Abstract
Estrogens play an important role in normal physiology and in a variety of pathological states involving diverse tissues including breast and bone. The mechanism by which estrogens exert cell type- and disease-specific effects, however, remains to be explained. We have compared the gene expression profile of the MCF7 breast cancer cell line with that of the osteoblast-like cell line U2OS-ERα by expression microarrays. We find that fewer than 10% of the 17β-estradiol (E2)-regulated genes are common to both cell types. We have validated this in primary calvarial osteoblasts. To dissect the mechanism underlying the cell type-specific E2 regulation of gene expression in MCF7 and U2OS-ERα cells, we compared the ERα binding sites on DNA in the two cell types by performing chromatin immunoprecipitation (ChIP) on genomic tiling arrays (ChIP-on-chip). Consistent with the distinct patterns of E2-regulated gene expression in these two cell lines, we find that the vast majority of ERα binding sites are also cell type specific and correlate both in position and number with cell type-specific gene regulation. Interestingly, although the forkhead factor FoxA1 plays a critical role in defining the ERα cistrome in MCF7 cells, it is not expressed in U2OS-ERα cells, and forkhead motifs are not enriched in the ERα cistrome in these cells. Finally, the ERα cistromes are correlated with cell type-specific epigenetic histone modifications. These results support a model for the cell type-specific action of E2 being driven primarily through specific ERα occupancy of epigenetically marked cis-regulatory regions of target genes.
ESTROGENS REGULATE FEMALE development and also play an important role in both males and females in the development and physiology of a variety of tissues including bone. The gene-regulatory effects of estrogens are mediated by two nuclear receptors: estrogen receptor-α (ERα) and ERβ. The biological responses to estrogens are diverse and include ductal outgrowth during normal mammary development, normal skeletal differentiation, the proliferation of ERα-positive breast cancer cells, and the prevention of osteoporosis, among many others (1,2,3).
ERα and ERβ can bind to DNA at specific DNA motifs termed estrogen response elements (EREs). In addition, ERα can indirectly activate transcription by binding to other DNA binding proteins such as Sp1 and c-fos or c-jun (4). We have defined the location of ERα binding to the genome in MCF7 breast cancer cells, the ERα cistrome, by combining chromatin immunoprecipitation (ChIP) with hybridization to whole-genome tiling arrays (5,6). We have defined more than 5000 high-confidence binding sites for ERα in MCF7 cells with the vast majority of sites located greater than 1 kb away from promoter proximal regions (7) consistent with these being primarily enhancer regions. Interestingly, the ERα cistrome defined in MCF7 cells represents only a small fraction of the potential high-affinity EREs in the genome (8). Whether these other potential ERα binding sites are used under different physiological conditions or in other cell types or are never occupied is unknown.
Tissue specificity of gene expression is determined by many factors, including DNA methylation, histone modifications, and transcription factor expression levels (9). Mechanisms of tissue specificity for 17β-estradiol (E2) signaling may include differences in ERα or ERβ levels, differential coactivator recruitment, and/or cell type-specific metabolism of estrogens (10,11,12). Recent work from our lab has suggested an important role for FOXA1 in defining ERα targets in MCF7 cells. In addition, we demonstrated that cell type-specific FOXA1 recruitment at enhancers is determined by a specific epigenetic signature of histone methylations at histone 3 lysine 4 (H3K4) and lysine 9 (H3K9) (6). FOXA1 is present at enhancers with H3K4 mono- and dimethylation and remodels the chromatin allowing for ERα recruitment in MCF7 cells vs. androgen receptor recruitment in LNCaP cells. However, it remains unknown what determines the ERα cistrome in other cell types including those lacking FOXA1.
E2 is critical for maintaining the appropriate ratio between bone-forming osteoblasts and bone-resorbing osteoclasts. E2 induces apoptosis in bone-resorbing osteoclasts (13,14,15), and we have shown that the mechanism of action of estrogen-induced apoptosis is due to an up-regulation of Fas ligand (FasL) in osteoblasts in response to E2, leading to apoptosis of osteoclasts (16). E2 also represses certain cytokines, such as IL-1 and IL-6 (reviewed in Ref. 17), that are osteoclastogenic. E2 has been shown to regulate osteoblast differentiation (18,19,20) and prevent osteoblast apoptosis (14). However, only a few estrogen-responsive genes in osteoblasts have been identified (15,21,22,23,24), and the tissue-specific transcriptional mechanisms remain unknown.
In this study, using expression microarrays, we first identified osteoblast-specific genes in U2OS-ERα cells and validated their expression profile in primary calvarial osteoblasts. We then compared the ERα binding sites in MCF7 and U2OS-ERα cells to determine whether differential ERα recruitment or other mechanisms play the primary role in defining the differences in E2-regulated gene expression in these two cell types.
RESULTS
Tissue-Specific Gene Expression Regulated by E2
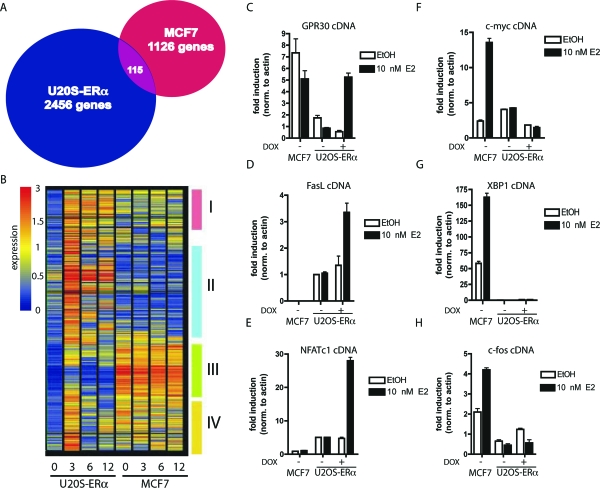
To characterize the regulation of genes by E2 in two tissue types, the osteoblast-like U2OS-ERα cell line and MCF7 breast cancer cell line were treated with 10 nm E2 for 0, 3, 6, and 12 h, and global gene expression profiles were determined by microarray analysis. The U2OS-ERα cell line is an osteosarcoma cell line that was stably transfected with a doxycycline (DOX)-inducible ERα. Upon treatment with DOX, the expression of ERα in U2OS-ERα cells is similar to MCF7 cells (23). The time-course experiments revealed that after 3 h E2 treatment, 2571 genes were induced at least 3-fold in U2OS-ERα cells (Fig. 1A). In contrast, only 1241 genes were up-regulated at 3 h in MCF7 cells. Surprisingly, only 115 genes (4.5% of the U2OS-ERα genes and 9.2% of the MCF7 genes) were induced by E2 in both cell lines. The number of genes regulated in common in the two cell types did not change significantly at 6 or 12 h (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). The number of genes that were commonly down-regulated in both cell types was approximately 11% of the genes down-regulated at 3 h by E2 (supplemental Fig. 1).
