Abstract
The structures of mitochondrial transporters and uncoupling proteins are 3-fold pseudosymmetrical, but their substrates and coupling ions are not. Thus, deviations from symmetry are to be expected in the substrate and ion-binding sites in the central aqueous cavity. By analyzing the 3-fold pseudosymmetrical repeats from which their sequences are made, conserved asymmetric residues were found to cluster in a region of the central cavity identified previously as the common substrate-binding site. Conserved symmetrical residues required for the transport mechanism were found at the water–membrane interfaces, and they include the three PX[DE]XX[RK] motifs, which form a salt bridge network on the matrix side of the cavity when the substrate-binding site is open to the mitochondrial intermembrane space. Symmetrical residues in three [FY][DE]XX[RK] motifs are on the cytoplasmic side of the cavity and could form a salt bridge network when the substrate-binding site is accessible from the mitochondrial matrix. It is proposed that the opening and closing of the carrier may be coupled to the disruption and formation of the 2 salt bridge networks via a 3-fold rotary twist induced by substrate binding. The interaction energies of the networks allow members of the transporter family to be classified as strict exchangers or uniporters.
Keywords: membrane proteins, substrate binding, amino acid sequence analysis, salt bridge networks, conformational changes
The members of the mitochondrial transporter or carrier family translocate nucleotides, amino acids, inorganic ions, keto acids, and vitamins across the mitochondrial inner membrane (1, 2). Uncoupling proteins, which generate heat by dissipating the proton electrochemical gradient, also belong to the family (3–5). The amino acid sequences have 3 homologous repeats (6) and a structure with pseudosymmetry (7). Each repeat is folded into 2 transmembrane α-helices linked by a short α-helix on the matrix side (8) and contains the signature motif PX[DE]XX[RK] (9). The proline residues kink the odd-numbered transmembrane α-helices H1, H3, and H5, and the charged residues form a salt-bridge network connecting the C-terminal end of the transmembrane α-helices, closing the transporter on the matrix side (8). During the transport cycle, the carriers form the cytoplasmic and matrix states in which the substrate-binding site of the carrier is open to the mitochondrial intermembrane space and matrix, respectively (10). According to the single binding center–gating pore mechanism, interconversion of the 2 conformational states via a transition intermediate leads to substrate translocation (11). In agreement, in the cytoplasmic state, a central substrate-binding site has been identified by applying chemical and distance constraints to comparative models (12, 13). The substrates bind to 3 major sites on the even-numbered α-helices, which are related by symmetry and located approximately in the middle of the membrane. In molecular dynamics simulations, ADP binds to the ADP/ATP carrier at the common substrate-binding site (14). The structural changes required for the translocation of the substrate are unknown, but docking of the substrate could disrupt the matrix salt bridge network, allowing the transporter to convert to the matrix state (12, 13). The yeast ADP/ATP carriers function as monomers (15), and other mitochondrial transporters are likely to operate in the same way.
Residues that are important for the transport mechanism are likely to be symmetrical, whereas residues involved in substrate binding will be asymmetrical reflecting the asymmetry of the substrates. By scoring the symmetry of residues in the sequence repeats, we have identified the substrate-binding sites and salt bridge networks that are important for the transport mechanism in family members. No preexisting molecular structure is required for the analysis, because only amino acid sequences are used. The symmetry analysis provides an assessment of the role of residues irrespective of the conformational state. The analysis also provides clues to the chemical identities of substrates of uncharacterized transporters.
Results
A symmetry score was developed to quantify the degree of similarity between symmetry-related residues (Fig. 1). The analysis was performed on subfamilies of mitochondrial transporters from fungi and metazoans, including uncoupling proteins and the transporters of CoA, Mg2+-ATP/Pi, thiamine pyrophosphate, ADP/ATP, pyrimidine nucleotides, NAD+, flavin nucleotides, peroxisomal adenine nucleotides, dicarboxylates, malate/oxoglutarate, oxaloacetate, citrate, succinate/fumarate, oxodicarboxylates, aspartate/glutamate, glutamate, S-adenosyl-methionine, ornithine, carnitine/acylcarnitine, phosphate, and GTP/GDP, and on the uncharacterized subfamilies SLC25A42, YPR011c, SLC25A43, YFR045w, YMR166c, Mtm1p, YDL119c, Ymc1p, AU042651, SLC25A45, C14ORF68, CACTL, Mrs3p, SLC25A44, and Yhm2p [references in refs. 1 and 2 or supporting information (SI) Text in SI Appendix]. The results for the fungal phosphate transporters (16, 17) and mammalian uncoupling proteins (3–5) are discussed as examples of the analysis.
