Abstract
Here, we describe a multi-parametric study of DNA hybridization to probes with 20–70% G + C content. Probes were designed towards 71 different sites/mutations in the phenylalanine hydroxylase gene. Seven probe lengths, three spacer lengths and six stringencies were systematically varied. The three spacer lengths were obtained by placing the gene-specific sequence in discrete steps along the 60-mer probes. The study was performed using Agilent 8 × 15 000 probes custom-made arrays and a home-built array washer providing different stringencies to each of the eight sub-arrays on the slides. Investigation of hybridization signals, specificity and dissociation curves indicated that probes close to the surface were influenced by an additional stringency provided by the microarray surface. Consistent with this, probes close to the surface required 4 × SSC, while probes placed away from the surface required 0.35 × SSC wash buffers in order to give accurate genotyping results. Multiple step dissociation was frequently observed for probes placed furthest away from surface, but not for probes placed proximal to the surface, which is consistent with the hypothesis that there is different stringency along the 60-mer. The results have impact on design of probes for genotyping, gene expression and comparative genome hybridization analysis.
INTRODUCTION
DNA microarrays technology has evolved rapidly in the last decade. The technique has been used as a platform for many different applications including gene expression profiling (1), microbial detection (2), single nucleotide polymorphism (SNP) genotyping (3), comparative genome hybridization (4), ChIP on chip analysis (5) and miRNA detection (6). Hybridization of complementary nucleic acids in solution is well described using thermodynamic nearest neighbor (NN) parameters (7). Although somewhat relevant, solution parameters used in thermodynamic models do not fully predict hybridization on microarray surface (7–9), because among others surface effects are normally not accounted for (10,11). An exception to this is the Hyther server (12) that makes linear correction of solution-based thermodynamic parameters, based on published microarray data obtained on a single substrate (13), and is therefore not directly applicable to many of the commonly used microarray substrates.
Constraints induced by tethering one end of the reactant (capture probe) to a solid support include the following: (i) the probe is not free to diffuse as it would be in solution, which reduces the reaction rate (14) and (ii) steric hindrance prevents the target in solution to make close approach/hybridize to the immobilized probe (15). Steric hindrance is however a general term that covers multiple factors negatively affecting the hybridization reaction between immobilized probe and target. Steric hindrance encompasses both physical constraints involving less accessibility at surface and electrostatic and physiochemical effects occurring between nucleic acids and surface. In the following, we will use the term surface effect, with disregard to physical constraints, to describe the factors affecting hybridization in a microarray spot. Surface effects are determined by electrostatic interactions between surface and probe (10,16), probe density (15,17,18) and physiochemical properties of the substrate, such as hydrophobicity/hydrophilicity. Significant electrostatic effects are predicted when using common glass substrate, with a negative potential at hybridization conditions (−30 mV to −80 mV). For short probe-surface distances these effects are most dramatic and may reduce the melting temperature (Tm) by tens of degree Celsius (10). The electrostatic effects from a microarray surface can be reduced, and higher hybridization signal achieved by using spacers molecules (15,19) and/or three-dimensional coatings (13,20–22), to position probes further away from the surface. Shchepinov et al. (15) found that the length as opposed to charge and hydrophobicity of spacer molecules was the most important factor for hybridization yield, and optimal spacer length was about 45–60 atoms, corresponding to about 8–10 nt. Using spotted arrays, Peplies et al. (23) tested different lengths (6-mer, 12-mer, 18-mer, 24-mer) of poly A spacer on probes (15–20-mer) targeting bacterial 16S rRNA. A linear relationship between spacer length and signal was observed when hybridizing to the ssDNA target (amplified from rRNA). However, when helper oligonucleotides (chaperones) were used to make target more accessible to probes, saturation in signal was observed already using shorter (6-mer or 18-mer) spacers. Based on these findings, the authors ascribed secondary structures in target molecules as the main factor affecting microarray hybridization signal in their study.
Probe density can also affect hybridization yield because high probe densities effectively cluster large number of negatively charged DNA together in the spot. This was confirmed experimentally using cleavable linkers where the highest hybridization yield was found at a probe density of about 50% of the maximum possible probe density (15). Electrostatic effect of probes and hybridized target was further demonstrated by Yao et al. (24) who examined hybridization of a DNA target (PCR amplicons) to oligonucleotide (DNA) or peptide nucleic acids (PNA) probes attached on streptavidin-modified gold surface via biotin/steptavidin interaction. Hybridization of targets to densely packed surfaces of DNA or PNA probes at low sodium concentration was postulated to result in the following effects: (i) less secondary structure of targets, thus better hybridization, (ii) more electrostatic interactions between hybridized DNA targets, decreasing hybridization and for DNA probes, (iii) electrostatic repulsion of negatively charged DNA probe and target. Hence, although PNA has a neutral backbone, electrostatic effects were observed between hybridized targets, and this effect could be decreased when the PNA probe layer was sufficiently diluted. DNA probes showed a more dramatic response to the ionic effect, and duplex formation decreased rapidly at rather high (150 mM) sodium concentration. Other factors influencing microarray hybridization signal include spot size (25), probe length and G + C content (2,26,27), target length (28), concentration (29) and complexity (30), labeling of target (31), temperature and composition of hybridization buffer (2,26) and/or stringency wash temperature (32) and buffer (Poulsen et al., unpublished data). Whereas signal intensities generally increase with probe length (2,29), there is an inverse relation between probe length and specificity (27,29).
Although the effect of probe length, spacer length and experimental conditions on microarray hybridization yield and specificity have been extensively reported (1,2,15,23,27), a larger study simultaneously modulating these factors has to our best knowledge not been reported. Especially, the stringency is seldom varied in probe characteristics studies (15,25,28,29,33,34) despite being a critical factor for obtaining signal and specificity. Consequently, investigation of factors that can influence hybridization signal and specificity can be under- or overestimated in significance because some combinations of these factors may have other assay optima than tested.
Here, we describe the use of high-density commercial arrays combined with an in-house built multi-stringency array washer (MSAW) that enabled systematic study of the importance of experimental parameters to hybridization signal and specificity. The parameters modulated were the length of the capture probe, the position of capture probe relative to the microarray surface and finally the stringency wash. We used a model system comprising different sites in the human phenylalaninehydroxylase (PAH) gene, covering a wide range (∼20–70%) of G + C content. We show how the interplay between the parameters investigated affected signal, specificity and melting curve shapes. Using linear regressions between experimental data and calculated melting temperature Tm and Gibbs free energy (ΔG), we demonstrate that the correlation to thermodynamic predictions is modulated by the distance between probe and surface and the experimental condition (stringency). Using a specific application comprising genotyping PAH mutations corroborates that distance from surface affects requirements for assay stringency and melting temperature. Finally, we discuss the implications of our results on probe design.
MATERIALS AND METHODS
Design and fabrication of the MSAW
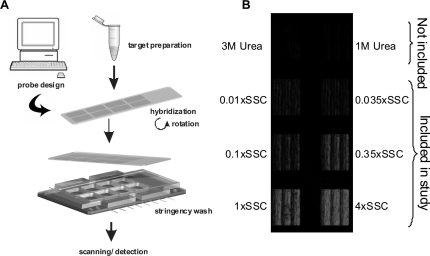
For wash of hybridized 8 × 15K Agilent arrays (Agilent Technologies, Palo Alto, CA, USA) a MSAW was designed and fabricated. The washer consisted of: (i) a bottom layer for buffer handling, (ii) an elastic layer defining the eight wash chambers and an alignment groove for the slide and (iii) a pressure lid (see Figure 1 and Supplementary Figure 1). For the bottom layer and pressure lid, polymethylmethacrylate (PMMA) sheets were micromachined on a laser ablation system (Synrad Inc., Mukilteo, WA, USA) controlled by computer assisted design (CAD) software (winMark Pro, Synrad). A schematic layout of the position of the respective sub-arrays provided by Agilent Technologies served as a guide when designing the washer. Flow pressure barriers (35) were implemented to ensure uniform flow in the large (10.7 × 14.6 mm) wash chambers. Bubble traps were incorporated in the inlet channel in the bottom PMMA layer (Supplementary Figure 1B) to prevent bubbles from reaching the arrays. The fabricated PMMA parts were aligned and heat bonded between glass blocks at 100°C for 90 min with a pressure of 100 cNm using a torque screw-driver (Lindstrom, Orange, CA, USA). The elastic polydimethylsiloxane (PDMS) layer was molded from a PMMA master structure with the inverse shape of the washing chambers, which was produced by computer-assisted micromilling (Folken Industries, Glendale, CA, USA). The master was fixed to the bottom of a small container; the PDMS prepolymer and catalyst (Sylgard 184, Dow Corning, Germany) were mixed and degassed under vacuum for 30 min, and poured onto the PMMA master. The PDMS was cured for 3 h at 60°C, after which the PMMA master was removed, which resulted in eight wash chambers and an alignment groove for the microarray slide. The MSAW device allowed each identical sub-arrays of the Agilent slide (see below) to be washed at different stringency, using wash buffers of varying ionic strength. Proper filling, bubble-free operation and no leakage were observed when pumping a water solution containing methylene blue through the device (Supplementary Figure 1A).
