Abstract
Molecular differentiation between races or closely related species is often incongruent with the reproductive divergence of the taxa of interest. Shared ancient polymorphism and/or introgression during secondary contact may be responsible for the incongruence. At loci contributing to speciation, these two complications should be minimized (1, 2); hence, their variation may more faithfully reflect the history of the species' reproductive differentiation. In this study, we analyzed DNA polymorphism at the Odysseus (OdsH) locus of hybrid sterility between Drosophila mauritiana and Drosophila simulans and were able to verify such a prediction. Interestingly, DNA variation only a short distance away (1.8 kb) appears not to be influenced by the forces that shape the recent evolution of the OdsH coding region. This locus thus may represent a test case of inferring phylogeny of very closely related species.
Species are delineated by shared reproductive physiology, development, sexual behavior, and morphology (3, 4). Divergence in these systems is manifested as hybrid sterility, hybrid inviability, premating isolation, and morphological differences, respectively. Races are less well defined but members often may cluster by morphological traits. One of the paradoxes concerning race or species differentiation is the common occurrences of ambiguity in distinguishing taxa by molecular means, even when grouping by reproductive or morphological traits is straightforward and clearcut. Human racial differentiation may be a most obvious example in which many morphological characters cluster by geographical origin, whereas almost all molecular polymorphisms are extensively shared among races (5). Morphological distinction among dog breeds is another example (6). In Drosophila, sexual isolation between the Zimbabwe and non-African races of Drosophila melanogaster is clearly determined by many genes spread over the autosomal genome (7), and yet, recent molecular data have failed to show much differentiation at autosomal loci (8, 9).
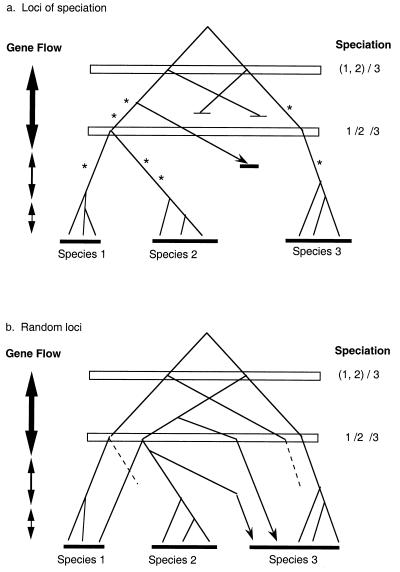
An explanation for the discordance between the “reproductive” and “molecular” phylogeny is that genomes may be mosaics with respect to molecular genealogy, as illustrated in Fig. 1. Most loci, chosen without regard to their roles in reproductive differentiation, may not reflect the biological divergence in their sequence polymorphism because of either shared ancient polymorphism or gene introgression through secondary contact (Fig. 1b). Ancient polymorphism may persist until present day in species with large population sizes (10, 11), and gene introgression, even at a very low level, may be sufficient to obliterate differentiation (12). In this context, we shall consider separately “speciation genes,” defined as loci that contribute directly to some aspects of biological divergence between closely related species (such as gametogenesis, behavior, or morphology).
Figure 1.
Contrasting gene genealogies at two types of loci. Speciation occurred first between species 3 and the ancestor of species 1 and 2, and then between the latter species. Gene flow across species boundaries diminished with time. (a) “Speciation loci.” Each favorable mutation (marked with an *) drives the spread of a single lineage, excluding other lineages (ending with a tick). This would result in the purge of shared ancient polymorphisms. In addition, any lineage introgressed from the other species (arrow) is quickly eliminated because of the incompatibility with the new genetic background. Monophyly by species and a clear species phylogeny are observed. (b) “Other loci.” Ancient polymorphism and introgression during secondary contact (arrow) lead to mixed genealogies between species. Dashed branches denote lineages lost because of genetic drift. Note that species 2 and 3 appear most closely related.
A hypothesis, proposed in various forms (1, 2, 13, 14), is that “speciation genes” may record a phylogenetic history more consistent with species' reproductive biology. This is because polymorphism and divergence at these loci should be relatively unaffected by shared polymorphisms or introgressions (see the legend of Fig. 1a). The cloning of the Odysseus (OdsH, H for homeodomain) locus of hybrid male sterility in the Drosophila simulans clade (15) therefore provides an opportunity to test this hypothesis. The sibling species of D. simulans, Drosophila mauritiana, and Drosophila sechellia often exhibit large intraspecific variation relative to interspecific divergence in their DNA (16, 17). On the other hand, these species do show within-species coherence and extensive between-species divergence with respect to reproductive and morphological characters (18, 19). Do the sequence polymorphisms of OdsH cluster by species? If they do, what would the phylogeny of the trio of species be? The latter question has attracted much attention (16, 17, 20).