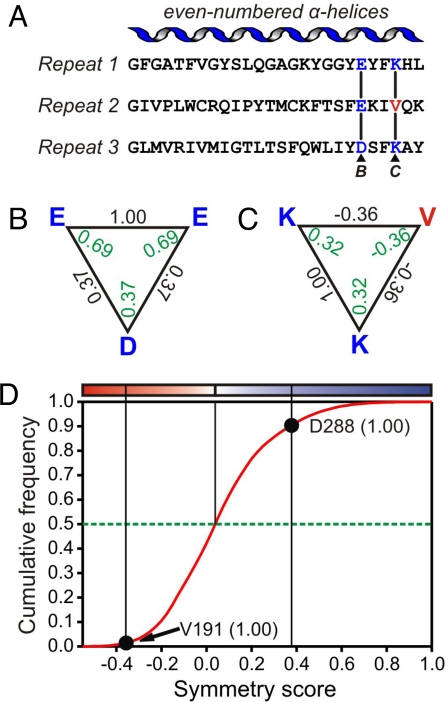
Fig. 1.
Scoring of symmetry in the yeast mitochondrial phosphate transporter ScPic2p. (A) The alignment of the even-numbered α-helices in ScPic2p. Highlighted in color are 2 examples of symmetry-related triplets of residues, as used in B and C. (B and C) The replacement scores (black) for each comparison in the triplet were obtained from the residue replace ability matrix (55). The symmetry score of a residue (green) is the average of the 2 replacement scores. (B) Triplet E-E-D with symmetric residues. (C) Triplet K-V-K with an asymmetric valine residue. (D) The cumulative frequency distribution of the symmetry scores for all possible triplets in the sequences of the subfamily of fungal Pic2p transporters. The symmetry scores are defined by a red-white-blue color scale, in which the median of the distribution is white and the minimal and maximal symmetry scores red and blue, respectively (Fig. 2 A and B). The symmetry scores of D288 (B) and V191 (C) (conservation scores in brackets) are shown.
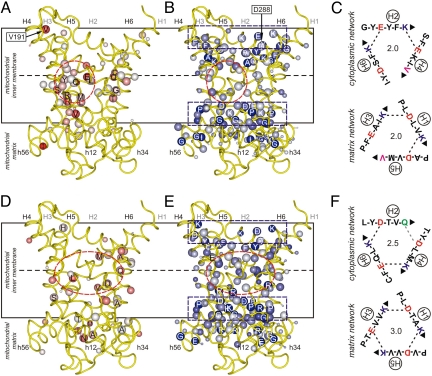
The majority of the conserved asymmetric residues in the fungal phosphate transporters cluster in a small region of the cavity corresponding to the middle of the membrane (Fig. 2A). The highly asymmetric residues Q86 and R175/Q176 in ScPic2p form contact points I and II of the common substrate-binding site, respectively (12). Asymmetric R272 is 1 α-helical turn away from the common contact point III (12). Thus, the interaction of the substrate with H6 may occur at a different position, as an adaptation to the size of the substrate. Upon binding of phosphate, the positively charged residues R175 and R272 could neutralize negative charges of phosphate and Q176 could form a hydrogen bond. The nearby asymmetric residue, E120, may be protonated to drive the import of phosphate against the membrane potential by cotransport with a proton. Another possible site for proton coupling is the asymmetric H33. The equivalent residues in the ortholog ScMir1p are essential for function (18).
Fig. 2.
Asymmetry and symmetry in fungal phosphate transporters and mammalian uncoupling proteins. Average symmetry and conservation scores in the subfamily of (A and B) the phosphate transporters and (D and E) the uncoupling proteins projected onto a model of the bovine ADP/ATP transporter (8). The positive conservation score and average symmetry score are represented by the size and color of the Cβ atom, respectively (Fig. 1D). (A and D) Asymmetric residues with negative average symmetry scores. Residues in the cavity are labeled. (B and E) Symmetric residues with positive average symmetry scores. Highly symmetric residues are labeled. Dashed circles and rectangles indicate the location of the substrate binding site and networks, respectively. Green spheres represent residues that are absent from the repeat. Also shown are V191 and D288 (Fig. 1D). For all analyzed sequences the average of the standard deviations for all of the average symmetry scores of the cavity residues was 0.10. (C and F) The residues of the cytoplasmic (Upper) and matrix network (Lower) in ScPic2p and HsUCP1, respectively. The positively and negatively charged residues of the salt bridges are shown in blue and red, respectively. Deviating polar, aromatic and apolar residues are shown in green, orange and pink, respectively. The interaction energies of the network are quantified as the number of salt bridges, taking hydrogen bonds and cation-π interactions as half the interaction energy of a salt bridge, and van der Waals interactions as negligible.
