Abstract
Cadmium is a highly toxic environmental contaminant implicated in various diseases. Our previous data demonstrated that Pca1, a P1B-type ATPase, plays a critical role in cadmium resistance in yeast S. cerevisiae by extruding intracellular cadmium. This illustrates the first cadmium-specific efflux pump in eukaryotes. In response to cadmium, yeast cells rapidly enhance expression of Pca1 by a post-transcriptional mechanism. To gain mechanistic insights into the cadmium-dependent control of Pca1 expression, we have characterized the pathway for Pca1 turnover and the mechanism of cadmium sensing that leads to up-regulation of Pca1. Pca1 is a short-lived protein (t½ < 5 min) and is subject to ubiquitination when cells are growing in media lacking cadmium. Distinct from many plasma membrane transporters targeted to the vacuole for degradation via endocytosis, cells defective in this pathway did not stabilize Pca1. Rather, Pca1 turnover was dependent on the proteasome. These data suggest that, in the absence of cadmium, Pca1 is targeted for degradation before reaching the plasma membrane. Mapping of the N terminus of Pca1 identified a metal-responding degradation signal encompassing amino acids 250–350. Fusion of this domain to a stable protein demonstrated that it functions autonomously in a metal-responsive manner. Cadmium sensing by cysteine residues within this domain circumvents ubiquitination and degradation of Pca1. These data reveal a new mechanism for substrate-mediated control of P1B-type ATPase expression. Cells have likely evolved this mode of regulation for a rapid and specific cellular response to cadmium.
Although several heavy metals are essential micronutrients, many of them are highly toxic with no physiological role. Genetic disorders in metal metabolism, such as hemochromatosis and Wilson disease, provide striking evidence for the toxicity of nutritional metal ions (1–3). Toxic heavy metals in the environment such as cadmium, lead, and mercury are linked to a number of human disorders, including cancer, organ damage, and reproductive defects (4–7). For instance, cadmium has a high affinity for sulfhydryl (-SH) groups in proteins and glutathione, a major cellular reducing buffer, which results in inactivation of proteins and oxidative stress (4, 9, 10). Cadmium markedly increases mutation rates by specifically inhibiting the DNA mismatch repair system (11). Estrogenic effects of cadmium also lead to infertility and cancer (12–14). Human exposure to cadmium through diet, smoking, pollution, and occupation is widespread (4, 5, 8).
Organisms have evolved systems for excretion and detoxification of nonphysiological metals and homeostatic acquisition of nutritional yet toxic metals. Metal transporters, cytosolic delivery molecules, and chelators have been identified in many organisms ranging from bacteria to humans (1, 3, 26–28). Given that cadmium is not a nutritional metal ion, organisms do not actively acquire cadmium. Rather, cadmium enters the body through nonspecific mechanisms and/or transporters involved in the acquisition of essential metal ions. DMT1 (DCT1, Nramp2) iron transporter, zinc transporters, and calcium channels are all involved in cadmium absorption (15–19). Metallothionein, a cysteine-rich low molecular weight metalchelating peptide, and GSH, a cysteine-containing tripeptide, sequester metal ions. In eukaryotes, this is considered the major mechanism for neutralizing toxic metals, including cadmium (20–23). Ace1 is a transcription factor inducing expression of copper defense genes in S. cerevisiae (24). Ycf1, a vacuolar membrane ATP-binding cassette transporter, sequesters glutathione-conjugated cadmium and other metals into the vacuole (25). Thus, Δace1 and Δycf1 strains are sensitive to copper and cadmium, respectively. Phytochelatin, a GSH polymer synthesized in plants and fission yeast, also chelates heavy metals (21). Several heavy metal efflux pumps, including P1B-type ATPases, ATP-binding cassette transporters, and cation diffusion facilitators play roles for extruding toxic heavy metals (3, 26–28). These metal transporters play vital roles for uptake, compartmentalization, and extrusion of nutritional and nonphysiological heavy metals. In bacterial cells, P1B-type ATPases are vital for extrusion of metals, including cadmium, copper, and lead (26, 27). However, the cadmium efflux systems in eukaryotes remain uncharacterized. Transcriptional control appears to be a major regulatory mechanism for regulation of P1B-type ATPases in bacteria and plants (26–28). In mammals copper controls subcellular trafficking of copper-transporting P1B-type ATPases from the trans-Golgi to the plasma membrane and cytoplasmic vesicle-like compartments (3, 29), which appears to be an important mechanism for excretion of excess copper.
In a previous study, we demonstrated that Pca1, a predicted P1B-type metal-transporting ATPase, is critical for cadmium defense in yeast Saccharomyces cerevisiae (30). Pca1 is rapidly up-regulated in response to cadmium without any change in its transcript levels (30), which suggests a new mode of substrate-mediated regulation for this family of proteins. To elucidate the mechanisms underlying the expressional control of Pca1 in response to cadmium, we have characterized the pathways for turnover of Pca1 and identified a metal-sensing cis-acting element that controls turnover of Pca1. Our data demonstrate that a degradation signal within the cysteine-rich N terminus rapidly targets Pca1 for ubiquitination and degradation by the proteasome. This occurs before Pca1 reaches the plasma membrane where it extrudes cadmium. However, direct cadmium sensing by the degradation signal prevents turnover, which leads to enhanced expression of Pca1. Cells have likely evolved this mode of regulation for a rapid and specific response against cadmium. Together, we have identified a new metal-binding domain and revealed a unique mode of regulation for a P1B-type ATPase by its own substrate.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Growth Conditions—A BY4741 haploid S. cerevisiae strain (MATa his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and null mutants, including ace1::KanMX6, ycf1:: KanMX6, pep4::KanMX6, vps4::KanMX6, and pdr5::KanMX6 strains, were purchased from OpenBiosystems. Gene deletion in each strain was confirmed by PCR using gene-specific primer sets. We received end4-1 (31), npi1 (32), and pre1-1 pre2-1 (33) strains from Drs. Caroline Philpott (National Institute of Health), Bruno André, (Université Libre de Bruxelles), and Dieter Wolf (Universität Stuttgart), respectively. Yeast cells were cultured with synthetic complete (SC)2 medium (2% dextrose, 0.2% amino acid mixture, and 0.67% yeast nitrogen base) lacking specific amino acids for plasmid selection. If no temperature is specified, the cells were cultured at 30 °C. For phenotypical analysis on solid medium (1.5% agar), 5 μl of yeast cells (A600 = 1.0) were spotted on selective medium supplemented with the indicated concentrations of cadmium (CdCl2) or copper (CuCl2) and incubated at 30 °C for 2 days before photography. Cycloheximide chases were performed by the addition of 100 μg/ml cycloheximide to logarithmically growing cultures prior to collection into equal volumes of ice-cold kill buffer (phosphate-buffered saline (PBS) containing 15 mm NaN3) at the indicated time points. For proteasome inhibition, a Δpdr5 strain that blocks proteasome inhibitor efflux (34), was cultured for 5 h in the presence of 40 μm MG132 (Calbiochem) in 0.1% dimethyl sulfoxide (Sigma).
