Abstract
Balancing selection is common on many defense genes, but it has rarely been reported for immune effector proteins such as antimicrobial peptides (AMPs). We describe genetic diversity at a brevinin-1 AMP locus in three species of leopard frogs (Rana pipiens, Rana blairi, and Rana palustris). Several highly divergent allelic lineages are segregating at this locus. That this unusual pattern results from balancing selection is demonstrated by multiple lines of evidence, including a ratio of nonsynonymous/synonymous polymorphism significantly higher than 1, the ZnS test, incongruence between the number of segregating sites and haplotype diversity, and significant Tajima's D values. Our data are more consistent with a model of fluctuating selection in which alleles change frequencies over time than with a model of stable balancing selection such as overdominance. Evidence for fluctuating selection includes skewed allele frequencies, low levels of synonymous variation, nonneutral values of Tajima's D within allelic lineages, an inverse relationship between the frequency of an allelic lineage and its degree of polymorphism, and divergent allele frequencies among populations. AMP loci could be important sites of adaptive genetic diversity, with consequences for host–pathogen coevolution and the ability of species to resist disease epidemics.
Keywords: Rana pipiens, Rana blairi, Rana palustris, antimicrobial peptide, balancing selection, fluctuating selection
Introduction
Genes encoding immune system proteins often harbor adaptive variation maintained by balancing selection (Garrigan and Hedrick 2003). For example, patterns of genetic diversity consistent with balancing selection have frequently been observed at the vertebrate major histocompatibility complex (Garrigan and Hedrick 2003; Piertney and Oliver 2006), immunoglobulin genes (Su and Nei 1999), plant R-genes (Stahl et al. 1999; Bakker et al. 2006), and other immunity genes (Bamshad et al. 2002; Jensen et al. 2008). Although extensive, these examples are not equally distributed among all classes of immunity genes. Nearly all immunogenetic adaptive variations have been found in detection and signaling proteins, not in the effector proteins that directly attack pathogens (Garrigan and Hedrick 2003; Lazzaro et al. 2004; Tiffin and Moeller 2006). The high polymorphism at many of these pathogen-detection genes, such as loci of the vertebrate adaptive immune system and plant loci involved in gene-for-gene interactions, is associated with either high specificity or a cost of resistance (Garrigan and Hedrick 2003; Tian et al. 2003; Bakker et al. 2006). When immunity mechanisms are highly specific, pathogens can easily counteract them by altering the specific molecular target, so it benefits hosts to carry a diversity of immunity molecules which are effective against different variants of the molecular target (Stahl et al. 1999; Tiffin et al. 2004). Also, if alleles vary in overall effectiveness but there is a cost to resistance, both resistance and susceptibility alleles can be maintained at a locus (Tian et al. 2003). Some effector molecules show high structural and functional diversity across taxa, suggestive of specific immunological roles, and some could impose a cost by damaging host cells (Hancock 2001). Thus, we hypothesize that balancing selection might be relatively common at certain effector genes, especially those that show high interspecies divergence and/or are potentially costly. Testing this hypothesis would help to indicate the conditions under which balancing selection occurs and further illuminate host–pathogen coevolution at the molecular level.
Several mechanisms of balancing selection have been proposed (Charlesworth 2006). Under overdominant selection, heterozygotes are consistently fitter than homozygotes. Under instantaneous frequency-dependent selection or minority advantage, the fitness of an allele is directly and inversely proportional to its frequency (Takahata and Nei 1990). Instantaneous frequency-dependent selection can arise due to behavioral interactions among individuals in a single species, but it is not thought to occur in host–parasite interactions (Seger 1988). Under time-delayed frequency-dependent selection, including the “trench warfare” model of host–pathogen coevolution, there is a lag between the change in host allele frequencies and the change in their fitnesses, owing to the need for the parasite to evolve first (Seger 1988; Stahl et al. 1999). Finally, under spatiotemporally varying selection, fitnesses of alleles vary over time and/or space due to variation in the presence or absence of pathogens which occurs independently of host allele frequencies (Hedrick 2002). These various mechanisms of balancing selection can be categorized according to whether or not allele frequencies change over time. Overdominance and instantaneous frequency-dependent selection are mathematically equivalent, and both are forms of stable balancing selection, predicting approximately constant intermediate allele frequencies over long periods of time (Takahata and Nei 1990). In contrast, the trench warfare model and spatiotemporally varying selection are forms of fluctuating selection, in which allele frequencies change frequency dynamically over time, becoming common when advantageous and rare when disadvantageous (Stahl et al. 1999; Tiffin et al. 2004).
Under stable balancing selection, allele frequencies should change more slowly over time than they would under neutral genetic drift, whereas under fluctuating selection, allele frequencies should change more quickly over time than they would under neutral genetic drift. Therefore, the two categories of balancing selection can be distinguished from each other by testing for evidence of substantial allele frequency change over time. When two or more allelic lineages are maintained by balancing selection, polymorphisms can be classified as either within-lineage variation or between-lineage variation (Innan and Tajima 1999). Under stable balancing selection, Tajima's D (Tajima 1989) for within-lineage variation should be close to 0. Under fluctuating selection, Tajima's D should be negative within lineages that have recently increased in frequency, which are analogous to growing populations, and positive for lineages that have recently decreased in frequency, which are analogous to populations going through a bottleneck. Similarly, stable balancing selection predicts highly similar allele frequencies among populations, high synonymous variation due to the antiquity of alleles at that locus, a correlation between the frequency of allelic lineages and the neutral variation they harbor, and a low probability of fixation for any allele. In contrast, fluctuating selection permits divergent allele frequencies among populations and predicts low synonymous variation due to periodic bottlenecks for every allelic lineage, no correlation between the frequency of allelic lineages and the neutral variation they harbor, and the occasional fixation of particular alleles.
Antimicrobial peptides (AMPs) of the innate immune system have only rarely been observed to be under pathogen-driven balancing selection (Tennessen 2005b). These cationic, amphipathic mature peptides are cleaved off of a larger protein and then bind to the cell membranes of bacterial, fungal, and enveloped viral pathogens, killing them (Hancock 2001). Research on AMPs has been substantial in recent years, due in part to an interest in developing them for therapeutic application (Hancock 2001). Positive selection on AMP genes is very common and has resulted in an enormous functional diversity of these molecules among species and among loci in many taxa (Tennessen 2005b). Some human and mussel AMP loci appear to be under balancing selection (Hollox and Armour 2008; Pahdi and Verghese 2008), but other studies of intraspecies genetic diversity at AMP genes have revealed no evidence for balancing selection (Clark and Wang 1997; Lazzaro and Clark 2003; Tennessen and Blouin 2007).