The uncoupling proteins have been proposed to transport protons (19) or fatty acids (20). The symmetry analysis shows they are highly symmetrical and have few conserved asymmetric residues in the cavity (Fig. 2D). In HsUCP1, the most noteworthy residues are the asymmetric Q85 and D28 (Fig. 2D) and the symmetric R84, R183 and R277 (Fig. 2E) at the contact points of the common substrate-binding site (12, 13). The asymmetric D28 is in the vicinity of the substrate-binding site and is involved in proton transport (21), whereas E191 (Fig. S4 in SI Appendix), which is further away, is involved in the pH control of nucleotide binding (22). The symmetry-related arginine residues are found also in the dicarboxylate (Fig. S5 in SI Appendix) and oxoglutarate transporters (Fig. S6 in SI Appendix). There are no patches of hydrophobic asymmetric residues in the cavities, such as those observed in carnitine/acylcarnitine transporters (Fig. S7 in SI Appendix), indicating that the activating fatty acids are not the intended substrates. Nor are there widespread asymmetric adaptations, as found in the nucleotide-binding sites of the ADP/ATP transporters (Fig. S8 in SI Appendix) and GTP/GDP transporters (Fig. S9 in SI Appendix). The positively charged arginine residues in the binding site could attract the negatively charged phosphate moieties of nucleotides or carboxylates of fatty acids nonspecifically. The lack of specific adaptations for the binding of nucleotides could explain why there is no specificity on the bases in binding. On this basis, the intended substrates of uncoupling proteins are likely to be small carboxylic or keto acids, transported in symport with protons.
Conserved and highly symmetric residues are found consistently on the matrix and cytoplasmic sides of the transporters at the water–membrane interface (Fig. 2 B and E and Figs. S1–S41 in SI Appendix).
Discussion
Location of the Substrate-Binding Sites.
The vast majority of conserved asymmetric residues cluster in the cavities approximately at the middle of the membrane, confirming the location of the common substrate-binding site for nucleotides, cofactors, inorganic ions, keto acids, and amino acids (Fig. S5–S41 in SI Appendix). The asymmetric residues in the substrate binding site fall into 3 categories that reflect their functional properties: (i) conserved residues interacting directly with the functional groups of the asymmetric substrate, (ii) residues under evolutionary pressure to maintain a neutral character so as to not interfere with substrate binding, and (iii) conserved residues involved in ion and proton coupling. The substrate-binding site contains also conserved symmetric residues that form contacts between the substrate and the even-numbered α-helices to couple the binding of the substrate to a symmetrical mechanism. The conserved asymmetric and symmetric residues form a single cluster, indicating that substrate binding occurs at a similar location in both conformational states, which is consistent with the single-binding center-gating pore model (11). There is a qualitative trend between the size of the substrate and the size of the cluster of asymmetric residues. For example, the mitochondrial dicarboxylate, phosphate, and oxaloacetate transporters have small clusters of asymmetric residues (Figs. S5, S10, and S11 in SI Appendix), whereas the mitochondrial ADP/ATP and GTP/GDP transporters have large ones (Figs. S8 and S9 in SI Appendix). The analysis confirms that neutralization of the negatively charged substrate by the protonated side chains of arginine or lysine residues is an important property of the substrate binding site (12, 13). In addition, negatively charged asymmetric residues are often present in the binding site, and they are likely to be involved in proton and ion coupling, as found in the uncoupling proteins (Fig. 2D) and the GTP/GDP, phosphate, aspartate/glutamate, oxodicarboxylate, Mg2+-ATP/Pi, ornithine, succinate/fumarate, and citrate transporters (Figs. S9, S10, and S12–S17 in SI Appendix) and the uncharacterized ScMrs3p-like transporters and ScYFR045w (Figs. S18 and S19 in SI Appendix).
Mitochondrial transporters have very few conserved asymmetric residues either on the outside facing the membrane or on the matrix α-helices that could form a plausible asymmetric protein–protein interface, as required for homodimerization of symmetrical proteins (Fig. 2 A and D and Figs. S5–S41 in SI Appendix). Thus, all mitochondrial transporters must be monomeric in structure and function.
Cytoplasmic and Matrix Network.