Figure 1.
The experimental setup and image of a processed slide. (A) Schematic overview of the experimental setup. Probe-design was performed in silico (computer) using NN algorithms to Tm-match PM/MM probe pairs. In situ synthesized microarrays, with eight identical sub-arrays were obtained from Agilent Technologies. Target amplification and labeling was performed by T7-IVT of pooled PCR products. Hybridization was performed in a dedicated hybridization station using bubble mixing during rotation. Hybridized arrays were subjected to stringency wash in an in-house MSAW, with eight different chambers, each processed with different stringency wash buffer. Finally, fluorescence detection of processed microarrays was performed in a confocal scanner. (B) A scanning image of a hybridized Agilent slide, containing eight identical sub-arrays that were processed at eight different stringencies (as indicated) in the MSAW. Data analysis did not include the two uppermost stringency conditions (wash buffers supplemented with 1 M or 3 M Urea) as the mechanism of dissociation differs when using a denaturant as compared to different sodium concentrations.
Spacer sequence selection
Effort was made to identify a spacer sequence (tag) that did not hybridize to the PAH target. Initial screening of published tags (36,37) and other sequences (Supplementary Table 1) was performed in silico using BlastN for short nearly exact matches, i.e. window/word size of 7. Candidates with E-values <25 were excluded, and the remaining candidates tested for hairpin structures using mFold with the following settings: nucleotide type = DNA and Na+ concentration = 37 mM. A total of 24 tags (Supplementary Table 1) including the most promising candidates from above screening and tags previously effectively immobilized to agarose (38) were experimentally tested. Spotted arrays of probes with the respective tag sequence in their 5′-end followed by a β-globin sequence 5′-GAAGTTGGTGGTGAGGCCC-3′ were individually hybridized with each amplified PAH exon to exclude tags that hybridized to PAH fragments. Furthermore the arrays were hybridized with target complementary to the β-globin sequence, and this hybridization served as positive control of spotting and immobilization of probes.
Selection of stringency buffers
From the selected spacer sequence above, probes were designed that targeted a GC-rich (c.734T > C) and an AT-rich (c.1315 + 1G > A) site in the PAH gene, of the lengths 13 nt, 17 nt or 25 nt. All probes contained the 25-nt spacer sequence in the 5′-end, and were spotted in identical sub-arrays, hybridized with target (see below) and subjected to wash with different buffers at 37°C.
DNA microarray probes
For the Agilent arrays, perfect match (PM) and mismatch (MM) probes were designed to target 71 different wild-type or mutant sites in the human PAH gene, respectively (Supplementary Table 2). Most MM probes had a single base substitution or deletion compared to the corresponding probe capturing PM sequence; however, MM probes targeting five sites had 2 or 3 point mismatches (deletions or substitutions). For each site, seven lengths of PM probes were designed, ranging from 13 nt to 25 nt with 2-nt increments. To form a probe pair with each length of PM probe, MM probes were designed to be as closely Tm matched with their respective PM probes as possible. This was performed using the OligoAnalyzer 3.0 (http://www.idtdna.com) using the settings: Target type = RNA, oligo concentration = 0.1 µM and Na+ concentration = 37 mM. Change in Gibbs free energy of hybridization at 37°C (ΔG°37) was calculated for probe capture sequences using the parameters published in ref. (39) for DNA/RNA hybridization at 1 M Na+. For the remaining of article, ΔG°37 will simply be referred to as ΔG. The default length of an Agilent microarray probe is 60 nt. We therefore divided each probe into three sections, proximal nearest surface, central and distal, furthest away from surface. Placement of the PAH-specific part of the 60-mer probe was obtained by varying the position of the selected spacer/tag sequence. The same tag sequence also served as fill sequence towards solution to obtain 60-mer probes. Full-length (25-mer), partial repetition or 3′-truncated sequence of the tag sequence was used to accommodate the desired length and placement of the PAH-specific probe sequence. The seven lengths of probes targeting 71 PAH sites were designed with three different spacer lengths: proximal probes (P) with no spacer, where the capture sequence was attached to the substrate and followed by a long (35–47-mer) fill sequence; central probes (C), where the capture sequence was positioned in the centre of the 60-mer probe, and on a short (18–24-mer) spacer and short (17–23-mer) fill sequence towards the liquid; and finally distal probes (D), with the capture sequence furthest away from the substrate, on a long spacer (35–47 nt). With the limitation of the 60-mer oligo, there was an inherent variation in spacer length and fill sequence towards liquid, e.g. short capture sequences of C and D probes had longer spacers than long capture sequence probes (Supplementary Figure 2). For negative control a probe consisting of repetitions of the spacer sequence was used.
Fabrication of DNA microarrays
Control experiments such as finding suitable washing buffers and spacer/tag were made using microarrays spotted on agarose coated slides (40) as previously described (32). For testing effects of probe and spacer length and different stringencies, we used custom-made (eArray 4.5) high-density Agilent arrays in the format 8 × 15K expression arrays. Triplicates of each probe were present in the eight identical sub-arrays.
DNA samples and target preparation
The DNA samples used in this study originated from 38 individuals that were heterozygous (n = 31) or homozygous (n = 7) for mutations in the PAH gene (Supplementary Table 3). The original molecular diagnosis was made by denaturing gradient gel electrophoresis (DGGE) (41) followed by direct DNA sequencing. The use of patient material carrying different mutations gave us information whether or not the probes were working selectively in the assay.
Exons 1–12 with flanking sequences of the PAH gene were individually amplified by PCR using the primer pairs in Supplementary Table 4. The PCR products ranged in size from about 150 bp to 330 bp. DNA samples were amplified in a total volume of 40 μl containing 0.67 μM of each primer (forward and reverse), 200 μM of each dNTP, 0.1 U/μl TEMPase Hot Start DNA polymerase (Ampliqon, Bie & Berntsen A/S, Rødovre, Denmark), 3 mM MgCl2 and 1 × TEMPase Buffer I provided with the enzyme. The reverse primers contained a T7 promoter sequence in the 5′-end and the resulting PCR fragments could thereby serve as DNA template for subsequent T7 RNA polymerase amplification. The PCR cycling conditions were 15 min at 95°C followed by 35 amplification cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final extension at 72°C for 7 min. PCR products were confirmed and quantified on an Agilent Bioanalyzer 2100 (Agilent Technologies) using a DNA 500 LabChip kit (Agilent Technologies). The PCR products were used directly (without purification steps) as a template for the T7 in vitro transcription (IVT).
For each of the 38 individuals, single-stranded RNA target was produced by T7 IVT in an 80-μl reaction mixture containing 47 μl of a mixture with equimolar concentrations of each PCR-amplified PAH fragment, 500 μM of each NTP, 12.5 μM (2.5%) Cy3-CTP (GE Healthcare Bio-Sciences Corp, Piscataway, NJ, USA), 0.0025 U/μl inorganic pyrophosphatase (Sigma-Aldrich, Brøndby, Denmark), 1 U/μl T7 RNA Polymerase-Plus™ (Ambion, Huntingdon, Cambridgeshire, UK) and 1 × transcription buffer provided with the enzyme. Transcription was performed overnight at 37°C.