Plasmid Construction and Manipulation—If no expression vector is specified, single copy yeast vectors (p416-GPD) (35) were used for glyceradehyde-3-phosphate dehydrogenase gene promoter-driven constitutive expression of Pca1, N-terminal truncated Pca1, CaCRP, Pca1-CaCRP fusion, N-terminal 250–350 peptide, Fet3 iron oxidase, CL1 degradation signal peptide, and Gap1 general amino acid permease. For the green fluorescent protein (GFP) or hemagglutinin (HA) epitope fusion, NotI-flanked GFP or three tandem HA epitopes were inserted into the NotI site, which was artificially generated after ATG translation start codon by PCR. Expression plasmids of CL1 artificial degradation signal fused with GFP were constructed as described (36, 45). Site-directed mutagenesis was conducted by the primer overlap extension method (37). We sequenced all constructs to confirm correct sequence and in frame fusion. Common molecular biology techniques, including plasmid amplification using Escherichia coli, and purification, followed previously established methods (38). Plasmid transformation into yeast was performed using the lithium acetate method (39).
Fluorescence Microscopy—Yeast cells were cultured in SC selective medium at 30 °C with agitation until mid-log phase. When indicated, metals (CdCl2 or CuCl2) were added to the culture medium prior to imaging. The cells were collected, resuspended in PBS, and imaged on a confocal microscope (Olympus FV500) as described (30).
Immunoblotting—Total protein extracts were prepared by breaking cells with glass beads in PBS containing protease inhibitor mixture (Complete Mini) (Roche Applied Science) and 1% Triton X-100. Protein concentrations were measured using the BCA kit (Pierce) according to the manufacturer's specifications. The lysates were denatured in SDS sample buffer containing dithiothreitol (25 mm) for 15 min at 37 °C, resolved by SDS-PAGE, and transferred to a nitrocellulose membrane. GFP fusion proteins were detected by rabbit anti-GFP polyclonal antibodies (Santa Cruz Biotechnology Inc.) and secondary goat anti-rabbit IgG antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology Inc.). HA-tagged proteins were probed with rabbit anti-HA polyclonal antibodies (Rockland) and secondary goat anti-rabbit IgG antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology Inc.). Loading control, 3-phosphoglycerate kinase (PGK), was detected with mouse monoclonal anti-PGK antibodies (Molecular Probes) and secondary sheep anti-mouse IgG antibodies conjugated to horseradish peroxidase (Amersham Biosciences). The signals were captured on film (Midwest Scientific) by chemiluminescence (SuperSignal West Pico) (Pierce).
Detection of Ubiquitination—The cells were broken by glass beads in PBS containing protease inhibitors (Complete Mini) (Roche Applied Science), 5 mm N-ethylmaleimide, 1 mm phenylmethylsulfonyl fluoride, and 1% Triton X-100. HA-tagged proteins were immunoprecipitated with a ProFound™ HA tag IP/Co-IP kit (Pierce) according to the manufacturer's specifications. The proteins were eluted by incubation of immobilized anti-HA agarose beads in 2× SDS sample buffer at 37 °C for 15 min and reduced in the presence of 0.1 m dithiothreitol for an additional 15 min at 37 °C. Ubiquitin-conjugated proteins were detected by immunoblotting using mouse monoclonal antibodies against ubiquitin (Covance).
RESULTS
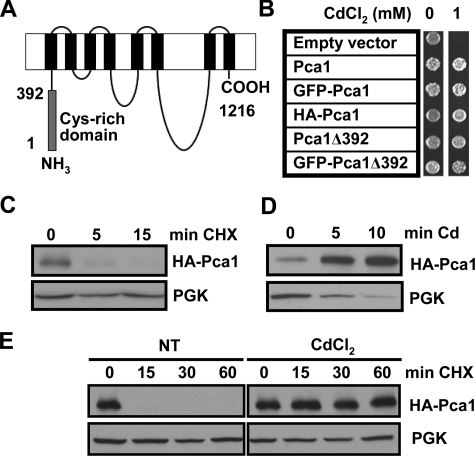
Cadmium Rapidly Induces Pca1 Expression by Increasing Stability—A unique feature of Pca1 is a cytosolic cysteine-rich N-terminal extension of nearly 400 residues (Fig. 1A). To investigate the mechanism underlying the post-transcriptional regulation and the role of the N-terminal extension in turnover of Pca1, we have constructed expression plasmids of full-length Pca1 and Pca1 with truncation of the first 392 amino acids. A GFP or triple HA epitope was fused at the N terminus. All of the Pca1 constructs were functional in conferring cadmium resistance (Fig. 1B). Western blot analysis of Pca1 showed that Pca1 is a short-lived protein (t½ < 5 min) (Fig. 1C). However, up-regulation of Pca1 is clearly visible 5 min after the addition of cadmium to culture medium (Fig. 1D), indicating a rapid cellular response to cadmium. Pca1 tagged with the GFP or c-Myc epitope at the N or C terminus, respectively, also showed similar regulation by cadmium (data not shown), suggesting that the effect of cadmium on Pca1 expression is independent of epitope or its location. This expression control by cadmium can be explained by enhanced translation or stability of Pca1 protein. To decipher these two possibilities, Pca1 turnover rate was examined by cycloheximide chases in the absence or presence of cadmium in culture media. Indeed, stability of Pca1 is dramatically increased in the presence of cadmium (Fig. 1E). These data suggest that the induction of Pca1 occurs through the ability of cadmium to prevent the rapid turnover of Pca1.
FIGURE 1.
Cadmium rapidly enhances Pca1 expression by increasing stability. A, schematic depiction of the Pca1 P1B-type ATPase. Gray and black rectangles represent the N-terminal cysteine-rich cytosolic domain encompassing the first 392 amino acids and eight predicted transmembrane domains, respectively. B, cadmium resistance in cells expressing Pca1 alleles. Pca1, N-terminal GFP-fused Pca1 (GFP-Pca1), three tandem HA epitope-tagged Pca1 (HA-Pca1), and Pca1 deleted of the N-terminal cysteine-rich domain (Pca1Δ392) with or without N-terminal GFP fusion were expressed in a BY4741 yeast strain in which endogenous Pca1 is nonfunctional (30). The cells (5 μl, A600 = 1.0) were spotted on SC solid medium containing the indicated concentration of CdCl2, and then cell growth was assessed after 2 days. C–E, HA-Pca1 expression was detected by Western blotting using anti-HA antibodies. The same blot was probed for PGK to determine equal loading. C, cycloheximide chase of yeast cells expressing HA-Pca1. Exponentially growing cells were co-cultured with cycloheximide (100 μg/ml) and then collected at the indicated time points for Western blot analysis. D, rapid up-regulation of Pca1 in response to cadmium. Cadmium (50 μm CdCl2) was added to exponentially growing yeast cultures, and then the cells were collected into ice-cold kill buffer (15 mm NaN3 PBS) at the indicated time points. E, cadmium-induced stabilization of Pca1 determined by cycloheximide chase. Yeast cells precultured for 15 min without (NT) or with cadmium (20 μm CdCl2) were collected at the indicated time points. Cycloheximide (100 μg/ml) was added to the culture medium after collection of time zero. Total protein extracts were subjected to Western blot analysis.