Triplets of symmetry-related residues with a high level of symmetry and conservation define members of the mitochondrial transporter family, and they are likely to be important for a common mechanism of transport (Fig. S1–S3 and SI Text in SI Appendix). There are 2 clusters of conserved and highly symmetric residues, which flank the common substrate binding site. The first cluster is found on the odd-numbered α-helices on the matrix side of the transporters, consisting of the PX[DE]XX[RK] motif (Fig. 2 C and F Lower and Fig. S1 in SI Appendix). As shown previously, the charged residues form a salt bridge network when the carrier is in the cytoplasmic state (8, 9). The second cluster has the consensus [FY][DE]XX[RK] motif and is found on the even-numbered α-helices on the cytoplasmic side (Fig. 2 C and F Upper and Fig. S3 in SI Appendix). The charged residues of this motif have no obvious function in the cytoplasmic state (8), but we propose they come together to form a salt bridge network when the transporter is in the matrix state with the substrate-binding site open to the mitochondrial matrix. According to their location, the salt bridge networks will be referred to as the matrix and cytoplasmic networks (Fig. 3). During the transport cycle, the matrix and cytoplasmic networks would form and disrupt to open and close the cavity to the mitochondrial matrix and the intermembrane space in an alternating fashion. Several observations support the existence of the cytoplasmic network. First, the residues are highly conserved throughout the mitochondrial transporter family (Fig. S3 in SI Appendix). Generally, charged or polar residues forming salt bridges or hydrogen bonds in the networks are strictly conserved, but hydrophobic substitutions, which break them, are not. Second, the residues are likely to be positioned at the water–membrane interface, as are the residues of the matrix network (Fig. 3). The networks would pose a barrier to protons and other ions at the interface, preventing the dissipation of the electrochemical gradients during the transport cycle. Third, the residues of the networks are sensitive to mutation and modification. Mutant oxoglutarate transporters, in which residues S203, E301, and N304 of the cytoplasmic network were replaced by cysteine, had transport activities of 15%, 3%, and 1% of the wild-type transporter, respectively (23). Modification by sulfhydryl reagents of single cysteine replacements in the carboxyl ends of the even-numbered α-helices render the transporter dysfunctional (23), suggesting they prevent formation of the network and the closure of the transporter. The mutations E95Q, K98A, E192Q, S195A, and K295A of the phosphate transporter ScMir1p (Fig. S10 in SI Appendix) reduced the transport activity to 72%, 3%, 39%, 3%, and 33% of the wild-type activity, showing that apolar substitutions render the transporter dysfunctional, whereas polar substitutions are less severe (18). Notably, mutations need not have a strong effect, because they may only change the transport mode from exchanger to uniporter or vice versa (see below). Fourth, the E601K mutation in the human aspartate/glutamate transporter HsAGC2 alters the cytoplasmic network on H6 and causes type II citrullinemia (24). The formation and disruption of the networks agrees fully with the accessibility and gating aspects of the single-binding center-gating pore model (11).
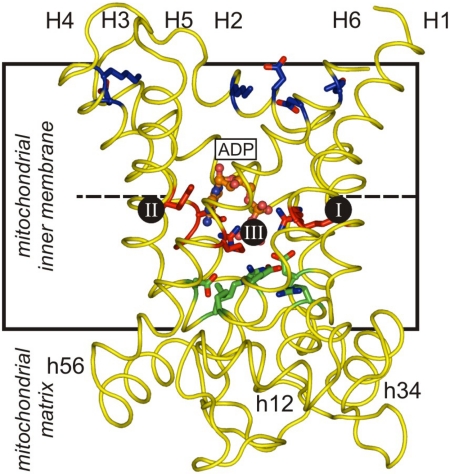
Fig. 3.
The substrate-binding site and salt bridge networks of the mitochondrial ADP/ATP carrier. An ADP molecule and a comparative model of the yeast ADP/ATP carrier ScAac2p (12) are shown in ball/stick and diagram representation, respectively. The residues involved in substrate binding are shown in red and the contact points of the common substrate-binding site are indicated by black circles and Roman numerals (12). The residues of the matrix and the cytoplasmic network are shown in green and blue, respectively.
Trigger for Substrate Translocation.
The networks pose an initial activation energy barrier that must be overcome by substrate binding to trigger the transport cycle. When a substrate enters the cavity from the intermembrane space and binds to the major contact points on the even-numbered α-helices, they might be pulled together to minimize bond distances, initiating the disruption of the matrix network, the transition to the matrix state, and the formation of the cytoplasmic network. When a substrate is translocated from the matrix to the cytoplasm, these events are reversed. Hydrophobic, aromatic, or polar residues in the networks lower the energy barrier for the translocation of substrates with low binding energies, as is the case for dicarboxylates, phosphate and some nucleotides (25). The phosphate anion has 2 negative charges for electrostatic interactions. Both networks of the phosphate transporter ScPic2p have a valine rather than an arginine or lysine residue (Fig. 2 A and C), so only 2 salt bridges remain. The binding of phosphate will provide sufficient energy to break the networks to trigger the structural changes required for the transition to the alternative conformational state. The observation that both networks in the phosphate transporter contain the conserved deviation provides further support for the formation of the 2 networks.
