation deconvolution software (supplemental Fig. S2).
Mass Spectrometry of B4M-PpsB—Whereas intact B4M-AcpM protein was analyzed directly by LC/MS, PpsB was first digested with trypsin/Glu-C before analysis using the method described previously (12). The digested protein samples were loaded onto a Zorbax 300SB-C18 column (3.5 μm, 150 × 0.3 mm; Agilent) and analyzed using a LTQ linear ion trap (Thermo Scientific). Solvent A was 3% acetonitrile and 0.1% formic acid in H2O, and solvent B was 0.1% formic acid in 100% acetonitrile. Samples were eluted using a flow rate of 15 μl/min with the following gradient profile: 0% B for 0–5 min, 0–15% B for 5–8 min, 15–45% B for 8–50 min, and 45–90% B for 50–55 min. Mass spectra were recorded over the m/z range of 200 to 2000 atomic mass units and processed using both Sequest (Thermo Scientific) and Mascot (Matrix Science) search engines. For the unmodified PpsB protein the sequence coverage was 84%, and the peptide fragment containing Ser-1458 (the site of phosphopantetheinylation) was clearly observed. The sequence coverage for B4M-AcpM was also high (70%). However, although the peptide containing Ser-1458 was now absent in the mass spectrum, a peptide with the expected mass increase resulting from covalent attachment of the B4M-phosphopantetheinyl group was not observed. This is possibly because of a reduction in ionization efficiency caused by the modification. Although the disappearance of the unmodified Ser-1458-containing peptide in the mass spectrum was indirect evidence that modification was successful, the ability of the B4M-PpsB to photocross-link KasA confirmed that Sfp was indeed able to load the B4M-phosphopantetheine onto PpsB.
Photo-cross-linking—Purified biotinyl C-12-KasA was combined with an equimolar amount of either B4M-AcpM or B4M-PpsB and incubated at 37 °C for 90 min. The cross-linking was performed by irradiating the sample with a handheld ultraviolet lamp (365 nm) for 15 min. The resulting cross-linked proteins were concentrated and analyzed by Western blot with either an anti-biotin or anti-His alkaline phosphatase antibody together with the BCIP/NBT phosphatase substrate.
In Vitro Reconstitution of PpsB Transfer to the FAS-II Pathway—Previous studies monitored the in vitro synthesis of radiolabeled fatty acids using cell lysates prepared from a strain of M. smegmatis that overexpressed KasA (25–27). The same procedure was used in the present work except that the addition of KasA (10 μm) to the reaction mixture replaced the use of the overexpressing KasA strain. Cell lysates were prepared from 200 ml of M. smegmatis cell culture grown to mid-log phase as described (26), and reactions were performed at pH 8.5 in 2-ml reaction mixtures containing cell lysate, 0.1 mm NADH, 0.1 mm NADPH, 500 mm α-cyclodextrin, 0.2 mm malonyl-CoA, and 150 μg AcpM. Fatty acid biosynthesis was initiated using either [1-14C]palmitoyl-PpsB or [1-14C]palmitoyl-KasA, and reactions were allowed to run at 37 °C overnight. Samples of the starting product, palmitoyl-[1-14C]PpsB, the elongation reaction after 1 h, and the elongation reaction after overnight incubation were analyzed by SDS-PAGE. Reactions were stopped with the addition of 2 ml of 20% tertbutylhydroxide, and the esters were saponified by heating at 120 °C overnight. Reactions were then cooled to room temperature, and the free fatty acids were methylesterified by adding an equal volume of a 40:1 CH2Cl2:MeI solution. The organic layer was separated and dried under air, and the [1-14C]methyl esters were recovered by extraction with ether. Samples were analyzed by C18 reverse-phase thin layer chromatography (TLC) using 1:1 acetonitrile:dioxane as the mobile phase. The separated [1-14C]methyl ester products on the TLC plate and the radiolabeled gel (which was dried using a Bio-Rad gel dryer) were visualized using a Typhoon 9400 PhosphorImager (GE Healthcare).
