Abstract
DNA bypass polymerases are utilized to transit bulky DNA lesions during replication, but the process frequently causes mutations. The structural origins of mutagenic versus high fidelity replication in lesion bypass is therefore of fundamental interest. As model systems, we investigated the molecular basis of the experimentally observed essentially faithful bypass of the guanine 10S-(+)-trans-anti-benzo[a]pyrene-N2-dG adduct by the Y-family human DNA polymerase κ, and the observed blockage of pol κ produced by the adenine 10S-(+)-trans-anti-benzo[a]pyrene-N2-dA adduct. These lesions are derived from the most tumorigenic metabolite of the ubiquitous cancer-causing pollutant, benzo[a]pyrene. We compare our results for the dG adduct with our earlier studies for the pol κ archaeal homolog Dpo4, which processes the same lesion in an error-prone manner. Molecular modeling, molecular mechanics calculations and molecular dynamics simulations were utilized. Our results show that the pol κ N-clasp is a key structural feature that accounts for the dA adduct blockage and the near-error-free bypass of the dG lesion. Absence of the N-clasp in Dpo4 explains the error-prone processing of the same lesion by this enzyme. Thus, our studies elucidate structure-function relationships in the fidelity of lesion bypass.
INTRODUCTION
Bulky lesions normally block the progress of processive high fidelity DNA polymerases (1–5). Subsequently, the replicative polymerase may be replaced by one or more translesion bypass polymerases (6–10). The error-free bypass of DNA lesions during translesion synthesis (TLS) is a repair pathway, which plays an important role in coping with highly blocking lesions in all three domains of life (6,10). However, translesion synthesis is often carried out by low-fidelity bypass DNA polymerases (11–14). In error-prone TLS, the mismatched nucleotides incorporated opposite the lesion introduce mutations into the genome that can result in the development of cancer (15,16). In humans, there are four known Y-family bypass DNA polymerases, pol η, pol ι, pol κ and Rev1 (7,17). Pol κ is the only Y-family polymerase found in humans which has homologs in both bacteria (DinB or Pol IV in Escherichia coli) and archaea (Dbh in Sulfolobus acidocaldarius and Dpo4 in Sufolobus solfataricus) (18).
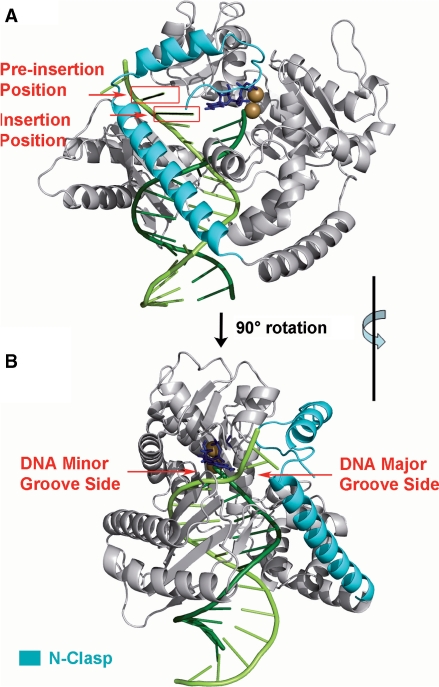
The structural features of low-fidelity bypass polymerases that allow for TLS of some lesions but not others, while high-fidelity polymerases are stalled by the same lesions, is a topic of significant interest (3,9,10). The structures of a number of Y-family polymerases have been investigated (10,19). A ternary complex crystal structure of pol κ lacking its C-terminal domain has recently been determined (20). This structure contains the major domains of pol κ including its signature N-terminal domain, namely, the ‘N-clasp’, an unmodified DNA primer/template complex, and an incoming dTTP Watson–Crick paired with the templating base adenine (20). The N-clasp in pol κ is at present unique, as it has not yet been observed in any other Y-family polymerase. With the N-clasp on the major groove side (Figure 1) of the nascent duplex, pol κ completely encircles a region of the DNA containing the template–primer junction at its active site. It is noteworthy that pol κ lacking the N-clasp (pol κ68–526) is severely impaired in its DNA synthesis activity (20).
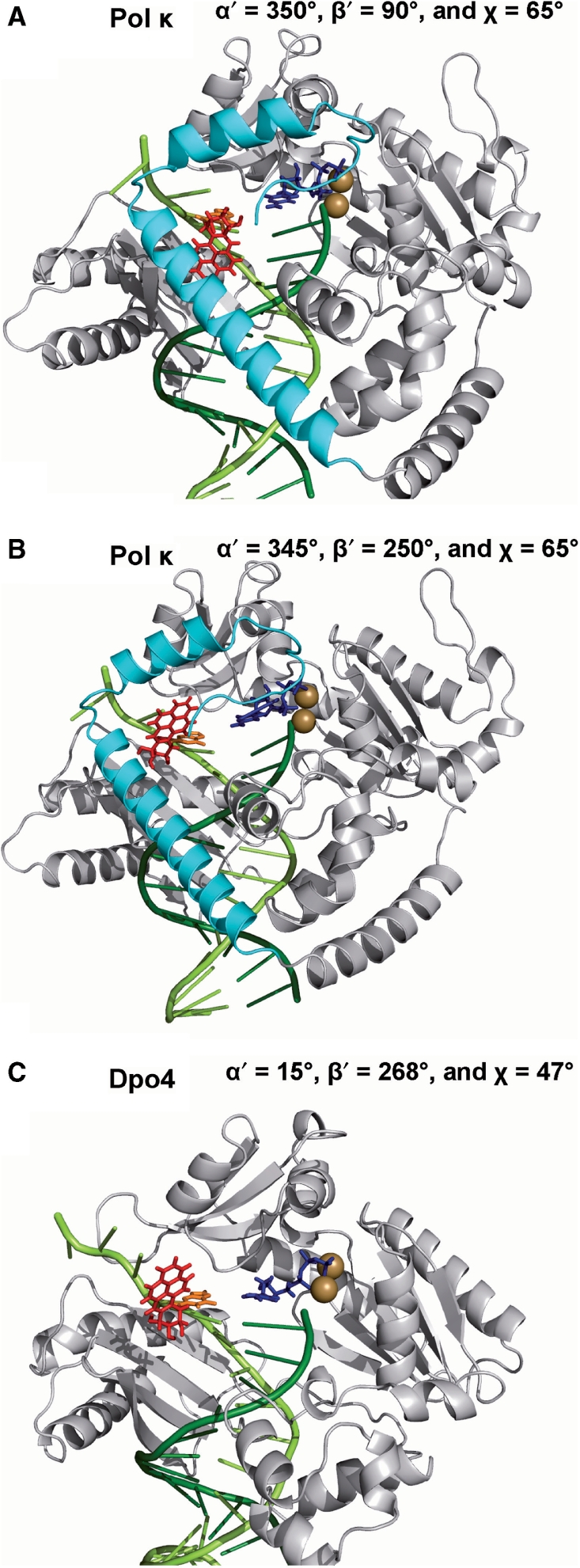
Figure 1.
(A) Definitions of preinsertion and insertion positions in the unmodified pol κ crystal structure (20) (PDB ID: 2OH2). (B) Denotation of DNA major and minor grooves shown through rotation of (A) by 90° along the vertical axis. Note that the N-clasp is on the major groove side. The enzyme is in gray with the N-clasp highlighted in cyan. The DNA template and primer strands are light and dark green, respectively. The incoming dNTP is blue. The Mg2+ ions are brown. Stereoviews are shown in Supplementary Figure S7.
The ability of pol κ to synthesize DNA both in the absence and presence of lesions has received considerable attention. In the case of unmodified DNA templates, pol κ synthesizes DNA with low fidelity (21), and it has also been shown that it is promiscuous in extending primer strands past mispaired primer–template termini (22). The processing of DNA lesions by pol κ, whether error-free or error-prone, has been shown to be highly lesion dependent. The cis-syn cyclobutane pyrimidine T–T dimer and the pyrimidine (6–4) pyrimidone T–T dimer, which are UV lesions, are complete blocks (23–25). The cisplatin lesion is also a complete block (24). The incorporation of correct or incorrect nucleotides opposite templating modified DNA bases catalyzed by pol κ is significantly slowed by apurinic sites (25), thymine glycol (26), N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) and N-(deoxyguanosin-8-yl)-2-aminofluorene (dG-AF) adducts (27), 1,N2-ethenoguanine (28), 1,N6-ethenodeoxyadenosine (29), O6-methylguanine (30) and the more bulky O6-alkylated guanine lesions (31), as well as by benzo[a]pyrene-derived lesions (25,32–37). On the other hand, 8-oxoguanine is bypassed efficiently (30). In certain cases, namely the cis-syn cyclobutane pyrimidine T–T dimer (22), O6-methylguanine (30), γ-hydroxy-1,N2-propano-2′-deoxyguanosine (γ-HOPdG) (38) and benzo[a]pyrene-derived lesions (36), it has been shown that pol κ extends efficiently past the lesion, while another polymerase is responsible for nucleotide incorporation opposite the lesion in vitro.