Figure 1.
ERα-Mediated Gene Expression in Different Cell Types
A, Venn diagram showing the number of genes regulated by 10 nm E2 for 3 h in MCF7 and U2OS-ERα cells. B, Cluster analysis of all of the genes up-regulated in U2OS-ERα cells at 3 h. Each column represents a time point (0, 3, 6, or 12 h) in U2OS-ERα or MCF7 cells and contains all of the genes up-regulated in U2OS-ERα cells at 3 h. Groups I–IV are described in the text. C–H, U2OS-ERα and MCF7 cells were deprived of E2 for 3 d in phenol red-free medium containing 5% CDT-FBS. U2OS-ERα cells were either treated with (to induce ERα) or without DOX for 24 h. They were then treated with 10 nm E2 for 3 h, and RNA was obtained. GPR30, FasL, NFATc1, c-myc, XBP1, and c-fos cDNA were analyzed by quantitative PCR. Error bars for all parts represent mean ± 1 sd.
Using a hierarchical clustering algorithm, we organized the 2571 up-regulated genes in U2OS-ERα on the basis of overall similarity in their gene expression patterns. Their relationships are clustered (Fig. 1B) to identify distinct U2OS-ERα expression patterns. Each column represents a time point (0, 3, 6, or 12 h) in U2OS-ERα or MCF7 cells and contains all of the genes significantly up-regulated in U2OS-ERα cells at 3 h. Four distinct clusters could be identified reflecting the relatedness of expression pattern within each cluster (Fig. 1B, I–IV). Group I contains genes that are up-regulated by E2 in U2OS-ERα cells at 3 h and remain elevated at 6 and 12 h. These genes are expressed in MCF7 cells but are not regulated by E2. Group II contains genes that are up-regulated by E2 in U2OS-ERα cells but are not expressed in MCF7 cells. Group III contains genes that are minimally up-regulated in U2OS-ERα cells at 3 h, and these same genes are highly expressed in MCF7 cells although not regulated by E2 in MCF7 cells. Genes in group IV are highly up-regulated by E2 only at 3 h in U2OS-ERα cells but not regulated in MCF7 cells and unlike group I do not appear to be elevated at both 6 and 12 h. Together, these groups illustrate global gene expression patterns that are tissue specific and time dependent.
To identify important genes uniquely up-regulated by E2 in U2OS-ERα cells, the hierarchical clustering and expression data were analyzed for bone-specific genes. Three representative genes with bone-specific functions were verified by quantitative PCR in Fig. 1, C–E [GPR30 (Fig. 1C), FASLG (FasL, Fig. 1D), and NFATC1 (Fig. 1E)]. G protein-coupled receptor 30 (GPR30), which binds E2 and regulates cellular calcium (25), is located at the growth plate, and its expression there changes during puberty (26). Notably, GPR30 is slightly down-regulated in MCF7 cells, whereas it is up-regulated in U2OS-ERα cells. FasL is important in the estrogen-mediated apoptosis of osteoclasts (15). Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 (NFATc1) is a critical transcription factor in osteoclasts but more recently has been shown to also have a role in osteoblasts (27), where it has been shown to regulate osteoblast-specific genes such as Osterix (28). Each of these genes showed no induction in the absence of ERα (Fig. 1). Furthermore, these genes are not up-regulated due simply to overexpression of ERα, because overexpression of ERα in another ERα-negative cell line (MDA-MB-231 cells) did not result in the regulation of genes such as NFATc1 or GPR30 (supplemental Fig. 2).
To verify that the genes up-regulated by E2 in MCF7 cells were not up-regulated in U2OS-ERα cells, several genes identified by the microarray analysis (and previously demonstrated to be up-regulated by E2 in MCF7 cells) were tested. c-Myc (MYC), X-box binding protein 1 (XBP1), and c-fos (FOS), as predicted, were induced by E2 in MCF7 cells but not in U2OS-ERα cells (Fig. 1, F–H), validating tissue specificity of these E2-regulated genes in both breast and osteoblast cells.
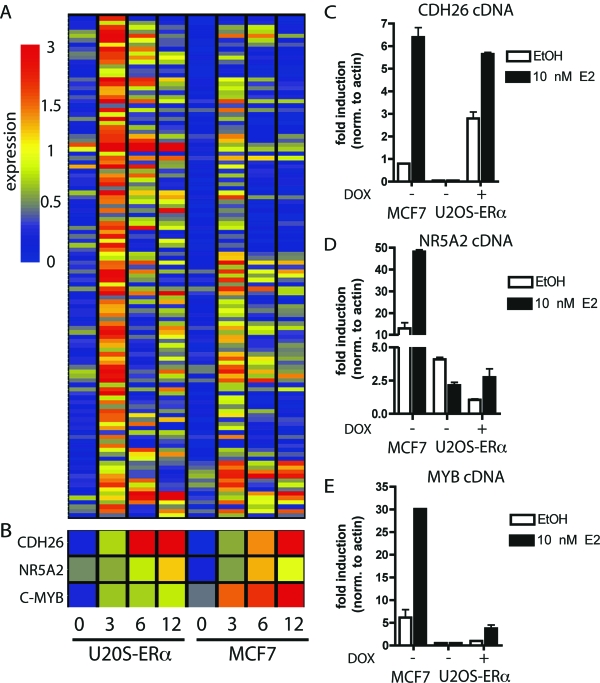
The 115 commonly regulated genes were also subjected to hierarchical clustering analysis (Fig. 2A). The majority of these genes peaked at 3 h in both MCF7 and U2OS-ERα cells, and expression decreased by 6 and 12 h E2 treatment. Interestingly, over half of the commonly regulated genes were expressed sequence tags (data not shown). To verify the microarray data, quantitative PCR was performed with cDNA from the two cell lines treated with E2 for 3 h. Three representative genes were chosen from the list of 115 genes, and their specific microarray profile is illustrated (Fig. 2B). Cadherin-like 26 (CDH26), nuclear receptor subfamily 5 member 2 (NR5A2/LRH1) and c-myb (MYB) were induced by E2 in both cell lines, although the basal levels of each of these genes were different (Fig. 2, C–E). CDH26, with similarities to cadherin proteins, has as of yet no known function. C-myb is a transcription factor that is highly correlated with ERα in breast cancers, but its role in osteoblasts is not defined. NR5A2 is a nuclear receptor that has been previously been shown to be regulated by E2 in breast cancer cells (29), but no known role for NR5A2 has been demonstrated in bone.
Figure 2.
Common E2-Regulated Genes in MCF7 and U2OS-ERα Cells
A, Cluster analysis of all of the genes up-regulated in both MCF7 and U2OS-ERα cells at 3 h. B, Representation of array data for CDH26, NR5A2, and MYB. C–E, U2OS-ERα and MCF7 cells were deprived of E2 for 3 d in phenol red-free medium containing 5% CDT-FBS. U2OS-ERα cells were either treated with (to induce ERα) or without DOX for 24 h. U2OS-ERα and MCF7 cells were treated with 10 nm E2 for 3 h, and RNA was obtained. CDH26, NR5A2, and MYB were analyzed by quantitative PCR. Error bars for all parts represent mean ± 1 sd.