Strict Equimolar Exchange of Substrates.
In most transporters, the cytoplasmic and matrix networks are complete, but in some, 1 or more salt bridges with interaction energies of 20–40 kJ/mol are replaced by hydrogen bond, cation-π or van der Waals interactions (Figs. S5–S41 in SI Appendix), which have interaction energies of 8–29, 8–16, and 1 kJ/mol, respectively. Thus, the interaction energies of the networks can be estimated (Fig. 2 C and F) and may determine whether interconversion of the transport states can occur in the absence of substrate, explaining why they operate as uniporters or exchangers. Strong networks are defined as having interaction energies of 2 to 3 salt bridges and weak ones as having equal to or less than half the interaction energies of complete networks. The division into 2 classes serves to illustrate the principles but is qualitative, because there will be a range of interaction energies, due to the type of substitutions, residue geometries, local structural environments, and physiological conditions.
Most mitochondrial transporters operate according to a strict exchange mechanism. When both networks are strong, interconversion between the conformational states may be prevented in the absence of substrate, because the networks will pose a considerable energy barrier with a low probability of disrupting spontaneously, and thus the exchange of the substrates will be equimolar (Fig. 4A). The dicarboxylate transporter, oxoglutarate carrier, ADP/ATP carrier, Mg2+-ATP/Pi carrier, succinate/fumarate carrier, and the pyrimidine nucleotide transporter are predicted to be strict exchangers (Figs. S5, S6, S8, S14, S16, and S24 in SI Appendix), as shown experimentally (26–33). Notably, the uncoupling proteins (Fig. 2 and Figs. S21, and S22 in SI Appendix) are predicted to be strict exchangers also. Although predominantly an exchanger, a small uniport activity was observed for the NAD+ transporters (34), which is not expected on the basis of the networks. NAD+ is recycled in mitochondria by oxidation/reduction reactions, and thus the only requirement for net import is to replenish the pools upon mitochondrial division, which could be achieved by exchange with mitochondrial AMP or GMP. The S-adenosylmethionine transporters are predicted to be strict exchangers, because the exchanged S-adenosylmethionine and S-adenosylhomocysteine are the substrate and product of methylation reactions. In agreement, the human version has no uniport activity (35), but the yeast version has some (36). Similarly, acylcarnitine/carnitine carriers are expected to be strict exchangers, because the exchanged substrates are the substrate and product of carnitine acyltransferase. The rat and yeast carriers have some uniport activity, albeit 10-fold lower than the rate of exchange (37, 38). The import of S-adenosylmethionine or carnitine, which is required to prime the equimolar exchange, could be carried out by related transporters ScYmc1p, ScYMR166c, and HsSLC25A29, which are predicted to have uniport activity (Figs. S33, S34, and S38 in SI Appendix).
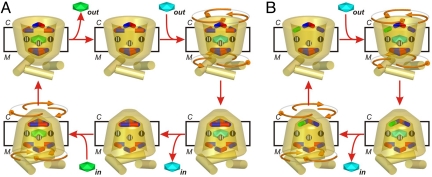
Fig. 4.
Dependency of the transport mode on the interaction energies of the salt bridge networks. Transport cycles showing (A) strict equimolar exchange and (B) uniport. The contact points of the common substrate binding site are shown as black spheres with Roman numerals. Negatively charged, positively charged and polar residues of the networks are shown as red, blue and green sticks, respectively. The exported and imported substrates are green and cyan, respectively. All transport steps are fully reversible, but the direction of transport (red arrows) is determined by the concentration gradients of the substrates and the membrane potential in the case of electrogenic transport. During substrate import, the matrix network on the odd-numbered α-helices is released, whereas the cytoplasmic network on the even-numbered α-helices is formed, and the reverse during substrate export. The structural changes might occur with 3-fold pseudosymmetry via a rotary twist (orange arrows).
Uniport of Substrates.