Hybridization and stringent wash
Prior to mixing, all components of the hybridization mix were heated at 95°C for 1 min. Amplified RNA target of 80 μl was mixed with 170 µl of MilliQ water and 250 µl of hybridization buffer (10 × SSC and 1% SDS) and centrifuged at 13 000 r.p.m. for 1 min. Hybridization was performed with bubble mixing in a hybridization chamber (Agilent Technologies) covering all eight sub-arrays at 37°C for 4 h according to manufacturers instructions. Disassembly of the microarray slide and gasket slide was performed in a 2 × phosphate buffered saline (PBS) buffer at room temperature (RT).
The dried microarray slide was positioned in the MSAW device (Figure 1 and Supplementary Figure 1A), and each sub-array was washed with different stringency wash buffers. All wash buffers contained 0.1% of SDS, but had different concentrations of SSC buffer (4 × SSC, 1 × SSC, 0.35 × SSC, 0.1 × SSC, 0.035 × SSC and 0.01 × SSC). This corresponded to a sodium concentration of approximately 660 mM, 165 mM, 58 mM, 17 mM, 6.5 mM, and 2.5 mM, respectively. During the wash, an equal flow rate 0.67 ml/min was obtained using an eight-channel peristaltic pump (Watson-Marlow Alitea, Stockholm, Sweden). The stringent wash was performed at 37°C (in a heat incubator) for 30 min, with wash buffers that were prewarmed to 37°C. Subsequently, the slide was dismounted from the MSAW, washed in 2 × PBS for 5 min at RT and dried.
Detection, quantification and data analysis
The processed microarrays were visualized on a Packard ScanArray Lite scanner (PerkinElmer Life Sciences, Boston, MA, USA) at a laser power of 65%, a PMT gain of 65%, and a resolution of 5 μm. The resulting images (TIFF) were quantified using the GenePix Pro 6.1 software (Molecular Devices, Sunnyvale, CA, USA). All sub-arrays were inspected visually and spots in areas with dust or other abnormalities were manually flagged. Data extracted with the software were imported into Microsoft Excel for further analysis. Local median background signal was subtracted from the mean spot intensity, and signal threshold was set at >2 SD of background signal. Subsequently, an average signal of the probe replicates in each sub-array was calculated if following criteria were fulfilled: (i) ≥2 out of three replicates had signal above threshold and (ii) SD of the replicates divided by the average signal was <0.5.
In order to determine the specificity of the PM probes and to genotype a subset of the PAH sites (Supplementary Table 2 and 3), a normalized ratio was calculated. This was done for each PM and MM probe pair in the separate stringency zones. This ratio was calculated by dividing the average signal from the PM probe by the sum of the signals from the average PM and average MM probes (SPM/(SPM + SMM)). This means, that at an appropriate stringency during the post-hybridization washing step, the normalized ratio of homozygous wild-type hybridization should approach an ideal value of 1.0, which represents a much more intense signal from the perfect match probe than from the mismatch probe. The ideal value for heterozygotes is 0.5, representing equal signal intensity for PM and MM probe, and about 0 for homozygous mutated, which represent a much more intense signal from the MM probe than the PM probe. For clearer measure of specificity of PM probes, the normalized ratio was transformed back into a a more clear signal ratio (SPM/SMM) using the following algorithm: Signal ratio = Normalized ratio/(1 − Normalized ratio).
Stripping procedure for hybridized slides
For reuse of slides, hybridized targets were stripped of using a modified version of the protocol of Hahnke et al. (42). Immediately after scanning the hybridized slides were washed in 0.05 N NaOH for 10 min at RT, followed by 1% SDS for 10 min at 70°C and finally neutralized in 0.1 × PBS for 5 min at RT. After the treatment, the microarray was scanned as described above. If no signal was observed by visual inspection of the image file, the slides were considered ready for reuse; otherwise the stripping procedure was repeated until no signal was observed.
RESULTS
Assay design
Screening of 24 different candidate sequences to serve as spacer and fill sequence for the high-density arrays, resulted in selection of a sequence (tag) 5′-AAGTATTCGTTCACTTCCGATATGC-3′ (36) that showed low cross-hybridization to the PAH targets. This was confirmed on the negative control spots on the Agilent arrays that had three repeats of the spacer (tag) sequence. Using this sequence in combination with sequences of 71 different sites in the PAH gene (Supplementary Table 2), 60-mer probes were designed. To obtain the appropriate length of the spacer/fill, repeats or truncated sequences of the above selected tag were used. In each probe, the PAH capture sequence was designed with seven different lengths, 13–25 nt in steps of 2 nt, and three different positions relative to the microarray surface [proximal (‘P’), central (‘C’) and distal (‘D’)]. The positioning of the PAH capture sequence was obtained by varying the location of the spacer and/or fill sequence (Supplementary Figure 2). The Agilent 8 × 15K array format was chosen for the experiment, because it allowed a multi-parametric test of probe design (about 15 000 probes per sub-array) in combination with multi-stringency wash of the eight identical sub-arrays (Figure 1), and thus making the experiment practically and economically feasible. To further increase data output, each microarray was re-used up to three times by the stripping of hybridized targets. Although the signal decreased for each successive re-hybridization, we were able to assign genotypes (data not shown).
The systematic study of the importance of experimental parameters to hybridization signal, specificity and dissociation curves, however, only included data from slides hybridized once (not re-used). The analysis included 64 of the 71 PAH sites, excluding seven PAH sites/mutations in proximity of common SNPs. With triplicates of each probe sequence the total number of analyzed probes in each sub-array was about 4100. The remaining probes, including probes with common SNPs and variant of PM probes, were not included in this analysis. The six analyzed sub-arrays of 13 hybridized microarray slides resulted in nearly 320 000 measurements.
To assess the homogeneity of the hybridization signal between each sub-array after a stringent wash, a control experiment was performed, where all six sub-arrays of a hybridized Agilent microarray were washed with the same buffer (0.1 × SSC, 0.1% SDS). This experiment showed a maximum of 20% variation in the average signal of all probes between the sub-arrays (data not shown).
Hybridization signal varies at different stringency and position of capture probe
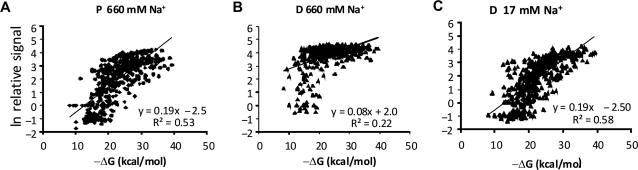
We hybridized high concentration of targets (ssRNA) at a low stringency condition (about 800 mM Na+ at 37°C) to the arrays, and subsequently washed off the hybridized targets using a spatial ionic strength buffer gradient. The relation between average signal intensity and calculated free energy of hybridization (ΔG) for the capture probes at different stringencies and positions relative to the surface showed the following: (i) low correlation (R2) when distal (Figure 2B) and central probes (data not shown) were washed at low stringency (660 mM Na+) and (ii) higher correlation for probes in the proximal position at low stringency (660 mM Na+) and for distal position at higher stringency (17 mM) (Figure 2B and C). It should be noted that the signal intensity for <1% of the probes showed evidence of detector saturation (signals above 60 000). These results indicated that probes placed in the distal and central position were not subjected to a sufficiently stringent wash at 660 mM Na+. That is, the low stringency wash was non-stringent for the majority of the probes at distal and central position. In contrast, probes at proximal position showed a relative good correlation between ΔG and signal at 660 mM Na+ indicating that the stringency at the proximal probe position was sufficient to resolve differences in ΔG in the probe set. It was therefore likely that the stringency at proximal probe position is higher than that at central and distal positions.
Figure 2.
Correlation between hybridization signals calculated Gibbs free energy (ΔG). For each of the six stringencies and position of probe capture sequence (proximal, central or distal) hybridization signals were plotted as function of calculated ΔG. Scatter plot showing the relationship between the natural logarithm (ln) of the relative hybridization signal, as percentage of probe with highest signal intensity, and calculated ΔG for (A) distal probes washed at 660 mM Na+, (B) proximal probes washed at 660 mM Na+ and (C) distal probes washed at 17 mM Na+.