RESULTS
Detection of Interactions between KasA and H37Rv Proteins Using a Bacteria Two-hybrid Screen—The full-length open reading frame of the kasA gene was cloned into the pBT bait plasmid so that KasA was expressed as a fusion protein with a DNA-binding domain (the bacterial phage λcl protein) (28). The M. tuberculosis library is composed of random genome fragments cloned into the pTRG target plasmid for expression as a C-terminal fusion with the α-subunit of RNA polymerase, which serves as an activation domain (28). Co-transformation of the pBT-kasA plasmid with the pTRG-MTB plasmids into E. coli yielded several colonies on double selective screening media, containing streptomycin and 3-AT, consistent with the activation of the two reporter genes, aadA and HIS3, by a KasA library protein-protein interaction. All library-target plasmids from these colonies were isolated, and the specificity of the interactions with KasA was verified by the absence of growth on double selective LB media upon co-transformation with other bait plasmids such as pBT-LGF2 and pBT-actin. Sequence analysis of the pTRG-MTB plasmids revealed two genes, ppsB and ppsD.
Interestingly, both ppsB and ppsD are part of a 50-kb pks gene cluster involved in the biosynthesis of the virulence lipid PDIM (7, 13, 15, 17). Both ppsB and ppsD encode large multidomain proteins that carry out multiple enzymatic reactions and include an ACP domain that carries the growing acyl product via a phosphopantetheinyl group (22, 29). The domain boundaries in the PpsB and PpsD proteins were determined utilizing the program NRPS-PKS (17, 30), and the region of interaction between KasA and PpsB or PpsD was localized to the C-terminal portion of each protein corresponding to the ACP domain. Because KasA binds substrates attached to the soluble AcpM protein, the observation of an interaction between KasA and the ACP domains of PpsB and PpsD suggested a mechanism for transferring acyl groups between the PDIM and FAS-II biosynthetic pathways.
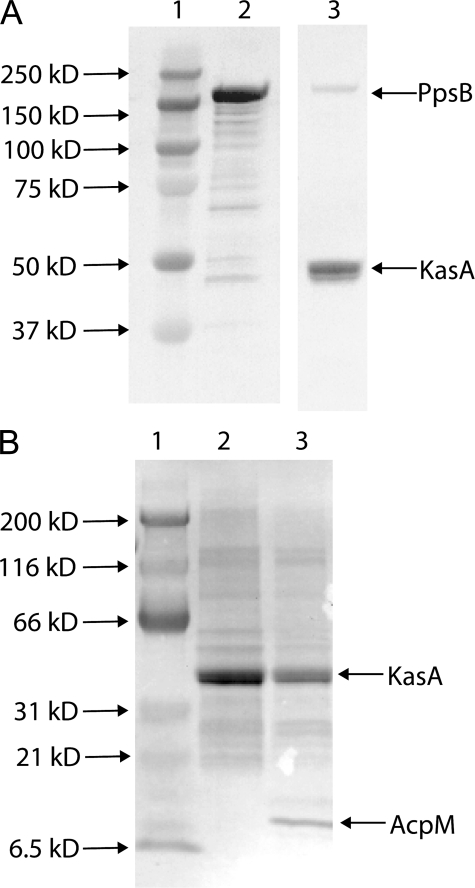
In Vitro Interaction of KasA and PpsB—To demonstrate the functional nature of the interaction between the FAS-II and PDIM pathways, we performed a series of experiments utilizing recombinant KasA and PpsB. KasA was purified from M. smegmatis using an acetamide-inducible mycobacterial expression vector (18), and the entire 1538-residue PpsB protein was expressed in E. coli to determine whether KasA was able to interact with the ACP domain in the full-length folded PpsB protein. Peptide digestion and mass spectrometry revealed that the PpsB protein was present in the apoform, indicating that it was not an efficient substrate for the endogenous E. coli phosphopantetheinyl transferase. Consequently, the malonyl-phosphopantetheine and biotinyl C-12-phosphopantetheine groups were subsequently loaded onto PpsB from their respective CoA using the promiscuous phosphopantetheinyl transferase protein, Sfp, from Bacillus subtilis (17, 21, 22). Phosphopantetheinylation and the subsequent transfer of the biotinyl C-12 acyl group to KasA were monitored by Western blot using anti-biotin antibodies. As shown in Fig. 4A, the purified biotinyl C-12 group can be transferred from PpsB to KasA (lanes 2 and 3) where it is presumably attached to KasA via the active site Cys-171 residue. Although acyl transfer to KasA involves direct acylation of Cys-171, the second half of the ping-pong reaction catalyzed by KasA involves condensation of the acyl group with malonyl-AcpM to generate a β-ketoacyl-AcpM. Thus, even though the above experiment demonstrates that PpsB can load acyl groups onto KasA, we were also interested in determining whether malonyl-ppsB could act as the second substrate in the KasA reaction. Consequently, malonyl-PpsB was incubated with biotinyl C-12-KasA. However, no biotinylation of the PpsB enzyme was detected, suggesting that the substrate transfer is unidirectional (data not shown). As a control, the ability of KasA to condense the biotinyl C-12 acyl group with malonyl-AcpM was confirmed by Western blot (Fig. 4B).