Benzo[a]pyrene (B[a]P) is a ubiquitous cancer-causing environmental pollutant, present in tobacco smoke and as a product of fossil fuel combustion (16). The most mutagenic and tumorigenic metabolite of B[a]P is the (+)-7r,8t-dihydrodiol-t9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene, (+)-anti-B[a]PDE (39,40). The (+)-anti-B[a]PDE molecule is unstable in aqueous environments and reacts readily with the exocyclic amino groups of adenine or guanine in DNA by the predominant trans opening of the epoxide, to form the 10S-(+)-trans-anti-B[a]P-N6-dA and –N2-dG adducts (B[a]P-dA and B[a]P-dG, respectively) (41) (Figure 2A). These adducts are the major adenine and guanine adducts (41–43) and have been extensively investigated in primer extension studies with a number of replicative and bypass polymerases (2,5,33,37,44–50). Interestingly, benzo[a]pyrene-derived bulky lesions to N2 of dG are bypassed by purified pol κ (25,33–36), or by a C-terminal truncated form of pol κ (37), with mostly correct insertion of dCTP opposite these lesions, and is thus error-free. Pol κ has also been implicated in the error-free bypass of these benzo[a]pyrene-derived DNA adducts in mouse embryonic cells (51), and in mouse embryo fibroblasts transfected with a gapped plasmid containing a site-specifically incorporated benzo[a]pyrene-derived N2-dG adduct (52).
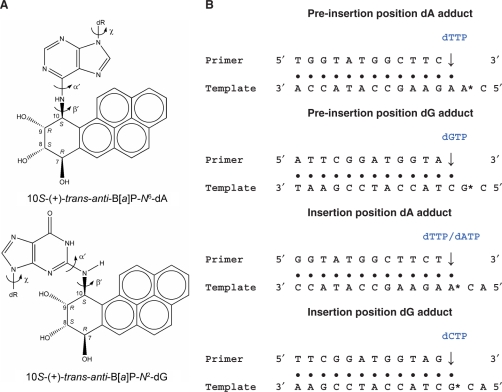
Figure 2.
(A) Structures of the 10S-(+)-trans-anti-B[a]P-N6-dA and the 10S-(+)-trans-anti-B[a]P-N2-dG adducts. The absolute configurations of the four chiral atoms C7, C8, C9 and C10 are indicated. Torsion angles are defined as χ = O4′(dR)−C1′ (dR)−N9−C4 (dR is deoxyribose.), α′ = N1−C6−N6−C10(B[a]P-dA) or N1−C2−N2−C10(B[a]P-dG), β′ = C6−N6−C10(B[a]P-dA)−C9(B[a]P-dA) or C2−N2−C10(B[a]P-dG)−C9(B[a]P-dG). (B) Base sequences of the four models. The incoming nucleotide dNTP is indicated in blue font. In the case of the dA adduct in the insertion position, dTTP was employed in the VDW energy survey model, while dATP was employed in the MD simulations (see text).
Primer extension studies by Rechkoblit et al. (33) using the (+)-trans-B[a]P-N2-dG and –N6-dA adducts, employing pol κ as the polymerase, have shown particularly interesting differences, namely, error-free bypass of B[a]P-dG and essentially full blockage of the polymerase by B[a]P-dA adducts under similar experimental conditions. Specifically, pol κ incorporates dCTP opposite the B[a]P-dG adduct, although at a slowed rate relative to unmodified template strands. However, the progress of pol κ is strongly impeded by the B[a]P-dA adduct, and it stalls at the step that involves nucleotide insertion opposite the lesion (33). The structural and mechanistic factors underlying this striking difference in primer extension efficiency are of great interest because they provide us with further insights into the abilities of Y-family polymerases to bypass some DNA lesions but not others. The availability of the crystal structure of pol κ (20) provides an excellent opportunity for examining the structural features of this polymerase that allow for the TLS of B[a]P-dG adducts, but not of the B[a]P-dA adducts, utilizing molecular modeling and dynamics approaches.
Our hypothesis is that the unique pol κ N-clasp plays an important role in the differential treatment of these bulky N2-dG and N6-dA adducts. To investigate this hypothesis, we modeled these B[a]P-dG and B[a]P-dA adducts on the templating strands in the pol κ active site region in two positions shown in Figure 1. The first position is termed the preinsertion position: in this case an unmodified template base is in the insertion position opposite incoming dNTP, while the lesion is adjacent on its 5′ side, in the single-strand overhang. For this position, we modeled dGTP paired with the unmodified template C, or dTTP paired with the unmodified template A. The second position we studied is the insertion position, in which the dNTP is paired with the B[a]P-dG or the B[a]P-dA adduct. For this position, we modeled dCTP paired with the dG adduct or dTTP paired with the dA adduct (Figure 2B). We then carried out an extensive automated van der Waals (VDW) energy survey with 373, 248 different conformations for each lesion; this produced a comprehensive topography of the energy as a function of the lesion conformation within the pol κ ternary complex. The survey was employed to locate structures with minimum steric contacts and thereby to provide initial structures for subsequent molecular dynamic (MD) simulations. Our results reveal that the N-clasp plays a critical role in the blockage by the B[a]P-dA adduct, which competes for space with the N-clasp and is always in close contact with it when in the preinsertion position. However, in the case of the dG adduct in this position, the B[a]P ring system is not near the N-clasp and is without steric close contacts. Moreover, at the insertion position, the B[a]P-dG can readily form Watson–Crick hydrogen bonds with incoming dCTP for error-free bypass. We compared our results for pol κ with previous modeling and experimental studies of the B[a]P-dG adduct for Dpo4 (47,53,54, Xu, P., Oum, L., Geacintov, N.E. and Broyde, S., manuscript in preparation), which lacks the N-clasp (Supplementary Figure S1). Dpo4 is relatively promiscuous in nucleotide selection and favors slippage in the bypass of this adduct. Our results describe how the structural differences between the two polymerases explain their differential processing of the same lesion.
METHODS
Construction of the initial models by molecular modeling
We utilized the ternary crystal structure of pol κ with DNA and incoming dTTP (PDB ID: 2OH2, complex B) (20) as the template to build our initial models. This pol κ structure did not resolve 57 residues (residue 225–281) in its palm subdomain. These residues are not present in the pol κ homolog Dpo4 and are called the flexible ‘dorsal tendril’ (55). We superimposed this pol κ complex with a Dpo4 ternary complex (PDB ID 2AGQ) (56) and found that the two enzymes share high structural similarity in the palm domain regardless of the unresolved residues in the pol κ structure (Supplementary Figure S1). Moreover, these residues are not located near the active site, and modeling them could lead to a problematic structure. Therefore, to obtain a reasonable model of a continuous pol κ structure, we closed the gap between residues 224 and 282 as a loop and discarded the missing 57 residues; the program MODELLER running at the ModLoop server was employed to model the loop containing residues 223, 224, 282 and 283 (57,58). Furthermore, this pol κ structure contains only one Mg2+ in its active site; however, a functional polymerase requires two Mg2+ ions to organize the active site (59,60). The nucleotide binding Mg2+ ion (MgA), already present in the structure, has normal octahedral coordination (61,62). We modeled the missing catalytic Mg2+ (MgB) (Supplementary Figure S2) in the pol κ active site following octahedral coordination criteria from a high resolution pol β crystal structure and a computational study with Dpo4 (61,62). The backbone torsions of the primer nucleotide were modestly adjusted (<30°) in order to achieve the coordination distance between O3′ and the catalytic Mg2+. Coordinates of all initial structures are provided in Supplementary data.