Why would the phylogeny of the three species of Drosophila be of general interest? The main reason is that this may be a test case revealing the complex forces that underlie the phylogenetic history of races or closely related species in general. These forces operate at the early stage of speciation (i.e., around the top nodes of Fig. 1), and the complex histories are therefore a manifestation of the population genetic dynamics of species formation.
Materials and Methods
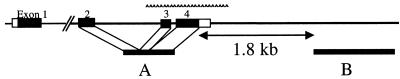
All of the D. melanogaster (Ore-R), D. simulans, and D. sechellia lines were obtained from the Bloomington Stock Center, Bloomington, IN. The seven D. mauritiana lines used in region A sequencing were obtained from the stock center, and 10 more lines from National Institute of Genetics (Mishima, Japan) were added to this collection in the study of region B. The regions we sequenced are diagrammed in Fig. 2, and details of the sequencing method were as described (9).
Figure 2.
Schematic drawing of the genomic region of OdsH. Exons are shown as solid boxes. Line segments A and B denote the regions sequenced in this study.
In total, we analyze the polymorphism and divergence data from eight gene regions: regions A and B of Fig. 2, asense, period, yolk protein-2, zeste, Cubitus interruptus, as listed in ref. 16, and Acp26Aa (unpublished observations). Cubitus interruptus shows virtually no polymorphisms. In Table 1, we consider only sites where more than 1 nt (for example, T and C) are present in more than one species. Although these are conventionally referred to as phylogenetically informative sites, such a term is more appropriate in dealing with only one sequence from each species. Because multiple sequences from three species (plus an outgroup) are analyzed here, these sites can be either ambiguous or unambiguous about species phylogeny. An unambiguous site is usually where two of the three species have the same derived, fixed nucleotide, whereas the third species has only the ancestral type (=outgroup). More generally, it is where (i) two species share a derived nucleotide and neither retains the ancestral one and (ii) the third species has the ancestral nucleotide without the derived one. All other configurations are ambiguous. Ambiguity can be caused by ancient polymorphism, introgression, reversion, or parallel mutations, although the latter two should contribute only a small fraction of shared polymorphisms between such closely related species.
Table 1.
The number of ambiguous and unambiguous sites in the coding region of OdsH and six other loci
| OdsH* | All other genes† | |
|---|---|---|
| Ambiguous sites | 2 | 30 |
| Unambigous sites | 7 | 1 |
| Sim − Mau‡ | 6 | 0 |
| Sec − Mau‡ | 1* | 0 |
| Sim − Sec‡ | 0 | 1 |
A high ratio of ambiguous/unambiguous sites indicates the influence of ancient polymorphism and/or secondary introgression. Unambiguous sites can be further classified by one of the three groupings they support.
This site is in the noncoding region of D. mauritiana, but coding region of the other two species because the former is 31 codons shorter than others.
† The chance of having an ambiguous site (because of shared polymorphism between species) may increase as the sample size increases. Since the OdsH data have fewer ambiguous sites but slightly larger sample for D. simulans and D. mauritiana (the number of D. sechellia sequences usually has no effect), the results are not biased by sampling. A complete profile of these sites in the four species will be given upon request.
‡ The closest pair of species grouped by the unambiguous sites. Sim, D. simulans; Mau, D. mauritiana; and Sec, D. sechellia.
Results
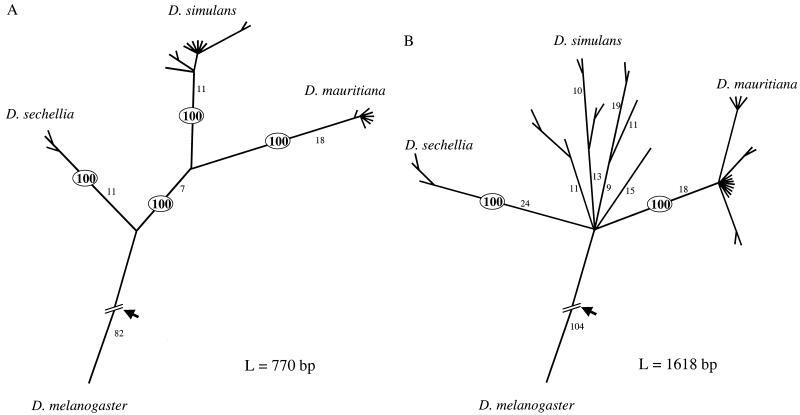
To find out whether the OdsH locus indeed behaves as expected, in Fig. 1a, we sequenced 11 samples from D. simulans, seven from D. mauritiana, three from D. sechellia, and one from D. melanogaster as shown in Fig. 2. Fig. 3A presents the genealogy of 770 bp of sequence spanning exons 2–4 of OdsH (15). Exon 1 is more than 10 kb away and is excluded from this analysis. As predicted, the genealogy based on the exons of OdsH is cleanly sorted by species (Fig. 3A). More importantly, this gene unambiguously groups two of the trio as each species' closest relative with a 100% bootstrapping value, unique among the eight gene regions that have polymorphism data in all three species. That D. mauritiana and D. simulans are most closely related is intriguing because reproductive incompatibility between them is much greater than between D. sechellia and D. simulans (1).