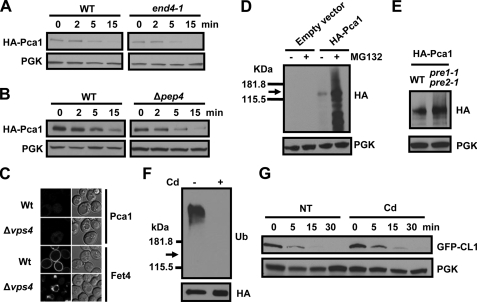
Pca1 Is Not Stabilized in a Strain Defective in Endocytosis—In yeast, several plasma membrane proteins are internalized into endocytic vesicles that enter the early endosome where they can be recycled back to the surface or are delivered to the vacuole for degradation (40, 41). We considered that Pca1 might be transiently localized at the cell surface before being rapidly internalized and degraded. In this pattern of trafficking, cadmium sensing would shift the distribution of Pca1 to the cell surface for cadmium efflux. If the internalization step were to be blocked, then Pca1 would be expected to accumulate at the plasma membrane in the absence of cadmium. To test this possibility, we investigated the subcellular localization of Pca1 fused with GFP (Fig. 1B) in a strain harboring a temperature-sensitive allele of END4. This strain exhibits conditional defects in both fluid phase and receptor-mediated endocytosis at the restrictive temperature (31). Confocal microscopy revealed that GFP-Pca1 expressed in the end4-1 mutant at the restrictive temperature did not accumulate at the plasma membrane (data not shown) and remained unstable as demonstrated by cycloheximide chase (Fig. 2A). The defect in endocytosis was confirmed by inhibition of internalization of the membrane impermeable dye, Lucifer Yellow CH (31) in the end4-1 strain at the restrictive temperature (data not shown). These results suggest that Pca1 turnover in the absence of cadmium occurs either at the plasma membrane or before sorting to the plasma membrane.
FIGURE 2.
Proteasome-dependent degradation of Pca1 and prevention of ubiquitination by cadmium. A, cycloheximide chase and Western blotting of HA-Pca1 expressed in end4-1 strain and isogenic parental strain (WT). The cells were cultured at 37 °C for 2 h to block endocytosis and collected at the indicated time points for Western blot analysis. B, cycloheximide chase of HA-Pca1 expressed in Δpep4 strain and isogenic parental (WT) strain. C, confocal microscopy of GFP-fused Pca1 or Fet4 expressed in Δvps4 or isogenic parental (Wt) strains. D, empty vector or HA-Pca1 was expressed in Δpdr5 cells in which proteasome inhibitor efflux is reduced (34). The cells were cultured in the presence (+) or absence (–) of proteasome inhibitor MG132 (40μm, 5 h). Total protein extracts were subject to Western blot analysis using anti-HA antibodies. E, HA-Pca1 was expressed in a temperature-sensitive pre1-1 pre2-1 proteasome defective mutant or isogenic parental strain (WT) at restrictive temperature (37 °C) for 2 h. Relative expression of HA-Pca1 was detected by anti-HA immunoblotting. F, cadmium inhibits Pca1 ubiquitination. Cells expressing HA-Pca1 were cultured with (+) or without (–) cadmium (50 μm CdCl2, 2 h). HA-Pca1 in total protein extracts was immunoprecipitated with beads conjugated with anti-HA antibodies. HA-Pca1 and ubiquitin-attached HA-Pca1 were detected by immunoblotting using anti-ubiquitin (Ub) and anti-HA antibodies. The arrow indicates the expected migration of nonubiquitinated HA-Pca1. G, cadmium does not stabilize a model substrate of the ubiquitination-proteasome degradation pathway. Expression levels of GFP-CL1 were detected in cells precultured for 1 h without (NT) or with cadmium (50 μm CdCl2). Cycloheximide (100 μg/ml) was added to the culture medium after collection of time zero. Total protein extracts prepared from cells collected at the indicated time points were subjected to Western blot analysis using anti-GFP antibodies. PGK was probed to determine equal loading.
Pca1 Degradation Is Dependent on the Proteasome But Not Vacuolar Proteases—Plasma membrane proteins can be directly sorted to the vacuole for degradation. For instance the Gap1 amino acid permease is directly targeted to the vacuole when preferred nitrogen sources are available (42). Perhaps in an analogous manner Pca1 may sense cadmium before trafficking to the plasma membrane or is otherwise degraded by the vacuole. To test this, we examined localization and turnover rate of Pca1 in a Δpep4 strain that is deficient in vacuolar protease activity (43). GFP-Pca1 did not accumulate in the vacuole of the Δpep4 strain, which was distinct from Fet3-GFP, a cell surface iron oxidase, previously shown to undergo vacuolar degradation (44) (data not shown). A cycloheximide chase confirmed that Pca1 undergoes degradation at a similar rate in the Δpep4 background compared with an isogenic wild type strain (Fig. 2B), suggesting no significant role for vacuolar proteases in degradation of Pca1. The dependence of Pca1 degradation through the vacuolar/endosomal pathway was further examined in a Δvps4 strain, which is defective in both anterograde (late endosome-to-vacuole) and retrograde (late endosome-to-Golgi) trafficking (74). Resident proteins of the late endosome are typically trapped in prevacuolar compartments/multivesicular bodies in Δvps4 cells. As shown in Fig. 2C, GFP-Pca1 does not accumulate in such compartments in the Δvps4 mutant, unlike Fet4-GFP, which undergoes vacuolar degradation. We conclude that Pca1 degradation is not dependent on the vacuolar degradation pathway.
Given that Pca1 turnover is not dependent on vacuolar proteases, we questioned whether the rapid degradation of Pca1 was dependent upon the proteasome. Indeed, treatment of cells with the protease inhibitor MG132 led to a significant increase in Pca1 expression compared with vehicle-treated controls (Fig. 2D). This experiment was performed in a Δpdr5 strain, previously shown to limit the efflux of protease inhibitors (34). We also examined the steady state expression of Pca1 in a pre1-1 pre2-1 double mutant, which is defective in proteasomal activity (33). Consistently, Pca1 is stabilized in this strain although to a much lesser extent, which is likely due to some remaining proteolytic activities in the pre1-1 pre2-1 strain that participates in Pca1 degradation (Fig. 2E). Consistent with the fact that ubiquitination is a prerequisite for proteasomal targeting, ubiquitination of Pca1 was readily detectible (Fig. 2F). However, it is diminished to nondetectable levels after cells are exposed to cadmium (Fig. 2F). This observation hints toward a mechanistic explanation for the role of cadmium in preventing the degradation of its own exporter. Rsp5 is an E3 ubiquitin-protein ligase that is known to be involved in regulation of several plasma membrane proteins, such as metal-dependent down-regulation of the Znt1 zinc transporter and Smf1 manganese transporter (40). However, an Rsp5-deficient strain did not stabilize Pca1 (data not shown), suggesting no role for this ubiquitin-protein ligase in Pca1 turnover.
