Abstract
Yersinia pestis, a Gram-negative bacterium that causes bubonic and pneumonic plague, is able to rapidly disseminate to other parts of its mammalian hosts. Y. pestis expresses plasminogen activator (PLA) on its surface, which has been suggested to play a role in bacterial dissemination. It has been speculated that Y. pestis hijacks antigen-presenting cells, such as macrophages (MΦs) and dendritic cells, to be delivered to lymph nodes to initiate dissemination and infection. Both alveolar MΦs and pulmonary dendritic cells express a C-type lectin receptor, DEC-205 (CD205), which mediates antigen uptake and presentation. However, no ligand has been identified for DEC-205. In this study, we show that the invasion of alveolar MΦsby Y. pestis depends both in vitro and in vivo on the expression of PLA. DEC-205-expressing MΦs and transfectants, but not their negative counterparts, phagocytosed PLA-expressing Y. pestis and Escherichia coli K12 more efficiently than PLA-negative controls. The interactions between PLA-expressing bacteria and DEC-205-expressing transfectants or alveolar MΦs could be inhibited by an anti-DEC-205 antibody. Importantly, the blockage of the PLA-DEC-205 interaction reduced the dissemination of Y. pestis in mice. In conclusion, murine DEC-205 is a receptor for PLA of Y. pestis, and this host-pathogen interaction appears to play a key role in promoting bacterial dissemination.
Yersinia pestis is the Gram-negative bacterium that causes bubonic and pneumonic plague. Y. pestis has evolved directly from Yersinia pseudotuberculosis within the last 10,000–20,000 years (1, 2). The Yersinia genus is composed of 12 species, and 3 species of Yersinia are pathogenic to humans (3, 4). Y. pseudotuberculosis and Yersinia enterocolitica cause mild enteric diseases, whereas the hallmark of Y. pestis is its rapid dissemination in the mammalian host resulting in bubonic and pneumonic plague and high mortality.
All three pathogenic Yersinia share a virulence plasmid, pCD1 (pYV), which is essential for pathogenesis (5, 6). This plasmid encodes a type III secretion system (7), YadA {an adherence and virulence surface molecule, although yadA is a pseudogene in Y. pestis (8, 9)}, and a number of secreted effector proteins that inhibit bacterial phagocytosis and specific innate immune responses (10).
Y. pestis harbors two additional plasmids, pPCP1 (9.6 kb) and pMT1 (pFra) (102 kb), which encode the plasminogen activator (PLA)3 and the F1 protein, respectively. The products of these genes are necessary for tissue invasion (11), capsule formation (12), and infection of the plague flea vector (13, 14). Capsule formation by Y. pestis has been proposed to confer anti-phagocytic capabilities (15, 16). PLA is an outer membrane protease that also plays a role in bacterial attachment and invasion of one eukaryotic epithelial cell line, but the invasive function does not involve its proteolytic activity (17). Recent studies have further confirmed the important roles of PLA in the progression of bubonic and pneumonic plague, in which PLA contributes to dissemination of Y. pestis, especially to bubonic plague (14, 18).
Lipopolysaccharide (LPS) of many Gram-negative bacterial pathogens mediates toxicity and resistance to serum killing and phagocytosis and generally consists of three structural regions as follows: (i) the lipid A backbone, (ii) an oligosaccharide core (core LPS), and (iii) O-antigen (19, 20). However, Y. pestis does not produce an O-antigen (21, 22). The shortened LPS is also referred to as lipo-oligosaccharide and is presumably exposed to the external environment. In general, Gram-negative bacteria are classified as smooth or rough based on the presence or lack of the O-antigen, respectively. An interesting observation supports the idea that Yersinia spp. expressing O-antigen blocks enzymatic activity of PLA (23). Therefore, it is also possible that expression of O-antigen in Y. pestis could physically shield interactions between PLA and potential host receptors.
Antigen-presenting cells (APCs) such as macrophages (MΦs) play an essential role in the host defense against invading pathogens and in the control and maintenance of innate and adaptive immunity (24, 25). Innate immune functions are initially carried out by the ability of APCs to phagocytose and kill invading pathogens or to deliver them to other types of host immune cells for further elimination of the pathogens. APCs express members of the C-type lectin family. For example, alveolar MΦs express a C-type lectin receptor, DEC-205 (CD205). Although DEC-205 participates in an important role in the antigen-presenting process (26, 27), no ligand for this receptor has yet been identified.
Some pathogens may pirate innate immune receptors for their dissemination. It has been speculated that after entering the host via a bite from an infected flea (10), Y. pestis may use innate immune receptors present on APCs to traffic to lymph nodes where they encounter host lymphocytes (10). This hypothesis is reminiscent of the mechanism HIV-1 uses to target APCs. HIV-1 may hijack another C-type lectin receptor, DC-specific intercellular adhesion molecule-grabbing non-integrin (DC-SIGN, CD209), so as to be captured and trafficked to target cells such as CD4 lymphocytes in lymph nodes (28–30). Human DC-SIGN is also expressed by a certain class of MΦs (31), including human alveolar MΦs.
Recently, we demonstrated that human DC-SIGN and mouse SIGN-R1 can be used as receptors for the core LPS of Y. pestis, promoting bacterial adherence and phagocytosis to human DCs and murine MΦs (20).4 Based on these studies, we have hypothesized that after Y. pestis enters the lungs through aspiration, it invades alveolar MΦs via core LPS-DC-SIGN-SIGN-R1 interaction (20).4 In this study, we explored whether Y. pestis can exploit this interaction to be disseminated to spleens in a mouse model. Unexpectedly, we found that in fact PLA can also mediate the binding of Y. pestis with host DEC-205, and the interaction may play a role in bacterial dissemination.
EXPERIMENTAL PROCEDURES
Bacterial Strains—Escherichia coli K12 strain CS180 contains core LPS but lacks O-antigen (33). CS1861 is an isogenic strain of CS180 harboring pSS37, a plasmid containing all the genes necessary for the expression of the Shigella dysenteriae 1 O-antigen (33–35) (Table 1). E. coli strains were cultured on Luria-Bertani medium (LB) supplemented with 1.5% agar at 37 °C overnight.
TABLE 1.
Bacterial strains used in this study
| Strains | Genotypes + phenotypes | Refs. |
|---|---|---|
| E. coli K-12 | ||
| CS180 | Original type (rough) | 33-35 |
| CS1861 | CS180 expressing O-antigen (smooth) | 33-35 |
| DH5α | Original type (rough), similar to CS180 | 14 |
| DH5α-PLA | DH5α expressing the PLA | 14 |
| Y. pestis | ||
| KIM6+ | The natural type (rough) | 38 |
| KIM10+ | KIM6+ with pPCP1 plasmid cured | 38 |
| KIM6- | KIM6+ with deletion of pgm locus | 38 |
| KIM10- | KIM6+ with pPCP1 plasmid cured and deletion of pgm locus | 38 |
| KIM6+-O+ | KIM6+ expressing the O-antigen (smooth) | 39 |
| KIM10+-PLA | KIM10+ expressing the PLA | 14, 38 |
| KIM10--Δail | KIM10+ with deletion of ailC | 40 |
| KIM10--Δail-PLA | KIM10--Δail expressing the PLA | 14, 38, 40 |
| KIM6+-pBR322 | KIM6+ carrying pBR322 plasmid | This study |
| KIM10+-pBR322 | KIM10+ carrying pBR322 plasmid | This study |
| Y. pseudotuberculosis | ||
| Y1, O:1Aa | Wild-type expressing the invasin, but with naturally pYV plasmid cured and deletion of ail (smooth) | 37 |
Y. pseudotuberculosis (Y1) is a serotype O:1a strain and lacks the virulence plasmid (pYV) and does not express the Ail protein.5 The strain was obtained from the CDC and used as a control strain for invasion (20, 36), because this bacterium invades almost all epithelial cell lines via an invasin-integrin interaction (37).