The DNA sequences were then modified to the ones used by Rechkoblit et al. (33) in conducting the primer extension studies. These sequences are shown in Figure 2B. To complete our initial models, the B[a]P-dA and -dG adducts were modeled at the preinsertion and insertion positions (Figure 1) by covalently linking the lesion, taken from an NMR solution structure of this adduct in an 11-mer B-DNA duplex (63), to the amino groups of adenine or guanine. Templating bases were Watson–Crick partnered with their complementary dNTPs by replacing the original thymine of dTTP in the crystal structure with the appropriate base (Figure 2B), without changing the glycosidic torsion. Four initial models (B[a]P-dA and B[a]P-dG adducts in preinsertion and insertion positions) were created for the energy survey.
Van de Waals energy surveys of the dA and dG adducts in pol κ
To obtain a VDW energy surface of pol κ containing the dA and dG adducts, we carried out an energy survey for the four pol κ models, utilizing the two torsion angles at the linkage site between the damaged base and B[a]P, α′ and β′, and the glycosidic linkage χ (Figure 2A), as variables. A torsion driver program was employed to rotate these dihedral angles at 5° intervals in combination over the whole 360°. Thus, we created 723 = 373, 248 structures for each model. The energy calculations were carried out by the SANDER module in AMBER 9.0 (64) with the Cornell et al. (65) force field and PARM99 (66), ff99SB (67) and parmbsc0 (68) parameter sets. The partial charges for the B[a]P-dG and -dA adducts were parametrized using quantum mechanical ESP calculations (HF/6-31G*, Gaussian 03) and RESP for fitting the charge to each atom center (69,70). The partial charges for the dNTPs and Mg2+ are taken from Perlow et al. (71,72). All of the added force field parameters, atom types, and topology assignments for the lesions, dNTPs and Mg2+ are listed in Supplementary Data (Tables S1–S7). In these single-point VDW energy calculations, we utilized an implicit solvent model with dielectric constant represented by the function ε = 4r (r is the distance between an atom pair). TECPLOT10 from Amtec Engineering Inc., was employed to generate 3D energy maps.
Molecular dynamics simulations of pol κ complexes
From our 3D VDW energy survey maps, we selected the lowest energy pol κ structure of each model as the initial structure for the MD simulations. The same force field was employed as in the energy survey study. In addition, we simulated a normal G • C pair control model without B[a]P modification utilizing the crystal structure (PDB ID: 2OH2) with sequence remodeled to that of the B[a]P-dG adduct in the insertion position (Figure 2B). Detailed MD simulation protocols are given in Supplementary Material. The number of counterions and water molecules added to the system and the sizes of the solvation boxes are given in Supplementary Table S8.
Stability of the molecular dynamics simulations
Plots of the time-dependent root-mean-square deviations (RMSD) of each ensemble member's binding pocket relative to the first structure of production MD are given in Supplementary Figure S3; the binding pocket is defined as any atom within 5 Å of the damaged nucleotide. This binding pocket for the dA adduct in the preinsertion position is unstable due to local crowding. The binding pocket generally fluctuates in a stable manner after 3.0 ns for the remaining three models. Our analyses were confined to the 5.0–8.0 ns time frame as a uniform window for all simulations.
Structural analyses
Snapshots of the structures during the simulations with solvent and counterions removed were obtained with the PTRAJ module of the AMBER 9.0 suite (64). We selected one best representative structure from each ensemble for graphic illustration. To pick the best representative structure, we extracted 100 structures at 30 ps intervals from the 5.0–8.0 ns time frame for each model. Each extracted structure was superimposed to the remaining 99 structures and the RMSDs were computed and summed. The structure with the smallest sum RMSD was selected as the most representative structure (Supplementary Table S9). This procedure essentially follows the philosophy in Moil-View (73), which, however, could not be utilized directly due to the size of our system. PTRAJ was also employed to determine the time-dependence of the RMSD, and the torsion angles α′, β′ and χ (Supplementary Figure S4). Detailed hydrogen bonding analyses were carried out with the CARNAL module in AMBER7.0. Criteria for hydrogen bonding are: donor heavy atom to acceptor heavy atom distance ⩽3.4 Å; donor heavy atom-hydrogen-acceptor heavy atom angle: 135° to 225°. To analyze the reaction-ready active site organization, we measured the Mg2+ octahedral coordination distances (Supplementary Figure S2), the in-line attack distance (O3′-Pα), and angle (O3′-Pα-O3α). To compute the occupancy, we established criteria, based on near reaction-ready pol β and Dpo4 structures (61,62), for a well-organized active site: in-line attack distance <3.5 Å; in-line attack angle 180 ± 30°; Mg2+ coordination distance <2.4 Å, except for the distance MgA-O1α: for this distance we accepted <3.0 Å, based on our simulation for the unmodified control and because one long distance has also been observed in several of our earlier simulations (47,74,75). The near reaction ready occupancy is the percent of time during the 5–8ns time frame of the simulation utilized for analysis that meets all these criteria simultaneously (Donny-Clark,K. Shapiro,R. and Broyde,S., submitted).
INSIGHT 2005 from Accelrys, Inc. and PyMOL 1.1 from DeLano Scientific LLC. were utilized for molecular modeling and visualization, respectively. Clustalw2 (76) running at Uniport servers, http://beta.uniprot.org, was employed for the multi-sequence alignment. MD simulations were carried out on our own cluster of Silicon Graphics Origin® and Altix® high performance computers.
RESULTS
The objective of our work was to structurally rationalize the origins of the observed blockage of the dA adduct, while the dG adduct is bypassed with near-high fidelity. We carried out a comprehensive molecular mechanics VDW energy survey for each adduct within the preinsertion and insertion positions of the polymerase–DNA complexes in order to search for feasible domains for the α′, β′ and χ flexible torsions (Figure 2A) that govern the lesion orientation (see Methods section). Structures with minimal steric collisions then provided initial models for subsequent MD simulations. Full energy spectra, in two-dimensional slices, of the results presented below are given in Supplementary Movies S1–S12.
The molecular mechanics energy surveys reveal that the dA adduct in the preinsertion position has close contacts with the pol κ N-clasp, while the dG adduct does not
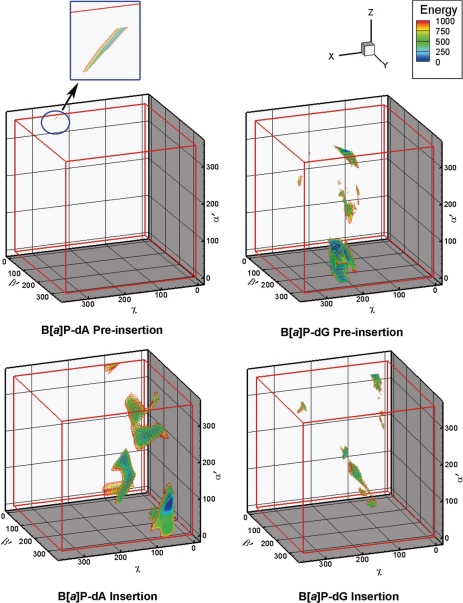
We determined 3D VDW energy topographies for the lesion at the pol κ preinsertion position (Figure 1). The VDW energy was computed for each conformer to evaluate steric hindrance between the lesion and the protein/DNA complex. Three-dimensional (α′, β′ and χ) energy contour maps for each model were then constructed based on the energy survey. Figure 3 shows the contour maps for these models. Energies are relative to the lowest energy conformer of each model (Supplementary Table S11). Our purpose here was to locate domains that were not severely forbidden. Therefore, at this stage, we visualized structural domains to an energy cutoff of 1000 kcal/mol to locate all potentially feasible structures, including those that could become viable with small protein movements; the steric term in the Lennard–Jones potential for the VDW energy increases very steeply, with the twelfth power of the interatomic distance, and small torsional movements can produce very large VDW energy increases. For the preinsertion position models, the maps reveal that the dA adduct is high energy essentially throughout the entire torsional space. There is only one domain which is not forbidden by our criteria. This domain, which contains the lowest energy structure, is extremely small (Figure 3, inset). However, the energy map for the dG adduct reveals two much larger such domains (Figure 3 and Supplementary Table S12).
Figure 3.