Figure 3.
Genealogies of region A or B (as shown in Fig. 2) among the four sibling species of Drosophila, as inferred by the maximum parsimony method (paup, version 3.1.1). Variation at other loci published so far exhibits genealogies similar to that of B (see text). Branch length is proportional to the inferred number of changes, which are given along the major branches. The short terminal branches often represent 0 changes. Adjacent nodes whose bootstrapping value is lower than 50% are compressed into a single node. The bootstrapping values of the four interspecific nodes are of special interest. In A, they are all 100%, as shown, but in B, two are less than 50% and are compressed into a star phylogeny. Arrow indicates the root. L, Number of nucleotides analyzed.
The pattern of Fig. 3A exhibits a resolution not observed in other single-copy genes published so far (16). We have followed the same procedure to construct the genealogies at six other loci where polymorphic data are available from D. simulans, D. mauritiana, D. sechellia, as well as the outgroup, D. melanogaster. We do not consider data sets that do not contain multiple sequences from both D. simulans and D. mauritiana. Among the six gene regions that have some degree of within-species variation, monophyletic clustering for all three species is not seen at any locus, even at a low stringency of 50% bootstrapping. As a consequence, the between-species phylogeny cannot be clearly inferred. (A visual representation of the genealogies of these genes resembles that of Fig. 3B; see below.) This analysis is consistent with earlier studies (16, 17). [Note that, in ref. 16, the five loci that have polymorphism data in all three species do not yield any conclusive grouping of species (their table 3). The grouping of D. mauritiana with D. sechellia is based on the three loci that have only one sequence from D. mauritiana.]
To reveal the differences in the phylogenetic information provided by OdsH vis-a-vis all other loci (which are not known to be associated with speciation), we shall distinguish between variant nucleotide sites that are phylogenetically ambiguous and unambiguous for the three species. Phylogenetically ambiguous sites designate shared variations across species, presumably resulting from ancient polymorphisms and/or subsequent introgressions. An example is the following nucleotide composition at a site: (G,C), (G,C), C, and G for D. simulans, D. mauritiana, D. sechellia, and D. melanogaster, respectively, where ( ) denotes polymorphism. In that case, all possible phylogenies among the three species are compatible with the data. An unambiguous site is, for example, G, G, T, T for the four species, respectively, where each species is fixed for a nucleotide. A precise definition of ambiguous vs. unambiguous sites is given in Materials and Methods. In Table 1, a vast majority of sites from other loci are ambiguous (30 of 31 sites), whereas, at OdsH, seven of the nine sites are unambiguous with six of them supporting the close kinship between D. mauritiana and D. simulans. The difference is highly significant (P < 0.001 by Fisher's exact test), suggesting a strong disparity in the impact of ancient polymorphism and/or introgression on the extant variations at speciation loci vis-a-vis others.
The next question, naturally, is how far away from OdsH would the pattern still resemble that of OdsH (Fig. 1a), as opposed to those of randomly chosen loci. A selection-driven genetic change on its way to fixation would affect the nearby region (21). If there is absolutely no recombination between a selected site and a linked neutral locus, the latter would also lose its ancient variation because of the fixation of the single haplotype that carries the selected variant. The process often is referred to as selective sweep (21, 22), which can be analyzed by examining the level and pattern of polymorphism (23, 24). Recombination, however, would decouple the dynamics of a nearby site from that of the selected variant. How far apart the two sites have to be for their dynamics to be completely decoupled depends on the time it takes for the favorable mutation to become fixed and the rate of recombination between the two sites. Recombination also can alter the effect of introgression. When there is introgression across species boundary, genes like OdsH would be excluded because they are incompatible with the new genetic background. Nearby sites may or may not escape the negative selection, depending on whether there is sufficient recombination to separate them from the locus of hybrid incompatibility after introgression (1).