New ERα Targets in Osteoblasts
Numerous ERα target genes have already been identified in breast cancer cells. In contrast, very little is known regarding E2 targets in osteoblasts. To find additional osteoblast-specific genes induced by E2, the U2OS-ERα expression microarray data set was then further mined for genes with a potential role in primary osteoblasts. These genes were then analyzed in primary murine osteoblast cultures treated in vitro with 10 nm E2 for 3 h.
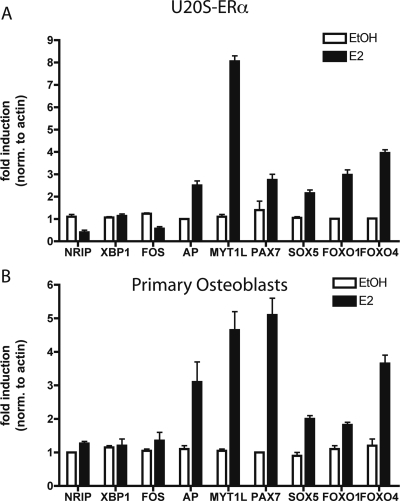
Alkaline phosphatase is the most common marker to identify osteoblasts. It is induced early in differentiation of osteoblasts and hydrolyzes phosphates for incorporation into hydroxyapatite (30). Alkaline phosphatase was up-regulated in U2OS-ERα cells and primary calvarial osteoblasts after treatment with E2 (Fig. 3, A and B).
Figure 3.
Novel E2 Targets in Bone
A, U2OS-ERα cells were treated with 10 nm E2 for 3 h, and RNA was obtained. Quantitative PCR was performed in triplicate on cDNA from three independent experiments. B, Primary calvarial osteoblasts were treated with 10 nm E2 for 3 h, and RNA was obtained. Quantitative PCR was performed in triplicate on cDNA from three independent experiments. Error bars for all parts represent mean ± 1 sd. AP, Alkaline phosphatase.
In addition, in primary osteoblasts, the transcription factors MYT1L, PAX7, SOX5, FOXO1, and FOXO4 were also induced by E2 (Fig. 3, A and B). Each of these is a transcription factor that is implicated in tissue-specific differentiation of a variety of tissues (31,32,33,34). These results identify new osteoblast-specific ERα targets that are regulated by E2 in primary calvarial osteoblast cultures.
Myt1L is a member of the myelin transcription factor 1 (Myt1) gene family that is comprised of three zinc finger genes [Myt1, Myt1L (Myt1-like) and NZF3]. Myt1 and MytlL recruit histone deacetylase proteins to regulate tissue specificity (32), although nothing is known about Myt1L in osteoblasts. Myt1L is induced by more than 8-fold in U2OS-ERα cells and by more than 7-fold in calvarial osteoblast cells (Fig. 3, A and B).
Pax7 is a member of the nine-member PAX family of genes with homology to Drosophila segmentation genes. Pax7 knockout mice have defects in facial skeletal structure, and thus PAX7 may be important in the skeleton (33). Pax7 has a role in muscle stem cells, indicating that Pax7 may be involved in the differentiation of a mesenchymal stem cell into either muscle or bone. This is the first demonstration that E2 regulates Pax7 in bone (Fig. 3, A and B).
Sox family members contain an SRY box, a region homologous to the DNA-binding domain of SRY, the mammalian sex-determining gene. Sox5 is important in mesenchymal stem cell differentiation (34) but has not been shown to be regulated by E2. There is a small but statistically significant induction of Sox5 by E2 in both U2OS-ERα cells and in calvarial osteoblast cells (Fig. 3, A and B).
Forkhead (Fox) family members, of which there are over 40 human proteins, are named for their forkhead (or winged helix) motif. Fox proteins transcriptionally regulate a wide variety of biological processes. Although FoxA1 is important in regulating E2-mediated transcription in MCF7 cells (35), U2OS-ERα cells do not express FoxA1. The U2OS-ERα expression data were screened for other potential Fox proteins that might be involved in osteoblast biology. We identified FoxO1 and FoxO4 as being regulated in both U2OS-ERα cells and osteoblast cells (Fig. 3, A and B).
As negative controls, we tested three genes that are induced by E2 in MCF7 cells (NRIP, XBP1, and FOS). These three genes were not regulated in U2OS-ERα cells or in primary osteoblasts.
The ERα Cistrome in U2OS-ERα Cells Is Highly Distinct from that in MCF7 Cells
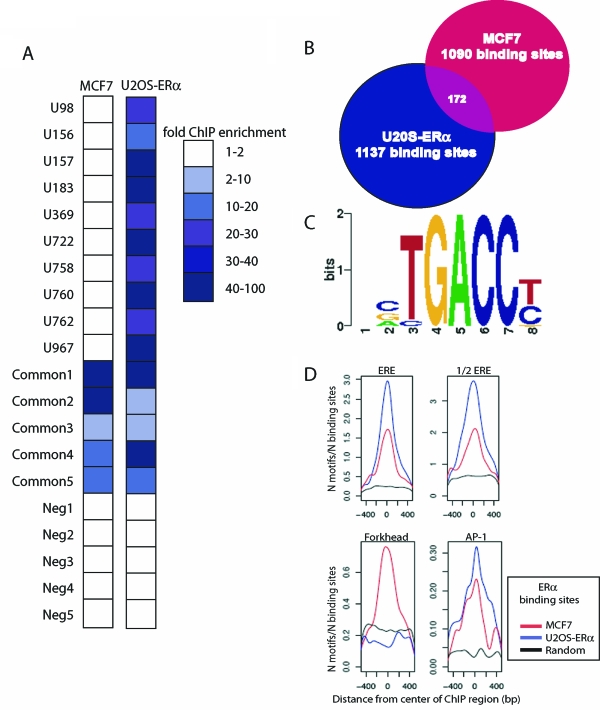
To understand how E2 regulates distinct sets of genes in MCF7 and U2OS-ERα cells, we hypothesized that the ERα binds to distinct enhancer elements in the two cell types. To test this, we performed ChIP-on-chip to begin to define the ERα cistrome in U2OS-ERα cells and compared it with the previously described ERα cistrome in MCF7 cells defined by our group (7). The partial ERα cistrome in U2OS-ERα cells was obtained using Affymetrix Human Tiling Array A. This array contains the nonrepetitive regions of chromosomes 1 and 6, representing one seventh of the genome, tiled at 35-bp resolution. We performed three biological replicates and identified enriched binding sites using MAT analysis (36). At a false discovery rate of 1%, we identified 1137 ERα binding sites on these two chromosomes in U2OS-ERα cells.