Three-dimensional van der Waals energy (kcal/mol) contour maps of the protein/DNA complex. Energies are relative to the lowest energy structure for each model. The energy map of the dA adduct in the preinsertion position shows only a very small contoured domain (volume less than 1 deg3, circled and exploded). Supplementary movies S1–S12 show the full energy spectra of the data in two-dimensional slices.
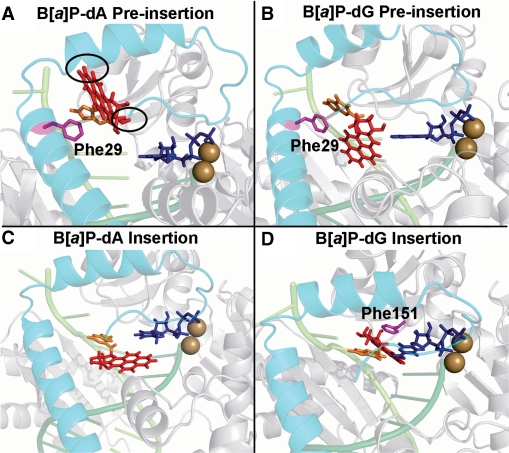
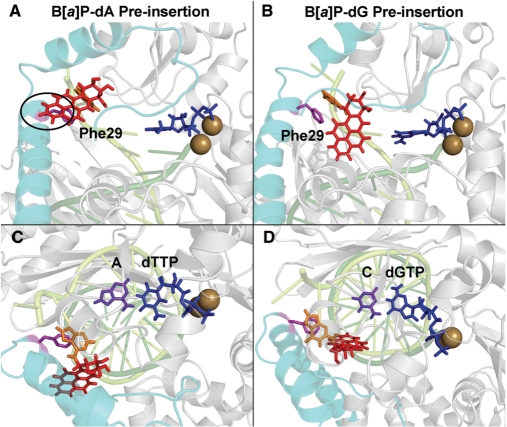
Figures 4A and B show the lowest energy structures of the dA and dG adducts in the pol κ preinsertion position. In the case of the dA adduct, the lowest energy conformation places the adduct in the anti conformation of the glycosidic torsion. However, there are still close contacts between the B[a]P ring system and the pol κ N-clasp (Figure 4), as the B[a]P rings are directed toward it. In the case of the dG adduct, the lowest energy conformation also has the adduct in the anti-conformation. Furthermore, while the B[a]P rings are linked to the N2 of dG, which is on the minor groove side of the guanine in duplex B-DNA, here the adduct is in a single-stranded region; this single-stranded region has a non-B-DNA backbone conformation, which places the B[a]P moiety on the major groove side of the evolving duplex (Figure 4B). There are no close contacts between the adduct and the enzyme, since the B[a]P rings are not near to the N-clasp. In addition, the aromatic ring of Phe29 on the N-clasp stacks with the modified guanine base better than it does with the modified adenine; this stacking helps stabilize the dG adduct in the preinsertion position (Figure 4).
Figure 4.
Pol κ/DNA complex models with lowest energy based on the VDW energy surveys. The complexes are shown in cartoon mode. The N-clasp is cyan; the DNA strands are light and dark green; the B[a]P lesion is red stick; the damaged adenine or guanine are in orange; the side chains of Phe29 and Phe151 are in magenta stick. (A) The dA adduct is in the preinsertion position. Note that the B[a]P rings are directed toward the N-clasp. The two black ovals indicate the crowding between the adduct and the N-clasp. (B) The dG adduct is in the preinsertion position. The adduct is not near to the N-clasp. Phe29 stacks with the damaged guanine on the DNA major groove side. (C) The dA adduct is in the insertion position opposite dTTP, but with syn glycosidic bond conformation that is not capable of Watson–Crick pairing. (D) The dG adduct is in the insertion position. The adduct is on the DNA minor groove side Watson–Crick paired with dCTP and B[a]P is stacked with Phe151. Stereoviews are shown in Supplementary Figure S8.
The molecular mechanics energy surveys show that the dG adduct in the insertion position favors the Watson–Crick capable anti-glycosidic conformation but the dA adduct prefers the syn domain
Here, we were motivated to elucidate the experimentally observed error-free insertion of dCTP opposite the dG adduct as well as the absence of dTTP incorporation opposite the dA adduct (33). The 3D energy maps (Figure 3) show relatively low energy domains for both dA and dG adducts in the pol κ insertion position, but the dG adduct prefers the anti-glycosidic bond conformation while the dA adduct favors the syn domain.
In the case of the dG adduct, the lowest energy anti-conformation is preferred by at least 89 kcal/mol over any syn conformers. The lowest energy structure places the dG adduct in the normal anti-conformation opposite the Watson–Crick partner dCTP, and positions the B[a]P in the pol κ on the minor groove side of the evolving DNA duplex, not near to the N-clasp. In addition, the B[a]P ring system stacks with Phe151 (Figure 4D). The B[a]P ring orientation for the dG adduct is different from that observed in NMR solution structures of this adduct in a full duplex (64), and a model template–primer complex containing a C opposite the lesion (77). In these structures, the B[a]P rings are oriented 5′ along the modified strand in the minor groove. In the present pol κ structures, the B[a]P rings are 3′ directed along the modified strand. While this is not energetically favored in solution (78), it is enforced in pol κ because steric collisions with the enzyme finger domain would occur with a 5′ orientation.
For the dA adduct, we also modeled the normal Watson–Crick partner, dTTP, as the incoming nucleotide. However, the energy survey revealed that the anti-glycosidic bond conformation is prohibited, and hence Watson–Crick pairing is impossible. On a structural level, this stems from the N6 linkage of the adenine to the B[a]P moiety which places the B[a]P ring system on the major groove side of the adducted adenine (Figure 4C); on this side the B[a]P rings collide with the enzyme active site or N-clasp or adjacent DNA and dNTP, depending on the α′ and β′ linkage site conformations. However, lower energy domains are obtained for the syn glycosidic region (Figure 3).
MD simulations of the lesions in the preinsertion position show crowding by the N-clasp in the dA but not the dG adduct
The energy survey provided lowest energy conformations as the initial structures for subsequent MD simulations. The MD simulations of the models with lesions in the preinsertion position show that the dG adduct can be housed in this site stably, as was indicated by the VDW energy survey. The dG adduct is not positioned near to the N-clasp, but is accommodated in the space between the N-clasp and the DNA (Figure 5B). In addition, the stacking interaction between the damaged guanine and Phe29 observed in the energy survey remains in place to help stabilize the adduct (Figure 5B). For the dA adduct, the MD simulation shows slight movement of the adduct and the N-clasp just sufficient to alleviate the close contacts (Figure 5A) between them that were observed in the energy survey. The B[a]P ring system continues to be directed toward the N-clasp during the simulation and is housed in the crowded environment created by it (Figure 6). There is some stacking between Phe29 and the modified adenine, but the stacking interactions are less favorable than with the dG adduct (Figure 5).
Figure 5.
MD simulation preinsertion position models for the dA and dG adducts. The best representative structures from the MD ensembles are shown. The color scheme is the same as in Figure 4. Views of (A) the dA adduct and (B) the dG adduct looking into the major groove side of the nascent DNA duplex. The adducts are positioned on the major groove side in both cases. Note the oval which highlights the crowding between the adduct and the N-clasp. Phe29 stacks partially with the damaged adenine but stacks very well with the damaged guanine. The (C and D) orientations are obtained from (A) and (B) through an ∼90° rotation to obtain views looking approximately down the DNA helix axis; these views show clearly the Watson–Crick base pairing alignment between the purple unmodified base 3′ to the lesion and its partner dNTP. Stereoviews are shown in Supplementary Figure S9. Movies are shown in Supplementary data.
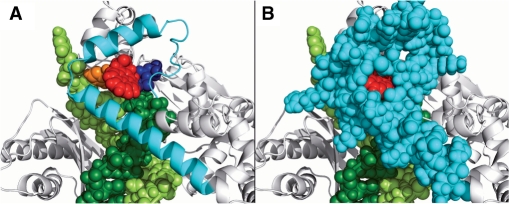
Figure 6.
Accommodation of the B[a]P ring system of the dA adduct in the preinsertion position. The adduct in the preinsertion position is directed toward and is crowded by the N-clasp. The color scheme is the same as in Figure 4. The protein is in cartoon mode and the DNA and incoming dNTP are in CPK. (A) The N-clasp is shown in cartoon in order to reveal the position of the B[a]P ring system. The (B) view is the same as (A), except that the N-clasp is shown in CPK. Stereoviews are shown in Supplementary Figure S10.