To measure the extent of hitchhiking, we surveyed the polymorphism in regions that are increasingly distant from the site of selection, i.e., the exons of OdsH. Region B of Fig. 2 is the region we surveyed from 11 D. simulans, 17 D. mauritiana, three D. sechellia, and one D. melanogaster lines. (Note that 10 more D. mauritiana lines are used for region B to increase the resolution.) To our surprise, this region appears to be completely unaffected by the events shaping the genealogy of the exons, even although the two regions are only 1.8 kb apart. Fig. 3 A and B contrast their genealogies. In region A, D. simulans alleles cluster and two of the three species (D. simulans and D. mauritiana) are unambiguously more closely related than each is to the third species.
Discussion
This study has several implications:
(i) The genome can indeed be a mosaic of regions of different genealogies among closely related species, because of shared ancient polymorphism and/or introgressions (1, 2, 13). Genomic regions not affected by either factor should be monophyletic by species and more faithfully representative of the biological species status. The coding sequence of OdsH appears to be such a region. As a consequence of monophyly by species, OdsH also provides a clearer resolution of phylogeny among species. The pattern is in contrast with the majority of variable sites in the genome, which are often phylogenetically ambiguous because of shared variants (see Table 1). The preponderance of ambiguous sites suggests that ancient polymorphism and/or introgression may play a very significant role in the earlier phase of speciation.
The phylogenetic pattern of Fig. 3A is corroborated by the joint analysis of 39 microsatellite loci from the three species, each with more than 20 individuals (20). To infer the phylogeny of closely related species accurately, polymorphism data from multiple loci generally are needed to overcome the noises of ancient polymorphisms (10, 11), but a single speciation locus may suffice.
(ii) Introgression can potentially bias phylogenetic inference, for example, when interspecific introgression is asymmetric. In that case, increasing the number of genes or individuals would not rectify the bias. Moreover, to bias the inference, introgression only needs to happen in the early stage of speciation (i.e., Fig. 1 Upper).
Is there evidence of introgression in the trio of Drosophila species? The cosmopolitan distribution of D. simulans and the fertility of all hybrid females suggest the possibility of unidirectional introgression from D. simulans into its island siblings. Indeed, it has been reported that 88% of D. mauritiana lines carry a D. simulans type mitochondrial molecule (25). That the sharing is because of introgression has been demonstrated by Ballard (26) who found only 1-bp difference in 15 kb between the two molecules from the two species. In contrast, another D. mauritiana mitochondrial allele (which presumably has diverged from its D. simulans counterpart since species divergence) differs from this introgressed type by more than 200 bp (1.5%), close to the level of divergence for most nuclear genes (16, 17). If mitochondrial DNA can still migrate across species boundary in the recent past, it is not farfetched to imagine more substantial gene flow earlier on. A previous analysis of DNA polymorphism on the fourth chromosome indeed suggests such a possibility (2). Given the large number of ambiguous sites in Table 1, introgression may have to be invoked in addition to the retention of ancient polymorphisms in the extant species. This is because the three species have diverged for 5–10 million generations since speciation (17), long enough for the majority of shared polymorphisms to have become fixed. Introgression thus may fill the gap in our account of Table 1. It also may explain why the Acp26Aa gene, which has been under selection and should have lost most shared ancient polymorphisms (9), yields only ambiguous sites.
How strongly a speciation gene's genealogical history contrasts with those of other loci depends on many variables, including the timing when the reproductive incompatibility caused by a specific genetic change evolved. If it evolved relatively late, introgression of this particular locus could happen during much of the species' history. For this reason, hybrid sterility because of OdsH most likely evolved early, a conjecture supported by the extensive amino acid differences between these species (15).
(iii) This present study also redresses a shortcoming in virtually all studies of the genetics of speciation. For technical reasons, such studies always have been done with only one or two representative lines from each species, but cloning has since made sampling many chromosomes feasible. By doing so, the results of Fig. 3A corroborate the conclusion that OdsH-induced hybrid sterility is a species phenomenon, not a peculiar property of a few lines.
(iv) Finally, the mixed genealogies near the OdsH locus suggest a molecular perspective on the species concept. What seems most surprising is the very different resolutions between the genealogical trees of regions of DNA less than 2 kb apart. The hitchhiking process, either in removing ancient polymorphisms or in excluding cointrogressions of tightly linked variations, must have been relatively ineffectual over a longer distance (see also ref. 27). This raises an intriguing possibility: diverging species that remain incompletely isolated reproductively (such as D. simulans and D. mauritiana) may be permeable to introgression over a large portion of their genomes. As only a small region near each locus of speciation is impermeable, the exchange may continue for some time until reproductive isolation is complete. During this period, regions of impermeability would only expand gradually because of the increase in the number of speciation loci. Whether this molecular perspective of “porous species,” suggested by the population genetics near OdsH, is general will have to await the cloning and characterization of other speciation loci.