FIGURE 4.
Transfer reactions involving the biotinyl C-12 acyl group. A, Western blot analysis of the transfer of the biotinyl C-12 acyl group from PpsB to KasA. Lane 1, Kaleidoscope Protein Standard (Bio-Rad); lane 2, biotinyl C-12-PpsB product after modification with Sfp using biotinyl C-12-CoA as the substrate; lane 3, transfer of biotinyl C-12 from PpsB to KasA. B, transfer of the biotinyl C-12 acyl group from KasA (lane 2) to AcpM (lane 3). Both Westerns were visualized with an anti-biotin alkaline phosphatase-conjugated antibody and developed with BCIP/NBT.
To ensure that the proteins interact directly in vitro in order to transfer the substrate, photo-cross-linking experiments were conducted. Using the Sfp phosphopantetheinyl transferase, the B4M group from B4M-CoA (Fig. 3) was transferred to the ACP domain of PpsB resulting in the generation of a photoreactive B4M-PpsB. Cross-linking was performed in the presence of biotinyl C-12-KasA using 365 nm light, and the biotinyl C-12-KasA·B4M-PpsB complex was detected by Western blot, generating a high molecular mass protein band of more than 200 kDa (Fig. 5A). This observation substantiates the hypothesis that PpsB is capable of transferring an acyl chain into the FAS-II pathway by directly interacting with KasA. Because KasA must interact with AcpM in order to initiate each cycle of elongation in the FAS-II cycle, as a control we also demonstrated that B4M-AcpM was able to photo-cross-link biotinyl C-12-KasA. Fig. 5B indicates the formation of a band on SDS-PAGE of 60 kDa following irradiation of the B4M-AcpM and biotinyl C-12-KasA reaction mixture.
FIGURE 5.
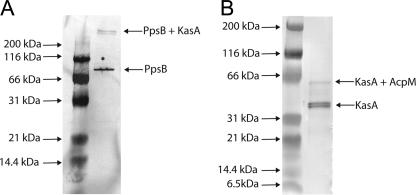
Photo-cross-linking reactions with B4M-PpsB and B4M-AcpM. A, Western blot analysis of B4M-PpsB before (163 kDa) and after photo-cross-linking in the presence of biotinyl C-12-KasA (205 kDa). This Western blot was developed with an anti-His alkaline phosphatase-conjugated antibody to detect the unbound and KasA-bound forms of PpsB. B, Western blot analysis of KasA before (44 kDa) and after the photo-cross-linking reaction with B4M-AcpM (60 kDa). To detect the unbound and AcpM bound form of KasA, this Western blot was developed with an anti-biotin alkaline phosphatase-conjugated antibody.
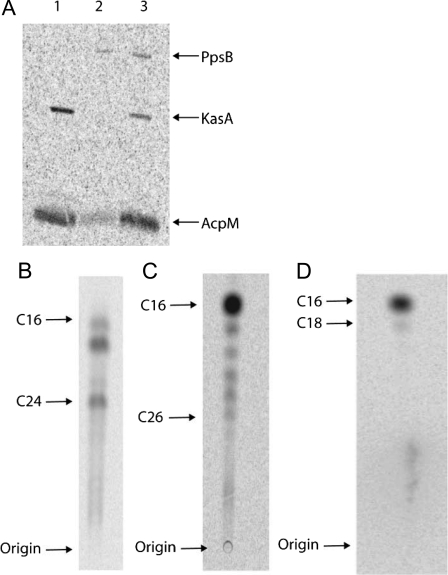
As we were able to demonstrate transfer of an acyl group from PpsB to KasA that could then be condensed with malonyl-AcpM, we wanted to determine whether acyl-PpsB could prime fatty acid synthesis in M. smegmatis whole cell lysate. Starting with purified [1-14C]palmitoyl-PpsB, we were able to detect the transfer of the [1-14C] palmitoyl group to both KasA and AcpM by phosphorimaging (Fig. 6A). The elongation of the C16 group by the FAS-II pathway was monitored by reverse-phase TLC and resulted in the same distinctive labeled fatty acid profile observed when [1-14C]palmitoyl-KasA was used as the starting material (Fig. 6, B and C) (26).