All strains of Y. pestis used in this study are derived from the KIM strain (38). There are nine derivatives of Y. pestis used in this study as follows. 1) KIM6+ was derived from wild-type KIM by curing the virulence plasmid (pCD1 or pYV). 2) KIM10+ was derived from KIM6+ by curing the pPCP plasmid. 3) KIM6– is a derivative of KIM6+ obtained after spontaneous deletion of the pgm (pigmentation) locus (38). 4) KIM10– is a derivative of KIM6+ obtained after curing the pPCP plasmid and deletion of the pgm locus. 5) KIM6+-O+ is KIM6+/pAY100.1 that expresses the O-antigen of Y. enterocolitica serotype O:3 from the O-antigen gene cluster cloned in plasmid pAY100.1 (39). 6) KIM10+-PLA is the KIM10+ containing a plasmid that expresses the PLA. 7) KIM10–-Δail is the KIM10–, whose ail gene is deleted (40). 8) KIM6+-pBR322 is KIM6+ that carries the pBR322 plasmid. 9) KIM10+-pBR322 is KIM10+ that carries pBR322 (Table 1). Strains were cultured at 26 °C on GC-based plates (Difco) supplemented with 1% hemoglobin (U. S. Biochemical Corp.) (20).
Mice—BALB/CJ mice were bred at the University of Illinois at Chicago-Rockford animal facilities and used in this study. BALB/CJ mice were selected, because of the expression of SIGN-R1 and DEC-205 in peritoneal MΦs and alveolar MΦs, respectively, in these mice (41–43).
Biological Reagents—Anti-mouse DEC-205, anti-mouse SIGN-R1, and anti-human DC-SIGN antibodies were purchased from SeroTec (Raleigh, NC). YTH71.3, a rat antibody that recognizes CEACAM1 (CD66a), CEACAM6 (CD66c), and CEACAM3 (CD66d), was purchased from Roche Applied Science.
Isolation of Mouse Peritoneal and Alveolar Macrophages—After euthanizing a mouse, its intact abdomen was exposed, cleaned with 70% ethanol, and opened. 5 ml of RPMI medium was injected into the intraperitoneal cavity. The mouse abdomen was gently massaged for 3 min, and then the lavage fluid was collected. The suspension containing the peritoneal MΦs was seeded in flasks and placed in a CO2 incubator for 2 h. The cell layers were washed three times to remove nonadherent cells. MΦs were removed from the plastic surface by incubating with citrate saline and re-seeded for interaction assays or stained with antibodies to check the expression level of receptors.
Alveolar MΦs were obtained using the following procedures. After euthanizing a mouse, its bronchial tract was opened, and 1 ml of RPMI medium was injected into the lungs via a tiny tube. The mouse chest was gently massaged for 3 min, and then the lavage fluid was collected. The purification processes followed the same process as the peritoneal MΦs.
Host Cell Lines—Mouse C-type lectin transfectants, CHO-mDC-SIGN, CHO-mSIGN-R1, CHO-mSIGN-R3, and CHO-mDEC-205 (CD205), were generated by transfecting CHO cells with mouse corresponding C-type lectin cDNAs, followed by selection for stable surface expression as originally described (41). CHO-NEO was used as a control cell line, which expresses the neomycin resistance gene only.
Adherence and Phagocytosis Assays—The assays for adherence and phagocytosis have been described previously (44, 45). Briefly, host cells (CHO, HeLa, dendritic cells, or macrophages) were plated in 24- or 96-well plates. Cells were suspended in RPMI medium with 2% FCS at a concentration of 4 × 105/ml. One-half ml of each of these cell suspensions were added to 24-well plates, and after addition of 50 μl of bacterial suspensions at a concentration of 1 × 107 colony-forming units (CFU)/ml, the cells were allowed to incubate for 2.5 h (2 h for dendritic cells and alveolar MΦs) at 37 °C in the presence of 5% CO2. The cell monolayers were then washed three times with phosphate-buffered saline. The number of associated bacteria (adherent and internalized) per cell was quantified by washing the cells three times with RPMI medium containing 2% FCS and plating the culture after the cells were lysed by the addition of 0.5% saponin (Calbiochem).
To determine the internalization of bacteria, gentamicin, which kills extracellular bacteria but cannot penetrate into host cells, was added into each well to a final concentration of 100 μg/ml, and the cultures were incubated for 60 min. Cells were washed twice to remove the antibiotic. Then the cells were suspended in phosphate-buffered saline containing 0.5% saponin, diluted, and plated on LB as well as the Y. pestis plates. The level of internalization of bacteria in these host cells was calculated by determining the CFU recovered from lysed cells. All experiments were performed in triplicate, and each experiment was repeated three times. The data were expressed as mean ± S.E. Statistical significance was calculated using the Student'st test.
For the inhibition assay, anti-DEC-205 antibody was added 20 min prior to the addition of bacteria at a concentration of 5 μg/ml. The concentration used was based on our preliminary data and was selected based on the fact that at these concentrations there was no influence on the survival of bacteria and host cells, as previously and recently shown (19, 20, 46).
Determination of Phagocytosis by Flow Cytometry—The following method was used to supplement the survival-based phagocytosis assay described above because APCs are known to kill some phagocytosed bacteria (20, 47). Briefly, bacteria were suspended in RPMI medium containing 5- and 6-carboxyfluorescein diacetate, succinimidyl ester (CFDA-S.E.; Molecular Probes, Eugene, OR), for 40 min and washed twice with RPMI medium to remove the excess dye. Labeled bacteria were added to MΦ cultures for 2 h. Cell cultures were washed twice to remove unbound bacteria. MΦs plus associated bacteria were fixed with 2% paraformaldehyde. Before flow cytometry, a 1:10 dilution of trypan blue (0.4%, Sigma) was added to the fixed cell cultures, and the mixture was incubated at ambient temperature for 10 min (47) to quench the fluorescence from extracellular labeled bacteria. Trypan blue blocks fluorescence but cannot penetrate host cells; therefore, fluorescence from internalized bacteria was not influenced by addition of trypan blue. The rate of bacterial internalization was determined by comparing the intensity of fluorescence-positive MΦs with various bacteria. The greater the fluorescence intensity, the more bacteria are phagocytosed by MΦs. All of the results were evaluated in triplicate.
In Vivo Phagocytosis Assays—After anesthetizing a mouse, its bronchial tract was opened, and about 20 μl of bacterial suspension (A600 = 0.1) was injected deep into the lungs using a tiny tube. After 2 h, mice were euthanized and alveolar MΦs were collected as described in the procedure for alveolar MΦs above. The numbers of cells in lavage fluid were counted for each collection. 1 × 105 mouse cells were seeded into each well of 96-well plates, containing RPMI medium with 2% FCS and gentamicin at a concentration of 100 μg/ml and were then incubated for 1.5 h to allow the MΦs to adhere to plates and kill the extracellular bacteria. Each well was washed three times with RPMI medium containing 2% FCS to remove nonadherent cells and lysed with saponin, following the same procedures as used in the in vitro phagocytosis assays.
Dissemination Assay—In this study, the dissemination was defined such that Y. pestis was transported from lungs to spleens. In detail, mice were inoculated with Y. pestis KIM6+pBR322, KIM6+-O+, KIM10+-pBR322 and KIM10+-PLA, at a concentration of A600 = 1.5 in phosphate-buffered saline (0.2 ml) via the intranasal route with a small tube after anesthesia (18, 48, 49). Mice were injected intravenously with ampicillin at the same concentration in each of following days. After the 1st day, mice were euthanized, and both the lungs and spleens were removed daily. Collected lungs and spleens were homogenized. The homogenized tissues were then lysed with 1% Triton X-100 to release the bacteria 10 min before they were plated onto agar plates containing ampicillin. The total isolated CFU per spleen was defined as the dissemination rate.
Three points should be noted. 1) 30 min before inoculation, mice were injected with ampicillin at a final concentration of 50 μg/g of mouse body weight to maintain the plasmid-based expression of O-antigen in KIM6+-O+ and PLA in KIM10+-PLA. The rationale for the high dose of bacteria (∼109/ml) in the inoculum is discussed in depth under “Discussion.” 2) To meet biosafety and regulatory requirements, the tetracycline marker on the original pBR322 has been deleted. 3) The Y. pestis strains used in this study were cultured at 26 °C; at this temperature, Y. pestis does not produce the F1 capsule, which is capable of blocking interactions with host cells (15, 16, 20).