We next compared the ERα cistrome for chromosomes 1 and 6 in U2OS-ERα cells with that in MCF7 cells. ERα binds to 1090 sites on chromosomes 1 and 6 in MCF7 cells compared with 1137 sites in U2OS-ERα cells. Strikingly, only 172 sites were common between MCF7 cells and U2OS-ERα cells, representing approximately 15% of the U2OS-ERα sites (Fig. 4B). Similar to what was observed for MCF7 cells, the majority of the ERα binding sites in U2OS-ERα cells are not found at proximal promoters (supplemental Fig. 3).
Figure 4.
Comparison of the ERα Cistromes in U2OS-ERα Cells and MCF7 Cells
A, Directed ChIP was performed on MCF7 and U2OS-ERα cells after 45 min E2 treatment. Quantitative PCR was performed on 10 ERα binding sites in U2OS-ERα cells, five common binding sites and five negative control sites. The 10 ERα binding sites are designated with a U, and the number of the binding site from the list of 1137 sites. The binding sites in both the MCF7 and the U2OS-ERα data sets are designated common. The five negative control sites (not found on either list) are designated Neg1–5. Data are presented as enrichment over background genomic DNA and are the average of three independent experiments. B, The number of ERα binding sites on chromosomes 1 and 6 in U2OS-ERα cells at a false discovery rate of 1% was compared with the number of ERα binding sites on chromosomes 1 and 6 in MCF7 cells under the same conditions. C, Identification of de novo enriched motifs within the U2OS-ERα ERα binding sites revealed a conical ERE half-site as the most enriched. D, Enrichment for the ERE, ERE half-site, forkhead, and activator protein-1 (AP-1) motifs in the center of the binding sites specific to MCF7 cells, U2OS-ERα cells, or a randomly generated data set. The occurrence of the motifs (N motifs) was normalized to the number of sites in each subset (N binding sites).
To verify validity of the ERα cistrome in U2OS-ERα cells and its cell type specificity, ChIP followed by quantitative PCR was performed at 10 sites identified in the U2OS-ERα data set as being unique as well as five sites that were found to be in common with MCF7 cells. None of the sites predicted to be unique to U2OS-ERα cells was found to be occupied in MCF7 cells, whereas all of these sites showed enrichment in U2OS-ERα cells that was greater than 10-fold over background genomic regions (Fig. 4A). The five sites identified as common ERα binding sites in MCF7 and U2OS-ERα cells, as expected, showed significant enrichment over the genomic background in both cell types (Fig. 4A). Finally, five sites containing predicted EREs that were not detected in either the MCF7 or U2OS-ERα cistromes were tested, and as expected, ERα was not recruited to any of these sites in either cell type (Fig. 4A).
We searched the ERα cistromes in MCF7 and U2OS-ERα cells for enriched transcription factor motifs by both de novo and candidate scanning approaches. This screen identified ERE half-sites as the most highly enriched transcription factor motif, as would be expected (Fig. 4, C and D). Activation protein-1 sites were also highly enriched in both cell types (Fig. 4D). Interestingly, the forkhead motif was enriched only in the MCF7 data set and not in the U2OS-ERα data set (7) (Fig. 4D). Importantly, FoxA1 is expressed in MCF7 cells and serves as a pioneer transcription factor for ERα in this cell type, opening chromatin to allow for ERα binding (35). FoxA1 is not expressed in U2OS-ERα cells (supplemental Fig. 4). However, like in MCF7 cells, ERα binds to only a small fraction of all available EREs or ERE half-sites. This suggests that U2OS-ERα cells must use an alternative mechanism that allows ERα to bind to a different subset of ERE-containing regulatory elements. Whether this is independent of (or involves) a different pioneer factor remains to be discovered.
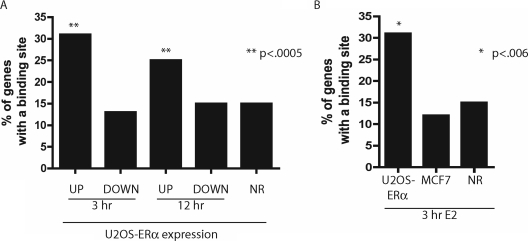
To determine whether the different ERα cistromes found in MCF7 and U2OS-ERα cells were linked to the differential gene regulation by E2, we analyzed the distribution of ERα recruitment sites relative to regulated genes in both cell lines. Correlation of ERα binding sites with E2-induced genes showed a bias of binding sites near both early (3 h up-regulated) and late (12 h up-regulated) E2-induced genes (P < 0.0005) as compared with nonregulated genes (Fig. 5A). Down-regulated genes showed no enrichment of binding sites when compared with the nonregulated genes. In addition, whereas the early up-regulated genes in U2OS-ERα cells had a bias of binding sites nearby, genes up-regulated in MCF7 at 3 h did not show any enrichment of binding sites from the U2OS-ERα data set over nonregulated genes (Fig. 5B). Thus, the ERα cistrome defined in U2OS-ERα cells predicts those genes up-regulated in this cell type and supports the notion that cell type-specific recruitment of ERα actively contributes to lineage-specific transcriptional response to E2.
Figure 5.
ERα Binding in U2OS-ERα Cells Best Predicts Genes Up-Regulated in U2OS-ERα Cells
A, The genes up-regulated or down-regulated at 3 and 12 h (or not regulated, NR) in U2OS-ERα cells were analyzed for the percentage with a binding site in the gene or 10 kb upstream of the transcriptional start site. **, P < 0.0005 compared with nonregulated genes. B, The genes up-regulated (or not regulated, NR) in U2OS-ERα cells and MCF7 cells were analyzed for the percentage with a binding site in the gene or 10 kb upstream of the transcriptional start site. *, P < 0.006 compared with nonregulated genes.
Alkaline Phosphatase Is a Direct Target of ERα
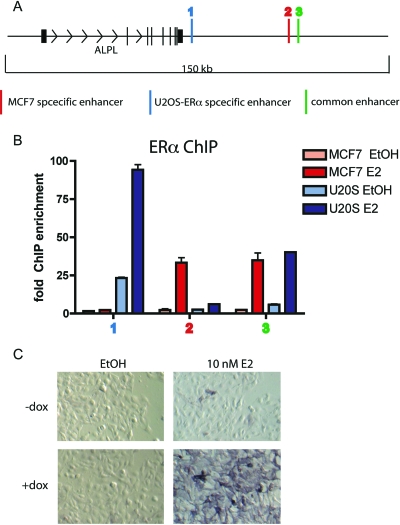
Alkaline phosphatase has been known to be up-regulated in the presence of E2, as compared with a vehicle control, in cell lines (37) and in animals (38). However, it was unknown whether alkaline phosphatase, as a marker of osteoblast differentiation, might be up-regulated indirectly, as a result of E2 up-regulation of other secondary genes responsible for osteoblast differentiation, or whether E2 directly induces ERα binding near the alkaline phosphatase gene. Here, we show for the first time by ChIP that ERα binds to DNA near the ALPL gene (alkaline phosphatase) after E2 treatment for 45 min (Fig. 6, A and B). In U2OS-ERα cells, ERα binds to two enhancers downstream of the ALPL coding region. In MCF7 cells, ERα binds to one of the same enhancers found in U2OS-ERα cells and one unique enhancer (Fig. 6A). After 3 h E2 treatment, ALPL mRNA increases only in U2OS-ERα cells (Fig. 3A) and not in MCF7 cells (data not shown). Together, these data for the first time show that ERα directly induces ALPL mRNA via binding to novel enhancers.