FIGURE 6.
Transfer reactions utilizing [1-14C]palmitate. A, SDS-PAGE confirming the transfer of [1-14C]palmitate from PpsB to KasA and AcpM in M. smegmatis whole cell lysate. Lane 1, final reaction mixture showing the complete transfer of [1-14C]palmitate from PpsB to KasA and AcpM; lane 2, [1-14C]palmitoyl-PpsB after purification; lane 3, sample taken from the elongation reaction 1 h after [1-14C]palmitoyl-PpsB was added; partial transfer of [1-14C]palmitate from PpsB to KasA and AcpM can be seen. B, TLC of fatty acid synthesis initiated by the addition of [1-14C]palmitoyl-PpsB to M. smegmatis whole cell lysate. Fatty acids increasing in chain length by 2 carbons can be observed from C16–C24. C, control TLC using [1-14C]palmitoyl-KasA as the starting material. D, control TLC using [1-14C]palmitoyl-CoA as the starting material. Without additional AcpM, NADH, and NADPH, the M. smegmatis lysate contains sufficient substrates and cofactors for only one round of elongation.
DISCUSSION
The FAS-II pathway in mycobacteria is essential for producing long chain fatty acids that serve as the precursors to mycolic acids. Several of the FAS-II components are essential to bacterial survival or replication and have been validated as legitimate drug targets. We have previously hypothesized that enzymes in the FAS-II pathway form a noncovalent complex in vivo (8), and we and others have previously demonstrated the existence of such interactions using two-hybrid techniques (10–12). The present studies were stimulated by the observation of an unexpected interaction between KasA and proteins involved in PDIM biosynthesis when the full-length KasA protein was used as a bait to screen a mycobacterial library.
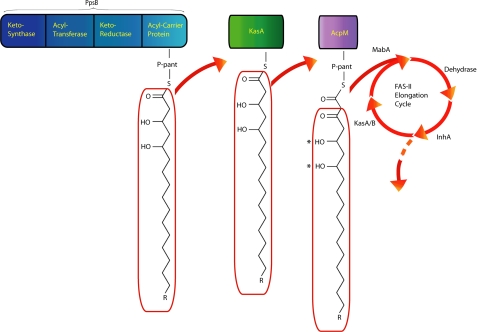
PpsB is a large, type I, modular polyketide synthase that accepts, elongates, and modifies the fatty acyl chain reaction product of the PpsA enzyme (Fig. 7). Neither PpsA nor PpsB contains an enoyl-reductase or a dehydrase domain, and therefore the product of PpsB contains two hydroxyl groups (a 1,3-diol) (17). The remaining Pps proteins, PpsC–PpsE, all contain both the enoyl-reductase and dehydrase domains and, in turn, are responsible for the addition of saturated alkyl units derived from malonyl or methyl-malonyl-CoA. The hydroxyl groups inserted into the phthiocerol chain by PpsA and PpsB are subsequently coupled with mycocerosic acids, which are in turn synthesized by the Mas polyketide synthase that extends C16–C18 fatty acids with methylmalonyl units, to give the final phthiocerol dimycocerosate (17, 31).
FIGURE 7.
The PpsB product and the proposed interaction between the PDIM and FAS-II pathways. The asterisk indicate the diol functionality present in the PpsB product that would be retained in fatty acids elongated by the FAS-II pathway.