The AMPs of leopard frogs (genus Rana; in this paper, we ignore the recent proposal by Frost et al. [2006] to revise the genus to Lithobates) are among the most well studied (Conlon et al. 2004). Most of them consist of an α-helix with a disulfide bridge forming a loop at the C-terminal end. They are functionally diverse, frequently with activity against both amphibian and human pathogens (Goraya et al. 2000; Chinchar et al. 2004; Rollins-Smith and Conlon 2005). Given the global crisis of amphibian population declines mediated by emerging infectious diseases and the immunological importance of AMPs, genetic diversity at AMP loci could be an important determinant of amphibian population stability (Daszak et al. 2003; Woodhams, Rollins-Smith, et al. 2006; Woodhams, Voyles, et al. 2006). Of the four AMP families secreted by the northern leopard frog, Rana pipiens (brevinin-1, ranatuerin-2, temporin-1, and esculentin-2), the brevinin-1 family is both the most diverse and the most active against microbes (Goraya et al. 2000; Tennessen and Blouin 2007). Previously, we investigated allelic variation at the Ranatuerin2 AMP locus in R. pipiens and found that a single haplotype had been fixed in the species by a positive selective sweep (Tennessen and Blouin 2007). We also found substantial diversity among AMP sequences at five loci of the brevinin-1 family in R. pipiens, but we were unable to assess how much of this variation was within versus between loci (Tennessen and Blouin 2007). In this paper, we test whether some of the brevinin-1 variants are allelic by designing primers that amplify a single brevinin-1 locus. We examine patterns of genetic diversity in over 400 individuals of R. pipiens. In order to assess the generality of our observations across species, we also sequence this locus in a smaller number of plains leopard frogs (Rana blairi) and pickerel frogs (Rana palustris). Several highly divergent allelic lineages are segregating at this locus, and we present evidence that the balanced maintenance of these alleles is due to a dynamic process of fluctuating natural selection.
Materials and Methods
Tissues
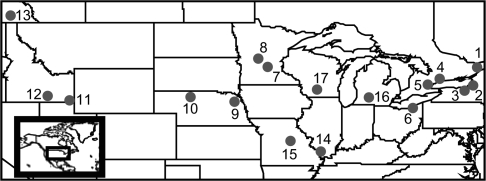
Samples of R. pipiens were collected from 13 sites throughout its range across northern North America as described previously (n = 20–46 individuals per site; supplementary table 1, Supplementary Material online; fig. 1; Hoffman et al. 2006). For one of the sites in New York, we also obtained a historical sample of R. pipiens collected in 1971 (approximately 15 generations earlier) as described previously (n = 25; Hoffman et al. 2006). We collected samples of R. palustris from two sites in Wisconsin and Michigan (n = 23 and 40, respectively) and samples of R. blairi from a single site in Illinois (n = 27; supplementary table 1, Supplementary Material online; fig. 1). Further samples of R. blairi were obtained from the Museum of Vertebrate Zoology (MVZ Herp 240161–240184), having all been originally collected from a single site in Missouri (n = 24; supplementary table 1, Supplementary Material online; fig. 1). The sample size at every site was at least 20 frogs (mean number of frogs per population = 28.8). All tissues consisted of toe clips preserved by desiccation in 1.5-ml tubes filled with Drierite desiccant (W. A. Hammond Drierite Co., Xenia, OH), except for the MVZ samples, which had been frozen. DNA was extracted as described previously (Hoffman and Blouin 2004) or using DNeasy Blood and Tissue Kits (Qiagen Inc., Valencia, CA).
FIG. 1.—
Map of sampling localities in the United States and Canada. Populations 1–13 are Rana pipiens, populations 14 and 15 are Rana blairi, and populations 16 and 17 are Rana palustris. Exact population locations are in supplementary table 1 (Supplementary Material online).
We carried out 25 μl polymerase chain reactions (PCRs) using standard buffer conditions, 1.5 mM MgCl2, 0.2 mM each deoxyribonucleotide triphosphate, approximately 100 ng DNA, and 0.5 Units Taq DNA polymerase. We visualized PCR products under ultraviolet light, purified them with the MoBio Ultraclean PCR cleanup kit (Solana Beach, CA), and sequenced them through the Nevada Genomics Center (Reno, NV).
Sequences
Previously, we had cloned two brevinin-1 sequences from a single R. pipiens individual that were similar in the intron but quite divergent in the mature peptide region (GenBank accession numbers DQ276967 and DQ276968; Tennessen and Blouin 2007). In this study, we tested whether they were allelic by assessing whether the genotype frequencies were in Hardy–Weinberg equilibrium within populations. To do so, we designed a primer (Brev1PF4; 5′-GAT GAC CCA ATA ATA ATT TTT C-3′) that would bind to these sequences but not to any other known brevinin-1 gene in R. pipiens. We used the primers Brev1PF4 and Brev1PR1 (Tennessen and Blouin 2007), which bind outside of the coding region, to amplify genomic DNA from R. pipiens, R. palustris, and R. blairi. PCR amplification conditions consisted of an initial denaturation step at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 45 s, annealing at 51 °C for 30 s, and extension at 72 °C for 1 min; and a final single extension step at 72 °C for 5 min. We sequenced the resultant PCR products with an internal primer (Brev1PF5; 5′-GAA AGC TCT GTG CCA TAG-3′).
Due to difficulties amplifying some R. blairi individuals with this primer pair, we designed a forward primer specific to R. blairi. We sequenced the primer-binding site in R. blairi using a cloning procedure described previously (Tennessen and Blouin 2007). Briefly, we amplified R. blairi genomic DNA using the more distal and degenerate forward primer Brev1PF3 with Brev1PR1 in PCRs with the high fidelity enzyme Pfu DNA polymerase (Promega, Madison, WI). The resultant PCR products were incubated with Taq polymerase in order to add 3′ adenines and cloned into Escherichia coli using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA). The recombinant locus was amplified from screened colonies using the primers T3 and T7, and the resultant PCR products were sequenced with T7. We used these sequences to design the primer Brev1BLF (5′-TAG ATG ACC TAA TAA TAA TTT TTC-3′), which binds in the intron. We amplified and sequenced DNA from all R. blairi individuals using both primer set (Brev1PF4 and Brev1PR1) and primer set (Brev1BLF and Brev1PR1).