RESULTS
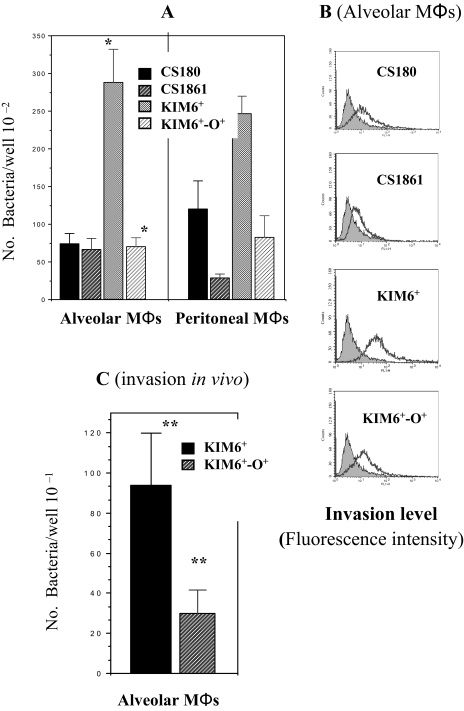
Y. pestis Invades Mouse Alveolar Macrophages—We recently showed that human DC-SIGN and mouse SIGN-R1 can be used as receptors for the core LPS of Y. pestis, promoting bacterial adherence and phagocytosis to human DCs and MΦs, including human alveolar MΦs and murine peritoneal MΦs (20).4 Therefore, we have speculated that Y. pestis might utilize the interaction of core-LPS and SIGN-R1 to invade murine alveolar MΦs in the case of the mouse model. To investigate this hypothesis, Y. pestis KIM6+ (a natural rough strain with the core LPS presumably exposed) and its isogenic derivative KIM6+-O+ (a smooth strain in which the outer core LPS is shielded by O-antigen) were examined for their ability to invade alveolar MΦs. Two corresponding E. coli K12 strains CS180 (rough) and CS1861 (CS180 expressing an O-antigen, smooth) were used as controls. We have used these sets of strains to demonstrate that exposure of the core-LPS of E. coli and Y. pestis is essential to initiate the interaction of core-LPS and DC-SIGN/SIGN-R1 (19, 20, 47, 50)4 (Table 1). In addition, mouse peritoneal MΦs were used as control host cells that can bind E. coli and Y. pestis through the interaction of core LPS and SIGN-R1.4 We reasoned that, like murine peritoneal MΦs, mouse alveolar MΦs might use the same mechanism to phagocytose Y. pestis. The results presented in Fig. 1 show three phenomena. 1) Y. pestis KIM6+ invades alveolar and peritoneal MΦs equally well (Fig. 1A). 2) Phagocytosis of Y. pestis strain KIM6+-O+ by peritoneal and alveolar MΦs was reduced compared with Y. pestis strain KIM6+ (Fig. 1, A and B), presumably as a result of expression of O-antigen. 3) The most interesting result was observed with our control infection using the E. coli strains. Peritoneal MΦs generated an effective phagocytic response to E. coli K12 CS180 (rough), but not to CS1861 (with expression of O-antigen), which was not the case when using alveolar MΦs. Both CS180 and CS1861 can only promote a limited phagocytic response in alveolar MΦs, indicating a clear difference in the interaction of rough Gram-negative bacteria with alveolar MΦs and peritoneal MΦs.
FIGURE 1.
Phagocytosis of Y. pestis by mouse alveolar macrophages occurs in vitro and in vivo. Gentamicin protection (A) and flow cytometry-based assays (B) were used to determine the invasion ability of Y. pestis KIM6+, KIM6+-O+, E. coli K12 CS180, and CS1681 (expression of O-antigen) into purified mouse alveolar MΦs and peritoneal MΦs, and the latter is shown Footnote 5. The invasive nature of the E. coli strains with human APCs has been described recently (19, 20, 47, 50). The in vivo phagocytosis of Y. pestis KIM6+ and KIM6+-O+ is shown in C.*, p < 0.001, calculated by Student's t test in the comparison of the invasion of alveolar MΦsby Y. pestis KIM6+ to KIM6+-O+. **, p < 0.005 in the in vivo comparison of invasion between these two strains. Labeled and unlabeled bacteria are indicated by open and filled symbols, respectively (B). The greater the fluorescence intensity, the more bacteria are phagocytosed by MΦs.
We have recently developed an in vivo phagocytosis assay using peritoneal MΦs,4 which lead us to examine the in vivo phagocytosis of Y. pestis by alveolar MΦs. The in vivo phagocytosis of KIM6+ and KIM6+-O+ (Fig. 1C) shows a similar pattern to the in vitro phagocytosis (Fig. 1A), although the numbers of recovered bacteria were much lower in vivo. We believe that the low recovery of bacteria in vivo is because of the technical difficulty in achieving high concentrations of bacteria in the lungs of live mice. It should be noted that the concentration of inoculating bacteria was 100 times higher, in comparison with in vitro invasion assay. For this experiment, we did not use CS180/CS1861, because E. coli K12 are very sensitive to serum killing. In short, the results indicate that the phagocytosis of Y. pestis by alveolar MΦs is involved with the molecule(s) other than core LPS on Y. pestis.
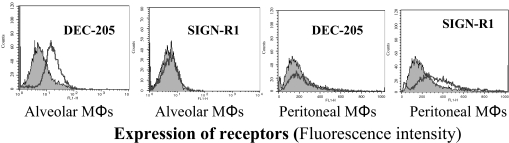
DEC-205, Expressed on Mouse Alveolar Macrophages, Is a Receptor for Y. pestis—The data presented in Fig. 1 indicate that the invasion of alveolar MΦsby Y. pestis might not be mediated by exposure of core LPS. This was initially surprising, as core LPS is a ligand for the SIGN-R1 receptor, expressed on peritoneal MΦs.4 However, studies from several other investigators have suggested that mouse alveolar MΦs in fact do not express SIGN-R1 (43) but may express another receptor, DEC-205 (51). To explain these contrasting experiments, we examined the expression of DEC-205 and SIGN-R1 on alveolar and peritoneal MΦs. Our results confirm that mouse alveolar MΦs and peritoneal MΦs predominantly express DEC-205 and SIGN-R1, respectively (Fig. 2). However, in comparison with its CHO transfectants (Fig. 3B), the expression of DEC-205 in nonactivated alveolar MΦs is moderate. Our data indicated that LPS-activated alveolar MΦs did not enhance the expression of DEC-205 and their ability to phagocytose the Y. pestis (data not shown).
FIGURE 2.
The expression of DEC-205 and SIGN-R1 in alveolar and peritoneal macrophages. Both alveolar and peritoneal MΦs were examined for their expression of DEC-205 and SIGN-R1 with flow cytometry. The level of expression for each receptor is shown with fluorescence intensity. The anti-human DC-SIGN antibody was used as a negative control, which is shown as filled symbols.
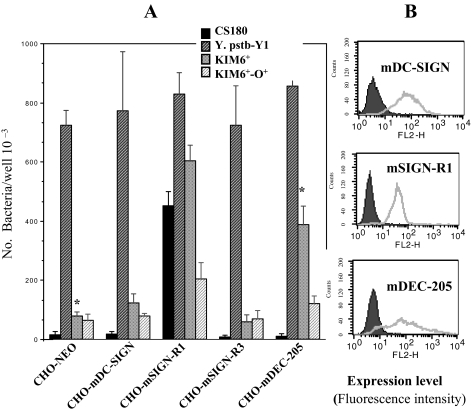
FIGURE 3.