Low interaction energies of the cytoplasmic and matrix networks would allow transporters to have both exchange and uniport activities, depending on whether substrates are present or not. If the cytoplasmic network is weak and the matrix network is strong, the protein could convert from the matrix to the cytoplasmic state in the absence of substrate, because a weak network has a higher probability of breaking spontaneously (Fig. 4B). Thus, net import of the substrate would occur, as predicted and shown experimentally for the transporters of thiamine pyrophosphate (Fig. S28 in SI Appendix) (39), oxaloacetate (Fig. S11 in SI Appendix) (40), citrate (Fig. S17 in SI Appendix) (41), aspartate/glutamate (Fig. S12 in SI Appendix) (42), and glutamate (Fig. S29 in SI Appendix) (43). The GTP/GDP carrier (Fig. S9 in SI Appendix) is predicted to be a net importer in agreement with its biological function, because the net influx of nucleotides is required for nucleic acid synthesis, yet no uniport activity has been observed (44). If the matrix network is weak and the cytoplasmic network is strong, net export would be anticipated when the substrate is absent from the intermembrane space. The peroxisomal AMP/ATP transporters (Fig. S20 in SI Appendix) have such an arrangement, but the orientation and transport mode in peroxisomal membranes are unknown. If both networks were weak, then the transporter would function as a uniporter, equilibrating the substrate pools on both sides of the membrane. The uncharacterized HsMCART1 and HsMCART2 (Fig. S30 in SI Appendix), HsMCART6 (Fig. S31 in SI Appendix), HsMTCH1, HsMTCH2, and TB1 (Fig. S4 in SI Appendix) have such an arrangement.
The predicted uniport or exchange transport modes are generally conserved among orthologs from different taxa, except for the oxodicarboxylates and ornithine transporters (Figs. S13 and S15 in SI Appendix), but the disparities may reflect different physiological functions in different organisms. There is also a good correlation between the predicted transport mode and the biological role of the transporter. For instance, most amino acid transporters are predicted to have net import activity, because amino acids are incorporated into proteins during synthesis in the mitochondrial matrix.
Mechanism of Transport.
Kinetic studies have suggested that most mitochondrial carriers carry out equimolar exchange with a sequential/simultaneous mechanism (45) in which the exchanged substrates bind to the transporter at the same time before transport (46). However, the single substrate-binding site is inconsistent with this mechanism and the transporters are more likely to have ping-pong mechanisms (45, 46) in which the transported substrate leaves the transporter before binding of the countersubstrate. A plausible explanation for equimolar exchange has been given by considering the interaction energies of the 2 networks without the need to invoke a dimer or 2 substrate binding sites in a monomer. The structural mechanism of the mitochondrial ADP/ATP transporters has been depicted as a rocking 2 domain structure with the translocation pathway in between monomers (11), like ABC transporters (47) and major facilitators (48, 49). Our symmetry analysis suggests that structural changes in mitochondrial transporters occur via a 3-fold symmetric mechanism instead. Symmetric motions of the α-helices combined with structural constraints suggest that the movements of the odd- and even-numbered α-helices could occur via a rotary twist relative to the matrix α-helices (Fig. 4).
Methods
Full-length fungal and metazoan protein sequences with the Pfam (50) mitochondrial transporter family signature (PF000153) were taken from UniProt (51). Additional sequences were identified from fungal genomes. Orthologous sequences were grouped by using a pairwise reciprocal procedure with BLASTP (52), clustering with CLUSTALW (53) and manual inspection with JalView (54). The upstream DNA sequences of proteins with truncated N termini were examined for alternative start codons. The sequence redundancy of orthologs was reduced by clustering at 90% sequence identity.
Amino acid residues related by 3-fold pseudosymmetry were grouped as triplets by using a multiple sequence alignment of the repeats (6) for a set of nonredundant transporters from fungi and metazoan. The symmetry-related triplets of transport proteins from Saccharomyces cerevisiae and Homo sapiens are in Fig. S1–S4 in SI Appendix. The triplets were numbered according to the residue number in the first repeat of bovine ADP/ATP carrier, e.g., triplet 27 corresponds to residues 27, 132, and 229 (Fig. S4 in SI Appendix). A symmetry score that quantifies the degree of similarity of residues in a triplet was calculated for each target residue by taking the average of the replacement scores in the residue replace ability matrix (55) for each of the other 2-aa of the triplet (Fig. 1). A single amino acid sequence can be used, but by using a set of orthologs, the average and standard deviation of the symmetry score were calculated for each amino acid. To account for sequence composition bias, a frequency distribution of the symmetry scores was determined by enumerating over all possible combinations of triplets in each sequence in the group of orthologs (Fig. 1D). The median value of the distribution was taken as the expected symmetry score for a triplet chosen randomly and was subtracted from the observed averaged score and color-coded (Fig. 1D). In addition, a conservation score was calculated for each aligned residue position in a set of orthologs by using a sum of pairs method with the residue replace ability matrix (55). With adaptations, these methods can be used to identify important residues in other transporters with pseudosymmetry, such as in members of the ABC transporter family (47) and the major facilitator superfamily (48, 49), whether a structure is available or not. PyMOL (56) was used for structure visualization.
Supplementary Material
Acknowledgments.