In order to compare patterns of genetic diversity at the brevinin-1 locus with other, putatively neutral, nuclear loci, we sequenced 1,246 bp of four nuclear genes in a subset of individuals: Arcadlin (primers ArcadF and ArcadR, 178 bp coding, 87 bp intron; Tennessen and Blouin 2007), Myosin (primers MyosinF and MyosinR, 65 bp coding, 85 bp intron; Tennessen and Blouin 2007), FIBI7 (primers FIBI7U and FIBI7L, 50 bp coding, 208 bp intron; Di Candia and Routman 2007), and Tyrosinase (primers Tyr1A and Tyr1G, 573 bp coding; Bossuyt and Milinkovitch 2000). These loci were chosen because we had preexisting protocols for amplifying them in Rana and because their polymorphisms are consistent with neutral evolution. We sequenced Arcadlin, Myosin, FIBI7, and Tyrosinase in all frogs from six populations: populations 1, 2, 14, 15, 16, and 17 (fig. 1). These six populations represented all R. palustris and R. blairi individuals but only a fraction of all R. pipiens individuals (59 frogs). In order to obtain a more representative sample of R. pipiens, we randomly chose two frogs from each of the remaining 11 contemporary R. pipiens populations and we sequenced Arcadlin, Myosin, FIBI7, and Tyrosinase in these 22 frogs.
Analysis
All sequences have been deposited in GenBank under accession numbers EU407141–EU407149, EU407151–EU407176, and EU769510–EU769553. All loci were nuclear and diploid, so two alleles were obtained from all individuals. Heterozygotes were identified by the presence of double peaks in electropherograms. To determine common haplotypes, we noted all homozygous genotypes and tested for recombination using the four-gamete test. Given evidence for minimal recombination, we determined the remaining haplotypes by subtracting out previously observed haplotypes from the heterozygous genotypes or, for some R. blairi genotypes, by using allele-specific PCR. To confirm that observed polymorphisms were allelic and not due to coamplification of duplicated loci, we tested whether each population was in Hardy–Weinberg equilibrium. It is highly unlikely that polymorphic sequences from multiple loci would conform to Hardy–Weinberg expectations.
We used PAUP* (version 4.0b10, Swofford 2002) to construct a maximum parsimony phylogeny of all brevinin-1 sequences. We used DnaSP (version 4.0, Rozas et al. 2003) to calculate standard statistics of selective neutrality, including ZnS (Kelly 1997), Tajima's D (Tajima 1989), haplotype diversity or expected heterozygosity (Hd), the number of haplotypes (h), genetic variation (π), and the number of segregating sites (S), and to test their significance using coalescent simulations (conditioned on S with no recombination and 1,000 replicates). To minimize the effects of population subdivision on these test statistics, we performed each test within individual populations. We used MEGA (version 2.1; Kumar et al. 2001) to test whether the ratio of nonsynonymous variation to synonymous variation (πn/πs) was significantly greater than 1. If observed πs was 0, we estimated πn/πs to be greater than what the value would be if a single synonymous polymorphism had been observed in a single sequence. Neutrality tests were conducted on the mature peptide region only because that is the region where nonneutral evolution most frequently occurs (Tennessen 2005b). We represented relationships among alleles with a haplotype network, as opposed to a phylogenetic tree, because networks show allele frequencies and because there is little evidence for recombination at this locus.
To distinguish between fluctuating and stable balancing selection, we performed several tests of allelic stasis. A model of stable balancing selection predicts that the silent variation at the selected locus should be higher than elsewhere in the genome because allelic lineages have been maintained at intermediate frequency for a very long time and many mutations have been able to accumulate over this long coalescence period (Charlesworth 2006). In contrast, under fluctuating selection each allelic lineage spends some time being rare when selection favors the other allelic lineage. Thus, the effective population size of the locus, estimated as the sum of the harmonic means of the effective sizes of all allelic lineages over time, is small. As a result, silent variation will be purged by genetic drift, making silent variation lower than silent variation at neutral loci (Tiffin et al. 2004). Thus, we calculated genetic variation (π) at silent sites (synonymous and noncoding) for Arcadlin, Myosin, FIBI7, Tyrosinase, and the brevinin-1 locus. We used equation 12.63 of Nei and Kumar (2000) to calculate the variance in π, and we used the square root of this variance as the standard error in t-tests comparing mean π values. Similarly, stable balancing selection predicts within-lineage Tajima's D values near 0, while fluctuating selection predicts high or low Tajima's D values for lineages that have been shrinking or growing, respectively. Thus, we calculated Tajima's D within each allelic lineage.
Our five easternmost populations of R. pipiens are the “Eastern” populations used by Hoffman et al. (2006) to generate a neutral distribution of the expected FST value, based on microsatellite markers and intersimple sequence repeat loci, using the method of Beaumont and Nichols (1996). Under stable balancing selection, FST would be lower than the neutral expectations. Under fluctuating selection, FST could be higher than the neutral expectation if selection coefficients varied substantially among populations or if the effective population size at the selected locus is substantially lower than at neutral loci. Therefore, we compared FST at the brevinin-1 locus with this distribution. Furthermore, we used the previously described genotype data from the R. pipiens individuals at nine microsatellite loci (Hoffman et al. 2006) to construct phylogenies of the R. pipiens populations east of the Mississippi River based on both the microsatellites and the brevinin-1 locus. We used the Fitch method based on the genetic distance of Reynolds et al. (1983) in PHYLIP (version 3.65; Felsenstein 1989). We only used the eastern populations because the high genetic divergence between eastern and western R. pipiens could result in substantial homoplasy at the microsatellite markers (Hoffman and Blouin 2004; Hoffman et al. 2006). Although microsatellites have a higher mutation rate than coding sequences, among closely related populations genetic distances based on both marker types should be correlated under neutrality (Richard and Thorpe 2001), and therefore differences between the brevinin-1 phylogeny and the microsatellite phylogeny could be due to selection. To formally evaluate the difference between interpopulation divergences at the brevinin-1 locus and at the microsatellite loci, we performed a Mantel test of pairwise genetic distances in FSTAT version 2.9.3 (Goudet 1995).
Populations with low genetic diversity at the brevinin-1 locus could have recently experienced a selective sweep, or they could simply have low genome-wide variation due to a low effective population size. To distinguish between these two hypotheses, we calculated the mean expected heterozygosity (He) for each R. pipiens population using the previously described genotypes at the nine microsatellite loci (Hoffman et al. 2006). This analysis was restricted to R. pipiens because we lacked microsatellite data for the other species. We compared population He values at the brevinin-1 locus with the population He values at the microsatellite loci, which we assumed to be selectively neutral. If populations with low brevinin-1 diversity also have low microsatellite diversity, genetic drift is probably responsible in both cases. If not, the low brevinin-1 diversity could be due to a recent or ongoing selective sweep.