Figure 6.
ERα Regulates Alkaline Phosphatase Transcription in U2OS-ERα Cells
A, Diagram of the ALPL locus. B, U2OS-ERα and MCF7 cells were treated with 10 nm E2 for 45 min, and ChIP was performed with an antibody to ERα. Quantitative PCR was performed with primers to the ALPL enhancers. Error bars represent mean ± 1 sd. C, U2OS-ERα cells were either treated with (to induce ERα) or without DOX for 24 h. The cells were then treated with 10 nm E2 for 24 h and stained for alkaline phosphatase.
To determine whether an increase in ALPL mRNA translates into an increase in alkaline phosphatase protein, U2OS-ERα cells were treated with or without DOX to induce ERα and then treated with 10 nm E2 for 24 h. In the presence of ERα and E2, U2OS-ERα cells stained positive for alkaline phosphatase, suggesting that U2OS-ERα cells are osteoblast-like. Taken together, these experiments show that alkaline phosphatase is directly regulated by E2, in addition to being regulated by 1,25-dihydroxyvitamin D3 (39,40).
E2-Regulated Genes Contain Multiple ERα Binding Sites
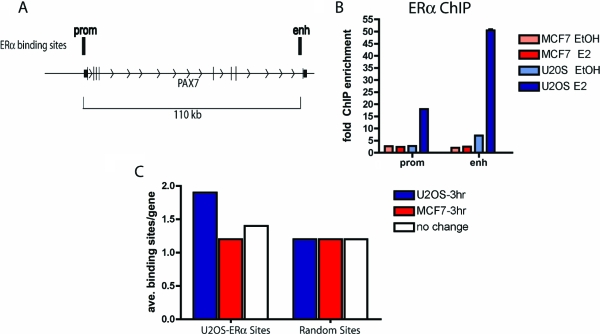
We observed that ERα binding sites often occurred in multiples near a given regulated gene. For example, PAX7 is up-regulated in U2OS-ERα cells (Fig. 3). There are ERα binding sites, identified by ChIP-on-chip, at the promoter of PAX7 and at the 3′ end of the gene (Fig. 7A). Quantitative PCR was performed to verify that ERα is recruited to each of these sites. Indeed, ERα is recruited only in U2OS-ERα to each of these sites (Fig. 7B). To systematically address the question of whether ERα binding occurs more frequently near an up-regulated gene on a genomic scale, we compared the average number of U2OS-ERα binding sites near the up-regulated genes in U2OS-ERα cells with the number of sites near nonregulated genes and genes up-regulated in MCF7 cells. There is an average of almost two binding sites in the genes up-regulated at 3 h in U2OS-ERα cells, compared with an average of 1.2 binding sites in the MCF7 up-regulated genes and 1.4 binding sites in the nonregulated genes in U2OS-ERα cells (Fig. 7C). A randomly generated set of binding sites occurred only an average of 1.2 times in the U2OS-ERα up-regulated genes, MCF7 up-regulated genes, or U2OS-ERα nonregulated genes (Fig. 7C). In the ERα cistrome defined in MCF7 cells, there are also multiple ERα binding sites near early up-regulated genes where there are approximately 2.3 binding sites per gene up-regulated by E2 at 3 h (supplemental Fig. 5).
Figure 7.
Genes Regulated by E2 at 3 h Have a Higher Number of ERα Binding Sites
A, Diagram of the PAX7 locus. B, U2OS-ERα and MCF7 cells were treated with 10 nm E2 for 45 min, and ChIP was performed with an antibody to ERα. Quantitative PCR was performed with primers to the PAX7 enhancer and promoter. Error bars represent mean ± 1 sd. C, Each of the genes up-regulated at 3 h by E2 in U2OS-ERα and MCF7 cells or nonregulated genes in U2OS-ERα cells was analyzed for the number of ERα binding sites in the gene and 10 kb upstream of the gene. A randomly generated set of binding sites was also compared with the genes up-regulated at 3 h in U2OS-ERα and MCF7 cells or nonregulated genes in U2OS-ERα cells.
Chromatin Modifications Correlate with Tissue-Specific Gene Expression
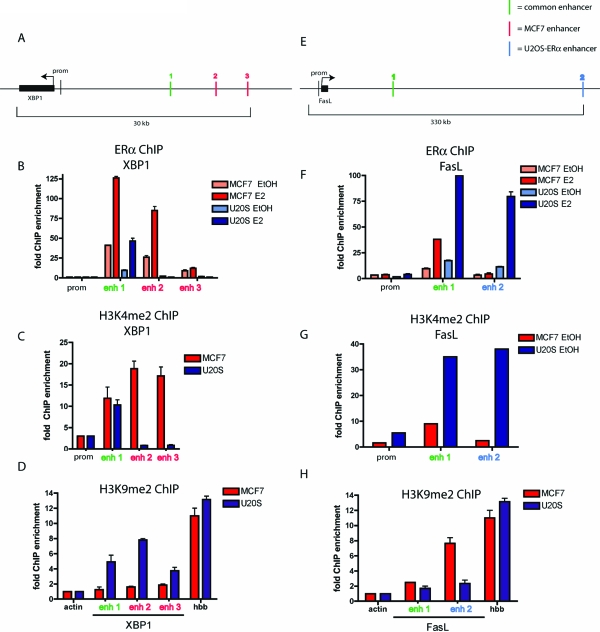
Specific histone modifications are correlated with either active or inactive transcriptional states. Mono- and dimethylation at lysine 4 of histone H3 (H3K4me1 and H3K4me2) are found predominantly near active genes (41,42), whereas dimethylation at lysine 9 of histone H3 (H3K9me2) associates with inactive genes (43). Using XBP1 as a model gene that is specifically induced upon E2 treatment in MCF7 cells and not in U2OS-ERα cells, we analyzed the three ERα-containing enhancers of XBP1 for an enrichment of the activating chromatin modification H3K4me2. ERα is recruited to all three enhancers upstream of the transcriptional start site of XBP1 in MCF7 cells. However, ERα is recruited only to the first enhancer (enhancer 1) in U2OS-ERα cells (Fig. 8B). To determine whether enhancers 2 and 3 had activating modifications in MCF7 cells but not in U2OS-ERα cells, ChIP was performed with an antibody specific to H3K4me2. Correlating with the presence of ERα recruitment, enhancer 1 of XBP1 was enriched for H3K4me2 in both cell types. Only in MCF7 cells are enhancers 2 and 3 enriched for H3K4me2 (Fig. 8C). The H3K4me2 is present before E2 treatment or ERα recruitment. We then analyzed these same enhancers for the repressive chromatin mark H3K9me2 by performing ChIP with an antibody specific for this modification. U2OS-ERα cells show enrichment of H3K9me2 at all three enhancers near XBP1 (Fig. 8D), suggesting that this region is less accessible in this cell type. The promoter of actin was used as a negative control for H3K9me2, because it is actively transcribed. The promoter of hemoglobin (HBB) is used as a positive control for H3K9me2, because it is not transcribed in MCF7 or in U2OS-ERα cells.