To exclude the possibility that stabilization of Pca1 by cadmium may reflect toxic effects of cadmium on the cellular systems for ubiquitination and proteasomal degradation, we tested the effects of cadmium on the expression of GFP-CL1 that has been utilized as a reporter for determining the activities of the ubiquitination-proteasome pathway (45). As shown in Fig. 2G, cadmium exposure at the same concentrations that markedly stabilize Pca1 (Fig. 1E) had no significant influence on the expression levels of GFP-CL1. Thus, cadmium inhibition of proteasomal degradation of Pca1 is specific. Our previous data also showed that Pca1 protein expression is enhanced when yeast cells are exposed to cadmium as low as 1 μm concentrations (30), which are similar to those found in plants where natural yeast cells grow (6).
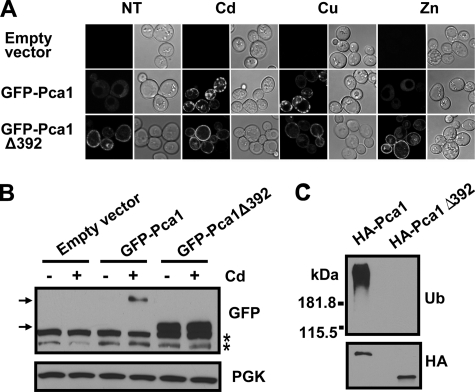
The Cysteine-rich Cytosolic Extension Contains a Degradation Signal—We postulated that the cysteinerich N-terminal extension of Pca1 might contain a metal-responding degradation signal. Indeed, whereas full-length Pca1 is hardly detectable in the absence of copper or cadmium, deletion of this domain (Pca1Δ392) resulted in constitutive expression at the cell surface even in the absence of cadmium (Fig. 3, A and B). These data strongly suggest that Pca1 turnover occurs via this domain. Consistent with the idea that Pca1 ubiquitination is dependent on a degradation signal within the N-terminal domain, ubiquitination was not detected in Pca1Δ392 (Fig. 3C). Although the Pca1(1–392) domain is necessary for Pca1 turnover, co-expression of this domain with Pca1Δ392 separately did not lead to enhanced turnover of Pca1Δ392 (data not shown). This suggests that to serve as a degradation signal for Pca1, this domain should be associated with the entire protein.
FIGURE 3.
The cysteine-rich N terminus of Pca1 contains a degradation signal. A, metal-dependent expression of GFP-Pca1 or GFP-Pca1Δ392. Cells expressing GFP-Pca1 or GFP-Pca1Δ392 were cultured in the absence (NT) or presence of metal ions (50μm CuCl2 (Cu), 50μm CdCl2 (Cd), or 15 mm ZnCl2 (Zn) for 1 h). GFP fluorescence in cells was visualized by confocal microscopy. A copper-sensitive Δace1 strain (24) was used for these experiments presented in both A and B. B, Western blotting of GFP-Pca1 and GFP-Pca1Δ392 in cells cultured with (+) or without (–) cadmium (50μm CdCl2 for 2 h) using anti-GFP antibodies. The asterisks indicate nonspecific bands. The top arrow indicates full-length GFP-Pca1, and the bottom arrow indicates GFP-Pca1Δ392. C, ubiquitination of Pca1 is dependent on the N-terminal domain. HA-Pca1 and HA-Pca1Δ392 were immunoprecipitated and then detected by Western blotting using anti-ubiquitin (Ub) and anti-HA antibodies.
To determine whether the N-terminal regulatory domain is sufficient for controlling the expression of another plasma membrane transporter, we fused the peptide comprising amino acid residues 1–392 of Pca1 to the N terminus of a copper-transporting P1B-type ATPase (CaCRP) identified from Candida albicans (46, 47) and then expressed in S. cerevisiae. The Pca1(1–392)-CaCRP fusion was fully functional in conferring copper tolerance to yeast cells indicating correct folding and trafficking (Fig. 4A). Confocal microscopy (Fig. 4B) and Western blot analysis (Fig. 4C) demonstrated that residues 1–392 in Pca1 control CaCRP expression specifically in a cadmium- and copper-dependent manner. Hence, this domain functions as an autonomous degradation signal that can be masked when cells are growing in medium supplemented with cadmium or copper.
FIGURE 4.
The N-terminal cytosolic extension of Pca1 is an auto-regulatory domain that can be masked by cadmium and copper. A, yeast cells expressing GFP-tagged wild type CaCRP or CaCRP fused with the first 392 amino acids of the N terminus of Pca1 (Pca1(1–392)) were spotted on solid medium containing toxic copper (CuCl2, 0.1 mm). B, expression of GFP-CaCRP and GFP-Pca1(1–392)-CaCRP in cells cultured with CuCl2 or CdCl2 (50μm, 2 h) were determined by visualization of GFP fluorescence with confocal microscopy. C, Western blot analysis of cells cultured as described above (B) using anti-GFP antibodies. A copper-sensitive Δace1 strain (24) was used for these experiments.
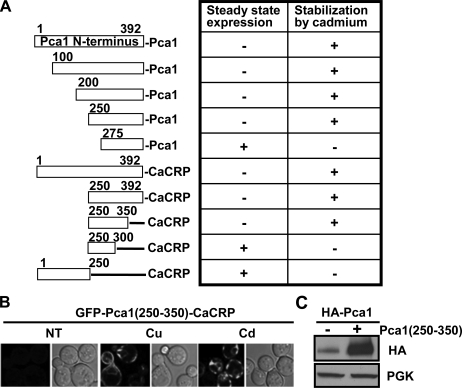
We aligned this domain with characterized P1B-type ATPases and other proteins. However, the Pca1 N-terminal domain does not possess any significant sequence similarity to known proteins. Therefore, we employed a mapping strategy to define a signal sequence of metal-dependent degradation. Serial deletions of the N terminus and fusion of various Pca1 N-terminal fragments to CaCRP were made (Fig. 5A) and then expressed in yeast cells. Characterization of expression levels and metal-dependent stabilization using confocal microscopy revealed that a region encompassing residues 250–350 (Pca1(250–350)) is necessary and sufficient for both rapid turnover and cadmium or copper-dependent stabilization (Fig. 5, A and B). Because the Pca1(250–350) degradation signal likely interacts with cellular factor(s) necessary for Pca1 degradation, we hypothesized that excess expression of this domain would compete for degradation of full-length Pca1. Indeed, co-expression of the Pca1(250–350) peptide with Pca1 leads to enhanced expression of Pca1 (Fig. 5C). Collectively, these data indicate that this Pca1(250–350) contains a degradation signal that can be masked by metal sensing.
FIGURE 5.
The N-terminal domain encompassing amino acids 250-350 is necessary and sufficient for the control of Pca1 and CaCRP expression in a metal-dependent manner. A, Pca1 with or without N-terminal truncations and Pca1 N-terminal fragments fused with CaCRP were expressed in yeast. A GFP was tagged in frame at the N terminus of these expression constructs for visualization. Steady state GFP fluorescence in cells growing exponentially was scored as weak (–) or strong (+) by confocal microscopy. Cadmium (50 μm CdCl2 for 2 h)-dependent enhancement (+) of GFP fluorescence was determined. B, confocal microscopy of GFP-Pca1(250–350)-CaCRP fusion in cells cultured in the absence (NT) or presence of metals (50 μm CuCl2 (Cu) or CdCl2 (Cd) for 2 h). C, determination of HA-Pca1 expression levels in cells with (+) or without (–) co-expression of the Pca1(250–350) peptide. HA-Pca1 was detected by immunoblotting with anti-HA antibodies. PGK in the same blot was probed as a loading control. A copper-sensitive Δace1 strain (24) was used for these experiments (A and B).