Results
Brevinin-1 Locus
We consistently obtained readable brevinin-1 sequences 236 bp in length, which consisted of a 98-bp partial intron followed by a 138-bp partial exon. The exon contained 61 bp of the C-terminal end of the propiece, the entire 72-bp mature peptide region, a stop codon, and 2 bp of postcoding sequence. Out of 517 frogs, 344 were homozygous. All common haplotypes (seen more than five times) were observed as homozygotes and showed no evidence of recombination. All remaining haplotypes (1% of the total) could easily be resolved, either because there was only one way to resolve the heterozygous genotypes such that at least one allele in every genotype matched an allele previously observed in that population or by using allele-specific PCR. It remains possible that some of these nonhomozygous haplotypes were erroneously resolved, which could bias our results, but they are so rare that they are unlikely to affect our conclusions substantially. All populations were found to be in Hardy–Weinberg equilibrium, and there was no evidence of more than two alleles in any one individual, confirming that only a single locus was amplified, here named the Brevinin1.1 locus.
Within R. pipiens, four common, highly divergent alleles were observed at the Brevinin1.1 locus: alleles Rp1, Rp2, Rp3, and Rp4 (fig. 2). These four alleles accounted for 97% of the sequences; all remaining minor variants were one or two steps away from one of them (fig. 3). Allele Rp1 encoded the previously described peptide brevinin-1Pa, and allele Rp2 encoded the previously described peptide brevinin-1Pg (Goraya et al. 2000; Tennessen and Blouin 2007). Alleles Rp3 and Rp4 both encoded a peptide, brevinin-1PLa, which previously had only been described in R. palustris (Basir et al. 2000). The distances among these four major R. pipiens alleles ranged from 1 to 13 substitutions (fig. 3). The species overall showed an excess of homozygotes, probably owing to population subdivision, because every individual population was in Hardy–Weinberg equilibrium (supplementary table 1, Supplementary Material online) and because this species shows substantial population structure at neutral markers (Hoffman and Blouin 2004; Hoffman et al. 2006). We observed similar patterns of diversity in R. blairi. A single R. blairi sample, MVZ Herp 240161, was homozygous for unique alleles at both Brevinin1.1 and Arcadlin and carried a unique allele at Myosin. Because this individual could be mislabeled or a migrant, we excluded it from all further analyses. Allele Rb1 accounted for 86% of the R. blairi sequences at the Brevinin1.1 locus (figs. 2 and 3). The remaining R. blairi alleles differed from allele Rb1 by 1–13 substitutions, including a single-nucleotide indel in the intron. In contrast to the other species, in R. palustris, we observed only two alleles which were separated by a single mutational step in the intron. Both coded for the peptide brevinin-1PLa (figs. 2 and 3). The more common allele (allele Rp4) was identical to an allele observed in R. pipiens.
FIG. 2.—
Alignment of Brevinin1.1 locus amino acid sequences from all species, the frequencies of these sequences in each species, the alleles encoding them (some rare alleles were not named), and the allelic lineage.
FIG. 3.—
Maximum parsimony haplotype network for a 236-bp segment of the Brevinin1.1 locus. White circles represent Rana pipiens haplotypes, gray circles represent Rana blairi haplotypes, and black circles represent Rana palustris haplotypes. Gray squares represent inferred nodes that were not observed. The size of the circles indicates the frequency of each allele in the respective species. Nonsynonymous substitutions are indicated by small black rectangular bars, and synonymous substitutions are indicated by small white rectangular bars; thus, branch lengths represent the total number of substitutions. Common alleles are labeled. Allelic lineages are surrounded by large rectangles. Highly divergent alleles, separated mostly by nonsynonymous substitutions, occur in both R. pipiens and R. blairi, causing several tests of selective neutrality to be significant (table 1). A haplotype is shared between R. pipiens and R. palustris, indicated by a white circle superimposed on a black circle. A pattern of both very common and very rare alleles can be seen, especially for the Lineage 1 lineage in R. pipiens, suggestive of a recent rapid increase in allele frequency.
Haplotype networks of the Brevinin1.1 locus alleles are shown in figure 3. There were three main allelic lineages, separated by substantial and mostly nonsynonymous divergence. There was no reciprocal monophyly between R. pipiens and either of the other two species, but R. palustris and R. blairi were reciprocally monophyletic with respect to each other. Although there were no shared Brevinin1.1 alleles between R. pipiens and R. blairi, both species had alleles belonging the same divergent allelic lineages, Lineage 1 and Lineage 2 (table 1 and fig. 3).
Table 1.
Population Genetic Parameters for the Brevinin1.1 Mature Peptide Region (72 bp) in Three Frog Species
| Species | 2Na | Lineage 1b | Lineage 2b | Lineage 3b | Sc | hd | Hde | πf | πsg | ZnSh | πn/πsi | Tajima's Dj |
| Rana pipiens | 848 | 60% | 34% | 6% | 16 | 8* | 0.55 | 0.06* | 0.009 | 0.32* | 8.67** | 2.23* |
| Rana blairi | 100 | 92% | 8% | 0% | 12 | 4** | 0.24** | 0.02 | 0.001 | 0.48* | 25.00** | −0.81 |
| Rana palustris | 86 | 0% | 0% | 100% | 0 | 1 | 0.00 | 0.00 | 0.000 | 0.00 | — | — |
NOTE.—For all statistical tests, *P < 0.05 and **P < 0.01.
2N = number of alleles = twice the number of individuals.
Lineages 1, 2, and 3 refer to the percentage of alleles in each species belonging to each of these three allelic lineages.
S, number of segregating sites.
h, number of haplotypes; tested whether h is significantly low given S.
Hd, haplotype diversity, equivalent to expected heterozygosity; tested whether Hd is significantly low given S.
π, mean number of pairwise differences among sequences; tested whether π is significantly extreme given S.
πs, mean number of pairwise differences among sequences at silent sites; tested whether πs is significantly extreme given silent S.
ZnS, linkage disequilibrium statistic of Kelly (1997); tested whether ZnS is significantly high given S.
πn/πs, ratio of nonsynonymous nucleotide variation to synonymous nucleotide variation; tested whether significantly different than 1.
Tajima's D, statistic of Tajima (1989); tested whether significantly different than 0.