Acknowledgments
We thank Shigeo Hayashi for providing the D. mauritiana stocks. We also thank Ian Boussy, Justin Fay, Mark Jensen, Eli Stahl, and Kevin Thornton for comments. This work was supported by grants from National Institutes of Health and National Science Foundation (to C.I.W.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF254805 for D. simulans, AF254806 for D. mauritiana, and AF254807 for D. sechellia).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090541597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090541597
References
- 1.Palopoli M F, Davis A W, Wu C-I. Genetics. 1996;144:1321–1328. doi: 10.1093/genetics/144.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilton H, Kliman R M, Hey J. Evolution (Lawrence, Kans) 1994;48:1900–1913. doi: 10.1111/j.1558-5646.1994.tb02222.x. [DOI] [PubMed] [Google Scholar]
- 3.Dobzhansky T. Genetics of the Evolutionary Process. New York: Columbia Univ. Press; 1970. [Google Scholar]
- 4.Mayr E. Animal Species and Evolution. Cambridge, MA: Belknap; 1963. [Google Scholar]
- 5.Nei M, Roychoudhury A K. Mol Biol Evol. 1993;10:927–943. doi: 10.1093/oxfordjournals.molbev.a040059. [DOI] [PubMed] [Google Scholar]
- 6.Vila C, Savolainen P, Maldonado J E, Amorim I R, Rice J E, Honeycutt R L, Crandall K A, Lundeberg J, Wayne R K. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 7.Hollocher H, Ting C-T, Wu M L, Wu C-I. Genetics. 1997;147:1191–1201. doi: 10.1093/genetics/147.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasson E, Wang I N, Zeng L W, Kreitman K, Eanes W F. Mol Biol Evol. 1998;15:756–769. doi: 10.1093/oxfordjournals.molbev.a025979. [DOI] [PubMed] [Google Scholar]
- 9.Tsaur S C, Ting C-T, Wu C-I. Mol Biol Evol. 1998;15:1040–1046. doi: 10.1093/oxfordjournals.molbev.a026002. [DOI] [PubMed] [Google Scholar]
- 10.Pamilo P, Nei M. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- 11.Wu C-I. Genetics. 1991;127:429–435. doi: 10.1093/genetics/127.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahata N. Genetics. 1991;129:585–595. doi: 10.1093/genetics/129.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R L, Wakeley J, Hey J. Genetics. 1997;147:1091–1106. doi: 10.1093/genetics/147.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avise J C. Molecular Markers, Natural History, and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 15.Ting C-T, Tsaur S C, Wu M-L, Wu C-I. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 16.Caccone A, Moriyama E N, Gleason J M, Nigro L, Powell J R. Mol Biol Evol. 1996;13:1224–1232. doi: 10.1093/oxfordjournals.molbev.a025688. [DOI] [PubMed] [Google Scholar]
- 17.Hey J, Kliman R M. Mol Biol Evol. 1993;10:804–822. doi: 10.1093/oxfordjournals.molbev.a040044. [DOI] [PubMed] [Google Scholar]
- 18.Wu C-I, Palopoli M F. Annu Rev Genet. 1994;28:283–308. doi: 10.1146/annurev.ge.28.120194.001435. [DOI] [PubMed] [Google Scholar]
- 19.True J R, Liu J, Stam L F, Zeng Z-B, Laurie C C. Evolution (Lawrence, Kans) 1997;51:816–832. doi: 10.1111/j.1558-5646.1997.tb03664.x. [DOI] [PubMed] [Google Scholar]
- 20.Harr B, Weiss S, Davis J R, Brem G, Schlotterer C. Curr Biol. 1998;8:1183–1186. doi: 10.1016/s0960-9822(07)00490-3. [DOI] [PubMed] [Google Scholar]
- 21.Maynard Smith J, Haigh J. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 22.Stephan W, Wiehe T H E, Lenz M W. Theor Popul Biol. 1992;41:237–254. [Google Scholar]
- 23.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y-X, Li W-H. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solignac M, Monnerot M. Evolution (Lawrence, Kans) 1986;40:531–539. doi: 10.1111/j.1558-5646.1986.tb00505.x. [DOI] [PubMed] [Google Scholar]
- 26.Ballard, J. W. O. (2000) J. Mol. Evol., in press.
- 27.Wang R L, Stec A, Hey J, Lukens L, Doebley J. Nature (London) 1998;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]