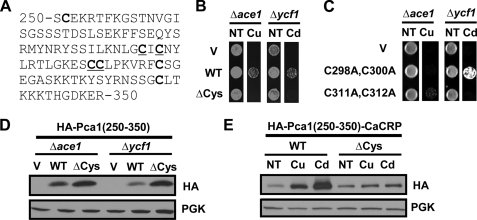
Cysteine Residues within the Pca1 Regulatory Domain Are Required for Metal Sensing—This 250–350-amino acid peptide contains seven cysteine residues (Fig. 6A). Given that cysteine is a known metal ligand, we predicted metal ion binding to this domain. A Pca1(250–350) peptide tagged with three tandem copies of HA epitope (HA-Pca1(250–350)) was expressed in Δace1 or Δycf1 strains that are sensitive to copper or cadmium, respectively (24, 25). We hypothesized that this peptide would bind and sequester metal ions like metallothionein (20), which would confer resistance to copper and cadmium. Growth assays on media containing copper or cadmium at toxic levels showed that cells expressing HA-Pca1(250–350) displayed cadmium and copper resistance compared with empty vector transformed cells (Fig. 6B). However, this peptide did not confer zinc tolerance in a zinc-sensitive strain (data not shown). These data are in accordance with previous observations in which copper and cadmium but not zinc stabilized Pca1 (Fig. 3A) and CaCRP fused with this regulatory domain (data not shown). The same peptide in which all cysteine residues are substituted to alanine (ΔCys) did not confer metal resistance (Fig. 6B), supporting roles for cysteine residues in metal binding. To ensure equal or greater levels relative to the wild type peptide (Fig. 6D), we expressed HA-Pca1(250–350)ΔCys peptide using a high copy plasmid.
FIGURE 6.
Cysteine residues within the regulatory domain of Pca1 are required for metal sensing. A, sequence of a metal-responsive degradation signal encompassing amino acids 250–350 in the N terminus of Pca1. Seven cysteine residues are in bold type. Cys298, Cys300, Cys311, and Cys312 are underlined. B, empty vector (V), a single-copy expression vector of N-terminal triple HA-tagged Pca1(250–350) (HA-Pca1(250–350), WT), and a multi-copy expression vector of HA-Pca1(250–350) with substitution of all seven cysteine residues to alanine (ΔCys) were transformed into copper-sensitive Δace1 (24) and cadmium-sensitive Δycf1 (25) strains. Cells growing exponentially were spotted on SC medium without (NT) or with supplementation of toxic copper or cadmium (50 μm CuCl2 (Cu) or CdCl2 (Cd)). Cell growth was assayed after 2 days. C, empty vector (V) and an expression vector of HA-Pca1(250–350) possessing specific cysteine mutations as indicated were transformed into cells. Cell growth was determined as described above (B). D, Western blotting of the HA-Pca1(250–350) peptides in total protein extracts of cells described above (B) with anti-HA antibodies. PGK was detected to determine equal loading. E, CaCRP fused with HA-Pca1(250–350) at the N terminus without (WT) or with site-directed mutagenesis of all seven cysteine residues to alanine (ΔCys) were expressed in a Δace1 strain. Total protein extracts of cells cultured without (NT) or with copper or cadmium (20 μm CuCl2 or CdCl2 for 1 h) were subject to Western blot analysis using anti-HA and anti-PGK antibodies.
We next examined which cysteine residues within this domain play roles for conferring resistance to cadmium and/or copper. Site-directed mutation of the CXC motif (C298A, C300A) abolished copper resistance but not cadmium resistance (Fig. 6C). Conversely, mutation of the CC motif (C311A,C312A) abolished cadmium resistance yet retained the ability to confer copper resistance (Fig. 6C). These data support distinct roles for these cysteine residues in copper or cadmium coordination.
We predicted that cysteine mutations would compromise metal-dependent masking of the degradation signal. Indeed, cadmium or copper could not stabilize CaCRP fused with HA epitope-tagged Pca1(250–350) where all seven cysteine residues were substituted to alanine (ΔCys) (Fig. 6E, upper panel, fifth and sixth lanes). However, steady state expression levels without metal addition to culture media (NT) were not significantly different between control (WT) and cysteine mutant (ΔCys) (Fig. 6E, upper panel, first lane versus fourth lane), suggesting that cysteine residues are specifically necessary for cadmium- or copper-induced stabilization but not turnover. Collectively, these data suggest that cysteine-based metal sensing within a cis-acting degradation signal serves as a molecular switch for rapid up-regulation of Pca1.
DISCUSSION
Our data reveal a new mode of expressional control of a metal-transporting P1B-type ATPase. In the absence of cadmium, Pca1 is rapidly targeted for degradation at the proteasome via an N-terminal degradation signal. However, cadmium sensing allows Pca1 to circumvent degradation, which is likely attributed to conformational changes within the degradation signal induced by metal binding. Hence, the cells are able to rapidly express Pca1 at the cell surface only when Pca1 is required for cadmium efflux.
Because cadmium is extremely toxic and cadmium levels in the environment fluctuate, transcriptional or translational regulation of a cadmium detoxification system may not be fast enough for cell protection. The constant synthesis of Pca1 would be an obvious advantage in coping with this problem. The underlying reason for rapid degradation of Pca1 could be explained by the fact that Pca1 expression is not necessary when cells are growing without cadmium stress. However, other factors may be important for keeping Pca1 levels at a minimum. For example, Pca1 could play another role other than cadmium efflux (e.g. efflux of nutritional metals), which would be deleterious in the absence of cadmium. Indeed, it was shown that cells overexpressing Pca1 grow slowly (48). If Pca1 transports nutritional metal ions because of broad substrate specificity, as shown for several other metal-transporting P-type ATPases (26–28), expression of Pca1 only when cadmium is in excess could prevent loss of essential metals that are limiting for growth. Although our previous data showed that Pca1 does not change cellular accumulation of zinc, manganese, cobalt, or iron (30), Pca1 may transport these or other metal ions under specific growth conditions. Second, it is not known what form(s) of cadmium and other metals are substrates of P1B-type ATPases. Ycf1, a vacuolar cadmium ATP-binding cassette transporter in yeast, transports both glutathione-conjugated cadmium and glutathione itself (25, 49). A recent report showed that cysteine dramatically increases the activities of CopA, ZntA, and HMA2 P1B-type ATPases (50). Pca1 may extrude cysteine, glutathione, or other ions such as sulfur with or without conjugation of cadmium. If so, constitutive extrusion of these cellular factors would be deleterious, because they are critical for redox homeostasis and metabolism (51, 52). Alternatively, cysteine or other cytosolic molecules may activate Pca1 as shown for ZntA and HMA2 (50), resulting in constitutive ATP hydrolysis in the absence of cadmium.