The RMSD plots (Supplementary Figure S3) of the active site region containing all atoms that are within 5 Å of any atom of the adducted nucleotide show that the dA lesion is unstable in the preinsertion position, while the dG adduct is stable. We also analyzed the time dependence for the adduct linkage site torsions α′ and β′, and the glycosidic torsion χ in the ensemble of structures. The ensemble average values and the SD for these torsion angles are listed in Supplementary Table S12, and the time dependences are given in Supplementary Figure S4. All torsions, but most notably the glycosidic torsion χ of the dA adduct have larger SD values than those of the dG adduct, showing that glycosidic torsional motions contribute significantly to the instability of the dA adduct.
The experimental data revealed that Watson–Crick paired nucleotide incorporation occurred at the unmodified bases 3′ to the dG and dA adduct in the preinsertion position (Figure 1). We therefore evaluated the near reaction-ready active site organization for the incoming dNTPs paired with these unmodified bases. Accordingly, we analyzed the Mg2+ coordination, the in-line attack distance (O3′-Pα) and angle (O3′-Pα-O3α) from the MD ensemble derived from the ternary complex, utilizing criteria for ideal active site organization involving Mg2+ coordination, in-line attack distance and attack angle which must be simultaneously met, as detailed in the Methods section. We obtained a near reaction-ready occupancy (the percent of time that the criteria are met) of 83.3% and 62.7% for the dG and dA adduct cases, respectively. By comparison, an unmodified control simulation had an occupancy of 71.1%. In addition, the dNTP is well aligned in a Watson–Crick paired orientation with the unmodified partner cytosine or adenine (Figure 5C and D, Supplementary Table S13). These results suggest that dNTP insertion is feasible when the adduct is in the preinsertion position for both the dG and dA adducts; this is considered further in relation to the experimental data in the Discussion section.
MD simulations of the lesions in the insertion position show that the dG adduct is well accommodated with Watson–Crick pairing to dCTP, while dNTP incorporation is disfavored in the dA adduct
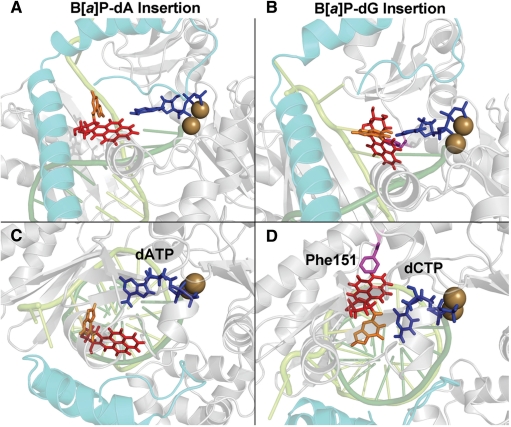
To further investigate the essentially error-free dCTP incorporation observed opposite the dG adduct, we modeled the adduct at the insertion position partnered with dCTP. The dG adduct forms a Watson–Crick base pair with the incoming dCTP (Figure 7D). The B[a]P ring system is stably accommodated in a shape complementary pocket on the evolving minor groove side of the DNA duplex (Figure 8). The stacking interaction between the B[a]P and Phe151, observed in the energy survey, remains stable during the simulation (Figure 7D). The active site RMSD and the SD values of the torsions α′, β′ and χ also show that the dG adduct is very stable in the insertion position (Supplementary Figures S3 and S4, Table S12). In addition, we obtained a near reaction-ready occupancy for the dG adduct of 66.3%.
Figure 7.
MD simulation insertion position models for the dA and dG adducts. The best representative structures from the MD ensembles are shown. The color scheme is the same as in Figure 4. Views of (A) the dA adduct and (B) the dG adduct looking into the major groove side of the nascent DNA duplex. The dA adduct is positioned on the major groove side, while the dG adduct is on the minor groove side. The damaged adenine is flipped out of the nascent DNA duplex. The (C and D) orientations are obtained from (A) and (B) through an ∼90° rotation to obtain views looking approximately down the DNA helix axis; these views show the alignment between the damaged base and the dNTP. The flipped-out damaged adenine mismatched with dATP cannot hydrogen bond with any dNTP. The damaged guanine is Watson–Crick paired with dCTP and the B[a]P has a stacking interaction with Phe151. Stereoviews are shown in Supplementary Figure S11. Movies are shown in Supplementary data.
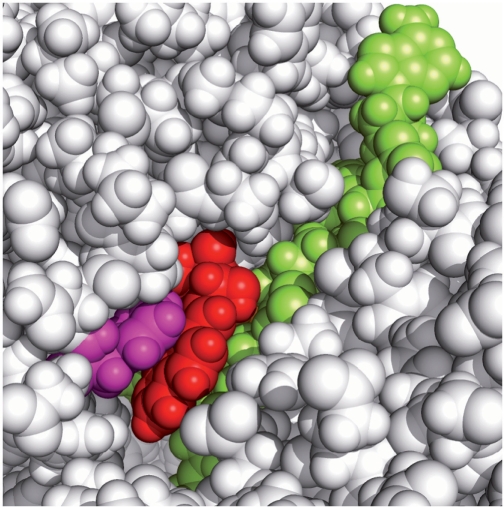
Figure 8.
CPK model of the B[a]P-dG adduct in the insertion position, looking into the minor groove of the nascent DNA duplex. The color scheme is the same as in Figure 4. The dG adduct is accommodated in a shape-complementary pocket on the evolving DNA minor groove side. The Phe151 residue in magenta stacks with the B[a]P ring system. This MD simulation frame (#7591) is among the optimal stacking structures of the ensemble. Stereoviews are shown in Supplementary Figure S12.
We wished to determine whether the aromatic Phe151 in pol κ, which stacks with the B[a]P to help stabilize its position for Watson–Crick pairing, is also present in the analogous position of Dpo4. For a broader perspective, we carried out multi-sequence alignment between all four human Y-family polymerases, as well as the archaeal enzymes Dpo4 and Dbh with Clustalw2 (76). The results show that only pol κ and Rev1 have a phenylalanine, while other polymerases have a nonaromatic residue in the analogous sequence position: pol η and pol ι have a leucine, and Dpo4 and Dbh have a lysine (Figure 9). These sequence aligned residues are located in analogous structural positions for pol κ and Dpo4, (see Discussion).
Figure 9.
Sequence alignment of S. acidocaldarius Dbh, S. solfataricus Dpo4, Homo sapiens Rev1, pol κ, pol η and pol ι. The red box shows that there is a phenylalanine present in pol κ and Rev1, while the other four polymerases have a non-aromatic residue at the analogous sequence position.
To investigate the very small amount of dATP incorporated opposite the dA adduct in the experiments (33), we modeled dATP as the incoming nucleotide for the MD simulation of this adduct (Supplementary Figure S5). The energy survey had revealed that the templating dA adduct favors the syn glycosidic conformation, and this is utilized as the initial model for the MD simulation. Since syn adenine prefers to hydrogen bond with purines in the anti-conformation (79,80), the incoming dATP was placed in the anti-glycosidic conformation (Supplementary Figure S5). Early in the MD simulation, the B[a]P ring system relocated to position the adduct on the DNA major groove side, between the DNA and the N-clasp, and away from the dATP (Figure 7A). This movement, seemingly independent of the dNTP, stems from rotations in the DNA backbone and glycosidic bond torsions (the latter from syn to high anti). This motion appears to be mandated by the proximity of the B[a]P ring system to the 3′-end primer thymine nucleobase in the lowest energy molecular mechanics structure utilized to initiate the MD (Supplementary Figure S6). This movement positions the damaged adenine away from the incoming dATP, thus preventing its hydrogen bonding interactions with any incoming dNTP (Figure 7C, Supplementary Table S13); stacking interactions between the B[a]P-modified adenine and the downstream templating base are also interrupted. The active site RMSD and the SD values of the torsions α′, β′ and χ, especially χ, show that the dA adduct is less stable than the dG adduct in the insertion position (Supplementary Figures S3 and S4, Table S12). The small amount of dATP incorporation observed in the experiments is considered in the Discussion.