Other Loci
For all individuals, preexisting protocols for amplifying and sequencing Arcadlin, Myosin, FIBI7, and Tyrosinase (Bossuyt and Milinkovitch 2000; Di Candia and Routman 2007; Tennessen and Blouin 2007) were successful. In total, we obtained 1,246 bp of nuclear sequence unlinked to Brevinin1.1, including 866 bp of coding sequence and 380 bp of intronic sequence. These loci showed moderate levels of variation (fig. 4 and table 2; supplementary table 2, Supplementary Material online). In R. pipiens and R. blairi, patterns of variation did not deviate from neutral expectations. In R. palustris, several neutrality tests indicated an excess of divergent, intermediate frequency haplotypes. Because these patterns were observed across all loci, they probably have demographic causes such as population subdivision or recent migration from genetically distinct populations and are not due to selection. There were no fixed differences between R. blairi and R. pipiens. There were eight fixed differences between R. palustris and R. blairi; of these, four were also fixed between R. palustris and R. pipiens.
FIG. 4.—
Maximum parsimony haplotype networks for four putatively neutral non-AMP nuclear loci unlinked to Brevinin1.1: Arcadlin, Myosin, FIBI7, and Tyrosinase. Symbols are the same as in figure 3. In contrast with the Brevinin1.1 locus (fig. 3), nonsynonymous substitutions (small black rectangular bars) are rarer than synonymous substitutions (small white rectangular bars). In both Rana pipiens (white circles) and Rana blairi (gray circles), non-AMP loci show intermediate frequency alleles and few missing transitional haplotypes (gray squares), in contrast with the Brevinin1.1 locus (fig. 3), which shows high- and low-frequency alleles forming divergent lineages separated by many missing transitional haplotypes. In Rana palustris (black circles), non-AMP loci show diverse, moderately divergent alleles, in contrast with the Brevinin1.1 locus (fig. 3), which shows very low diversity.
Table 2.
Population Genetic Parameters for Four Putatively Neutral Nuclear Loci Unlinked to Brevinin1.1 (1,246 bp) in Three Frog Species (Arcadlin, Myosin, FIBI7, and Tyrosinase)
| Species | 2Na | Locus | Lengthb | Sc | hd | Hde | πf | πsg | ZnSh | πn/πsi | Tajima's Dj |
| Rana pipiens | 162 | Arcadlin | 265 | 6 | 9 | 0.75 | 0.01 | 0.010 | 0.06 | 0.04** | 0.54 |
| Myosin | 150 | 7 | 10 | 0.76 | 0.01 | 0.020 | 0.12 | 0.00* | 1.27 | ||
| FIBI7 | 258 | 12 | 9 | 0.57 | 0.01 | 0.010 | 0.24 | 0.00 | 0.12 | ||
| Tyrosinase | 573 | 17 | 17 | 0.84 | 0.00 | 0.016 | 0.06 | 0.08* | −0.41 | ||
| Mean ± standard deviation | 312 ± 182 | 10.5 ± 5.1 | 11.3 ± 3.9 | 0.73 ± 0.11 | 0.01 ± 0.00 | 0.014 ± 0.005 | 0.12 ± 0.08 | 0.03** | 0.38 ± 0.71 | ||
| Rana blairi | 100 | Arcadlin | 265 | 2 | 3 | 0.46 | 0.00 | 0.004 | 0.02 | 0.00* | 0.35 |
| Myosin | 150 | 2 | 3 | 0.50 | 0.00 | 0.006 | 0.04 | 0.00 | 0.66 | ||
| FIBI7 | 258 | 2 | 3 | 0.10 | 0.00 | 0.000 | 0.00 | 0.00 | −1.15 | ||
| Tyrosinase | 573 | 3 | 4 | 0.56 | 0.00 | 0.000 | 0.02 | >8.7 | 0.07 | ||
| Mean ± standard deviation | 312 ± 182 | 2.3 ± 0.5 | 3.3 ± 0.5 | 0.40 ± 0.21 | 0.00 ± 0.00 | 0.002 ± 0.003 | 0.02 ± 0.02 | 0.14 | −0.02 ± 0.79 | ||
| Rana palustris | 86 | Arcadlin | 265 | 4 | 4 | 0.56 | 0.00 | 0.009 | 0.33 | 0.00** | 1.09 |
| Myosin | 150 | 4 | 2* | 0.34 | 0.01 | 0.010 | 1.00* | 0.64 | 1.39 | ||
| FIBI7 | 258 | 14 | 7 | 0.72 | 0.02** | 0.024** | 0.37* | 0.00* | 2.53** | ||
| Tyrosinase | 573 | 7 | 5 | 0.75 | 0.00 | 0.011 | 0.38 | 0.07** | 0.56 | ||
| Mean ± standard deviation | 312 ± 182 | 7.3 ± 4.7 | 4.5 ± 2.1** | 0.59 ± 0.19 | 0.01 ± 0.01** | 0.014 ± 0.007** | 0.52 ± 0.32** | 0.13** | 1.39 ± 0.83** |
NOTE.—For all statistical tests, *P < 0.05 and **P < 0.01.
2N = number of alleles = twice the number of individuals.
Length, locus length in base pairs.
S, number of segregating sites.
h, number of haplotypes; tested whether h is significantly low given S.
Hd, haplotype diversity, equivalent to expected heterozygosity; tested whether Hd is significantly low given S.
π, mean number of pairwise differences among sequences; tested whether π is significantly extreme given S.
πs, mean number of pairwise differences among sequences at silent sites; tested whether πs is significantly extreme given silent S.
ZnS, linkage disequilibrium statistic of Kelly (1997); tested whether ZnS is significantly high given S.
πn/πs, ratio of nonsynonymous nucleotide variation to synonymous nucleotide variation; tested whether significantly different than 1.
Tajima's D, statistic of Tajima (1989); tested whether significantly different than 0.