Effects of capture sequence and spacer length on hybridization signal
An average hybridization signal was calculated for each probe length, at the tested spacer lengths and stringencies. For each of the six different stringencies, the average signal intensity was plotted against probe length. Selected signals from wash with a low stringent (660 mM Na+) and high stringent buffer (17 mM Na+) are shown in Figure 3A. The low stringency was used to determine maximum hybridization signal and the high stringency was used to determine the hybridization signal at conditions giving specificity. An increase in the hybridization signal was observed at all stringency conditions with increasing capture sequence length (Figure 3A and data not shown). A further increase in signal was observed when the probes were positioned distal to the surface as compared to proximal probes (Figure 3A and data not shown). At low stringency, the increase in hybridization signal with probe length was most prominent for probe sequences proximal to the surface. In contrast, the relative gain in signals decreased with probe length for probes with short or long spacer (central and distal placed probes, Figure 3A). At high stringency there was a linear relationship between probe length and signal, and a distinct gain of signal for probes with long spacers as opposed to short (or no) spacer. Hence, the effect of capture sequence and spacer length on hybridization signal changes with assay conditions and became more pronounced at higher stringencies.
Figure 3.
The effect of capture sequence and spacer length on hybridization signal. (A) The average signal intensity in arbitrary units (a.u.) of 13 hybridizations to 64 different PM probes as a function of capture probe length. Error bars are SEM. Diamonds represent proximal probes (P), squares represent central probes (C) and triangles represent distal probes (D). Filled symbols are probes washed in a low stringency buffer containing 660 mM Na+ (4× SSC + 0.1% SDS), and open symbols are probes washed with a high stringency buffer containing 17 mM Na+ (0.1× SSC + 0.1% SDS). (B) The percentage of the 832 hybridized probes (13 × 64) having signal over background and filtration settings (see Materials and methods section) as function of capture probe length.
The percentage of hybridized PM probes yielding signal above cut-off (threshold) (see Material and methods section) at high stringency (17 mM Na+) as function of the length of capture sequence and spacer is shown in Figure 3B. For short capture sequences (13-mers) only about 20% of the proximal probes yielded signal compared to 50% for central and distal probes. This proportion rapidly increased with increasing probe length. About 90% of the probes yielded hybridization signals when the probe length was equal to or larger than 19 nt (central probes), 21 nt (distal probes) or 23 nt (proximal probes).
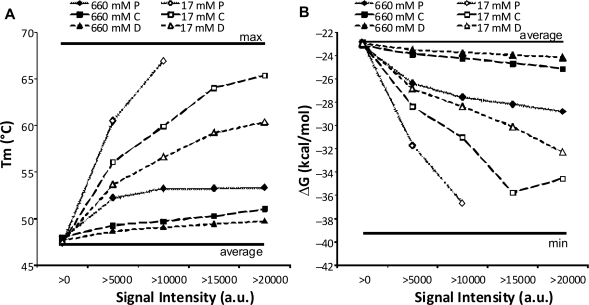
Calculated Tm and ΔG of probes at different positions
The average calculated Tm and ΔG for the probes giving signal were calculated for each position and stringency. The signals were furthermore categorized into probes giving >0 (above cut-off) in signal, and in steps of 5000, probes giving >20 000 fluorescent arbitrary units (Figure 4). Applying an increasingly higher signal threshold generally increased the difference in calculated Tm and ΔG between probes placed at different positions. Differences in Tm and ΔG (ΔTm and ΔΔG) were also highly dependent on the stringency applied (Figure 4, Supplementary Figures 3 and 4). At low stringency, ΔTm between proximal and distal probes was about 3–4°C irrespective of signal threshold except for threshold >0 that included nearly all probes. This shows that in order to give signal in the assay, proximal probes should have 3–4°C higher calculated Tm than probes placed in the distal position. In contrast, ΔTm increased with increased stringency and signal threshold applied. Spots giving intensity values >5000 at proximal position had calculated Tm that were about 7°C higher than distal-placed probes. At signal threshold >10 000, the difference increased to 12°C. Centrally placed probes had always calculated Tm between distal- and proximal-placed probes, indicating a decreasing requirement of selecting probes with high calculated Tm with distance from the surface. Furthermore, in order to yield high signals at high stringency, there was a general selection for probes with higher Tm compared to probes giving signal at low stringency (Figure 4A and Supplementary Figure 3). For instance, at a signal threshold of >5000, distal-placed probes had 6°C higher calculated Tm at high stringency compared to low stringency. Again, this difference increased with increasing requirements of signal in the assay where >20 000 threshold for distal-placed probes at high stringency required probes with calculated Tm that was 11°C higher compared to low stringency (Figure 4). In conclusion, these result showed that: (i) to expect increasingly high signals in the assay, probes should have increasingly higher calculated Tm which corroborates previous findings (43), (ii) displacing the capture probe from the surface decreased the need for selecting probes with high calculated Tm and (iii) high stringency in the assay increases the need to use probes with high calculated Tm.
Figure 4.
Tm and ΔG of probes with signal. (A) Average calculated Tm and (B) ΔG of probes with signal intensity (in arbitrary units, a.u.) above the limit indicated. The vertical lines indicate the average Tm and ΔG, and maximum Tm and minimum (most negative) ΔG of the probe set. Symbols are as in Figure 3A.
Calculated ΔG followed the same trends as Tm: probes functioning in the assay had increasingly lower calculated ΔG values with increasing stringency applied in the assay (Figure 4B, Supplementary Figure 4), displacing the probe from the surface lessened the requirement to select probes with low ΔG, and increasing the requirement for the signal strength of the probes increased the requirement to select probes with lower ΔG. Both analysis of Tm and ΔG supported that proximal probes need to be strong binders (have higher calculated Tm and ΔG), particularly at high stringencies applied to obtain high signal. At many stringencies, several proximal-placed probes did not give any signal at all (or signals <5000). In contrast, distally placed probes could be poorer binders and still function adequately at high stringencies.
Salt dissociation curves
For each PM probe the average wild-type hybridization signal was plotted as a function of the sodium concentration. With 64 different PAH sites, 7 probe lengths and 3 spacer lengths this resulted in more than 1300 dissociation curves. As examples of probes with extreme %G + C content, dissociation curves for 25-mer probes at a site with high G + C (64%) or low G + C (24%) content are shown (Figure 5A and B). For both sites, the signals were highest when the probes were located on long spacers (distal) and lowest for probes without spacer (proximal). Within the stringency buffers tested, the AT-rich probe did not reach a plateau in maximum signal (Figure 5B). On the contrary, the probe for the GC-rich site (with or without spacer) reached a plateau in maximum signal (Figure 5A). A surprising finding was the multi-state dissociation curve of the GC-rich 25-mer distal probe. The multi-state dissociation curves differed from the simple two-state dissociation model (bound or unbound), in that dissociation seem to happen in several steps/with more than one plateau. It however appeared that, this type of multi-state dissociation curve was common (e.g. 55% of 25-mers) for distal probes with long capture sequence (Figure 5C). For all mutation sites, it applied that if multi-state dissociation was observed at one capture sequence length, then it was also observed for the corresponding longer sequences. For example, a multi-state dissociation observed for 19 nt capture sequence, was also observed for 21 nt, 23 nt and 25 nt. A lower proportion (12% of 25-mers) of multi-state dissociation was also observed in central-placed capture sequence probes. No multi-state dissociation was observed for proximal-placed probes. Probes giving rise to multi-state dissociation curves had a significantly (P < 0.001) higher calculated Tm than probes giving rise to two-state dissociation curves (Figure 5D). Likewise, the presence or absence of multi-state dissociation curves was dependent on the calculated Gibbs free energy (ΔG) where probes with multi-state dissociation curves had a significantly lower calculated ΔG (P < 0.01) than probes displaying two-state dissociation (Supplementary Figure 5).
Figure 5.
Dissociation of probe–target hybrids. Dissociation curves were made for probes washed at six different sodium concentrations ranging from 2.5 mM to 600 mM Na+. Average signal of 25-mer PM probe as function of sodium concentration for (A) a G + C-rich PAH site (c.60+5G>T) and (B) a A + T-rich PAH site (c.1315+1G>A). (C) The share (percentage) of 64 different PM probes with multi-state dissociation curves as function of capture sequence length and (D) the average calculated Tm of the probes with multi-state or two-state dissociation curves. Error bars are SD. In (A–C), diamonds or hatched bars represent proximal probes (P), squares or open bars represent central probes (C) and triangles or filled bars represent distal probes (D). In Figure D, probes with two-state dissociation curves are represented with open bars and probes with multi-state dissociation curves are represented with filled bars.