Y. pestis but not E. coli K12 invades CHO-mDEC-205. A, invasion of E. coli K12 CS180, Y. pseudotuberculosis (Y1), and Y. pestis KIM6+ and KIM6+-O+ into CHO cell lines transfected by the mouse C-type lectins; CHO-mDC-SIGN, CHO-mSIGN-R1, CHO-mSIGN-R3 and CHO-mDEC-205 (CD205) were quantified by incubating the cell lines for 2.5 h with the bacterial strains and by killing the extracellular bacteria with 100 μg/ml of gentamicin. The number of phagocytosed bacteria was determined by counting CFU recovered following gentamicin treatment. The expression levels of each transfectant are shown in B. C-type lectin and CHO-NEO transfectants are shown in the open and filled curves, respectively. The level of expression for each receptor is shown with fluorescence intensity. Based on the Student's t test, *, p < 0.001, comparing the interaction of CHO-mDEC-205 with KIM6+ to the interaction of CHO-NEO with the same bacterium.
Because alveolar MΦs do not express SIGN-R1, we examined whether the invasion of Y. pestis into alveolar MΦs results from the interaction between Y. pestis and DEC-205 (Fig. 2). Four CHO transfectants stably expressing the mouse C-type lectin receptors mDC-SIGN, mSIGN-R1, mSIGN-R3, and mDEC-205 (CD205) (41, 42) were infected with Y. pestis KIM6+ and KIM6+-O+. CHO-NEO was used as a negative control cell line. CS180 and Y. pseudotuberculosis (Y1) were used as the positive control strains, because Y. pseudotuberculosis (Y1) grown at 26 °C invades most epithelial cell lines, including CHO (36) via the invasin protein (52), and CS180 was shown to interact with mCHO-SIGN-R1 via its core LPS.4 In addition, in comparison with other strains, only half the numbers of Y. pseudotuberculosis were loaded, because of its ability to promote an aggressive invasion into epithelial cells. The expression of each lectin on CHO cells is shown in Fig. 3B.4 Several phenomena can be observed from these results (Fig. 3A). Y. pestis KIM6+ interacts with CHO-mDEC-205 but not the CHO-NEO, showing that DEC-205 is a receptor for Y. pestis. The interactions of CHO-mSIGN-R1 with Y. pestis KIM6+ and CS180 are most likely because of the exposure of core LPS on the bacteria as the isogenic derivative of both KIM6+ and CS180 expressing an O-antigen no longer effectively interact with this transfectant, as also shown recently.4 We observed that the expression of O-antigen on KIM6+ was still able to block the interaction of this Y. pestis with CHO-mDEC-205, indicating that other potential ligands may also be shielded by expression of O-antigen.
It should be noted that our preliminary data showed that there is also a SIGN-R1- or DEC-205-independent interaction, which can be inhibited by addition of heparin, a synthetic form of heparan sulfate. We present this information, because heparin was included in the assays measuring bacteria-CHO-transfectant interaction to reduce some non-DEC-205 or SIGN-R1-specific interaction between Y. pestis and epithelial cells.4 The rationale for the addition of heparin is addressed in detail under “Discussion.”
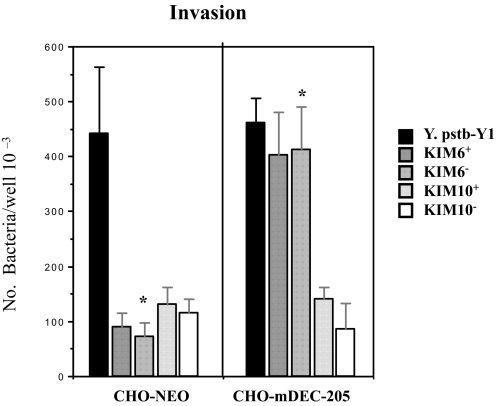
Plasmid pPCP1 Is Involved in the Interaction of Y. pestis with DEC-205 Receptor—As suggested above, core LPS, a ligand for mouse SIGN-R1,4 may not participate in the interaction of KIM6+ Y. pestis with alveolar MΦs, which express the DEC-205. It should be recognized that Y. pestis KIM6+ also contain the pPCP1 plasmid as well as the pgm locus. This led us to speculate that the ligands for the DEC-205 receptor might be encoded either on the pPCP1 plasmid, which encodes nine expressible genes, or the pgm locus containing many important genes that are related to pathogenesis of Y. pestis (53–55). Four strains, KIM6+, KIM6– (lacking the pgm locus), KIM10+ (lacking pPCP plasmid), and KIM10– (lacking both pgm locus and pPCP plasmid), were examined for their ability to interact with CHO-mDEC-205. Y. pseudotuberculosis (Y1) was used as a positive control. As shown in Fig. 4, Y. pestis KIM6+ and KIM6– but not the KIM10+ and KIM10– interact with CHO-DEC-205, indicating that pPCP1 rather than the pgm locus is necessary for the interaction with DEC-205. Again, heparin was present in this assay. Our unpublished data5 further show that KIM10+ binds to CHO cells better than KIM6+ in the absence of heparin, indicating that the presence of the pPCP1 plasmid may inhibit other adherence factors.
FIGURE 4.
Plasmid pPCP1 plays a role in mediating the interaction of Y. pestis with mDEC-205. KIM6+, KIM6– (lacking the pgm locus), KIM10+ (lacking pPCP plasmid), and KIM10– (lacking both pgm locus and pPCP1 plasmid) were tested for their ability to invade alveolar MΦs(left) and CHO-mDEC-205 (right), to determine the involvement of plasmid pPCP1 or pgm locus. The invasion assay followed the same procedures as for the transfectants and MΦs in Figs. 1 and 3. *, both p < 0.001, calculated by Student'st test in the comparison of the invasion of MΦs(left) and CHO-mDEC-205 (right) by KIM6+ to KIM10+. It should be noted that the inoculum of Y. pseudotuberculosis was one-third that of the other strains.
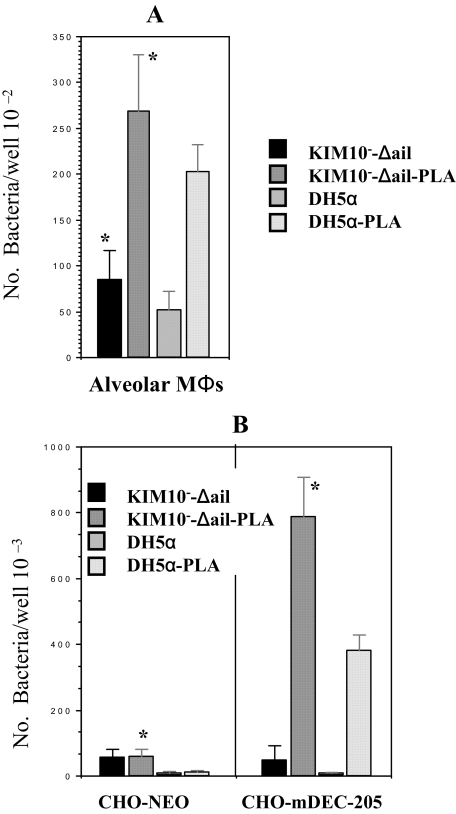
Plasminogen Activator of Y. pestis Is a Ligand for DEC-205 and Plays a Role in Interaction with Alveolar Macrophages—Although the pPCP1 plasmid contains nine potential expressible genes, PLA is the most studied and notable component. In addition, work from other investigators indicates that expression of O-antigen in Yersinia spp. blocks the activity of PLA encoded by the pPCP1 plasmid (23). The data described in Fig. 3A also show that expression of O-antigen inhibits the interaction of Y. pestis KIM6+ containing pPCP1 with CHO-mDEC-205. Therefore, KIM10–-Δail, KIM10–-Δail-PLA (40), E. coli K12 DH5α, and E. coli K12 DH5α-PLA were tested for their interactions with CHO-mDEC-205 and alveolar MΦs. CHO-NEO was used as the negative control. We find that removal of the ail gene reduces the non-C-type lectin-mediated interaction5 as described and discussed under “Experimental Procedures” and “Discussion.” Fig. 5 shows that KIM10–-Δail-PLA and E. coli K12 DH5α-PLA, but not the KIM10–-Δail and E. coli K12 DH5α, interact with CHO-DEC-205, demonstrating that PLA of Y. pestis is a ligand for DEC-205. In addition, expression of PLA in KIM10–-Δail and DH5α enhances the ability to interact with alveolar MΦs, indicating that PLA participates in the interaction of Y. pestis with alveolar MΦs.