Neutrality Tests
To test whether the high allelic variation at the Brevinin1.1 locus in R. pipiens and R. blairi was maintained by balancing selection, we conducted multiple tests of selective neutrality (table 1; supplementary table 1, Supplementary Material online). Neutrality tests were not conducted on R. palustris because that species had no variation in the mature peptide region. The ratio of mature peptide region nonsynonymous variation to synonymous variation (πn/πs) was significantly greater than 1 in most populations as well as in R. pipiens overall and R. blairi overall (table 1; supplementary table 1, Supplementary Material online; fig. 5). For both species, nonsynonymous variation in the mature peptide region was also significantly higher than synonymous variation at Arcadlin, Myosin, FIBI7, and Tyrosinase (fig. 5). Coalescent simulations of several population genetic parameters in DnaSP demonstrated that polymorphisms were structured into divergent allelic lineages, with skewed frequencies, more so than would be expected under neutrality given the number of segregating sites (table 1; supplementary table 1, Supplementary Material online; fig. 3). Given the number of segregating sites, the number of haplotypes (h) was too low for all populations that had any variation and the haplotype diversity (Hd) was too low for nearly all populations. Linkage disequilibrium (ZnS) was significantly higher than neutral expectations for all populations that had any variation. In R. pipiens, the presence of several common, divergent alleles made pairwise variation (π) significantly higher than expected given the number of segregating sites, resulting in significantly positive values of Tajima's D. In contrast, Tajima's D was negative in R. blairi, owing to rare but divergent alleles.
FIG. 5.—
Genetic diversity (π) at the Brevinin1.1 locus compared with neutral π for three frog species. “Non-AMP silent” is synonymous and noncoding π at four putatively neutral nuclear loci: Arcadlin, Myosin, FIBI7, and Tyrosinase. “AMP silent” is synonymous and noncoding π at Brevinin1.1. “AMP nonsynonymous” is nonsynonymous π in the mature peptide region of Brevinin1.1. Asterisks represent values that are significantly different from the corresponding non-AMP silent value (P < 0.05). For Rana pipiens and Rana palustris, Brevinin1.1 silent sites show lower variation than is seen at non-AMP silent sites. For R. pipiens and Rana blairi, Brevinin1.1 shows higher nonsynonymous variation than is seen at silent sites; R. palustris has no nonsynonymous variation at Brevinin1.1.
Multiple tests rejected neutrality in both R. pipiens and R. blairi, providing strong evidence for balancing selection. In R. pipiens, neutrality was also rejected if the sequences from any one allelic lineage were removed (significant ZnS and h values, πn/πs > 1). This result suggested that all three allelic lineages are adaptively divergent R. pipiens. Similarly, in R. blairi, if Lineage 2 sequences were removed, multiple tests still rejected neutrality (significant ZnS and h values, πn/πs > 1). This result suggested that alleles Rb1 and Rb2, while both in Lineage 1, might be adaptively divergent.
Distinguishing Stable from Fluctuating Selection
The first indication about the type of balancing selection acting was that allele frequencies were not intermediate but skewed. In R. pipiens, Lineage 1 alleles were 10 times more common than Lineage 3 alleles. In R. blairi, allele frequencies were even more skewed, with 86% of the sequences consisting of the same allele, and other alleles being quite rare. In R. palustris, only a single allelic lineage was observed. The other allelic lineages could be so rare they were not sampled in R. palustris or they might not exist in this species. No species showed a pattern of equally frequent allelic lineages consistent with symmetrical models of stable balancing selection. The observed skew was more consistent with fluctuating selection or highly asymmetrical overdominance.
To formally distinguish between stable balancing selection and fluctuating selection, we compared silent variation at four non-AMP nuclear loci (Arcadlin, Myosin, FIBI7, and Tyrosinase) with silent variation at the Brevinin1.1 locus (fig. 5). Silent variation at the Brevinin1.1 locus was lower than at all the non-AMP loci for both R. pipiens (Brevinin1.1 πs = 0.005; P = 0.08) and R. palustris (Brevinin1.1 πs = 0.002; P = 0.02). Silent variation was extremely low at all loci for R. blairi (Brevinin1.1 πs = 0.002; P > 0.1). These results were consistent with fluctuating selection, which predicts low synonymous diversity, but not with stable balancing selection, which predicts high synonymous diversity. Although demographic processes appeared to have caused nonneutral patterns at the non-AMP loci R. palustris, these processes could not explain the difference in genetic diversity between Brevinin1.1 and the other loci, which therefore is more likely to be due to selection.
We calculated values of Tajima's D within allelic lineages (table 3). In R. pipiens, Tajima's D was significantly negative within the common Lineage 1 (D = −1.84; P < 0.01). Because we did not sequence non-AMP loci in all R. pipiens individuals, we cannot completely understand possible demographic factors that could affect Tajima's D, so this result should be interpreted with caution. In both R. pipiens and R. blairi, there was a nonsignificant trend of other common allelic lineages having negative Tajima's D values and rare allelic lineages having positive Tajima's D values consistent with fluctuating selection (table 3). Because population subdivision can affect Tajima's D, we also calculated within-lineage Tajima's D values for individual populations of R. pipiens and R. blairi. For all populations, including those shown to have neutral Tajima's D values at non-AMP loci (populations 1, 2, 14, and 15; supplementary table 2, Supplementary Material online), the trend was the same as for the whole species (table 3). Only one allelic lineage with a single polymorphism occurred in R. palustris, so within-lineage Tajima's D could not be meaningfully estimated.
Table 3.
Values of Tajima's D (Tajima 1989) within Allelic Lineages within Species
| Species | Lineage | Da | Frequencyb | Nc | Population D (Mean)d | Population D (range)e |
| Rana pipiens | 1 | −1.84** | 60% | 7 | −1.11 | −1.69 to −0.53 |
| 2 | −0.79 | 34% | 3 | −0.95 | −1.09 to −0.84 | |
| 3 | 0.62 | 6% | 1 | 1.45 | 1.45 | |
| Rana blairi | 1 | −1.26 | 87% | 1 | −0.42 | −0.42 |
| 2 | 0.16 | 8% | 0 | — | — |
NOTE.—Only lineages displaying variation for the species considered are shown. In both R. pipiens and R. blairi, common lineages have negative Tajima's D values and rare lineages have positive Tajima's D values, consistent with fluctuating selection. This trend is statistically significant for Lineage 1 in R. pipiens. Only a single polymorphism occurs in Rana palustris, so Tajima's D cannot be meaningfully estimated.
D, within-lineage Tajima's D; tested whether significantly different than 0; **P < 0.01.
Frequency, frequency of each allelic lineage in each species.
N, number of populations in each species displaying sufficient variation in that allelic lineage such that within-lineage Tajima's D could be calculated.
Population D (mean), mean within-lineage Tajima's D value among individual populations.
Population D (range), range of within-lineage Tajima's D values among individual populations.