Correlation between experimentally determined Sh and calculated Tm
The sodium concentration, at which half the signal was dissociated (Sh) was manually read from nearly 500 dissociation curves, encompassing 23 different PAH sites (Supplementary Table 2). These sites were selected to include the average and extreme sequences (with respect to %G + C). The probes investigated had an average calculated Tm of about 47°C, a Tm range of 25–67°C and an average G + C content of 48% with G + C content ranging between 22% and 73%. Sh was determined at half of the maximum signal observed for probes without a maximum plateau in signal. For short probes (13 nt) Sh could only be read for 20% of the probes, due to low signal. However, for longer capture probes (19–25 nt) Sh was read for 80–95% of probes, not including probes with low signal or extreme multi-state dissociation profiles. For other probes with multi-state dissociation, Sh was read as half of the maximum signal even if this could give rise to some additional variation in results.
For each spacer length, the natural logarithm of the experimentally determined Sh was plotted against calculated Tm of each probe (Figure 6), showing that experimentally determined Sh to some extent correlated with calculated Tm (R2 between 0.42 and 0.64), the best correlation found for distal probes. At first glance there appeared to be a little difference between the relationships in the different positions. However, since Tm of the probes was calculated using 37 mM Na+ and the assay was processed at 37°C, it was expected that the regression lines would cross this point if there was good concordance between observed and calculated melting point (Tm). For example, if there was solution like environment in the spot, probes with higher calculated Tm than 37°C should dissociate at lower Na+ concentration than at 37 mM. However, plotting a line at 37 mM Na+ showed that most probes melted at higher Na+ concentration than 37 mM (Figure 6). Centrally placed probes dissociated, in general, at lower Na+ concentration than proximally placed probes and distally placed probes dissociated, in general, at higher salt concentration than centrally placed probes (Figure 6). However, even distally placed probes melted at lower Na+ concentrations than expected.
Figure 6.
Experimentally determined Sh as function of the calculated Tm. The natural logarithm (ln) of the sodium (Na+) concentration at which half of the signal is dissociated (Sh) as function of the calculated melting temperature (Tm). (A) proximal probes (P), (B) central probes (C) and (C) distal probes (D). Sh was read from seven capture lengths of PM probes targeting 23 different PAH sites (Supplementary Table 2), when appropriate dissociation curves were at hand (not too low signal or extreme multi-state dissociation). On each graph, the equation and correlation (R2) for the linear regression line is drawn.
Effects of capture sequence and spacer length on hybridization specificity
For each probe the maximum specificity (signal ratio, see Materials and methods section) within the stringency gradient was found, and an average signal ratio was calculated for probes with same capture sequence length and spacer (Supplementary Figure 6A). Maximum specificity was over all similar for probes with or without spacer, and showed a general decrease in specificity with increasing probe (capture sequence) length. The exception was short capture sequences (13-mer) that had higher specificity using probes with spacer (C and D). Probes without spacer (P) obtained their maximum specificity at capture sequences of 15 nt. Maximum specificity was between signal ratios of 3 and 17. Decrease in specificity with increasing probe length was also observed for probes with more than one (2–3 nt) mismatches, although the specificity (signal ratio) was much higher (Supplementary Figure 6B). Maximum specificity ranged in signal ratio from 6 to 300. Also in this case, the spacer had very little effect on the specificity.
Next, we investigated the relation between hybridization signal and specificity. For each capture sequence length and stringency an average specificity (signal ratio) and PM probe signal was calculated for the 64 different PAH sites and plotted (Supplementary Figure 7). Irrespective of spacer length, the highest specificity was observed as the signal approached zero. The best correlation between signal and specificity was observed for distal probes with signals spreading over a large signal range. In contrast, proximal probes mainly had signals near zero, and a poorer correlation (Supplementary Figure 7A and C). Furthermore, distally placed probes had higher specificity at high signal. For instance, at a signal of 10 000 fluorescent units, distal probes had an average signal ratio of ∼3.5, while central and proximal probes had about 3 and 2.5, respectively (Supplementary Figure 7). It should be noted that even if specificity of wild-type duplexes was investigated and shown (Supplementary Figures 6 and 7), the selectivity of the assay was ensured using heterozygotes (see genotyping below). In other words, a patient heterozygous for a particular mutation resulted in little specificity of probes covering the particular mutation, while at the same time giving rise to specificity at sites where the patient had wild-type sequence. The specificity observed in the assay is therefore selective and not a general effect of changing the experimental conditions.
Application-specific assay recommendations
As an example of a specific microarray application, we performed genotyping of PAH mutations/sites with available mutated patient samples (Supplementary Tables 2 and 3). To score genotypes, a normalized signal ratio (see the Materials and methods section) was calculated for each probe pair. In order to determine if a probe pair resulted in sufficient separation of the three possible genotypes homozygous wild-type (Wt/Wt), heterozygote (Wt/Mt) or homozygous mutant (Mt/Mt), and hence successful genotyping of a mutation, the following criteria were used: difference between minimum Wt/Wt ratio and maximum Wt/Mt ratio, as well as difference between minimum Wt/Mt ratio and maximum Mt/Mt ratio should be >0.05. For each site, the minimum and maximum wild-type ratios were determined from all wild-type hybridizations (>30 hybridized slides). The ratio of heterozygotes and potentially also homozygous mutated for each site derived from one or more hybridized samples (Supplementary Table 3). As most microarray assays are processed at one condition (stringency) and frequently have melting temperature (Tm) matched probe sets, we identified the optimal parameter settings of Tm and stringency from our results. Including only probe pairs within a fixed range of Tm in the assay, the stringency resulting in most sites being successfully genotyped was identified for each probe position relative to the surface. The fixed Tm of 46°C ± 3°C was chosen as it encompassed the average Tm of probe pairs of ∼47.5°C, and because the highest share of sites (95%) were represented with probe pairs in this Tm-range. Likewise, the Tm range giving the highest share of sites being successfully genotyped at a fixed stringency condition (0.35 × SSC) was determined (Table 1). A similar analysis was performed on probe pairs resulting in specificity above a certain threshold. Here, an adequate specificity of each probe pair was defined as 2.3 times more signal from the PM probe as from its corresponding MM probe (SPM/SMM > 2.3) (Table 1). The PAH sites and patients included in the signal and specificity analysis are found in Supplementary Tables 2 and 3. It should be noted that the identified stringency and Tm were assessed differently for the genotyping and specificity application (Table 1). As mentioned above, the optimal assay parameters (stringency or Tm) for genotyping at fixed conditions were found as the ones giving highest number of successfully genotyped mutations (% sites). In case of the specificity assay, the optimal settings were identified at probe-pair level. That is, the parameter settings yielding the highest share of probe pairs (% probes) with sufficient specificity were found by comparison between the number of probes with specificity >2.3 and the total number of probes with signal above zero at the fixed setting.
Table 1.
Optimal assay parameters at fixed probe-pair Tm or stringency wash
| Genotyping |
Specificity (SPM/SMM) > 2.3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Tm = 46° ± 3°C |
Stringency 0.35 × SSC |
Tm = 46° ± 3°C |
Stringency 0.35 × SSC |
|||||||
| Position form surface | Stringency | Sites (%) | Tm(°C) ± 3°C | Sites (%) | Stringency | Probes (%) | SPM/SMM | Tm(°C) ± 3°C | Probes (%) | SPM/SMM |
| Proximal | 4 × SSC | 60 | 52 | 66 | 0.1 × SSC | 85 | 4.9 | 46 | 83 | 4.9 |
| Central | 1 × SSC | 75 | 48 | 77 | 0.01 × SSC | 76 | 4.3 | 46 | 76 | 3.8 |
| Distal | 0.35 × SSC | 72 | 46 | 72 | 0.01 × SSC | 89 | 5.7 | 42 | 89 | 4.6 |
Genotyping of 42 different PAH mutations was performed using 38 patients samples (Supplementary Tables 2 and 3). For probe pairs within a fixed range of melting temperature (46°C ± 3°C) the stringency (×SSC), giving the highest share of mutations/sites (% sites) being successfully genotyped, is noted for each position from surface. Likewise, at a fixed stringency (0.35 × SSC) the Tm range (Tm ± 3°C) giving highest percentage of sites being successfully genotyped is shown. For the 64 PAH sites and 13 hybridizations included in signal and specificity analysis (Supplementary Tables 2 and 3) the same analysis was performed. However, the parameter settings were assessed as the ones giving the highest share of probes (% probes) giving specificity (signal ratio) >2.3, as compared with probes within the fixed setting giving signal. Signal ratio SPM/SMM was calculated as described in the Materials and methods section.