DISCUSSION
The pol κ N-clasp plays a central role in determining lesion blockage or bypass
Running start primer extension studies using site specifically modified DNA containing the (+)-trans-B[a]P-N2-dG and N6-dA adducts showed that pol κ incorporates mostly dCTP opposite the dG adduct and continues to extend past the lesion. However, for the dA adduct, the polymerase is mainly stopped just before the lesion site; a minor amount of adenine incorporation is observed opposite the adduct after long (30-min) reaction time and there is no further extension. In both the dG and dA adduct cases, nucleotide incorporation opposite unmodified bases 3′ to the lesion is efficient. However, the polymerase stalled after this dNTP incorporation in the dG adduct case. Our modeling studies provide structural insights into these observations.
Well-organized active site for dNTP incorporation opposite the 3′ lesion-flanking base
Our MD simulation of the preinsertion position models (Figure 1B) revealed that the active site is well organized for incorporation of the incoming dNTP opposite the unmodified base 3′ to the lesion, in both the dA and dG adduct cases (Figure 5C and D): Mg2+ coordination, O3′ in-line attack distances and angles, and Watson–Crick alignment between the dNTP and its partner base (Supplementary Table S13) are appropriate for a near-reaction ready structure based on criteria derived from a high resolution polymerase crystal structure (61) and QM/MM calculations (62). These results are in agreement with the experimental observation that these dNTP insertions are efficient.
The B[a]P-dA adduct is blocked by the N-clasp at the preinsertion position
Following nucleotide insertion, further progression of the polymerase is mostly blocked by the dA adduct (33,50). The underlying reasons for this blockage are apparent from our energy survey and subsequent MD simulation for the dA adduct in the preinsertion position: our models show that the B[a]P ring system, linked to the N6-group of adenine positioned in the major groove, is in steric conflict with the pol κ N-clasp (Figure 6). While nucleotide incorporation opposite the unmodified base 3′ to the lesion is efficient, the steric hindrance posed by the N-clasp would very likely present a barrier to translocation, which would account for the observed blockage. The small amount of dATP incorporation observed experimentally (33), suggests that the translocation impeded by the N-clasp may be relieved by rare enzyme movements that allow the dA adduct to proceed to the insertion position. Our MD simulations for the dA adduct opposite dATP in the insertion position showed that the modified adenine flips out of the duplex (Figure 7A and C), suggesting that the dATP incorporation might be untemplated. This would be analogous to the incorporation of dATP opposite an abasic site in pol κ (24) and pol β (81), i.e. mechanisms that follow the ‘A-rule’ (82). Mechanisms similar to those proposed here for the B[a]P-N6-dA lesions may also be plausible for other bulky lesions that are linked on the major groove side and that also block pol κ, at least partially. These lesions include the 2-aminofluorene (AF)- and 2-(acetylamino) fluorene (AAF)-C8-dG adducts (27).
The B[a]P-dG adduct can progress to the insertion site to form a Watson–Crick pair with dCTP
In the case of the dG adduct, following the facile nucleotide incorporation opposite the unmodified base 3′ to the lesion, the experiments show that the polymerase stalls, but then continues to insert dCTP opposite the lesion in the insertion position. Our energy survey and MD simulation show that in the preinsertion position, the B[a]P rings are not close to the N-clasp. A non-B-DNA conformation in this single-stranded preinsertion position places the B[a]P-dG adduct on the major groove side of the evolving duplex (Figure 5B), while in a full duplex of B-DNA the B[a]P moiety is on the minor groove side (64). Our subsequent studies for the insertion position show that the B[a]P-dG adduct is Watson–Crick-paired with dCTP, with the B[a]P ring system positioned on the minor groove side of the nascent duplex (Figure 7B). There is no steric hindrance in either position. The polymerase pause following nucleotide incorporation opposite the undamaged base 3′ to the lesion in the preinsertion position, may result from the following features: an energy barrier must be overcome to achieve the rearrangement of the lesion from the major to the minor groove side, as it translocates from the preinsertion, to the insertion position. However, once this barrier is overcome, the adduct is well positioned for Watson–Crick pairing of the modified dG with dCTP and for nucleotide incorporation (Figure 7D, Supplementary Table S13). The arrangements of the interacting partners at this active site are well poised for the nucleotidyl transfer reaction, which is consistent with the experimentally observed essentially high fidelity bypass of this lesion.
It is noteworthy that there are other bulky N2-dG adducts that are bypassed by pol κ, primarily in an error-free manner (32,83,84); these may assume conformations similar to those of the B[a]P- N2-dG adduct in the preinsertion and insertion sites of pol κ. Such adducts include the minor groove conformation of the N2-(2-hydroxyestron-6(α,β)-yl)-2′-deoxyguanosine in duplex DNA suggested by a modeling study (85), as well as the 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene adduct in double-stranded DNA positioned in the minor groove as shown by NMR solution (86) and modeling studies (87).
Error-free bypass of the B[a]P-N2-dG adduct is supported by specific structural features of pol κ, notably the N-clasp, not present in the error-prone Dpo4 polymerase
It is noteworthy that pol κ bypasses the B[a]P-dG lesion in an essentially error-free manner, while the bypass of the same lesion catalyzed by its DinB prokaryotic homolog Dpo4 is error-prone. In the latter, various mismatches and a prevalence of deletions have been observed, with the sequence context in which the lesion is embedded playing an important role in the specific mutagenic outcome (47,53,54,88). Our modeling and MD simulations for the pol κ insertion position provide some structural insights for this striking difference.
In pol κ, the anti glycosidic bond conformation of the damaged templating dG, needed for Watson–Crick pairing at the insertion position, is strongly preferred over the syn conformation because the pol κ N-clasp has close contacts with the syn-oriented B[a]P-dG moiety (Figure 10A and B). In contrast, in Dpo4 both syn and anti B[a]P-dG conformations are feasible (47,53, 88,89, Xu, P., Oum, L., Geacintov, N.E. and Broyde, S., manuscript in preparation). The syn B[a]P-dG adduct is comfortably accommodated on the major groove side, which does not have an N-clasp and is therefore open to the solution environment (Figure 10C). Furthermore, modeling studies show that the syn conformation, which is incapable of forming Watson–Crick hydrogen-bonding with the dNTP, can readily support mismatches (47,53,54, Xu, P., Oum, L., Geacintov, N.E. and Broyde, S., manuscript in preaparation), thus providing a rationale for the observed low fidelity of Dpo4.
Figure 10.
(A and B) The syn B[a]P-dG adduct structures have close contacts with the pol κ N-clasp. The lesion is positioned on the major groove side of the nascent DNA duplex. Two structures from the energy survey are shown. (A) The syn adduct structure with globally lowest VDW energy. (B) To facilitate comparison, a second syn structure is presented with adduct oriented as in the Dpo4 model shown in (C); the lowest VDW energy structure in this conformational domain for pol κ was selected. (C) A model derived from MD simulation of B[a]P-dG in Dpo4 (53). Dpo4 has an open space on the DNA major groove side because it lacks the N-clasp, and can therefore accommodate the syn B[a]P-dG. In all cases the syn adduct is positioned opposite dCTP. The color scheme is the same as in Figure 4. Stereoviews are shown in Supplementary Figure S13.
In addition, the aromatic Phe151 residue of pol κ, positioned in the nascent minor groove of the DNA duplex, stacks with the B[a]P ring system (Figure 8) which helps to stabilize the adducted guanine in an orientation which supports Watson-Crick pairing with dCTP. In contrast, Dpo4 has a lysine residue in the analogous position (Figure 11), which does not contribute to a stabilization of the aromatic ring system. Thus, even when the anti-conformation that is capable of Watson–Crick pairing is assumed, in Dpo4 the adducted guanine would be conformationally more flexible and hence more likely to pair with other dNTPs.
Figure 11.
Superimposition of the anti B[a]P-dG adduct in the pol κ insertion position with the Dpo4 ternary complex (PDB ID: 2AGQ). The most representative structure from the MD ensemble is shown. Pol κ is in gray cartoon; and Dpo4 is in blue cartoon; B[a]P rings are red; the damaged guanine is orange. The magenta Phe151 stacks with the B[a]P ring system. However, the analogous position in Dpo4 is occupied by a green lysine. Superimpositions are performed with the PyMol Align function which overlays the backbone Cα atoms. Stereoviews are shown in Supplementary Figure S14.