We are very grateful to John Walker for editing the manuscript. This work was supported by the Medical Research Council U.K. and the European Union EDICT grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809580105/DCSupplemental.
References
- 1.Palmieri F. The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflügers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 2.Palmieri F, et al. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta. 2006;1757:1249–1262. doi: 10.1016/j.bbabio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 3.Fleury C, et al. Uncoupling protein-2: A novel gene linked to obesity and hyperinsulinemia. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 4.Aquila H, Link TA, Klingenberg M. The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. EMBO J. 1985;4:2369–2376. doi: 10.1002/j.1460-2075.1985.tb03941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boss O, et al. Uncoupling protein-3: A new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 6.Saraste M, Walker JE. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144:250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- 7.Kunji ERS, Harding M. Projection structure of the atractyloside-inhibited mitochondrial ADP/ATP carrier of Saccharomyces cerevisiae. J Biol Chem. 2003;278:36985–36988. doi: 10.1074/jbc.C300304200. [DOI] [PubMed] [Google Scholar]
- 8.Pebay-Peyroula E, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 9.Nelson DR, Felix CM, Swanson JM. Highly conserved charge-pair networks in the mitochondrial carrier family. J Mol Biol. 1998;277:285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan BB, et al. Antibody evidence for different conformational states of ADP, ATP translocator protein isolated from mitochondria. Proc Natl Acad Sci USA. 1976;73:2280–2284. doi: 10.1073/pnas.73.7.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingenberg M. In: A Study of Enzymes. Kuby SA, editor. Vol II. Boca Raton, FL: CRC; 1991. pp. 367–390. [Google Scholar]
- 12.Robinson AJ, Kunji ERS. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci USA. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunji ERS, Robinson AJ. The conserved substrate binding site of mitochondrial carriers. Biochim Biophys Acta. 2006;1757:1237–1248. doi: 10.1016/j.bbabio.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc Natl Acad Sci USA. 2008;105:9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bamber L, et al. The yeast mitochondrial ADP/ATP carrier functions as a monomer in mitochondrial membranes. Proc Natl Acad Sci USA. 2007;104:10830–10834. doi: 10.1073/pnas.0703969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runswick MJ, Powell SJ, Nyren P, Walker JE. Sequence of the bovine mitochondrial phosphate carrier protein: Structural relationship to ADP/ATP translocase and the brown fat mitochondria uncoupling protein. EMBO J. 1987;6:1367–1373. doi: 10.1002/j.1460-2075.1987.tb02377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stappen R, Kramer R. Kinetic mechanism of phosphate/phosphate and phosphate/OH- antiports catalyzed by reconstituted phosphate carrier from beef heart mitochondria. J Biol Chem. 1994;269:11240–11246. [PubMed] [Google Scholar]
- 18.Wohlrab H, Annese V, Haefele A. Single replacement constructs of all hydroxyl, basic, and acidic amino acids identify new function and structure-sensitive regions of the mitochondrial phosphate transport protein. Biochemistry. 2002;41:3254–3261. doi: 10.1021/bi0117551. [DOI] [PubMed] [Google Scholar]
- 19.Klingenberg M, Echtay KS. Uncoupling proteins: The issues from a biochemist point of view. Biochim Biophys Acta. 2001;1504:128–143. doi: 10.1016/s0005-2728(00)00242-5. [DOI] [PubMed] [Google Scholar]
- 20.Jezek P, Garlid KD. Mammalian mitochondrial uncoupling proteins. Int J Biochem Cell Biol. 1998;30:1163–1168. doi: 10.1016/s1357-2725(98)00076-4. [DOI] [PubMed] [Google Scholar]
- 21.Echtay KS, Winkler E, Bienengraeber M, Klingenberg M. Site-directed mutagenesis identifies residues in uncoupling protein (UCP1) involved in three different functions. Biochemistry. 2000;39:3311–3317. doi: 10.1021/bi992448m. [DOI] [PubMed] [Google Scholar]
- 22.Echtay KS, Bienengraeber M, Klingenberg M. Mutagenesis of the uncoupling protein of brown adipose tissue. Neutralization of E190 largely abolishes pH control of nucleotide binding. Biochemistry. 1997;36:8253–8260. doi: 10.1021/bi970513r. [DOI] [PubMed] [Google Scholar]