Under asymmetrical stable balancing selection, such as asymmetrical overdominance, the more favored allelic lineage will be more common and harbor most of the neutral genetic variation, owing to its larger effective size (Innan and Tajima 1999; Stahl et al. 1999). We therefore compared levels of silent variation among allelic lineages. Within R. pipiens, values of silent π were 0.0004 ± 0.0011 for Lineage 1, 0.0002 ± 0.0008 for Lineage 2, and 0.0038 ± 0.0037 for Lineage 3. Within R. blairi, values of silent π were 0.0018 ± 0.0018 for Lineage 1 and 0.0053 ± 0.0042 for Lineage 2. Thus, for both species, genetic variation was relatively low in the more common allelic lineages and much higher in the rarest allelic lineages.
We tested for excessive differentiation among populations, a signature of fluctuating selection. The FST value among the five easternmost populations of R. pipiens was 0.20, and the expected neutral value of FST at the same level of heterozygosity was approximately 0.06 (fig. 2A in Hoffman et al. [2006]). Thus, the observed FST value at the Brevinin1.1 locus was higher than the mean neutral expectation but not significantly so. The high FST value is primarily due to population 2, where allele Rp1 predominates, in contrast to the other four populations, where allele Rp2 predominates (supplementary table 1, Supplementary Material online). In addition, FST between populations 1 and 2 was 0.02 at the four non-AMP loci sequenced in this study and 0.04 at the microsatellites, but 0.34 at Brevinin1.1. The high divergence of population 2 at Brevinin1.1 but not at microsatellites is shown in figure 6. The matrix of pairwise microsatellite differences among eastern populations was not correlated with the corresponding matrix of Brevinin1.1 pairwise differences (Mantel test; P > 0.1). Therefore, the high genetic divergence between population 2 and the other eastern populations is inconsistent with neutral expectations and could be due to spatially differing selective pressures.
FIG. 6.—
Distance-based phylogenies of contemporary Rana pipiens populations east of the Mississippi River, populations 1–6 (fig. 1; supplementary table 1, Supplementary Material online). (A) Phylogeny based on genetic distance at nine microsatellite loci. All populations show intermediate divergence from each other. (B) Phylogeny based on the Brevinin1.1 locus. All populations show intermediate divergence from each other except for population 2, which has highly different allele frequencies. Very little branching is apparent because almost all alleles at the Brevinin1.1 locus in these populations are either allele Rp1 or allele Rp2, so populations are effectively differentiated from each other in a single dimension (i.e., frequency of allele Rp1). Because the high divergence between population 2 and the other populations is only apparent at Brevinin1.1, not at the microsatellites, it is likely adaptive.
The two R. pipiens populations with the lowest microsatellite He values were fixed for the allele Rp1, in contrast to the high genetic variation seen in other populations (supplementary table 1, Supplementary Material online). This result was consistent with the hypothesis that these two populations had lower effective sizes than the others, in which case genetic drift would be more likely to fix alleles in these two populations. After these two fixed populations, the R. pipiens population with the lowest Brevinin1.1 Hd was population 2 (supplementary table 1, Supplementary Material online), which had slightly higher than average microsatellite He (0.84), and therefore its effective population size did not appear to be substantially lower than the other populations. Because population 2 had a very different allele frequency pattern than its closest geographic neighboring populations (fig. 6), and because it showed an unusually low Brevinin1.1 Hd value for its microsatellite He value, population 2 was the population most likely to have undergone a recent shift in allele frequencies at the Brevinin1.1 locus due to selection. However, allele frequencies were nearly identical between the 1971 sample and the 2001 sample, both at the Brevinin1.1 locus (FST = 0.0; supplementary table 1, Supplementary Material online) and at the nine microsatellite loci (FST = 0.0).
Discussion
Patterns of genetic diversity at the Brevinin1.1 locus are highly unusual in three species of leopard frogs, and multiple lines of evidence suggest that strong balancing selection is maintaining adaptive variation in the mature peptide region. Evidence for nonneutrality includes high ratios of nonsynonymous/synonymous variation, unusual values of Hd, h, and ZnS given the number of segregating sites, and significant values of Tajima's D (table 1; supplementary table 1, Supplementary Material online). Because we did not sequence non-AMP loci from all R. pipiens individuals, we cannot rule out the possibility that demographic factors have contributed to some nonneutral patterns; however, in populations 1 and 2, neutrality is strongly rejected the Brevinin1.1 locus but not at other loci (supplementary tables 1 and 2, Supplementary Material online), suggesting that our results are not an artifact of demography. Likewise, we cannot rule out the possibility that the high variation at the Brevinin1.1 locus arose not merely through point mutations but also through gene conversion from other loci, which can contribute to a pattern of strikingly divergent alleles (Storz et al. 2007). Several Lineage 1 alleles, including allele Rp1, are identical in the mature peptide region to other brevinin-1 loci that exist in leopard frog genomes (Tennessen and Blouin 2007). Lineage 2 and Lineage 3 sequences have not been observed at other brevinin-1 loci (Tennessen and Blouin 2007). However, even if demographic processes or gene conversion have enhanced the diversity at this locus, they must have acted in concert with selection as this is the only explanation for some nonneutral patterns such as an elevated πn/πs ratio. The role played by these peptides in amphibian immunity indicates that pathogens are the most likely selective pressure acting on this locus. Perhaps this locus is specialized on a particular pathogen that is coevolving with leopard frogs, and alleles differ in their ability to defend against different strains. Alternatively, perhaps some alleles are more effective than others overall but there is a cost to resistance.
Stable balancing selection includes both overdominance and instantaneous frequency-dependent selection, while fluctuating selection can be caused by time-delayed frequency-dependent selection (Seger 1988) or spatiotemporally varying selection (Hedrick 2002). Although we cannot determine with absolute certainty which type of balancing selection is acting, our data support the fluctuating selection hypothesis more strongly than the stable balancing selection hypothesis. First, allele frequencies are skewed instead of intermediate in all three species. Second, Tajima's D for within-lineage variation is negative for common lineages and positive for rare lineages in both R. blairi and R. pipiens (table 3). This result suggests that now-common alleles were recently rare and have rapidly increased in frequency. Third, synonymous variation is lower at the Brevinin1.1 locus than at unlinked, putatively neutral loci, the opposite of the prediction from stable balancing selection; for R. palustris, this difference is statistically significant, consistent with a recent selective sweep fixing Lineage 3 in this species (fig. 5). This effect holds despite evidence that amphibian mature peptide regions feature an elevated mutation rate and/or positive selection on synonymous sites, which would tend to cause the opposite pattern (Tennessen 2005a). Fourth, the allelic lineages with the most variation are not the most common, again suggesting that allele frequencies were quite different in the recent past. Finally, genetic distance at Brevinin1.1 is exceptionally high among some R. pipiens populations, and stable balancing selection would predict a trend in the opposite direction (fig. 6).