FIGURE 5.
PLA-expressing Y. pestis and E. coli invade CHO-mDEC-205 and alveolar macrophages. KIM10–-Δail, KIM10–-Δail-PLA, E. coli K12 DH5α, and E. coli K12 DH5α-PLA were examined for their invasion into alveolar MΦs (A) and CHO-mDEC-205 (B). The invasion assay followed the same procedures as for transfectants and MΦs, described in Figs. 1 and 3. *, both are p < 0.001, calculated by Student'st test in the comparison of the invasion of MΦs(A) and CHO-mDEC-205 (B) by KIM10–-Δail to KIM10–-Δail-PLA.
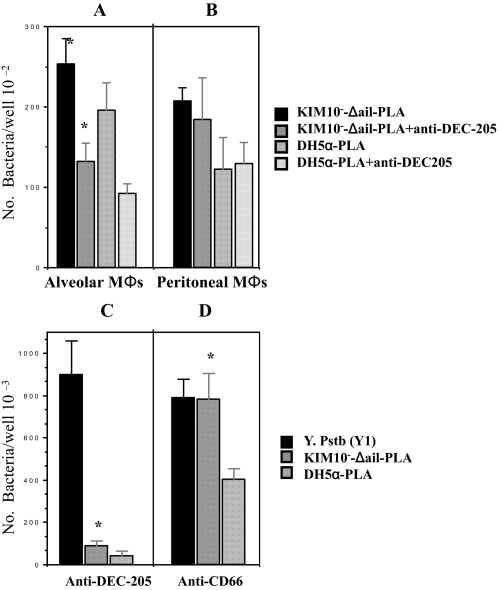
Interactions of CHO-DEC-205 and Alveolar Macrophages with PLA-expressing Y. pestis and E. coli Is Inhibited by DEC-205 Antibody—To verify the specificity of the Y. pestis-alveolar MΦ interaction, we examined whether the interaction of PLA-expressing Y. pestis KIM10–-Δail and E. coli DH5α with CHO-DEC-205 and alveolar MΦs could be inhibited by anti-DEC-205 antibody. Anti-CD66 antibody was employed as a control antibody. Again, we used Y. pseudotuberculosis serotype O:1a grown at 26 °C, which mediates a DEC-205-independent interaction, as a control strain. Fig. 6 shows that anti-DEC-205 antibody blocks the invasion of the PLA-KIM10–-Δail and -DH5α to CHO-DEC-205. Although this antibody reduced the phagocytosis of PLA-KIM10–-Δail and DH5α by alveolar MΦs (Fig. 6A), the blockage was not complete, indicating that other receptors on alveolar MΦs might be involved in this interaction. Also, the dynamic nature of APCs, which presumably phagocytose most antigens, could lead to specific or non-specific phagocytosis. It should be noted that the concentrations of antibody used in this study did not influence the viability of either bacteria or host cells (19) as also found with our control strain Y. pseudotuberculosis (20). Together, these data indicate that there is a specific interaction between DEC-205 of alveolar MΦs and PLA of Y. pestis.
FIGURE 6.
Inhibition of mDEC-205-mediated phagocytosis of PLA-expressing Y. pestis and E. coli K12 by anti-mDEC-205. The KIM10–-Δail-PLA and E. coli K12 DH5α-PLA were incubated with alveolar MΦs(A) and peritoneal MΦs(B) for 1.5 h in the presence of anti-DEC-205 (5 μg/ml). CHO-mDEC-205 cells were also incubated with the KIM10–-Δail-PLA, E. coli K12 DH5α-PLA, and Y. pseudotuberculosis for 2 h in presence or absence of anti-DEC-205 (5 μg/ml) (C) and anti-CD66 (5 μg/ml, the control antibody) (D). All reagents were added to media for 20 min before addition of bacteria. The concentration of each reagent used in this experiment was based on previously published data n (19, 20, 47, 50). The phagocytosis rate of Y. pestis was evaluated by the recovery of bacteria following gentamicin protection. Y. pseudotuberculosis serotype O:1a were used as control strains to show PLA-DEC-205-independent interaction with CHO. Based on the Student'st test, *, p < 0.001, compared the DEC-205-PLA interaction in the presence of anti-DEC-205 antibody to the interaction with the presence of anti-CD66 antibody.
Expression of PLA and O-antigen Promotes and Inhibits Early Dissemination of Y. pestis, Respectively—PLA plays an important role in promoting dissemination of Y. pestis (14, 18). We have presented data showing that DEC-205 is a receptor for the PLA ligand. Expression of O-antigen by Y. pestis inhibits with the PLA-DEC-205 interaction. The obvious question is whether the PLA-DEC-205 interaction enhances the dissemination of Y. pestis and whether blockage of this interaction would reduce the dissemination. Therefore, Y. pestis KIM6+, KIM10+, and KIM10+-PLA (PLA-expressing Y. pestis KIM10+) were examined for their ability to translocate from lungs to spleens. Briefly, mice were infected with these strains via nasal inoculation to mimic pneumonic plague (18, 48, 49). Then mice lungs and spleens were isolated, homogenized, and spread onto Y. pestis plates. Our results (Fig. 7, A and B) show that both KIM6+ and KIM10+ could be transmitted to spleens, but the KIM6+ is more effective, as KIM10+ was retained in the lung more. The most interesting result is that the single gene-expressing KIM10+-PLA partially regained its ability to be transmitted to spleens, but this only occurred on day 2. Furthermore, the retention rates in the lung for KIM10+-PLA were dramatically reduced after day 1. Finally, our data also show that both KIM6+ and KIM10+ can persistently be disseminated to spleens for up to 9 days although the isolation rates are very low in the later stages (data not shown), indicating that the nonvirulent Y. pestis can stay in the lungs for a reasonably long period. However, if they are phagocytosed, these nonvirulent Y. pestis can only live about 2–3 days. In short, the results suggest that PLA plays a role in early Y. pestis dissemination during pneumonic plague.
FIGURE 7.
Expression of PLA and O-antigen enhances and reduces the dissemination of Y. pestis, respectively. After nasal inoculation of KIM6+, KIM10+ (lacking the pPCP plasmid), and KIM10+-PLA, mice were euthanized. Both lungs (A) and spleens (B) were separated, homogenized, and spread on Y. pestis plates. The dissemination rate is the CFU recovered from whole spleens or lungs. C and D, KIM6+ and KIM6+-O+ (expression of O-antigen) were inoculated to mice following the same procedures as in A and B. The data from day 4 were shown, and note the difference in scale between lungs and spleens.
On the other hand, we also speculated that the PLA-DEC-205 interactions, if blocked by expression of O-antigen, could reduce the dissemination. To investigate this hypothesis, Y. pestis KIM6+ and its isogenic derivative KIM6+-O+ were examined for their ability to disseminate to mouse spleens. Four days after inoculation, the rate of dissemination was determined using the same procedures shown in Fig. 7, A and B. A high number of Y. pestis KIM6+ (rough) but not KIM6+-O+ (smooth) were isolated from spleens (Fig. 7, C and D). In contrast, KIM6+-O+ bacteria were retained in the lungs in much higher numbers than KIM 6+ bacteria. It is our presumption that the reduction in dissemination of Y. pestis KIM6+-O+ might be due to its inability to interact with DEC-205.
Because expression of O-antigen inhibits dissemination and the interaction between PLA and DEC-205, we suggest that the PLA-promoted dissemination is based on the ability of PLA to interact with DEC-205. Because KIM10+ is still able to be disseminated, other factors must also have participated in the Y. pestis dissemination from lungs to spleens.