For both the genotyping application and investigation of probe specificity it was observed that surface proximal probes required lower stringency and higher melting points to function optimally in the assay. However, successful genotyping required less stringent assay condition and higher Tm of the probes, than necessary to obtain specificity >2.3 (Table 1). Neither application resulted in 100% of sites or probe pairs successfully genotyped or with sufficient specificity, respectively using fixed Tm and stringency.
DISCUSSION
Although microarray technology is a promising, high-throughput technique with several biomedical and environmental applications, more systematic studies are required to parameterize new models of molecular interactions at microarray surfaces (7,44). In contrast to hybridization in the solution, microarray hybridization occurs at a solid phase and solution interface, which significantly alters the reaction of duplex formation. In microarray hybridizations, the effects of counter ions (cations e.g. Na+), in the hybridization and post-stringency wash buffer, on the stability of nucleic acid duplexes are manifold. These include shielding the negatively charged phosphate groups in nucleic acid and hence reducing the electrostatic repulsion between immobilized probes, probe and target and hybridized targets (24). Cations also shield the often negatively charged glass surface by forming a layer of counter ions. The Debye layer is defined as the distance from the surface where effect of surface charge diminishes significantly due to accumulation of counter ions. The length of the Debye layer ranges from less than a nanometer to tens of nanometers depending on the temperature and the counter ion concentration (10). Theoretical studies suggest that surface effects can reduce Tm of hybrids significantly (10), which we can corroborate experimentally in this report. We observed that probes at proximal position yielding signal had up to 10°C higher calculated Tm than probes in distal and central position (Figure 4). Furthermore, proximal-placed probes yielding signal had calculated ΔG that were up to 8 kcal/mol lower than central- and distal-placed probes. This indicates that probes close to the surface are exposed to an additional stringency provided by the surface and therefore, in order to give signals, the probes need to have higher affinity (higher Tm and a lower ΔG) (Figure 4 and Supplementary Figures 3 and 4). This, as yet undefined stringency, apparently selects for probes with higher calculated Tm and lower ΔG. Several observations in the present study support that there is a difference between surface proximal and distal probes: (i) the signal from proximally placed probes are equal to the signal from distally placed probes when washed at 30-fold higher Na+ concentration (Figure 3A), (ii) the fraction of probes giving signal is generally higher for distally placed probes compared to the corresponding proximal-placed probes (Figure 3B), (iii) the dissociation curves switch from being multi-state to two-states by placing probes closer to the surface (Figure 5) and (iv) proximal probes typically melted at higher Na+ concentrations than the corresponding distally placed probes (Figure 6). These differences can be explained by surface effects as discussed below.
It is quite common that probes immobilized on surfaces have a much lower observed Tm compared to calculated Tm presumably because of effect from the surface (13,45–47). This is consistent with our findings, since we observe an increasingly large surface effect on probes placed in the distal, central and proximal position, respectively. That proximally placed probes require to be exceptionally strong binding capture molecules to give signal (Figure 4) and that proximally placed probes melting at lower than expected stringency (Figure 6) explains previous findings indicating that the first 10 nt of 60-mer probes only function as spacers and not as capture sequence [(1) and reviewed in (48)]. However, it appears that even the distal position is affected by repulsive forces in the spot, because at this position probes melted at higher Na+ concentration than expected if we assume that there is solution like condition at the distal position (Figure 6). The results strongly indicate that large-scale multi-parametric studies to measure the actual Tm (and ΔG) at different positions in the DNA polymer employing thermal melting in conjunction with varying the ionic strength in the buffers is highly needed. Such studies would give accurate Tm and ΔG compensations factors for different ionic strength buffers and spacer lengths and could be used to make reliable microarray correction of the commonly used solution phase NN model.
As extensively reported, the hybridization yield increased with the length of the capture sequence (2,26,27) and when using spacer to distance the probes from the surface (15). Unlike the previous reports, we investigated effects of spacers in a stringency gradient and found that the relative effect of probe length and spacer was modulated by the applied stringency. At low Na+ concentrations, there was a greater effect of spacer length and also a linear relationship between probe length and hybridization signal. At high Na+ the hybridization signal from distal probes did not increase proportionally with the probe length, which is likely due to the non-stringent condition (660 mM and 165 mM Na+) applied for central and distal probes. Such non-stringent conditions will result in probes with different calculated Tm and ΔG yielding the same signals due to: (i) the spots being saturated or (ii) probes with different ΔG and Tm do not display differences in hybridizing or dissociating, respectively. When the sodium concentration is reduced, there is more electrostatic repulsion between probe–target, hybridized targets (16,24) and surface and nucleic acids (10), which explains why a longer spacer becomes increasingly important for duplex formation with decreasing cation concentration. In summation, it is strongly indicated, that the conditions for forming hybrids over the 60-mer probe molecule differs with distance from the surface. We generally observed larger differences between the proximal and central/distal position than between central and distal position. This is in accordance with the nature of the repulsive effect of the surface which declines exponentially with the distance from the surface (10). Based on previous work (10) we calculated that, at the highest stringency (thickest Debye layer), surface-repulsive effects are affecting about 30-nt counting from the surface. This corresponds approximately to the position where centrally placed probes are centered around. Many have speculated that steric factors are inhibiting hybridization close to surfaces (15,17,34). We hypothesize that the decrease in hybridization signal obtained on probes closer to the surface is at least partly due to the higher stringency closer to the surface.
It should be noted that, in our experiment (and most microarrays experiments) the spacers used (18–47 nt) are shorter than the ssRNA target (150–330 nt). Other reports addressing the effect of spacers used short synthetic target (15,17). However, as reviewed elsewhere (49) the use of spacers becomes more important when long targets are hybridized to arrays. One possible reason is that target molecules have long dangling ends that are facing down to the surface after hybridization, thereby giving large electrostatic or physical constraints due to the proximity to the surface as compared with targets with shorter dangling ends (50). However, Peplies et al. (23) demonstrated that the requirements of spacers decreased if helper oligonucleotides were used to break hair pin structures of target. Hence, the relative effect of spacers on signal is in addition to the parameters shown in this study (capture probe length and assay condition) also affected by other assay parameters, such as probe density (16,17), the length of target and possible secondary structures (16,23) and most likely also the type of solid support and chemistry.
A notable characteristics of especially distal-placed probes with high relative Tm and low ΔG, is that these probes display evidence of multi-state dissociation (Figure 5D and Supplementary Figure 5), which never was observed for proximal-placed probes. The reason for the multi-state dissociation in probes at distal position could be that secondary structures (hair pins) in the capture probe sequence can be more readily formed at distal position compared with central and proximal positions. The mechanism of why central and proximal probes have more difficulties to form hair pins could be either by steric hindrance or that hair pin formation is suppressed by the higher stringency existing closer to the surface of the slide (see above). Another possible explanation for multi-state dissociation curves is binding of non-targets (unspecific binding) to some of the probe molecules in the spot (51), which dissociate at lower stringency than perfectly matched duplexes. Melting curves obtained by dynamic allele-specific hybridization (DASH), a platform with targets tethered to solid support and probes in solution, showed a similar multi-state melting when hybridizing with many (>250 different) probes, which was ascribed to cross-hybridization (52). The authors did not provide details if this melting profile was associated with certain targets (%G + C content and Tm), but merely noted that the cross-hybridized probes melted of at relatively low st Despite the frequently observed multi-state dissociation curves, distal probes showed a better correlation between Sh and calculated Tm than central and proximal probes (Figure 6). Proximal probes might be more affected by the sequence-specific positional effect of the probe, which has been reported previously (53), than distally placed probes. Taking into the account that surface effect decays exponentially with distance from the surface, it is likely that probes at distal positions are less affected by strong surface effects than the proximal probe. Probes close to the surface are sensitive to even short displacements from the surface where single base differences in spacer length leads to changed hybridization properties (53). At this point, it is unclear if distal-placed probes are subjected to any base-to-base positional effects or only segment-to-segment positional effects as documented here. The poorer correlation of proximal probes might also to some extent be due to technical difficulties to read the Sh from the dissociation curves starting a low signal (Figure 5). At present, we however reject the idea that the multi-state dissociation curves are caused by failures in the synthesis of the array. The reason is that synthesis failures are random giving a distribution of different capture sequence with a distribution of melting points. Synthesis failures are therefore less likely to explain the observed discrete ‘steps’ in the melting curves (Figure 5).