Internalization followed by degradation at the vacuole is a major pathway for turnover of plasma membrane proteins. For example, Zrt1 zinc transporter, Fet3/Ftr1 iron transporter complex, and Smf1 manganese transporter are subject to this mode of post-translational regulation (40, 44, 53, 54). Given that these transporters play roles in metal ion uptake, they are down-regulated when cells are exposed to excess metal ions. Substrate-dependent mono or poly-ubiquitination of membrane transporters is often a signal that triggers turnover of these transporters by endocytosis followed by vacuolar degradation (40). However, Pca1 regulation is distinct when compared with other metal ion transporters in that it is rapidly up-regulated by substrate-dependent rescuing from degradation. Anti-ubiquitin immunoblotting suggests that Pca1 is poly-ubiquitinated to multiple species in the absence of cadmium. Because a stable Pca1 deleted of the first 392 amino acids is not ubiquitinated, this modification is likely the signal for degradation. However, given the fact that Pca1 functions at the plasma membrane to extrude cadmium, proteasome-dependent turnover of Pca1 is a unique example. To the best of our knowledge, HMG-CoA reductase, which resides within the endoplasmic reticulum membrane is the only known polytopic membrane protein that is degraded by the proteasome in a regulated manner (75). Cellular sterol metabolic status regulates the turnover of this rate-limiting enzyme of ergosterol synthesis (75). In addition, several mutated polytopic membrane proteins are removed from the endoplasmic reticulum via the endoplasmic reticulum-associated degradation system in which the proteasome is involved (76). This mechanism of turnover is known as a protein quality control pathway that plays an important role for the elimination of terminally misfolded proteins. However, Pca1 is a naturally expressed and functional protein that is regulated according to cellular needs. It is interesting to note that cellular trafficking of Arn1, a transporter of an iron-siderophore complex, is regulated by its substrate. Intracellular sensing triggers plasma membrane sorting of Arn1, and its cycling between the plasma membrane and endosomes is induced by substrate binding at the cell surface (55, 56). A similar mechanism may control the sorting of Pca1 to the plasma membrane after substrate sensing. We are currently pursuing the identification of trans-acting molecular factors involved in this process.
Most P1B-type ATPases possess N-terminal metal-binding domains (MBDs) that are thought to play roles in catalysis and/or regulation. The conserved MBD (∼70 amino acids, βαββαβ fold) contains a GMXCXXC sequence in which two cysteine residues coordinate a metal ion (3, 29). Although this domain is conserved in many P1B-type ATPases, the numbers of it vary, ranging from one to six. The N terminus of Pca1 contains only one conserved MBD in which two cysteine residues are essential for function (30). Human copper transporting ATPases possess six MBDs at the N terminus. The Atx1 metallochaperone delivers copper through direct docking to MBDs with different preferences among them (57–59), suggesting distinct functions of these domains. The structure and metal coordination of MBDs are relatively well characterized; however, the roles for these domains in ATPase function are not yet clear. Copper binding was initially proposed as a substrate recognition step followed by transfer of metal to the translocation pore. However, site-directed mutagenesis and deletion of MBDs suggest a regulatory role. For example, the fifth or sixth MBD of human ATP7a is necessary and sufficient for copper-induced redistribution (60). An inhibitory role of the N terminus of a copper transporting ATPase has been proposed as well (61). Interestingly, a recent report (62) suggested the presence of either additional regulatory copper-binding site(s) within the protein or another unknown mechanism for copper sensing. Collectively, whereas significant progress in the characterization of conserved MBDs in N terminus of P1B-type ATPases has been made, the functional roles of these domains remain elusive.
We have identified a distinct MBD in the N terminus of Pca1 that is necessary for extremely efficient control of Pca1 expression in response to cadmium. Several atypical MBDs have been identified in other P1B-type ATPases as well. The core sequence of N-terminal MBDs of Arabidopsis thaliana HMA2–4 is substituted by a degenerate sequence, GICC(T/S)SE (63). HMA2 and HMA4 zinc ATPases possess C-terminal MBDs (several di-cysteine pairs and an 11-histidine stretch) that regulate enzyme activities (63, 64). A CHHC predicted metal-binding sequence was identified at the C terminus of the CopA copper ATPase in Archaeoglobus fulgidus (65). The histidine-rich N-terminal metal-binding domain of the CopB copper ATPase in A. fulgidus appears to have a regulatory role affecting metal transport rates by controlling metal release (66). In addition to heavy metal-transporting P1B-type ATPases, it was also shown that Ca2+-transporting P-type ATPases carry inhibitory domains (67). Cytosolic regulators and phosphorylation of C-terminal residues control activities of H+-transporting ATPases in plants (68). Hence, auto-regulatory mechanisms may be widespread in P-type ATPases.
We propose that direct metal binding to the Pca1(250–350) domain induces conformational changes that would mask a signal recognized by the protein degradation machinery. Expression of Pca1(250–350) itself confers both cadmium and copper tolerance, which supports the conclusion that this domain directly binds metals. Substitution of cysteine residues within this domain abolished metal resistance, suggesting a role for cysteines in metal coordination. Site-directed mutation of cysteine residues suggests that binding site(s) of cadmium and copper do not overlap directly. Given that Pca1 does not transport copper (30), the physiological significance of the stabilization of Pca1 by copper is not obvious at this moment. It is necessary to point out that in a copper-sensitive Δace1 strain, Pca1 stabilization by copper is observed at similar concentrations to that of cadmium; however, in wild type cells a relatively high copper concentration (∼2 mm) is required for Pca1 stabilization (data not shown). CUP1- and CRS5-encoded metal chelators that are up-regulated by Ace1 likely have higher affinity to copper than the N terminus of Pca1.
Fusion of the Pca1(250–350) domain with a stable protein leads to rapid turnover and metal-dependent stabilization, demonstrating a role as an autonomous and metal-responsive degradation signal. However, this domain does not carry significant sequence identity with characterized degradation signals or known proteins. Given that Pca1(250–350) is located in the middle of the N terminus, the N-end rule pathway of protein degradation is not likely involved in Pca1 degradation (69). Pro, Glu, Ser, and Thr amino acid-enriched sequence (PEST sequence) is a proteolytic signal (70). However, Pca1 250–350 is not predicted to contain a PEST signal. It has been proposed that exposure of hydrophobic residues could recruit molecular chaperones, which results in refolding to a functional protein or targeting for degradation. Structural characterization of this domain in apo and cadmium-bound forms would reveal what characteristics lead to degradation of Pca1 and how metal binding masks this degradation signal. We are currently pursuing characterizations of structure and metal binding of this domain in detail; however, purification of ample quantities of the Pca1(250–350) peptide has been a technical challenge.3
It is interesting to note that the N terminus of a human copper ATPase interacts with several proteins, although the physiological significance of these interactions remains to be elucidated. For example, MURR1/COMMD1 binds with the N terminus of ATP7b, which decreases the stability of newly synthesized ATP7b (71). A hepatocytic isoform of the PLZF (promyelocytic leukemia zinc finger) protein interacts with the C terminus of ATP7b, which positively regulates extracellular signal-regulated kinase (ERK) signal transduction (72). Glutaredoxin interacts with the N terminus of ATP7a and ATP7b (76), and the dynactin subunit p62 interacts with the N terminus of ATP7b (73). These protein-protein interactions may play critical roles for the function, regulation of catalytic activities, and/or subcellular trafficking of ATP7b and possibly other P1B-type ATPases. The regulatory domain identified at the N terminus of Pca1 may interact with accessory protein(s) that target Pca1 to the ubiquitination-proteasome pathway.