Modeling pol κ with bulky DNA lesions: computational considerations
Finally, we wish to discuss our computational approaches. We utilized molecular dynamics simulations with AMBER 9.0 (64) in order to obtain ensembles of structures to aid in interpreting the experimental data. Simulations of such large systems still pose challenges and require necessary approximations. The first challenge is to obtain initial structures for the subsequent MD simulations. Here, we had to first complete the pol κ crystal structure, which had 57 residues far from the active site and DNA-binding region missing. To avoid possibly serious problems with modeling these residues de novo, we elected to close the gap with a loop as a better option. We also modeled the missing catalytic Mg2+ in the active site based on ideal octahedral coordination from a high resolution pol β crystal structure (61) and a QM/MM computational study of Dpo4 (62).
In this connection, it must be mentioned that the proper handling of magnesium ions in force fields such as AMBER remains a research frontier. This was pointed out by David Case who heads the AMBER development team ‘there is no simple way to handle divalent ions in (current) molecular mechanics force fields that is fully satisfactory’ (http://archive.ambermd.org/200507/0033.html). While various approaches are being explored, none appears yet ready for general use by the AMBER community. Furthermore, in relation to organization of DNA polymerase active sites, as benchmarked by a high resolution crystal structure (61) and by QM/MM calculations from our group (62), the AMBER force field is actually performing rather well in reproducing the octahedral coordination of the two active site magnesium ions; this was demonstrated in the present study as well as in earlier work from our group (74). However, there is no doubt that advances are needed in this area.
We then utilized a rational and unbiased approach to obtain initial conformations of the lesions in pol κ: we evaluated the VDW energy utilizing an implicit solvation model for 373, 248 different conformers, in an evenly and tightly spaced (5° intervals in combination) grid search of three flexible torsions governing the lesion orientation; this was done for each of the four models. We then selected the structure with the lowest VDW energy for each case to initiate the MD simulations with explicit aqueous solvent and Na+ counterions. A question arises here: if neighboring structures in the lowest energy region were utilized to initiate the MD, would such structures produce different outcomes in the MD simulations? While the sampling problem in MD simulations (90–92) remains demanding, our sterically very confined pol κ system has extremely limited opportunities for housing the bulky lesions we investigated, and these were ferreted out in the energy survey. Examination of the energy spectra (Figure 3 and Movies S1–S12 in Supplementary Material) reveals the very narrow conformational domains that could be feasible. For the dA adduct, it is very clear that the preinsertion position blocks accommodation of the lesion by collision with the N-clasp, which in and of itself explains the experimental data that this adduct mainly blocks the polymerase. For the dG adduct, the dominant experimental observation to be explained is the virtually error-free incorporation of dCTP opposite the lesion by pol κ. This structural requirement for Watson–Crick base pairing in the experimental data is directly rationalized from the energy survey, as it was the lowest VDW energy structure.
Overall, approximations dictated by the state-of-the-art were required in order to provide structural insights on the unexplained differential processing of the dA and dG adducts by pol κ, since pol κ crystal structures with these lesions are not available. However, the unique structural features of pol κ enforce very strong steric constraints that make our structural interpretation of the data plausible.
CONCLUSIONS
Pol κ with its unique N-clasp fully encircles the DNA and appears to be structured for near error-free bypass of minor groove bulky lesions such as the B[a]P-N2-dG adduct. These adducts are readily accommodated in the preinsertion and insertion positions of the enzyme and are not obstructed by the N-clasp. At the insertion position, Phe151 stacks with the B[a]P ring system on the minor groove side of the evolving DNA double-strand, which helps to stabilize Watson–Crick base pairing that, in turn, facilitates quite faithful lesion bypass. However, the major groove B[a]P-N6-dA adduct is located in a region that is sterically crowded with residues of the N-clasp in the preinsertion position, and is therefore largely blocked from entering into the insertion position. Structural comparisons of pol κ and Dpo4 that lacks the N-clasp and contains a lysine in place of Phe151, help explain why Dpo4 is error-prone in bypassing the B[a]P-dG lesion, while pol κ bypasses this lesion in an essentially error-free manner: the N-clasp in pol κ prevents the adoption of the mismatch-supporting syn conformation of this lesion. Thus, our results provide structural explanations for understanding the observed error-free and error-prone translesion synthesis associated with a pair of DinB bypass polymerases that are based on specific differences in their structural properties.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health, National Cancer Institute (CA28038 to S.B. and CA099194 to N.E.G). Partial support for computational infrastructure and systems management was also provided by Grant CA75449 to S.B. Computational support was provided in part by the National Science Foundation Partnerships for Advanced Computational Infrastructure. Funding for open access charge: National Institute of Health, National Cancer Institute CA 28038 to S.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We wish to thank a dedicated referee for sage advice.
REFERENCES
- 1.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 2.Xu P, Oum L, Beese LS, Geacintov NE, Broyde S. Following an environmental carcinogen N2-dG adduct through replication: elucidating blockage and bypass in a high-fidelity DNA polymerase. Nucleic Acids Res. 2007;35:4275–4288. doi: 10.1093/nar/gkm416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broyde S, Wang L, Rechkoblit O, Geacintov NE, Patel DJ. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 2008;33:209–219. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 5.Hanrahan CJ, Bacolod MD, Vyas RR, Liu T, Geacintov NE, Loechler EL, Basu AK. Sequence specific mutagenesis of the major (+)-anti-benzo[a]pyrene diol epoxide-DNA adduct at a mutational hot spot in vitro and in Escherichia coli cells. Chem. Res. Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr. Opin. Genet. Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 10.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 12.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 13.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 15.Clapp RW, Jacobs MM, Loechler EL. Environmental and occupational causes of cancer: new evidence 2005-2007. Rev. Environ. Health. 2008;23:1–37. doi: 10.1515/reveh.2008.23.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann AR. Translesion synthesis in mammalian cells. Exp. Cell Res. 2006;312:2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Jarosz DF, Godoy VG, Walker GC. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle. 2007;6:817–822. doi: 10.4161/cc.6.7.4065. [DOI] [PubMed] [Google Scholar]
- 19.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 20.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28:4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc. Natl Acad. Sci. USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J. Biol. Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki N, Ohashi E, Hayashi K, Ohmori H, Grollman AP, Shibutani S. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase kappa and Escherichia coli DNA polymerase IV. Biochemistry. 2001;40:15176–15183. doi: 10.1021/bi010702g. [DOI] [PubMed] [Google Scholar]
- 28.Choi JY, Zang H, Angel KC, Kozekov ID, Goodenough AK, Rizzo CJ, Guengerich FP. Translesion synthesis across 1,N2-ethenoguanine by human DNA polymerases. Chem. Res. Toxicol. 2006;19:879–886. doi: 10.1021/tx060051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine RL, Miller H, Grollman A, Ohashi E, Ohmori H, Masutani C, Hanaoka F, Moriya M. Translesion DNA synthesis catalyzed by human pol eta and pol kappa across 1,N6-ethenodeoxyadenosine. J. Biol. Chem. 2001;276:18717–18721. doi: 10.1074/jbc.M102158200. [DOI] [PubMed] [Google Scholar]