Specificity is another important feature of microarray hybridization, and was assessed by comparing the hybridization signal of perfect match and mismatch probe pairs. The maximum specificity of probes at a given stringency condition was generally independent of spacer length, but responded more dramatically to the length of the probes and the number of mismatches (2 nt or 3 nt instead of single nucleotide) (Supplementary Figure 6), the latter in agreement with previous findings (2,54). With distal and central probes it was possible to simultaneously have high signal and discrimination/specificity. Unlike the results presented here, several studies on commercial (Agilent and NimbleGen) and spotted arrays describe better discrimination of mismatches (higher specificity) at the distal end of the probe as compared with the proximal end of the probe. This difference in specificity is also denoted by 5′-end bias, provided that the 3′-end is attached to surface (1,27,29,55). A simple reason for the discrepancies in results can be that the reported assay conditions were optimized for signal and hence selection for hybrids in the distal end (1,27,29,55), whereas we find the optimal specificity by choosing between six different conditions. Taken into account that the stringency is higher in the proximal position (see above) than the distal positions, an assay run at one stringency for accessing positional effects is suboptimal for probes placed close to the surface. Furthermore, our experiment deviates from the above studies, because we placed the capture probe in three different positions relative to the microarray surface, hence our 60-mer probe (polynucleotide) was designed to be only partially hybridized to the target. Hybridization to the entire 60-nt sequence, which is common for gene expression analysis (1) will in contrast most likely select for binding at distal position, and it is therefore understandable that mismatches have larger effect when placed in the distal end.
The increased stringency in the surface proximal probe position, described above, was also demonstrated using a specific application, comprising genotyping of PAH mutations. Here, surface proximal probes required less stringent assay condition or higher calculated melting temperature (Tm) of the probes than distally placed probes. These results were also corroborated when finding optimal assay parameters for probe pairs with specificity above a defined threshold (2.3) (Table 1). However, optimal parameters differed between the two applications as the genotyping requiring lower stringency and higher calculated Tm of the probes than required to get specificity alone (Table 1). We speculate that this is because the specificity is maximized when unspecific binding is minimized, hence at high stringency and/or using weakly binding probes (as indicted by low calculated Tm, Table 1). In contrast, successful genotyping relies on specific hybridization to both the perfect match probe and its corresponding mismatch probe. Moreover, the genotyping application included hybridization data from slides re-hybridized up to three times. Successive loss of signal due to re-hybridizations, could lead to a selection of less stringent assay condition and/or probes with higher melting temperature. The inability to identify common assay parameters for all PAH sites and probe pairs (Table 1) was probably a consequence of the large spread in G + C content of 20–70% in the investigated probe sets (Poulsen et al., unpublished data). We have previously noted that even Tm matched probe set can be challenging to function optimally at only one stringency (32).
Our results also suggest improvements to probe design, when the whole 60-mer oligonucleotide probe is complementary to target, which is often the case for probes used in gene expression profiling and comparative genomic hybridization. Based on the information gained in our study, a 60-mer polynucleotide should be divided into three independent sections (proximal, central and distal) when designing the complete probe. First, the section close to the surface needs to have higher binding strength than the central and distal parts in order to provide hybridization to the complete oligo. In specific, probe segments closest to the surface should have 2–4°C and 4–6°C higher calculated melting temperature compared with the central and distal segments, respectively (Table 1). Furthermore, to avoid hair pins or partial hybridization, central and distal segments need to have relatively low melting temperature and binding strength, with Tm and ΔG of about ≤45°C and ≥−22 kcal/mol, respectively (Figure 5 and Supplementary Figure 5). The two criteria for probe choice are compatible with each other since proximal probes never show multi-state hybridization (Figure 5) allowing them to be designed with higher calculated Tm, which is needed for optimal performance (Table 1). It should however be noted, that the specified optimal assay parameters (Table 1) are only valid for the given stringency (0.35 × SSC) and assay setup, and will be modified by stringency, probe density, immobilization chemistry and microarray substrate. In order to get a complete list of thermodynamic correction factors usable for probe design, larger systematic studies investigating the parameter listed above are needed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Technical University of Denmark (to L.P.) Aase and Ejnar Danielsen Foundation. Funding for open access publication charges: Technical University of Denmark.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 2.Letowski J, Brousseau R, Masson L. Designing better probes: effect of probe size, mismatch position and number on hybridization in DNA oligonucleotide microarrays. J. Microbiol. Methods. 2004;57:269–278. doi: 10.1016/j.mimet.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 4.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 5.Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZDD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Ann. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 8.Wick LM, Rouillard JM, Whittam TS, Gulari E, Tiedje JM, Hashsham SA. On-chip non-equilibrium dissociation curves and dissociation rate constants as methods to assess specificity of oligonucleotide probes. Nucleic Acids Res. 2006;34:e26. doi: 10.1093/nar/gnj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozhitkov A, Noble PA, Domazet-Loso T, Nolte AW, Sonnenberg R, Staehler P, Beier M, Tautz D. Tests of rRNA hybridization to microarrays suggest that hybridization characteristics of oligonucleotide probes for species discrimination cannot be predicted. Nucleic Acids Res. 2006;34:e66. doi: 10.1093/nar/gkl133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vainrub A, Pettitt BM. Surface electrostatic effects in oligonucleotide microarrays: control and optimization of binding thermodynamics. Biopolymers. 2003;68:265–270. doi: 10.1002/bip.10271. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Wu C, Carta R, Zhao H. Free energy of DNA duplex formation on short oligonucleotide microarrays. Nucleic Acids Res. 2007;35:e18. doi: 10.1093/nar/gkl1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bommarito S, Peyret N, SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotin AV, Drobyshev AL, Proudnikov DY, Perov AN, Mirzabekov AD. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 1998;26:1515–1521. doi: 10.1093/nar/26.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Wolf LK, Georgiadis RM. Secondary structure effects on DNA hybridization kinetics: a solution versus surface comparison. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shchepinov MS, CaseGreen SC, Southern EM. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halperin A, Buhot A, Zhulina EB. Hybridization at a surface: the role of spacers in DNA microarrays. Langmuir. 2006;22:11290–11304. doi: 10.1021/la0616606. [DOI] [PubMed] [Google Scholar]