DISCUSSION
Y. pestis, the causative agent of pneumonic plague, is able to rapidly disseminate from the lungs to other organs. Investigators have shown that PLA plays a crucial role in promoting Y. pestis dissemination. It is speculated that Y. pestis hijacks APCs to be delivered to lymph nodes, resulting in this dissemination. In this study, we found that the PLA of Y. pestis is a ligand for DEC-205. Our results also suggest that the PLA-DEC-205 interaction may mediate the dissemination of Y. pestis.
The DEC-205 receptor is expressed on many APCs in both humans and mice (56, 57). The main function of DEC-205 receptor is related to antigen presentation (26, 27, 58). Therefore, the DEC-205 molecule has been used to enhance host responses for vaccine development against the HIV GAG protein and V-antigen from Y. pestis (59–61). On the other hand, an important corollary to the ligand-receptor interaction is that some pathogens, including Y. pestis, have evolved mechanisms of exploiting those very host defense molecules designed to eliminate them, with the result of an expanded ability to disseminate in the host, as explored in this study. However, there are still several technical and scientific issues, which need to be addressed to validate our findings and conclusions.
In this study, nine Y. pestis KIM variants were chosen for investigating various aspects of the bacterial-host interaction, including dissemination; however, none of the selected strains were fully virulent (Table 1). This raises the issue of whether the conclusions we have reached are significant in understanding the pathogenic process of plague. Because of the virulent nature of Y. pestis, this pathogen is considered a “select agent,” and its use in experiments are tightly restricted by IRBs and federal regulations; thus, it was not possible for us to use a virulent strain in our experiments. However, over the entire course of the dissemination experiments, the mouse fatality rate was zero. In fact, we did not even observe that there were any symptoms of discomfort, such as fever among the inoculated mice, suggesting that the innate immune system is very effective against these invading pathogens. These observations clearly confirm the reduced pathogenicity of these mutant strains.
Nevertheless, an advantage of using nonvirulent strains is that they allow us to apply genetically defined mutants to decode the complexity of host-pathogen interactions during Y. pestis dissemination and infection. For example, we have used nonvirulent strains to examine the role of specific ligand-receptor pairs in the dissemination of Y. pestis to mouse spleens from the lungs in experiments mimicking pneumonic plague. If a virulent strain had been used, it would have been impossible to define the dissemination rate; these bacteria are fully capable of replicating in spleens. Also, we observed that the number of KIM10+-PLA CFU retained in the lungs is dramatically reduced by day 2, with a corresponding rise in the isolation of KIM10+-PLA from spleens. These results suggest that the high expression of PLA by Y. pestis induces the rapid phagocytosis by alveolar MΦs, resulting in bacterial clearance or dissemination and that the mutant strains can assist in modeling various stages of Y. pestis pathogenicity. As shown in Figs. 3 and 5, KIM10-PLA invades alveolar MΦs and CHO-DEC-205 more effectively than the KIM6+, suggesting that the expression of PLA in KIM10-PLA might be higher than in KIM6+.
For the same reason, we speculate that Y. pestis KIM6+ bacteria can only survive for about 2–3 days after being phagocytosed or if they have initiated the dissemination process. Fig. 7 shows that after a short increase in dissemination on day 2, the recovery rates of KIM10+-PLA from both lungs and spleens are dramatically reduced. The data thus suggest that the dissemination is initiated after bacteria are phagocytosed. We would also like to mention that we were unable to demonstrate spleen dissemination by KIM6– (data not shown) but do show for KIM6+ in this study. We believe that the pgm locus may help Y. pestis to survive in the MΦs during infection, as shown in a recent study (55).
The overall dissemination rate is very low (Fig. 7). In whole spleens, an average of 500 KIM6+ CFU was recovered at the peak of dissemination. Based on the study of Lathem et al. (18), the number of Y. pestis CO92 could reach 108–109 in spleens before these mice succumbed to the infection. It is possible that the nonvirulent strains are more susceptible to killing in the host, thus affecting overall dissemination numbers. Alternatively, it is possible that only a small fraction of the inoculum was able to reach the lungs. Two observations should be recognized here. First, the only difference between wild-type Y. pestis and KIM6+ is the lack of the pYV plasmid in the latter, possibly affecting its ability to disseminate and/or survive. Second, under the experimental conditions described in this study, we were unable to isolate bacteria from blood samples of the infected mice (data not shown). We speculate that the bacteria transit in the bloodstream at concentrations too low or too rapidly for detection. It is also possible that the bacteria isolated from spleens have not entered there via the bloodstream but via an as yet unknown mechanism.
We also utilized the Y. pestis strain KIM10–-Δail, which is highly attenuated. The deleted components of this strain include the pgm locus, pPCP1 plasmid, the pYV plasmid (encoding type III secretion system and other cytotoxic components), and the ail gene (40). The reason for selecting this strain was to reduce the interference of other known virulence components, which might mask aspects of the pathogen-host interaction under study. For example, although Y. pestis does not express invasin, it directly interacts with epithelial cells such as HeLa cells (52). We have observed that the interaction of Y. pestis with epithelial cells might involve the binding of the Ail protein with cell surface heparan sulfate proteoglycans.5 As shown in Fig. 5, KIM10–-Δail or KIM10–-Δail-PLA has lost their ability to interact with epithelial cells in the absence of either SIGN-R1 or DEC-205.
Another issue of concern is why we used such a high concentration (OD = 1.5) of Y. pestis CFU as an inoculum. For the nonvirulent strains of Y. pestis, this large inoculum was required for us to recover any bacteria disseminating to the spleen. It is known that Y. pestis is an extremely virulent pathogen. Several experiments have indicated that LD50 of Y. pestis can be as low as 1 (62). However, to dissect the course of the bacterial infection, the use of attenuated Y. pestis mutants is more suitable.
In our recent studies (20),4 we have utilized the anti-human DC-SIGN and anti-mouse SIGN-R1 antibodies to inhibit the interactions of Y. pestis with human monocyte-derived DCs and mouse MΦs, but neither of these antibodies was effective. In fact, we must combine both anti-human DC-SIGN and -human Langerin (CD207) (another host receptor for Y. pestis)5 antibodies to show a moderate reduction in this interaction (20). As shown in Fig. 6A in this study, interaction of Y. pestis with alveolar MΦs was reduced by half following a single anti-DEC-205 antibody treatment, indicating that DEC-205 is a major receptor for Y. pestis and plays a specific role during the phagocytosis of Y. pestis by alveolar MΦs. However, this inhibition by anti-DEC-205 antibody was not complete, indicating that alveolar MΦs likely possess an additional receptor(s) for Y. pestis.
Both Welkos et al. (63) and Lathem et al. (18) indicated that PLA might not play a significant role in dissemination if the strain CO92 was infected via aerosol inhalation to non-human primates and intranasal route to mice, respectively. CO92, the most virulent strain, was used in their studies. Therefore, we suggest that binding of the bacteria to APCs, although actively blocking endocytosis via the type III secretion system, may facilitate the transport of viable bacteria to lymph nodes. As shown in Fig. 7, the KIM10+, without expression of PLA, was still able to be disseminated, although not as efficiently at the KIM6+.
What are the other possible mechanisms for promoting dissemination of Y. pestis? This study focused on the interactive roles between PLA of Y. pestis and DEC-205 of alveolar MΦs. However, it is likely that mouse pulmonary DCs, which express both DEC-205 and Langerin (CD207) rather than alveolar MΦs, are the cells responsible for migrating between tissues (64–66). Therefore, we would like to propose another model for pneumonic plague. After entering the lungs via aspiration, Y. pestis uses its Ail protein (40, 67) to bind to cell surface heparan sulfate proteoglycans and invade epithelial layers of bronchial tracts in the lungs. Then the Y. pestis encounters the pulmonary DCs via the SIGN-R1, DEC-205, or Langerin, which lead to the dissemination.
We have presented evidence in vitro and in vivo showing that Y. pestis may use the PLA-DEC-205 interaction to promote its invasion and possible dissemination. However, the fundamental question still remains as to whether the PLA-DEC-205 interaction plays a role in an in vivo infection.