- 30.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc. Natl Acad. Sci. USA. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 32.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 33.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Kolbanovskiy A, Wu X, Zhang Y, Wang Z, Zhuang P, Amin S, Geacintov NE. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol kappa. Biochemistry. 2003;42:2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Wu X, Guo D, Rechkoblit O, Geacintov NE, Wang Z. Two-step error-prone bypass of the (+)- and (-)-trans-anti-BPDE-N2-dG adducts by human DNA polymerases eta and kappa. Mutat. Res. 2002;510:23–35. doi: 10.1016/s0027-5107(02)00249-x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair. 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki N, Ohashi E, Kolbanovskiy A, Geacintov NE, Grollman AP, Ohmori H, Shibutani S. Translesion synthesis by human DNA polymerase kappa on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N(2)-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 38.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol. Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buening MK, Wislocki PG, Levin W, Yagi H, Thakker DR, Akagi H, Koreeda M, Jerina DM, Conney AH. Tumorigenicity of the optical enantiomers of the diastereomeric benzo[a]pyrene 7,8-diol-9,10-epoxides in newborn mice: exceptional activity of (+)-7beta,8alpha-dihydroxy-9alpha,10alpha-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. Proc. Natl Acad. Sci. USA. 1978;75:5358–5361. doi: 10.1073/pnas.75.11.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 41.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem. Res. Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 42.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 43.Meehan T, Straub K. Double-stranded DNA steroselectively binds benzo(a)pyrene diol epoxides. Nature. 1979;277:410–412. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 44.Christner DF, Lakshman MK, Sayer JM, Jerina DM, Dipple A. Primer extension by various polymerases using oligonucleotide templates containing stereoisomeric benzo[a]pyrene-deoxyadenosine adducts. Biochemistry. 1994;33:14297–14305. doi: 10.1021/bi00251a043. [DOI] [PubMed] [Google Scholar]
- 45.Perlow RA, Broyde S. Extending the understanding of mutagenicity: structural insights into primer-extension past a benzo[a]pyrene diol epoxide-DNA adduct. J. Mol. Biol. 2003;327:797–818. doi: 10.1016/s0022-2836(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 46.Perlow-Poehnelt RA, Likhterov I, Wang L, Scicchitano DA, Geacintov NE, Broyde S. Increased flexibility enhances misincorporation: temperature effects on nucleotide incorporation opposite a bulky carcinogen-DNA adduct by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:1397–1408. doi: 10.1074/jbc.M606769200. [DOI] [PubMed] [Google Scholar]
- 47.Xu P, Oum L, Geacintov NE, Broyde S. Nucleotide selectivity opposite a benzo[a]pyrene-derived N2-dG adduct in a Y-family DNA polymerase: a 5′-slippage mechanism. Biochemistry. 2008;47:2701–2709. doi: 10.1021/bi701839q. [DOI] [PubMed] [Google Scholar]
- 48.Hruszkewycz AM, Canella KA, Peltonen K, Kotrappa L, Dipple A. DNA polymerase action on benzo[a]pyrene-DNA adducts. Carcinogenesis. 1992;13:2347–2352. doi: 10.1093/carcin/13.12.2347. [DOI] [PubMed] [Google Scholar]
- 49.Chary P, Latham GJ, Robberson DL, Kim SJ, Han S, Harris CM, Harris TM, Lloyd RS. In vivo and in vitro replication consequences of stereoisomeric benzo[a]pyrene-7,8-dihydrodiol 9,10-epoxide adducts on adenine N6 at the second position of N-ras codon 61. J. Biol. Chem. 1995;270:4990–5000. doi: 10.1074/jbc.270.10.4990. [DOI] [PubMed] [Google Scholar]
- 50.Frank EG, Sayer JM, Kroth H, Ohashi E, Ohmori H, Jerina DM, Woodgate R. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase iota. Nucleic Acids Res. 2002;30:5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl Acad. Sci. USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 53.Perlow-Poehnelt RA, Likhterov I, Scicchitano DA, Geacintov NE, Broyde S. The spacious active site of a Y-family DNA polymerase facilitates promiscuous nucleotide incorporation opposite a bulky carcinogen-DNA adduct: elucidating the structure-function relationship through experimental and computational approaches. J. Biol. Chem. 2004;279:36951–36961. doi: 10.1074/jbc.M404332200. [DOI] [PubMed] [Google Scholar]
- 54.Oum L. Base-sequence effects on in vitro translesion bypass catalyzed by A- and Y-family polymerases. PhD Thesis. 2007 [Google Scholar]
- 55.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 59.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 60.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Yu X, Hu P, Broyde S, Zhang Y. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl Acad. Sci. USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, et al. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 65.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 66.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 67.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 70.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the multimolecule and multiconformational resp methodology to biopolymers - charge derivation for DNA, Rna, and proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 71.Perlow RA, Broyde S. Evading the proofreading machinery of a replicative DNA polymerase: induction of a mutation by an environmental carcinogen. J. Mol. Biol. 2001;309:519–536. doi: 10.1006/jmbi.2001.4674. [DOI] [PubMed] [Google Scholar]
- 72.Perlow RA, Broyde S. Toward understanding the mutagenicity of an environmental carcinogen: structural insights into nucleotide incorporation preferences. J. Mol. Biol. 2002;322:291–309. doi: 10.1016/s0022-2836(02)00751-9. [DOI] [PubMed] [Google Scholar]
- 73.Simmerling C, Kollman P. MOIL-View: a program for visualization of structure and dynamics of biomolecules. Abstracts of Papers of the American Chemical Society. 1996;211 92-Comp. [Google Scholar]
- 74.Wang L, Broyde S. A new anti conformation for N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (AAF-dG) allows Watson-Crick pairing in the Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) Nucleic Acids Res. 2006;34:785–795. doi: 10.1093/nar/gkj479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L, Rechkoblit O, Wang L, Patel DJ, Shapiro R, Broyde S. Mutagenic nucleotide incorporation and hindered translocation by a food carcinogen C8-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): modeling and dynamics studies. Nucleic Acids Res. 2006;34:3326–3337. doi: 10.1093/nar/gkl425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 77.Cosman M, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Structural alignments of (+)- and (-)-trans-anti-benzo[a]pyrene-dG adducts positioned at a DNA template-primer junction. Biochemistry. 1995;34:15334–15350. doi: 10.1021/bi00046a043. [DOI] [PubMed] [Google Scholar]
- 78.Xie XM, Geacintov NE, Broyde S. Origins of conformational differences between cis and trans DNA adducts derived from enantiomeric anti-benzo[a]pyrene diol epoxides. Chem. Res. Toxicol. 1999;12:597–609. doi: 10.1021/tx990021a. [DOI] [PubMed] [Google Scholar]
- 79.Brown T, Hunter WN, Kneale G, Kennard O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc. Natl Acad. Sci. USA. 1986;83:2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel DJ, Kozlowski SA, Ikuta S, Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG X dA mismatch site. Biochemistry. 1984;23:3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- 81.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase beta violates the “A-rule”. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 82.Strauss BS. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair. 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- 83.Poon K, Itoh S, Suzuki N, Laxmi YRS, Yoshizawa I, Shibutani S. Miscoding Properties of 6α- and 6β-Diastereoisomers of the N2-(Estradiol-6-yl)-2′-deoxyguanosine DNA Adduct by Y-Family Human DNA Polymerases. Biochemistry. 2008;47:6695–6701. doi: 10.1021/bi7022255. [DOI] [PubMed] [Google Scholar]
- 84.Yasui M, Dong H, Bonala RR, Suzuki N, Ohmori H, Hanaoka F, Johnson F, Grollman AP, Shibutani S. Mutagenic properties of 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene, a persistent acetylaminofluorene-derived DNA adduct in mammalian cells. Biochemistry. 2004;43:15005–15013. doi: 10.1021/bi048279+. [DOI] [PubMed] [Google Scholar]
- 85.Wang L, Hingerty BE, Shapiro R, Broyde S. Structural and stereoisomer effects of model estrogen quinone-derived DNA adducts: N6-(2-hydroxyestron-6(alpha,beta)-yl)-2′-deoxyadenosine and N2-(2-hydroxyestron-6(alpha,beta)-yl)-2′- deoxyguanosine. Chem. Res. Toxicol. 2004;17:311–324. doi: 10.1021/tx034218l. [DOI] [PubMed] [Google Scholar]
- 86.Zaliznyak T, Bonala R, Johnson F, de Los Santos C. Structure and stability of duplex DNA containing the 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG(N2)-AAF) lesion: a bulky adduct that persists in cellular DNA. Chem. Res. Toxicol. 2006;19:745–752. doi: 10.1021/tx060002i. [DOI] [PubMed] [Google Scholar]
- 87.Grad R, Shapiro R, Hingerty BE, Broyde S. A molecular mechanics and dynamics study of the minor adduct between DNA and the carcinogen 2-(acetylamino)fluorene (dG-N2-AAF) Chem. Res. Toxicol. 1997;10:1123–1132. doi: 10.1021/tx970092e. [DOI] [PubMed] [Google Scholar]
- 88.Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl Acad. Sci. USA. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 90.Roitberg A, Simmerling C. Foreword. J. Mol. Graph Model. 2004;22:317. doi: 10.1016/j.jmgm.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 91.Cheatham TE., III Simulation and modeling of nucleic acid structure, dynamics and interactions. Curr. Opin. Struct. Biol. 2004;14:360–367. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 92.Adcock SA, McCammon JA. Molecular dynamics: survey of methods for simulating the activity of proteins. Chem. Rev. 2006;106:1589–1615. doi: 10.1021/cr040426m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.