- 23.Cappello AR, et al. Functional and structural role of amino acid residues in the even-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol. 2006;363:51–62. doi: 10.1016/j.jmb.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 24.Palmieri F. Diseases caused by defects of mitochondrial carriers: A review. Biochim Biophys Acta. 2008;1777:564–578. doi: 10.1016/j.bbabio.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Cappello AR, et al. Functional and structural role of amino acid residues in the odd-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol. 2007;369:400–412. doi: 10.1016/j.jmb.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 26.Indiveri C, Capobianco L, Kramer R, Palmieri F. Kinetics of the reconstituted dicarboxylate carrier from rat liver mitochondria. Biochim Biophys Acta. 1989;977:187–193. doi: 10.1016/s0005-2728(89)80070-2. [DOI] [PubMed] [Google Scholar]
- 27.Palmieri L, et al. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 1997;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- 28.Fiermonte G, et al. Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2004;279:30722–30730. doi: 10.1074/jbc.M400445200. [DOI] [PubMed] [Google Scholar]
- 29.Pfaff E, Klingenberg M, Heldt HW. Unspecific permeation and specific exchange of adenine nucleotides in liver mitochondria. Biochim Biophys Acta. 1965;104:312–315. doi: 10.1016/0304-4165(65)90258-8. [DOI] [PubMed] [Google Scholar]
- 30.Marobbio CM, Di Noia MA, Palmieri F. Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: Bacterial expression, reconstitution and functional characterization. Biochem J. 2006;393:441–446. doi: 10.1042/BJ20051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmieri F, Prezioso G, Quagliariello E, Klingenberg M. Kinetic study of the dicarboxylate carrier in rat liver mitochondria. Eur J Biochem. 1971;22:66–74. doi: 10.1111/j.1432-1033.1971.tb01515.x. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri F, Quagliariello E, Klingenberg M. Kinetics and specificity of the oxoglutarate carrier in rat-liver mitochondria. Eur J Biochem. 1972;29:408–416. doi: 10.1111/j.1432-1033.1972.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 33.Indiveri C, Palmieri F, Bisaccia F, Kramer R. Kinetics of the reconstituted 2-oxoglutarate carrier from bovine heart mitochondria. Biochim Biophys Acta. 1987;890:310–318. doi: 10.1016/0005-2728(87)90158-7. [DOI] [PubMed] [Google Scholar]
- 34.Todisco S, Agrimi G, Castegna A, Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- 35.Agrimi G, et al. Identification of the human mitochondrial S-adenosylmethionine transporter: Bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem J. 2004;379:183–190. doi: 10.1042/BJ20031664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marobbio CM, Agrimi G, Lasorsa FM, Palmieri F. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 2003;22:5975–5982. doi: 10.1093/emboj/cdg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Indiveri C, Tonazzi A, Palmieri F. Characterization of the unidirectional transport of carnitine catalyzed by the reconstituted carnitine carrier from rat liver mitochondria. Biochim Biophys Acta. 1991;1069:110–116. doi: 10.1016/0005-2736(91)90110-t. [DOI] [PubMed] [Google Scholar]
- 38.Palmieri L, et al. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 1999;462:472–476. doi: 10.1016/s0014-5793(99)01555-0. [DOI] [PubMed] [Google Scholar]
- 39.Marobbio CM, et al. Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 2002;21:5653–5661. doi: 10.1093/emboj/cdf583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmieri L, et al. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J Biol Chem. 1999;274:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- 41.De Palma A, Scalera V, Bisaccia F, Prezioso G. Citrate uniport by the mitochondrial tricarboxylate carrier: A basis for a new hypothesis for the transport mechanism. J Bionenerg Biomembr. 2003;35:133–140. doi: 10.1023/a:1023794019331. [DOI] [PubMed] [Google Scholar]
- 42.Cavero S, et al. Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol Microbiol. 2003;50:1257–1269. doi: 10.1046/j.1365-2958.2003.03742.x. [DOI] [PubMed] [Google Scholar]
- 43.Fiermonte G, et al. Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem. 2002;277:19289–19294. doi: 10.1074/jbc.M201572200. [DOI] [PubMed] [Google Scholar]
- 44.Vozza A, Blanco E, Palmieri L, Palmieri F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- 45.Cleland WW. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. Biochim Biophys Acta. 1963;67:104–137. doi: 10.1016/0006-3002(63)91800-6. [DOI] [PubMed] [Google Scholar]
- 46.Palmieri F. Mitochondrial carrier proteins. FEBS Lett. 1994;346:48–54. doi: 10.1016/0014-5793(94)00329-7. [DOI] [PubMed] [Google Scholar]
- 47.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 48.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y, et al. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 50.Finn RD, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apweiler R, et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 55.Cserzo M, Bernassau JM, Simon I, Maigret B. New alignment strategy for transmembrane proteins. J Mol Biol. 1994;243:388–396. doi: 10.1006/jmbi.1994.1666. [DOI] [PubMed] [Google Scholar]
- 56.DeLano WL. PyMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.