Figure 8.
Histones in U2OS-ERα and MCF7 Cells Are Differentially Methylated
A, Diagram of the XBP1 locus with the three enhancers shown as vertical lines. B, U2OS-ERα and MCF7 cells were treated with 10 nm E2 for 45 min, and ChIP was performed with an antibody to ERα. Quantitative PCR was performed with primers to the XBP1 enhancers and promoter. C, ChIP was performed as in B but with an antibody to H3K4me2. D, ChIP was performed as in B but with an antibody to H3K9me2. Quantitative PCR was performed with primers to the XBP1 enhancers, actin promoter, and hemoglobin (HBB) promoter. E, Diagram of the FasL locus with the two enhancers. F, ChIP was performed as in B. Quantitative PCR was performed with primers to the FasL enhancers and promoter. G, ChIP was performed as in B but with an antibody to H3K4me2. Quantitative PCR was performed with primers to the FasL enhancers and promoter. H, ChIP was performed as in B but with an antibody to H3K9me2. Quantitative PCR was performed with primers to the FasL enhancers, actin promoter, and hemoglobin (HBB) promoter. Error bars for all parts represent mean ± 1 sd.
Conversely, a representative gene up-regulated by E2 in U2OS-ERα cells, but not in MCF7 cells, was analyzed for tissue-specific histone modifications. FasL is specifically up-regulated in osteoblasts and U2OS-ERα cells (Fig. 1). ERα is recruited to two enhancers near FasL in U2OS-ERα cells (Fig. 8E). ERα is recruited to only one of these enhancers in MCF7 cells (Fig. 8F). Correlating with ERα recruitment, H3K4me2 is enriched at enhancer 1 in both cell types but only at enhancer 2 in U2OS-ERα cells (Fig. 8G). Conversely, H3K9me2 is high at enhancer 2 in MCF7 cells (Fig. 8H), where there is no ERα recruitment. Taken together, these data demonstrate a link between cell-specific histone modifications and ERα recruitment at enhancers and ultimately differential gene regulation.
DISCUSSION
Defining the mechanisms underlying the cell type-specific regulation of gene expression by E2 is central to understanding its biology. Here we have explored gene regulation by E2 in two cell types. MCF7 cells were used as a model of E2 action in breast epithelial cells, and U2OS-ERα cells were used as model of E2 action in osteoblasts, providing us with new insights into the mechanism of differential transcriptional regulation.
We find very different E2-directed programs in the different cell types. Fewer than 10% of the genes induced in MCF7 cells are induced in U2OS-ERα cells. This is consistent with the very different biological activity of E2 in these two cell types; E2 induces proliferation in breast cancer cells and differentiation in osteoblasts. We found that critical genes involved in proliferation are induced by E2 in MCF7 cells including c-myc and cyclin D1 (44), whereas osteoblast-specific differentiation genes were induced by E2 in U2OS-ERα cells including NFATc1 and alkaline phosphatase. These and other E2-regulated genes found in U2OS-ERα cells were validated in primary calvaria osteoblast cultures.
Tissue specificity in response to E2 in MCF7 and U2OS-ERα cells was not found to be due to differences in ERα expression levels (23) or to differences in response kinetics (supplemental Fig. 1). However, we found by defining the ERα cistromes in these two cell types that fewer than 15% of the binding sites are common to both cell types, consistent with their unique gene expression programs. We show that genes uniquely induced by E2 in U2OS-ERα cells are regulated primarily by cell type-specific binding of ERα.
In addition to finding different ERα cistromes in the two cell types, we find that transcriptional activation may require multiple ERα binding events near a regulated gene. Most of the nuclear receptors have previously been studied in detail on only one or two target genes that are highly regulated. For example, the androgen receptor regulates PSA, the vitamin D receptor regulates cyp24, and ERα regulates pS2. Each of these receptors has been shown to bind to at least three enhancers near its respective classical target gene (45,46,47,48). Here we demonstrate that multiple recruitment sites for ERα at induced target genes is true on the genomic scale. For example, ERα is recruited to three enhancers near the XBP1 gene in MCF7 cells, and transcription is subsequently induced. In U2OS-ERα cells, ERα is not recruited to two of those enhancers, and transcription remains unchanged in the presence of E2. Reciprocally, ERα is recruited to two enhancers near the FasL gene in U2OS-ERα cells but only one of those enhancers in MCF7 cells.
To understand the mechanism of how ERα distinguishes between the binding sites in the two cell types, we analyzed the chromatin modifications at these putative enhancers. It has been shown that the methylation pattern at H3K9 and H3K4 correlates with the binding of FoxA1 in MCF7 (and LNCaP) cells and that FoxA1 acts as a pioneer factor in the recruitment of ERα (6). We have shown here that before E2 treatment, some enhancers have active chromatin marks (dimethylation at histone 3 lysine 4) or heterochromatic marks (dimethylation at histone 3 lysine 9) and that these correlate with ERα binding to DNA. However, unlike in MCF7 cells, the FoxA factors are expressed at low levels in U2OS-ERα cells, and there is no enrichment for the forkhead motif in the ERα cistrome in U2OS-ERα. Thus, whether ERα recruitment is independent of or involves a different pioneer factor in osteoblasts and other estrogen-responsive cell types remains to be determined.
In summary, we have shown that E2 regulates very different transcriptional programs in breast epithelial cells and osteoblasts. Interestingly, we find that the vast majority of the differential gene regulation can be explained by cell type-specific recruitment of ERα to multiple cis-regulatory regions of target genes, primarily enhancers. The activation state of these enhancers is defined by the presence of cell type-specific epigenetic histone modifications. These findings reveal important new insights into the complex biology governed by estrogen and extend the question of cell type-specific estrogen regulation to how the epigenetic signature of active enhancers is determined during development.
MATERIALS AND METHODS
Reagents
17β-Estradiol (E2) and DOX were purchased from Sigma-Aldrich Co. (St. Louis, MO). The following antibodies were used: ERα (HC-20 from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, and Ab-10 from Thermo Scientific, Fremont, CA), H3K4me2 (ab7766 from Abcam Inc., Cambridge, MA), and H3K9me2 (Upstate, Lake Placid, NY).