Although some interactions involved in both FAS-II and PDIM biosynthetic pathways have been identified previously, this is the first evidence of a protein-protein interaction linking the two systems (10, 11, 32, 33). It is clear from these experiments that the interaction between KasA and PpsB is specific and results in the transfer of substrate from PpsB to KasA. Although the experiments conducted here were conducted with palmitate, the ability of PpsB to load acyl groups onto KasA would result in the incorporation of a dihyroxy group into the long chain fatty acids produced by the mycobacterium. We propose that this is an additional mechanism for increasing the diversity of lipids in the cell and note that there are drastic alterations in the cell wall during the infection process resulting in part from a shift in lipid production (34–37). Knowledge of the mechanism of cell wall biosynthesis and the connectivity between the pathways involved in the production of mycobacterial will provide additional therapeutic targets for drug discovery.
Supplementary Material
Acknowledgments
M. tuberculosis genomic DNA was obtained through the CSU-Tuberculosis Research Materials and Vaccine Testing, National Institutes of Health, NIAID Contract N01-A1-75320.
This work was supported, in whole or in part, by National Institutes of Health Grant AI44639 (to P. J. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: FAS-I, fatty-acid synthase type I; FAS-II, fatty acid synthase type II; PDIM, phthiocerol dimycocerosate; ACP, acyl carrier protein; AcpM, mycobacterial acyl carrier protein; biotinyl C-12, 12-N-biotinyl(aminododecanoyl); B4M, benzophenone-4-maleimide; BCIP/NBT, 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium; HPLC, high pressure liquid chromatography; LC/MS, liquid chromatography/mass spectrometry; Sfp, B. subtilis phosphopantetheinyl transferase.
References
- 1.World Health Organization (1993) Treatment of Tuberculosis: Guidelines for National Program, 3rd Ed., World Health Organization, Geneva, Switzerland
- 2.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, Tekaia, F., Badcock, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R., Devlin, K., Feltwell, T., Gentles, S., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., Krogh, A., McLean, J., Moule, S., Murphy, L., Oliver, K., Osborne, J., Quail, M. A., Rajandream, M. A., Rogers, J., Rutter, S., Seeger, K., Skelton, J., Squares, R., Squares, S., Sulston, J. E., Taylor, K., Whitehead, S., and Barrell, B. G. (1998) Nature 393 537-544 [DOI] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A., and Yuan, Y. (1998) Prog Lipid Res. 37 143-179 [DOI] [PubMed] [Google Scholar]
- 4.Barry, C. E., III, and Mdluli, K. (1996) Trends Microbiol. 4 275-281 [DOI] [PubMed] [Google Scholar]
- 5.Banerjee, A., Dubnau, E., Quemard, A., Balasubramanian, V., Um, K. S., Wilson, T., Collins, D., de Lisle, G., and Jacobs, W. R., Jr. (1994) Science 263 227-230 [DOI] [PubMed] [Google Scholar]
- 6.Asselineau, J., and Lederer, E. (1950) Nature 166 782-783 [DOI] [PubMed] [Google Scholar]
- 7.Kolattukudy, P. E., Fernandes, N. D., Azad, A. K., Fitzmaurice, A. M., and Sirakova, T. D. (1997) Mol. Microbiol. 24 263-270 [DOI] [PubMed] [Google Scholar]
- 8.Rawat, R., Whitty, A., and Tonge, P. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13881-13886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odriozola, J. M., Ramos, J. A., and Bloch, K. (1977) Biochim. Biophys. Acta 488 207-217 [DOI] [PubMed] [Google Scholar]
- 10.Veyron-Churlet, R., Bigot, S., Guerrini, O., Verdoux, S., Malaga, W., Daffe, M., and Zerbib, D. (2005) J. Mol. Biol. 353 847-858 [DOI] [PubMed] [Google Scholar]
- 11.Veyron-Churlet, R., Guerrini, O., Mourey, L., Daffe, M., and Zerbib, D. (2004) Mol. Microbiol. 54 1161-1172 [DOI] [PubMed] [Google Scholar]