Acknowledgments
We thank Drs. D. Kornitzer, C. Philpott, B. André, and D. Wolf for providing CaCRP plasmid, the end4-1, npi1, and pre1-1 pre2-1 yeast strains, respectively. We also thank Lee lab members, including H. Kim, W. Wei, A. Adle, J. Bies, L. Volentine, N. Smith, and X. Wu, for technical assistance and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK79209 (to J. L.) and P20RR-17657 (to the Nebraska Redox Biology Center). This work was also supported by funds provided through the Hatch Act in the University of Nebraska Research Division. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SC, synthetic complete; GFP, green fluorescent protein; HA, hemagglutinin; PBS, phosphate-buffered saline; PGK, phosphoglycerate kinase; E3, ubiquitin-protein isopeptide ligase; WT, wild type; MBD, metal-binding domain.
D. J. Adle and J. Lee, unpublished data.
References
- 1.De Domenico, I., McVey Ward, D., and Kaplan, J. (2008) Nat. Rev. Mol. Cell. Biol. 9 72–81 [DOI] [PubMed] [Google Scholar]
- 2.Tao, T. Y., and Gitlin, J. D. (2003) Hepatology 37 1241–1247 [DOI] [PubMed] [Google Scholar]
- 3.Lutsenko, S., Barnes, N. L., Bartee, M. Y., and Dmitriev, O. Y. (2007) Physiol. Rev. 87 1011–1046 [DOI] [PubMed] [Google Scholar]
- 4.Jarup, L., Berglund, M., Elinder, C. G., Nordberg, G., and Vahter, M. (1998) Scand. J. Work Environ. Health 24 (Suppl. 1) 1–51 [PubMed] [Google Scholar]
- 5.Agency for Toxic Substance and Disease Registry (1999) Toxicological Profile for Cadmium CAS Number 7440-43-9 [PubMed]
- 6.Chaney, R. L., Malik, M., Li, Y. M., Brown, S. L., Brewer, E. P., Angle, J. S., and Baker, A. J. (1997) Curr. Opin. Biotechnol. 8 279–284 [DOI] [PubMed] [Google Scholar]
- 7.Satarug, S., and Moore, M. R. (2004) Environ. Health Perspect. 112 1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alloway, B. J., Jackson, A. P., and Morgan, H. (1990) Sci. Total Environ. 91 223–236 [DOI] [PubMed] [Google Scholar]
- 9.Ercal, N., Gurer-Orhan, H., and Aykin-Burns, N. (2001) Curr. Top. Med. Chem. 1 529–539 [DOI] [PubMed] [Google Scholar]
- 10.Brennan, R. J., and Schiestl, R. H. (1996) Mutat. Res. 356 171–178 [DOI] [PubMed] [Google Scholar]
- 11.Jin, Y. H., Clark, A. B., Slebos, R. J., Al-Refai, H., Taylor, J. A., Kunkel, T. A., Resnick, M. A., and Gordenin, D. A. (2003) Nat. Genet. 34 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henson, M. C., and Chedrese, P. J. (2004) Exp. Biol. Med. (Maywood) 229 383–392 [DOI] [PubMed] [Google Scholar]
- 13.Johnson, M. D., Kenney, N., Stoica, A., Hilakivi-Clarke, L., Singh, B., Chepko, G., Clarke, R., Sholler, P. F., Lirio, A. A., Foss, C., Reiter, R., Trock, B., Paik, S., and Martin, M. B. (2003) Nat. Med. 9 1081–1084 [DOI] [PubMed] [Google Scholar]
- 14.Safe, S. (2003) Nat. Med. 9 1000–1001 [DOI] [PubMed] [Google Scholar]
- 15.Bressler, J. P., Olivi, L., Cheong, J. H., Kim, Y., and Bannona, D. (2004) Ann. N. Y. Acad. Sci. 1012 142–152 [DOI] [PubMed] [Google Scholar]
- 16.Dalton, T. P., He, L., Wang, B., Miller, M. L., Jin, L., Stringer, K. F., Chang, X., Baxter, C. S., and Nebert, D. W. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3401–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes, D. S., Fragoso, L. C., Riger, C. J., Panek, A. D., and Eleutherio, E. C. (2002) Biochim. Biophys. Acta 1573 21–25 [DOI] [PubMed] [Google Scholar]
- 18.Leslie, E. M., Liu, J., Klaassen, C. D., and Waalkes, M. P. (2006) Mol. Pharmacol. 69 629–639 [DOI] [PubMed] [Google Scholar]
- 19.Perfus-Barbeoch, L., Leonhardt, N., Vavasseur, A., and Forestier, C. (2002) Plant J. 32 539–548 [DOI] [PubMed] [Google Scholar]
- 20.Klaassen, C. D., Liu, J., and Choudhuri, S. (1999) Annu. Rev. Pharmacol. Toxicol. 39 267–294 [DOI] [PubMed] [Google Scholar]
- 21.Cobbett, C., and Goldsbrough, P. (2002) Annu. Rev. Plant. Biol. 53 159–182 [DOI] [PubMed] [Google Scholar]
- 22.Singhal, R. K., Anderson, M. E., and Meister, A. (1987) FASEB J. 1 220–223 [DOI] [PubMed] [Google Scholar]
- 23.Wimmer, U., Wang, Y., Georgiev, O., and Schaffner, W. (2005) Nucleic Acids Res. 33 5715–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, P., and Thiele, D. J. (1993) Biofactors 4 105–115 [PubMed] [Google Scholar]
- 25.Li, Z. S., Lu, Y. P., Zhen, R. G., Szczypka, M., Thiele, D. J., and Rea, P. A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silver, S., and Phung, L. T. (1996) Annu. Rev. Microbiol. 50 753–789 [DOI] [PubMed] [Google Scholar]
- 27.Nies, D. H. (1999) Appl. Microbiol. Biotechnol. 51 730–750 [DOI] [PubMed] [Google Scholar]
- 28.Williams, L. E., and Mills, R. F. (2005) Trends Plant. Sci. 10 491–502 [DOI] [PubMed] [Google Scholar]
- 29.Lutsenko, S., and Petris, M. J. (2003) J. Membr. Biol. 191 1–12 [DOI] [PubMed] [Google Scholar]
- 30.Adle, D. J., Sinani, D., Kim, H., and Lee, J. (2007) J. Biol. Chem. 282 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raths, S., Rohrer, J., Crausaz, F., and Riezman, H. (1993) J. Cell Biol. 120 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springael, J. Y., and Andre, B. (1998) Mol. Biol. Cell. 9 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinemeyer, W., Gruhler, A., Mohrle, V., Mahe, Y., and Wolf, D. H. (1993) J. Biol. Chem. 268 5115–5120 [PubMed] [Google Scholar]
- 34.Fleming, J. A., Lightcap, E. S., Sadis, S., Thoroddsen, V., Bulawa, C. E., and Blackman, R. K. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mumberg, D., Muller, R., and Funk, M. (1995) Gene (Amst.) 156 119–122 [DOI] [PubMed] [Google Scholar]