The most straightforward approach would be to test whether a DEC-205 knock-out mouse model would be resistant to plague or reduce the dissemination of Y. pestis, an experiment that we are pursuing. The knock-out mouse model has been used successfully to identify viral receptors. For example, the CEACAM1 (CD66a) receptor knock-out mice have an increased resistance to mouse hepatitis viral infection (68), because mouse CEACAM1 is a receptor for mouse hepatitis virus (69). However, there are potential limitations of this approach for studying bacteria-host cell interactions. Strangely, there are no credible receptor knock-out models that are more resistant to bacterial infection. The reason might be simple; bacterial infections are more multifactorial than viral infections. Knock-out of one receptor might not be enough to determine the fate of a bacterial infection, a fact that might contribute to the failure of CCR5 knock-mice to resist Y. pestis infection (32, 70), even if CCR5 is a receptor for Y. pestis. Thus the mouse knock-out model may only provide unequivocal results if DEC-205 is the only receptor for Y. pestis. Unfortunately, many pathogens do not depend on only one receptor in their interactions with host cells, as we have indicated in our published or unpublished data5 that Y. pestis interacts with human DC-SIGN, human Langerin, mouse SIGN-R1, mouse Langerin, cell surface heparan sulfate proteoglycan, and of course mouse DEC-205.
Taken together, this study has demonstrated that DEC-205 functions as a cellular receptor for Y. pestis, with PLA serving as a ligand. The data presented in this study indicate that interactions between PLA and DEC-205 may play a role in the dissemination of Y. pestis.
Acknowledgments
We are indebted to Dr. Ralph Steinman at the Rockefeller University for insightful advice and generous support.
This work was supported, in whole or in part, by National Institutes of Health grants (USPHS). This work was also supported in part by a grant from the University of Illinois, College of Medicine, Rockford (to T. C.), and work in the Skurnik laboratory was supported by Academy of Finland Grant 114075. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PLA, plasminogen activator; MΦ, macrophage; APC, antigen-presenting cell; DC, dendritic cell; LPS, lipopolysaccharide; CFU, colony-forming unit; FCS, fetal calf serum; CHO, Chinese hamster ovary; HIV-1, human immunodeficiency virus, type 1.
S. Zhang, C. G. Park, S. S. Bartra, A. Shetty, P. Zhang, G. Zheng, S. Bulgheresi, Z. Yu, G. V. Plano, J. Klena, M. Skunilk, J. Hinnebusch, and T. Chen, submitted for publication.
T. Chen, unpublished data.
References
- 1.Achtman, M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A., and Carniel, E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., Morelli, G., Zhu, P., Wirth, T., Diehl, I., Kusecek, B., Vogler, A. J., Wagner, D. M., Allender, C. J., Easterday, W. R., Chenal-Francisque, V., Worsham, P., Thomson, N. R., Parkhill, J., Lindler, L. E., Carniel, E., and Keim, P. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17837–17842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vazquez-Torres, A., and Fang, F. C. (2000) Curr. Opin. Microbiol. 3 54–59 [DOI] [PubMed] [Google Scholar]
- 4.Isberg, R. R., and Barnes, P. (2001) J. Cell Sci. 114 21–28 [DOI] [PubMed] [Google Scholar]
- 5.Brubaker, R. R. (1983) Rev. Infect. Dis. 5 Suppl. 4, 748–758 [DOI] [PubMed] [Google Scholar]
- 6.Portnoy, D. A., and Martinez, R. J. (1985) Curr. Top. Microbiol. Immunol. 118 29–51 [DOI] [PubMed] [Google Scholar]
- 7.Cornelis, G. R., and Wolf-Watz, H. (1997) Mol. Microbiol. 23 861–867 [DOI] [PubMed] [Google Scholar]
- 8.Rosqvist, R., Skurnik, M., and Wolf-Watz, H. (1988) Nature 334 522–524 [DOI] [PubMed] [Google Scholar]
- 9.Skurnik, M., and Wolf-Watz, H. (1989) Mol. Microbiol. 3 517–529 [DOI] [PubMed] [Google Scholar]
- 10.Viboud, G. I., and Bliska, J. B. (2005) Annu. Rev. Microbiol. 59 69–89 [DOI] [PubMed] [Google Scholar]
- 11.Lahteenmaki, K., Virkola, R., Saren, A., Emody, L., and Korhonen, T. K. (1998) Infect. Immun. 66 5755–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlyshev, A. V., Galyov, E. E., Smirnov, O., Guzayev, A. P., Abramov, V. M., and Zav'yalov, V. P. (1992) FEBS Lett. 297 77–80 [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch, B. J., Rudolph, A. E., Cherepanov, P., Dixon, J. E., Schwan, T. G., and Forsberg, A. (2002) Science 296 733–735 [DOI] [PubMed] [Google Scholar]
- 14.Sebbane, F., Jarrett, C. O., Gardner, D., Long, D., and Hinnebusch, B. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5526–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weeks, S., Hill, J., Friedlander, A., and Welkos, S. (2002) Microb. Pathog. 32 227–237 [DOI] [PubMed] [Google Scholar]
- 16.Du, Y., Rosqvist, R., and Forsberg, A. (2002) Infect. Immun. 70 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahteenmaki, K., Kukkonen, M., and Korhonen, T. K. (2001) FEBS Lett. 504 69–72 [DOI] [PubMed] [Google Scholar]
- 18.Lathem, W. W., Price, P. A., Miller, V. L., and Goldman, W. E. (2007) Science 315 509–513 [DOI] [PubMed] [Google Scholar]
- 19.Zhang, P., Snyder, S., Feng, P., Azadi, P., Zhang, S., Bulgheresi, S., Sanderson, K. E., He, J., Klena, J., and Chen, T. (2006) J. Immunol. 177 4002–4011 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, P., Skurnik, M., Zhang, S., Zheng, G., Kalyanasundaram, R., Bulgheresi, S., He, J., Klena, J., Hinnebusch, J., and Chen, T. (2008) Infect. Immun. 76 2070–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skurnik, M., Peippo, A., and Ervela, E. (2000) Mol. Microbiol. 37 316–330 [DOI] [PubMed] [Google Scholar]
- 22.Prior, J. L., Parkhill, J., Hitchen, P. G., Mungall, K. L., Stevens, K., Morris, H. R., Reason, A. J., Oyston, P. C., Dell, A., Wren, B. W., and Titball, R. W. (2001) FEMS Microbiol. Lett. 197 229–233 [DOI] [PubMed] [Google Scholar]
- 23.Kukkonen, M., Suomalainen, M., Kyllonen, P., Lahteenmaki, K., Lang, H., Virkola, R., Helander, I. M., Holst, O., and Korhonen, T. K. (2004) Mol. Microbiol. 51 215–225 [DOI] [PubMed] [Google Scholar]
- 24.Medzhitov, R., Preston-Hurlburt, P., and Janeway, C. A., Jr. (1997) Nature 388 394–397 [DOI] [PubMed] [Google Scholar]
- 25.Takeda, K., Kaisho, T., and Akira, S. (2003) Annu. Rev. Immunol. 21 335–376 [DOI] [PubMed] [Google Scholar]
- 26.Bonifaz, L., Bonnyay, D., Mahnke, K., Rivera, M., Nussenzweig, M. C., and Steinman, R. M. (2002) J. Exp. Med. 196 1627–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonifaz, L. C., Bonnyay, D. P., Charalambous, A., Darguste, D. I., Fujii, S., Soares, H., Brimnes, M. K., Moltedo, B., Moran, T. M., and Steinman, R. M. (2004) J. Exp. Med. 199 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geijtenbeek, T. B., Kwon, D. S., Torensma, R., van Vliet, S. J., van Duijnhoven, G. C., Middel, J., Cornelissen, I. L., Nottet, H. S., KewalRamani, V. N., Littman, D. R., Figdor, C. G., and van Kooyk, Y. (2000) Cell 100 587–597 [DOI] [PubMed] [Google Scholar]