- 12.Kruh, N. A., Rawat, R., Ruzsicska, B. P., and Tonge, P. J. (2007) Protein Sci. 16 1617-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan, P. J., and Nikaido, H. (1995) Annu. Rev. Biochem. 64 29-63 [DOI] [PubMed] [Google Scholar]
- 14.Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B., and Guilhot, C. (1999) Mol. Microbiol. 34 257-267 [DOI] [PubMed] [Google Scholar]
- 15.Cox, J. S., Chen, B., McNeil, M., and Jacobs, W. R., Jr. (1999) Nature 402 79-83 [DOI] [PubMed] [Google Scholar]
- 16.Daffe, M., and Laneelle, M. A. (1988) J. Gen. Microbiol. 134 2049-2055 [DOI] [PubMed] [Google Scholar]
- 17.Trivedi, O. A., Arora, P., Vats, A., Ansari, M. Z., Tickoo, R., Sridharan, V., Mohanty, D., and Gokhale, R. S. (2005) Mol. Cell 17 631-643 [DOI] [PubMed] [Google Scholar]
- 18.Changsen, C., Franzblau, S. G., and Palittapongarnpim, P. (2003) Antimicrob. Agents Chemother. 47 3682-3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parish, T., and Stoker, N. G. (2001) Mycobacterium tuberculosis Protocols, p. 47, Humana Press, Totowa, NJ
- 20.Schaeffer, M. L., Agnihotri, G., Volker, C., Kallender, H., Brennan, P. J., and Lonsdale, J. T. (2001) J. Biol. Chem. 276 47029-47037 [DOI] [PubMed] [Google Scholar]
- 21.Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., LaCelle, M., Marahiel, M. A., Reid, R., Khosla, C., and Walsh, C. T. (1996) Chem. Biol. 3 923-936 [DOI] [PubMed] [Google Scholar]
- 22.Quadri, L. E., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., and Walsh, C. T. (1998) Biochemistry 37 1585-1595 [DOI] [PubMed] [Google Scholar]
- 23.Schaeffer, M. L., Agnihotri, G., Kallender, H., Brennan, P. J., and Lonsdale, J. T. (2001) Biochim. Biophys. Acta 1532 67-78 [DOI] [PubMed] [Google Scholar]
- 24.La Clair, J. J., Foley, T. L., Schegg, T. R., Regan, C. M., and Burkart, M. D. (2004) Chem. Biol. 11 195-201 [DOI] [PubMed] [Google Scholar]
- 25.Marrakchi, H., Laneelle, G., and Quemard, A. (2000) Microbiology 146 289-296 [DOI] [PubMed] [Google Scholar]
- 26.Slayden, R. A., and Barry, C. E., III (2002) Tuberculosis (Edinb.) 82 149-160 [DOI] [PubMed] [Google Scholar]
- 27.Slayden, R. A., Lee, R. E., and Barry, C. E., III (2000) Mol. Microbiol. 38 514-525 [DOI] [PubMed] [Google Scholar]
- 28.Dove, S. L., Joung, J. K., and Hochschild, A. (1997) Nature 386 627-630 [DOI] [PubMed] [Google Scholar]
- 29.Walsh, C. T., Gehring, A. M., Weinreb, P. H., Quadri, L. E., and Flugel, R. S. (1997) Curr. Opin. Chem. Biol. 1 309-315 [DOI] [PubMed] [Google Scholar]
- 30.Ansari, M. Z., Yadav, G., Gokhale, R. S., and Mohanty, D. (2004) Nucleic Acids Res. 32 W405-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azad, A. K., Sirakova, T. D., Fernandes, N. D., and Kolattukudy, P. E. (1997) J. Biol. Chem. 272 16741-16745 [DOI] [PubMed] [Google Scholar]
- 32.Jain, M., and Cox, J. S. (2005) PLoS Pathog. 2005 1 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao, A., and Ranganathan, A. (2004) Mol. Genet. Genomics 272 571-579 [DOI] [PubMed] [Google Scholar]
- 34.Jain, M., Petzold, C. J., Schelle, M. W., Leavell, M. D., Mougous, J. D., Bertozzi, C. R., Leary, J. A., and Cox, J. S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5133-5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., Dolganov, G., Efron, B., Butcher, P. D., Nathan, C., and Schoolnik, G. K. (2003) J. Exp. Med. 198 693-704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh, A., Gupta, R., Vishwakarma, R. A., Narayanan, P. R., Paramasivan, C. N., Ramanathan, V. D., and Tyagi, A. K. (2005) J. Bacteriol. 187 4173-4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talaat, A. M., Lyons, R., Howard, S. T., and Johnston, S. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4602-4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.