- 36.Gilon, T., Chomsky, O., and Kulka, R. G. (1998) EMBO J. 17 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overton, M. C., and Blumer, K. J. (2002) J. Biol. Chem. 277 41463–41472 [DOI] [PubMed] [Google Scholar]
- 38.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman J. G., Smith, J. A., and Struhl, K. (1987) Current Protocols in Molecular Biology, Greene Publishing Associates and Wiley-Interscience, New York
- 39.Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995) Yeast 11 355–360 [DOI] [PubMed] [Google Scholar]
- 40.Horák, J. (2003) Biochim. Biophys. Acta 1614 139–155 [DOI] [PubMed] [Google Scholar]
- 41.Lemmon, S. K., and Traub, L. M. (2000) Curr. Opin. Cell Biol. 12 457–466 [DOI] [PubMed] [Google Scholar]
- 42.Roberg, K. J., Rowley, N., and Kaiser, C. A. (1997) J. Cell Biol. 137 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ammerer, G., Hunter, C. P., Rothman, J. H., Saari, G. C., Valls, L. A., and Stevens, T. H. (1986) Mol. Cell. Biol. 6 2490–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felice, M. R., De Domenico, I., Li, L., Ward, D. M., Bartok, B., Musci, G., and Kaplan, J. (2005) J. Biol. Chem. 280 22181–22190 [DOI] [PubMed] [Google Scholar]
- 45.Zhang, F., Su, K., Yang, X., Bowe, D. B., Paterson, A. J., and Kudlow, J. E. (2003) Cell 115 715–725 [DOI] [PubMed] [Google Scholar]
- 46.Weissman, Z., Berdicevsky, I., Cavari, B. Z., and Kornitzer, D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3520–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riggle, P. J., and Kumamoto, C. A. (2000) J. Bacteriol. 182 4899–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rad, M. R., Kirchrath, L., and Hollenberg, C. P. (1994) Yeast 9 1217–1225 [DOI] [PubMed] [Google Scholar]
- 49.Rebbeor, J. F., Connolly, G. C., Dumont, M. E., and Ballatori, N. (1998) J. Biol. Chem. 273 33449–33454 [DOI] [PubMed] [Google Scholar]
- 50.Yang, Y., Mandal, A. K., Bredeston, L. M., González-Flecha, F. L., and Argüello, J. M. (2007) Biochim. Biophys. Acta 1768 495–501 [DOI] [PubMed] [Google Scholar]
- 51.Vido, K., Spector, D., Lagniel, G., Lopez, S., Toledano, M. B., and Labarre, J. (2001) J. Biol. Chem. 276 8469–8474 [DOI] [PubMed] [Google Scholar]
- 52.Fauchon, M., Lagniel, G., Aude, J. C., Lombardia, L., Soularue, P., Petat, C., Marguerie, G., Sentenac, A., Werner, M., and Labarre, J. (2002) Mol. Cell 9 713–723 [DOI] [PubMed] [Google Scholar]
- 53.Gitan, R. S., Shababi, M., Kramer, M., and Eide, D. J. (2003) J. Biol. Chem. 278 39558–39564 [DOI] [PubMed] [Google Scholar]
- 54.Liu, X. F., and Culotta, V. C. (1999) J. Biol. Chem. 274 4863–4868 [DOI] [PubMed] [Google Scholar]
- 55.Kim, Y, Lampert, S. M., and Philpott, C. C. (2005) EMBO J. 24 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim, Y., Yun, C. W., and Philpott, C. C. (2002) EMBO J. 21 3632–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker, J. M., Tsivkovskii, R., and Lutsenko, S. (2002) J. Biol. Chem. 277 27953–27959 [DOI] [PubMed] [Google Scholar]
- 58.Walker, J. M., Huster, D., Ralle, M., Morgan, C. T., Blackburn, N. J., and Lutsenko, S. (2004) J. Biol. Chem. 279 15376–15384 [DOI] [PubMed] [Google Scholar]
- 59.Achila, D., Banci, L., Bertini, I., Bunce, J., Ciofi-Baffoni, S., and Huffman, D. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5729–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strausak, D., La Fontaine, S., Hill, J., Firth, S. D., Lockhart, P. J., and Mercer, J. F. (1999) J. Biol. Chem. 274 11170–11177 [DOI] [PubMed] [Google Scholar]
- 61.Huster, D., and Lutsenko, S. (2003) J. Biol. Chem. 278 32212–32218 [DOI] [PubMed] [Google Scholar]
- 62.Cater, M. A., La Fontaine, S., and Mercer, J. F. (2007) Biochem. J. 401 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verret, F., Gravot, A., Auroy, P., Preveral, S., Forestier, C., Vavasseur, A., and Richaud, P. (2005) FEBS Lett. 579 1515–1522 [DOI] [PubMed] [Google Scholar]
- 64.Eren, E., Kennedy, D. C., Maroney, M. J., and Argüello, J. M. (2006) J. Biol. Chem. 281 33881–33891 [DOI] [PubMed] [Google Scholar]
- 65.Mandal, A. K., and Argüello, J. M. (2003) Biochemistry 42 11040–11047 [DOI] [PubMed] [Google Scholar]
- 66.Mana-Capelli S., Mandal, A. K., and Argüello, J. M. (2003) J. Biol. Chem. 278 40534–40541 [DOI] [PubMed] [Google Scholar]
- 67.Toyoshima, C., and Inesi, G. (2004) Annu. Rev. Biochem. 73 269–292 [DOI] [PubMed] [Google Scholar]
- 68.Portillo, F. (2000) Biochim. Biophys. Acta 1469 31–42 [DOI] [PubMed] [Google Scholar]
- 69.Varshavsky, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rechsteiner, M., and Rogers, S. W. (1996) Trends Biochem. Sci. 21 267–271 [PubMed] [Google Scholar]
- 71.De Bie, P., van de Sluis, B., Burstein, E., van de Berghe, P. V., Muller, P., Berger, R., Gitlin, J. D., Wijmenga, C., and Klomp, L. W. (2007) Gastroenterology 133 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ko, J. H., Son, W., Bae, G. Y., Kang, J. H., Oh, W., and Yoo, O. J. (2006) J. Cell. Biochem. 99 719–734 [DOI] [PubMed] [Google Scholar]
- 73.Lim, C. M., Cater, M. A., Mercer, J. F., and La Fontaine, S. (2006) Biochem. Biophys. Res. Commun. 348 428–436 [DOI] [PubMed] [Google Scholar]
- 74.Babst, M., Sato, T. K., Banta, L. M., and Emr, S. D. (1997) EMBO J. 16 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hampton, R. Y. (2002) Annu. Rev. Cell Dev. Biol. 18 345–378 [DOI] [PubMed] [Google Scholar]
- 76.Sayeed, A., and Ng, D. T. (2005) Crit. Rev. Biochem. Mol. Biol. 40 75–91 [DOI] [PubMed] [Google Scholar]