- 17.Peterson AW, Wolf LK, Georgiadis RM. Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 18.Fixe F, Dufva M, Telleman P, Christensen CBV. Functionalization of poly(methyl methacrylate) (PMMA) as a substrate for DNA microarrays. Nucleic Acids Res. 2004;32:e9. doi: 10.1093/nar/gng157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lytton-Jean AKR, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J. Am. Chem. Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 20.Dorris DR, Nguyen A, Gieser L, Lockner R, Lublinsky A, Patterson M, Touma E, Sendera TJ, Elghanian R, Mazumder A. Oligodeoxyribonucleotide probe accessibility on a three-dimensional DNA microarray surface and the effect of hybridization time on the accuracy of expression ratios. BMC Biotechnol. 2003;3:6. doi: 10.1186/1472-6750-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weckx S, Carlon E, De Vuyst L, Van Hummelen P. Thermodynamic behavior of short oligonucleotides in microarray hybridizations can be described using Gibbs free energy in a nearest-neighbor model. J. Phys. Chem. B. 2007;111:13583–13590. doi: 10.1021/jp075197x. [DOI] [PubMed] [Google Scholar]
- 22.Hong BJ, Sunkara V, Park JW. DNA microarrays on nanoscale-controlled surface. Nucleic Acids Res. 2005;33:e106. doi: 10.1093/nar/gni109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peplies J, Glockner FO, Amann R. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 2003;69:1397–1407. doi: 10.1128/AEM.69.3.1397-1407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao D, Kim J, Yu F, Nielsen PE, Sinner EK, Knoll W. Surface density dependence of PCR amplicon hybridization on PNA/DNA probe layers. Biophys. J. 2005;88:2745–2751. doi: 10.1529/biophysj.104.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dandy DS, Wu P, Grainger DW. Array feature size influences nucleic acid surface capture in DNA microarrays. Proc. Natl Acad. Sci. USA. 2007;104:8223–8228. doi: 10.1073/pnas.0606054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pullat J, Fleischer R, Becker N, Beier M, Metspalu A, Hoheisel JD. Optimization of candidate-gene SNP-genotyping by flexible oligonucleotide microarrays; analyzing variations in immune regulator genes of hay-fever samples. BMC Genomics. 2007;8:282. doi: 10.1186/1471-2164-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relogio A, Schwager C, Richter A, Ansorge W, Valcarcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu WT, Guo H, Wu JH. Effects of target length on the hybridization efficiency and specificity of rRNA-based oligonucleotide microarrays. Appl. Environ. Microbiol. 2007;73:73–82. doi: 10.1128/AEM.01468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki S, Ono N, Furusawa C, Kashiwagi A, Yomo T. Experimental optimization of probe length to increase the sequence specificity of high-density oligonucleotide microarrays. BMC Genomics. 2007;8:373. doi: 10.1186/1471-2164-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carletti E, Guerra E, Alberti S. The forgotten variables of DNA array hybridization. Trends Biotechnol. 2006;24:443–448. doi: 10.1016/j.tibtech.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Naef F, Lim DA, Patil N, Magnasco M. DNA hybridization to mismatched templates: a chip study. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2002;65:040902. doi: 10.1103/PhysRevE.65.040902. [DOI] [PubMed] [Google Scholar]
- 32.Petersen J, Poulsen L, Petronis S, Birgens H, Dufva M. Use of a multi-thermal washer for DNA microarrays simplifies probe design and gives robust genotyping assays. Nucleic Acids Res. 2008;36:e10. doi: 10.1093/nar/gkm1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moiseev L, Unlu MS, Swan AK, Goldberg BB, Cantor CR. DNA conformation on surfaces measured by fluorescence self-interference. Proc. Natl Acad. Sci. USA. 2006;103:2623–2628. doi: 10.1073/pnas.0511214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Giallo ML, Lucarelli F, Cosulich E, Pistarino E, Santamaria B, Marrazza G, Mascini M. Steric factors controlling the surface hybridization of PCR amplified sequences. Anal. Chem. 2005;77:6324–6330. doi: 10.1021/ac0506175. [DOI] [PubMed] [Google Scholar]
- 35.Petronis S, Stangegaard M, Christensen CB, Dufva M. Transparent polymeric cell culture chip with integrated temperature control and uniform media perfusion. Biotechniques. 2006;40:368–376. doi: 10.2144/000112122. [DOI] [PubMed] [Google Scholar]
- 36.Hirschhorn JN, Sklar P, Lindblad-Toh K, Lim YM, Ruiz-Gutierrez M, Bolk S, Langhorst B, Schaffner S, Winchester E, Lander ES. SBE-TAGS: an array-based method for efficient single-nucleotide polymorphism genotyping. Proc. Natl Acad. Sci. USA. 2000;97:12164–12169. doi: 10.1073/pnas.210394597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li JG, Liljedahl U, Heng CK. Tag/anti-tag liquid-phase primer extension array: a flexible and versatile genotyping platform. Genomics. 2006;87:151–157. doi: 10.1016/j.ygeno.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Dufva M, Petersen J, Stoltenborg M, Birgens H, Christensen CB. Detection of mutations using microarrays of poly(C)10-poly(T)10 modified DNA probes immobilized on agarose films. Anal. Biochem. 2006;352:188–197. doi: 10.1016/j.ab.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 40.Dufva M, Petronis S, Bjerremann Jensen L, Krag C, Christensen C. Characterization of an inexpensive, non-toxic and highly sensitive microarray substrate. Biotechniques. 2004;37:286–296. doi: 10.2144/04372MT02. [DOI] [PubMed] [Google Scholar]
- 41.Guldberg P, Henriksen KF, Guttler F. Molecular analysis of phenylketonuria in Denmark: 99% of the mutations detected by denaturing gradient gel electrophoresis. Genomics. 1993;17:141–146. doi: 10.1006/geno.1993.1295. [DOI] [PubMed] [Google Scholar]
- 42.Hahnke K, Jacobsen M, Gruetzkau A, Gruen JR, Koch M, Emoto M, Meyer TF, Walduck A, Kaufmann SH, Mollenkopf HJ. Striptease on glass: validation of an improved stripping procedure for in situ microarrays. J. Biotechnol. 2007;128:1–13. doi: 10.1016/j.jbiotec.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Wei H, Kuan PF, Tian S, Yang C, Nie J, Sengupta S, Ruotti V, Jonsdottir GA, Keles S, Thomson JA, et al. A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets. Nucleic Acids Res. 2008;36:2926–2938. doi: 10.1093/nar/gkn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajiyama T, Miyahara Y, Kricka LJ, Wilding P, Graves DJ, Surrey S, Fortina P. Genotyping on a thermal gradient DNA chip. Genome Res. 2003;13:467–475. doi: 10.1101/gr.790603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urakawa H, El Fantroussi S, Smidt H, Smoot JC, Tribou EH, Kelly JJ, Noble PA, Stahl DA. Optimization of single-base-pair mismatch discrimination in oligonucleotide microarrays. Appl. Environ. Microbiol. 2003;69:2848–2856. doi: 10.1128/AEM.69.5.2848-2856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee I, Dombkowski AA, Athey BD. Guidelines for incorporating non-perfectly matched oligonucleotides into target-specific hybridization probes for a DNA microarray. Nucleic Acids Res. 2004;32:681–690. doi: 10.1093/nar/gkh196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao XL, Gulari E, Zhou XC. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579–596. doi: 10.1002/bip.20005. [DOI] [PubMed] [Google Scholar]
- 49.Halperin A, Buhot A, Zhulina EB. On the hybridization isotherms of DNA microarrays: the Langmuir model and its extensions. J. Phys. Condens. Matter. 2006;18:S463–S490. [Google Scholar]
- 50.Stedtfeld RD, Wick LM, Baushke SW, Tourlousse DM, Herzog AB, Xia YM, Rouillard JM, Klappenbach JA, Cole JR, Gulari E, et al. Influence of dangling ends and surface-proximal tails of targets on probe-target duplex formation in 16S rRNA gene-based diagnostic arrays. Appl. Environ. Microbiol. 2007;73:380–389. doi: 10.1128/AEM.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koltai H, Weingarten-Baror C. Specificity of DNA microarray hybridization: characterization, effectors and approaches for data correction. Nucleic Acids Res. 2008;36:2395–2405. doi: 10.1093/nar/gkn087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jobs M, Howell WM, Stromqvist L, Mayr T, Brookes AJ. DASH-2: flexible, low-cost, and high-throughput SNP genotyping by dynamic allele-specific hybridization on membrane arrays. Genome Res. 2003;13:916–924. doi: 10.1101/gr.801103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat. Biotechnol. 2003;21:818–821. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- 54.Karaman MW, Groshen S, Lee CC, Pike BL, Hacia JG. Comparisons of substitution, insertion and deletion probes for resequencing and mutational analysis using oligonucleotide microarrays. Nucleic Acids Res. 2005;33:e33. doi: 10.1093/nar/gni034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaing C, Gardner S, McLoughlin K, Mulakken N, Alegria-Hartman M, Banda P, Williams P, Gu P, Wagner M, Manohar C, et al. A functional gene array for detection of bacterial virulence elements. PLoS ONE. 2008;3:e2163. doi: 10.1371/journal.pone.0002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.