Cell Culture
MCF7 cells and MDA-MB-231 cells were obtained from American Type Culture Collection (Manassas, VA). MDA-MB-231 cells stably transfected with ERα were previously described (39). U2OS-ERα cells, kindly provided by Dr. Thomas Spelsberg, were maintained as described (21). Twenty-four hours before treatment with E2, ERα expression in U2OS-ERα cells was induced by treatment with 100 ng/ml DOX.
Primary Osteoblast Cells
All animal work was approved by the Animal Care and Use Committee at Dana-Farber Cancer Institute. Neonatal BALB/c calvaria were obtained 2 d after birth and incubated for 40 min in αMEM/1.0 mg/ml collagenase P/1.25% trypsin at 37 C. These were washed in αMEM and transferred to αMEM/1.0 mg/ml collagenase P/1.25% trypsin for 1 h at 37 C (49). Digestion was stopped by addition of αMEM/10% fetal bovine serum (FBS). The cells from the second digest were allowed to attach for 48 h and then differentiated in mineralization medium with media replacement every 3 d. Differentiation was confirmed by quantitation of bone sialoprotein and osteocalcin mRNA, alkaline phosphatase positivity, and Von Kossa staining for mineralization.
RNA and Quantitative PCR
Total RNA was converted to cDNA with Superscript III First-Strand Synthesis Kit according to the manufacturer’s instructions (Invitrogen Corp., Carlsbad, CA). Primers were selected using Primer3 (50), and the sequences are listed in supplemental Tables 1–3 (published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). cDNA was subjected to quantitative PCR using the Applied Biosystems (Foster City, CA) SYBR Green Mastermix. Each RNA sample was collected in triplicate, and each PCR was amplified in triplicate.
Expression Microarrays
MCF7 and U2OS-ERα cells were deprived of hormones as previously described (15) and stimulated with 10 nm E2 for 0, 3, 6, or 12 h, after which total RNA was collected using the RNEasy Kit (QIAGEN, Valencia, CA). Expression microarrays were Affymetrix U133Plus2.0, and all experiments were performed in triplicate. Data were analyzed using GeneSpring (Agilent Technologies, Santa Clara, CA). The data have been submitted to the Gene Expression Omnibus (GEO).
ChIP
Cells were hormone deprived by culture for 3 d in phenol red-free medium (Invitrogen) supplemented with 5% charcoal-dextran-treated (CDT)-FBS. Cells were treated with 10 nm E2 or ethanol as a vehicle control for 45 min, and ChIP was performed as described (7,35). Immunoprecipitated DNA was amplified by quantitative PCR using the Applied Biosystems SYBR Green Mastermix. Each ChIP was performed in triplicate, and each PCR was amplified in triplicate. Error bars represent mean and 1 sd.
ChIP-on-Chip
ChIP-on-chip experiments using Array A of the Affymetrix (Santa Clara, CA) Human Tiling 2.0R Array Set were performed as previously described (6,7) with three independent biological replicates conducted. Analyses were performed using MAT (36). We used a statistical false discovery rate of 1% as a cutoff in all analyses. ChIP-quantitative PCR experiments were performed as described previously (7). All ChIP-on-chip data used in this study can be accessed at http://research.dfci.harvard.edu/brownlab/datasets/. De novo motif searches were performed on sequences ±100 bp from the center of ERα ChIP regions in U2OS-ERα cells as previously described (35). Location analysis of binding sites was performed using the cis-regulatory element annotation system (51).
Association of Trends in Gene Expression with Transcription Factor Binding Sites
Genes close to a ChIP region were defined as those having such a binding site 10 kb upstream of the transcription start site or within the exons or introns of the gene. Fisher’s exact test was used to assess the statistical significance of the association between close genes and up-regulated genes. The number of binding sites per gene was performed using the ChIP regions and 10 kb upstream of the transcription start site or within the exons or introns of the gene.
Alkaline Phosphatase
U2OS-ERα cells were either treated with (to induce ERα) or without DOX for 24 h. The cells were then treated with 10 nm E2 for 24 h, fixed with 3.7% formaldehyde, and stained for alkaline phosphatase (Sigma).
Supplementary Material
Footnotes
This work was supported by a postdoctoral fellowship from the Susan G. Komen for the Cure Foundation to S.A.K. and by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK074967 to M.B.), the National Cancer Institute (P01 CA8011105 and the DF/HCC Breast Cancer SPORE Grant to M.B.), and the Dana-Farber Cancer Institute Women’s Cancers Program.
Disclosure Statement: S.A.K., G.A.M.-C., M.L., J.E., and J.S.C. have nothing to disclose. M.B. has received sponsored research support from and is a consultant and scientific advisor to Wyeth Pharmaceuticals.
First Published Online September 25, 2001
Abbreviations: CDT, Charcoal-dextran-treated; ChIP, chromatin immunoprecipitation; DOX, doxycycline; E2, 17β-estradiol; ERα, estrogen receptor; ERE, estrogen response element; FasL, Fas ligand; FBS, fetal bovine serum; GPR30, G protein-coupled receptor 30; Myt1, myelin transcription factor 1; NFATc1, nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1; NR5A2, nuclear receptor subfamily 5 member 2.