- 29.Engering, A., Van Vliet, S. J., Geijtenbeek, T. B., and Van Kooyk, Y. (2002) Blood 100 1780–1786 [DOI] [PubMed] [Google Scholar]
- 30.McDonald, D., Wu, L., Bohks, S. M., KewalRamani, V. N., Unutmaz, D., and Hope, T. J. (2003) Science 300 1295–1297 [DOI] [PubMed] [Google Scholar]
- 31.Granelli-Piperno, A., Pritsker, A., Pack, M., Shimeliovich, I., Arrighi, J. F., Park, C. G., Trumpfheller, C., Piguet, V., Moran, T. M., and Steinman, R. M. (2005) J. Immunol. 175 4265–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mecsas, J., Franklin, G., Kuziel, W. A., Brubaker, R. R., Falkow, S., and Mosier, D. E. (2004) Nature 427 606. [DOI] [PubMed] [Google Scholar]
- 33.Schnaitman, C. A., and Klena, J. D. (1993) Microbiol. Rev. 57 655–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klena, J., Ashford, R. S., II, and Schnaitman, C. A. (1992) J. Bacteriol. 174 7297–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klena, J. D., and Schnaitman, C. A. (1993) Mol. Microbiol. 9 393–402 [DOI] [PubMed] [Google Scholar]
- 36.Chen, T., Belland, R., Wilson, J., and Swanson, J. (1995) J. Exp. Med. 182 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isberg, R., and Leong, J. (1990) Cell 60 861–871 [DOI] [PubMed] [Google Scholar]
- 38.Fetherston, J. D., Schuetze, P., and Perry, R. D. (1992) Mol. Microbiol. 6 2693–2704 [DOI] [PubMed] [Google Scholar]
- 39.Oyston, P. C., Prior, J. L., Kiljunen, S., Skurnik, M., Hill, J., and Titball, R. W. (2003) J. Med. Microbiol. 52 289–294 [DOI] [PubMed] [Google Scholar]
- 40.Bartra, S. S., Styer, K. L., O'Bryant, D. M., Nilles, M. L., Hinnebusch, B. J., Aballay, A., and Plano, G. V. (2008) Infect. Immun. 76 612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang, Y. S., Yamazaki, S., Iyoda, T., Pack, M., Bruening, S. A., Kim, J. Y., Takahara, K., Inaba, K., Steinman, R. M., and Park, C. G. (2003) Int. Immunol. 15 177–186 [DOI] [PubMed] [Google Scholar]
- 42.Takahara, K., Yashima, Y., Omatsu, Y., Yoshida, H., Kimura, Y., Kang, Y. S., Steinman, R. M., Park, C. G., and Inaba, K. (2004) Int. Immunol. 16 819–829 [DOI] [PubMed] [Google Scholar]
- 43.Wieland, C. W., Koppel, E. A., den Dunnen, J., Florquin, S., McKenzie, A. N., van Kooyk, Y., van der Poll, T., and Geijtenbeek, T. B. (2007) Microbes Infect. 9 134–141 [DOI] [PubMed] [Google Scholar]
- 44.Chen, T., Grunert, F., Medina-Marino, A., and Gotschlich, E. (1997) J. Exp. Med. 185 1557–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, T., Bolland, S., Chen, I., Parker, J., Pantelic, M., Grunert, F., and Zimmermann, W. (2001) J. Biol. Chem. 276 17413–17419 [DOI] [PubMed] [Google Scholar]
- 46.Chen, T., and Gotschlich, E. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14851–14856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, P., Schwartz, O., Pantelic, M., Li, G., Knazze, Q., Nobile, C., Radovich, M., He, J., Hong, S. C., Klena, J., and Chen, T. (2006) J. Leukocyte Biol. 79 731–738 [DOI] [PubMed] [Google Scholar]
- 48.Overheim, K. A., Depaolo, R. W., Debord, K. L., Morrin, E. M., Anderson, D. M., Green, N. M., Brubaker, R. R., Jabri, B., and Schneewind, O. (2005) Infect. Immun. 73 5152–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lathem, W. W., Crosby, S. D., Miller, V. L., and Goldman, W. E. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17786–17791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klena, J., Zhang, P., Schwartz, O., Hull, S., and Chen, T. (2005) J. Bacteriol. 187 1710–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ordway, D., Harton, M., Henao-Tamayo, M., Montoya, R., Orme, I. M., and Gonzalez-Juarrero, M. (2006) J. Immunol. 176 4931–4939 [DOI] [PubMed] [Google Scholar]
- 52.Sikkema, D. J., and Brubaker, R. R. (1987) Infect. Immun. 55 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fetherston, J. D., Bertolino, V. J., and Perry, R. D. (1999) Mol. Microbiol. 32 289–299 [DOI] [PubMed] [Google Scholar]
- 54.Bearden, S. W., and Perry, R. D. (1999) Mol. Microbiol. 32 403–414 [DOI] [PubMed] [Google Scholar]
- 55.Pujol, C., Grabenstein, J. P., Perry, R. D., and Bliska, J. B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12909–12914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato, M., McDonald, K. J., Khan, S., Ross, I. L., Vuckovic, S., Chen, K., Munster, D., MacDonald, K. P., and Hart, D. N. (2006) Int. Immunol. 18 857–869 [DOI] [PubMed] [Google Scholar]
- 57.Pack, M., Trumpfheller, C., Thomas, D., Park, C. G., Granelli-Piperno, A., Munz, C., and Steinman, R. M. (2008) Immunology 123 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dudziak, D., Kamphorst, A. O., Heidkamp, G. F., Buchholz, V. R., Trumpfheller, C., Yamazaki, S., Cheong, C., Liu, K., Lee, H. W., Park, C. G., Steinman, R. M., and Nussenzweig, M. C. (2007) Science 315 107–111 [DOI] [PubMed] [Google Scholar]
- 59.Bozzacco, L., Trumpfheller, C., Siegal, F. P., Mehandru, S., Markowitz, M., Carrington, M., Nussenzweig, M. C., Piperno, A. G., and Steinman, R. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1289–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trumpfheller, C., Finke, J. S., Lopez, C. B., Moran, T. M., Moltedo, B., Soares, H., Huang, Y., Schlesinger, S. J., Park, C. G., Nussenzweig, M. C., Granelli-Piperno, A., and Steinman, R. M. (2006) J. Exp. Med. 203 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Do, Y., Park, C. G., Kang, Y. S., Park, S. H., Lynch, R. M., Lee, H., Powell, B. S., and Steinman, R. M. (2008) Eur. J. Immunol. 38 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perry, R. D., and Fetherston, J. D. (1997) Clin. Microbiol. Rev. 10 35–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Welkos, S., Pitt, M. L., Martinez, M., Friedlander, A., Vogel, P., and Tammariello, R. (2002) Vaccine 20 2206–2214 [DOI] [PubMed] [Google Scholar]
- 64.Legge, K. L., and Braciale, T. J. (2005) Immunity 23 649–659 [DOI] [PubMed] [Google Scholar]
- 65.Hammad, H., and Lambrecht, B. N. (2007) Adv. Immunol. 93 265–278 [DOI] [PubMed] [Google Scholar]
- 66.Hao, X., Kim, T. S., and Braciale, T. J. (2008) J. Virol. 82 4908–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kolodziejek, A. M., Sinclair, D. J., Seo, K. S., Schnider, D. R., Deobald, C. F., Rohde, H. N., Viall, A. K., Minnich, S. S., Hovde, C. J., Minnich, S. A., and Bohach, G. A. (2007) Microbiology 153 2941–2951 [DOI] [PubMed] [Google Scholar]
- 68.Hemmila, E., Turbide, C., Olson, M., Jothy, S., Holmes, K. V., and Beauchemin, N. (2004) J. Virol. 78 10156–10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan, K., Zelus, B. D., Meijers, R., Liu, J. H., Bergelson, J. M., Duke, N., Zhang, R., Joachimiak, A., Holmes, K. V., and Wang, J. H. (2002) EMBO J. 21 2076–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elvin, S. J., Williamson, E. D., Scott, J. C., Smith, J. N., Perez De Lema, G., Chilla, S., Clapham, P., Pfeffer, K., Schlondorff, D., and Luckow, B. (2004) Nature 430 417. [DOI] [PubMed] [Google Scholar]