References
- Yager JD, Davidson NE 2006 Estrogen carcinogenesis in breast cancer. N Engl J Med 354:270–282 [DOI] [PubMed] [Google Scholar]
- Bodine PV, Henderson RA, Green J, Aronow M, Owen T, Stein GS, Lian JB, Komm BS 1998 Estrogen receptor-α is developmentally regulated during osteoblast differentiation and contributes to selective responsiveness of gene expression. Endocrinology 139:2048–2057 [DOI] [PubMed] [Google Scholar]
- Simoncini T, Mannella P, Fornari L, Caruso A, Varone G, Genazzani AR 2004 Genomic and non-genomic effects of estrogens on endothelial cells. Steroids 69:537–542 [DOI] [PubMed] [Google Scholar]
- Castro-Rivera E, Samudio I, Safe S 2001 Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J Biol Chem 276:30853–30861 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Brown M 2006 Estrogen receptor target gene: an evolving concept. Mol Endocrinol 20:1707–1714 [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008 FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M 2006 Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S 2004 Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18:1411–1427 [DOI] [PubMed] [Google Scholar]
- Vanyushin BF 2005 Enzymatic DNA methylation is an epigenetic control for genetic functions of the cell. Biochemistry (Mosc) 70:488–499 [DOI] [PubMed] [Google Scholar]
- Simpson ER, Clyne C, Speed C, Rubin G, Bulun S 2001 Tissue-specific estrogen biosynthesis and metabolism. Ann NY Acad Sci 949:58–67 [DOI] [PubMed] [Google Scholar]
- Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC 2005 Estrogen receptor α and β heterodimers exert unique effects on estrogen- and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol 19:1555–1568 [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M 2002 Molecular determinants for the tissue specificity of SERMs. Science 295:2465–2468 [DOI] [PubMed] [Google Scholar]
- Kameda T, Mano H, Yuasa T, Mori Y, Miyazawa K, Shiokawa M, Nakamaru Y, Hiroi E, Hiura K, Kameda A, Yang NN, Hakeda Y, Kumegawa M 1997 Estrogen inhibits bone resorption by directly inducing apoptosis of the bone-resorbing osteoclasts. J Exp Med 186:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousteni S, Chen JR, Bellido T, Han L, Ali AA, O'Brien CA, Plotkin L, Fu Q, Mancino AT, Wen Y, Vertino AM, Powers CC, Stewart SA, Ebert R, Parfitt AM, Weinstein RS, Jilka RL, Manolagas SC 2002 Reversal of bone loss in mice by nongenotropic signaling of sex steroids. Science 298:843–846 [DOI] [PubMed] [Google Scholar]
- Krum SA, Miranda-Carboni GA, Hauschka PV, Carroll JS, Lane TF, Freedman LP, Brown M 2008 Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J 27:535–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum SA, Brown M 2008 Unraveling estrogen action in osteoporosis. Cell Cycle 7:1348–1352 [DOI] [PubMed] [Google Scholar]
- Zallone A 2006 Direct and indirect estrogen actions on osteoblasts and osteoclasts. Ann NY Acad Sci 1068:173–179 [DOI] [PubMed] [Google Scholar]
- Syed F, Khosla S 2005 Mechanisms of sex steroid effects on bone. Biochem Biophys Res Commun 328:688–696 [DOI] [PubMed] [Google Scholar]
- Qu Q, Perala-Heape M, Kapanen A, Dahllund J, Salo J, Vaananen HK, Harkonen P 1998 Estrogen enhances differentiation of osteoblasts in mouse bone marrow culture. Bone 22:201–209 [DOI] [PubMed] [Google Scholar]
- Kousteni S, Almeida M, Han L, Bellido T, Jilka RL, Manolagas SC 2007 Induction of osteoblast differentiation by selective activation of kinase-mediated actions of the estrogen receptor. Mol Cell Biol 27:1516–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem M, Harris SA, Spelsberg TC, Riggs BL 1996 Estrogen inhibits interleukin-6 production and gene expression in a human osteoblastic cell line with high levels of estrogen receptors. J Bone Miner Res 11:193–199 [DOI] [PubMed] [Google Scholar]
- Chen XW, Garner SC, Anderson JJ 2002 Isoflavones regulate interleukin-6 and osteoprotegerin synthesis during osteoblast cell differentiation via an estrogen-receptor-dependent pathway. Biochem Biophys Res Commun 295:417–422 [DOI] [PubMed] [Google Scholar]
- Monroe DG, Getz BJ, Johnsen SA, Riggs BL, Khosla S, Spelsberg TC 2003 Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERα or ERβ. J Cell Biochem 90:315–326 [DOI] [PubMed] [Google Scholar]
- Rickard DJ, Hofbauer LC, Bonde SK, Gori F, Spelsberg TC, Riggs BL 1998 Bone morphogenetic protein-6 production in human osteoblastic cell lines. Selective regulation by estrogen. J Clin Invest 101:413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER 2005 A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science 307:1625–1630 [DOI] [PubMed] [Google Scholar]
- Chagin AS, Savendahl L 2007 GPR30 estrogen receptor expression in the growth plate declines as puberty progresses. J Clin Endocrinol Metab 92:4873–4877 [DOI] [PubMed] [Google Scholar]
- Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, Crabtree GR 2006 Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Dev Cell 10:771–782 [DOI] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H 2005 NFAT and Osterix cooperatively regulate bone formation. Nat Med 11:880–885 [DOI] [PubMed] [Google Scholar]
- Annicotte JS, Chavey C, Servant N, Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F, Maudelonde T, Lazennec G, Cavailles V, Fajas L 2005 The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene 24:8167–8175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcerzak M, Hamade E, Zhang L, Pikula S, Azzar G, Radisson J, Bandorowicz-Pikula J, Buchet R 2003 The roles of annexins and alkaline phosphatase in mineralization process. Acta Biochim Pol 50:1019–1038 [PubMed] [Google Scholar]
- Huang H, Tindall DJ 2007 Dynamic FoxO transcription factors. J Cell Sci 120:2479–2487 [DOI] [PubMed] [Google Scholar]
- Romm E, Nielsen JA, Kim JG, Hudson LD 2005 Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem 93:1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Stoykova A, Torres M, Gruss P 1996 Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development 122:831–838 [DOI] [PubMed] [Google Scholar]
- Prior HM, Walter MA 1996 SOX genes: architects of development. Mol Med 2:405–412 [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M 2005 Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006 Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA 103:12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheven BA, Damen CA, Hamilton NJ, Verhaar HJ, Duursma SA 1992 Stimulatory effects of estrogen and progesterone on proliferation and differentiation of normal human osteoblast-like cells in vitro. Biochem Biophys Res Commun 186:54–60 [DOI] [PubMed] [Google Scholar]
- Plant A, Tobias JH 2001 Characterisation of the temporal sequence of osteoblast gene expression during estrogen-induced osteogenesis in female mice. J Cell Biochem 82:683–691 [DOI] [PubMed] [Google Scholar]
- Rodan SB, Wesolowski G, Hilton DJ, Nicola NA, Rodan GA 1990 Leukemia inhibitory factor binds with high affinity to preosteoblastic RCT-1 cells and potentiates the retinoic acid induction of alkaline phosphatase. Endocrinology 127:1602–1608 [DOI] [PubMed] [Google Scholar]
- van Leeuwen JP, van Driel M, van den Bemd GJ, Pols HA 2001 Vitamin D control of osteoblast function and bone extracellular matrix mineralization. Crit Rev Eukaryot Gene Expr 11:199–226 [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002 Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas 3rd EJ, Gingeras TR, Schreiber SL, Lander ES 2005 Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169–181 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007 High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M 2006 A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 20:2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J 1996 Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem 271:6379–6388 [DOI] [PubMed] [Google Scholar]
- Carlberg C, Dunlop TW, Saramaki A, Sinkkonen L, Matilainen M, Vaisanen S 2007 Controlling the chromatin organization of vitamin D target genes by multiple vitamin D receptor binding sites. J Steroid Biochem Mol Biol 103:338–343 [DOI] [PubMed] [Google Scholar]
- Martinez E, Givel F, Wahli W 1987 The estrogen-responsive element as an inducible enhancer: DNA sequence requirements and conversion to a glucocorticoid-responsive element. EMBO J 6:3719–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J 1997 An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol 11:148–161 [DOI] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G 1999 A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev 13:1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000 Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS 2006 CEAS: cis-regulatory element annotation system. Nucleic Acids Res 34:W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.