Our results provide unique insight into the evolution of immunity genes. Nearly all examples of pathogen-induced balancing selection are detection and signaling proteins, such as those of the vertebrate adaptive immune system (Garrigan and Hedrick 2003). The Brevinin1.1 locus represents a rare example of an effector gene harboring an adaptive polymorphism. Loci encoding AMPs in mussels and humans also show high variation consistent with balancing selection (Hollox and Armour 2008; Pahdi and Verghese 2008). Drosophila AMPs are not under balancing selection (Clark and Wang 1997; Lazzaro and Clark 2003), possibly because fly AMPs, which also show little evidence for positive selection, may have less specific targets than AMPs in other taxa (Sackton et al. 2007). The AMP locus Ranatuerin2 has undergone a selective sweep in R. pipiens and shows no evidence of balancing selection (Tennessen and Blouin 2007), but this pattern is also consistent with fluctuating selection if the other alleles are extremely rare or have recently been eliminated. Fluctuating selection in particular has only occasionally been convincingly demonstrated for any immunity locus, effector or otherwise. Many of the best examples of fluctuating selection on defense genes are from plants (Stahl et al. 1999; Tiffin et al. 2004; Tiffin and Moeller 2006), but a few animal examples also exist (Jensen et al. 2008). Even for well-studied loci under balancing selection like the genes of the major histocompatibility complex, the importance of stable versus fluctuating selection is not clear (Piertney and Oliver 2006); the universally observed high allelic diversity at these loci suggests that if fluctuating selection occurs, the fluctuations are nearly instantaneous and, therefore, closer to stable balancing selection than they are at the Brevinin1.1 locus. Microbes can evolve resistance to detection mechanisms by simply substituting one or more amino acid residues on their surface proteins. In contrast, microbial resistance to AMPs involves changing the biochemistry of the cell membrane or producing AMP-degrading enzymes, adaptations which might involve several coordinated changes across multiple genes. Thus, we hypothesize that it is more difficult for microbes to evolve resistance to AMPs than to detection mechanisms, resulting in longer lag periods between host evolution and pathogen evolution and therefore greater fluctuations in allele frequencies, including occasional allele fixation. Fluctuating selection may be difficult to distinguish from positive selection if divergent, low-frequency alleles are not sampled, which may be partly why documented examples of balancing selection on effector molecules are rare. Overall, our results help to illustrate when and how balancing selection can act on effector loci.
Allele frequency differences among populations are either due to genetic drift or different selective pressures in different habitats. In R. pipiens, allele frequencies differ substantially between populations east and west of the Mississippi River (supplementary table 1, Supplementary Material online), but this differentiation is consistent with genetic drift, given the high east–west mitochondrial DNA divergence also observed in this species (Hoffman and Blouin 2004). The high differentiation among the eastern populations at Brevinin1.1, despite low differentiation at neutral markers, is more likely to be due to selection (fig. 6). Population 2 is the most likely candidate for a recent population-specific shift in allele frequencies because it is quite divergent from other populations nearby and because its haplotype diversity is low despite having normal levels of neutral genetic diversity. Allele frequencies in this population have not changed noticeably between 1971 and 2001, so if there is a difference in selective pressures between population 2 and its neighbors, it is more than a few decades old.
Variation at AMP loci is likely to be an important contributor to the ability of amphibian species to adapt to novel disease threats. Agents of emerging infectious diseases are causing precipitous declines, and in some cases extinction, in many amphibian species (Carey et al. 1999; Daszak et al. 2003). It is thought that AMPs are an important defense against these pathogens (Chinchar et al. 2004; Rollins-Smith and Conlon 2005; Woodhams, Rollins-Smith, et al. 2006; Woodhams, Voyles, et al. 2006). Several epizootics of the fungal pathogen Batrachochytrium dendrobatidis and iridovirus have caused declines of R. pipiens populations in recent decades (Carey et al. 1999; Green et al. 2002; Greer et al. 2005). Although these epizootics could be selective agents on the Brevinin1.1 locus, we did not observe any historic allele frequency change in population 2, and allelic lineages are too divergent to have arisen within a few decades. Therefore, nonneutral patterns of genetic diversity are probably primarily due to long-term allelic fluctuations caused by unidentified native diseases, with recent disease outbreaks possibly shaping this variation.
Our study adds to the growing body of knowledge on diversifying selection among AMPs by demonstrating that AMPs encoded by alleles at the same locus can show evidence of major adaptive differences. We have shown that genes encoding immune effector proteins can harbor balanced polymorphisms comparable to many pathogen-detection genes. Researchers developing AMPs for therapeutic applications (Hancock 2001) should consider examining multiple individuals of the same species to encounter functionally novel allelic variants. Our data are consistent with a model of fluctuating selection, likely caused by pathogens. However, because the results of many of our tests were merely suggestive, we are reluctant to completely reject the hypothesis of stable balancing selection. Further study is required to determine the specific functional difference among these peptides, the particular pathogens that are driving selection, and the precise mechanism maintaining variation. We are currently examining the antimicrobial effects of these peptides in vitro (Tennessen JA, Woodhams DC, Reinert LK, Blouin MS, and Rollins-Smith LA, unpublished data). Loci encoding AMPs, such as Brevinin1.1, can be important sites of adaptive genetic diversity and major players in the coevolutionary arms race between hosts and pathogens.
Supplementary Material
Supplementary tables 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors are grateful to Eric Hoffman, Eric Tobin, Mike Redmer, Stephan Swanson, Mike Foy, and the Museum of Vertebrate Zoology for help obtaining samples. Becky Cooper provided invaluable laboratory assistance. Hitoshi Araki, Kaitlin Bonner, and Mark Christie commented on earlier versions of the manuscript. We also thank three anonymous reviewers for their helpful comments. This research was supported by an Environmental Protection Agency Science to Achieve Results fellowship to J.A.T. and an Idea Network of Biomedical Research Excellence grant (2 P20 RR016463) to the Nevada Genomics Center.
References
- Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, et al. (11 co-authors) A strong signature of balancing selection in the 59 cis-regulatory region of CCR5. Proc Natl Acad Sci USA. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basir YJ, Knoop FC, Dulka J, Conlon JM. Multiple antimicrobial peptides and peptides related to bradykinin and neuromedin N isolated from skin secretions of the pickerel frog, Rana palustris. Biochim Biophys Acta. 2000;1543:95–105. doi: 10.1016/s0167-4838(00)00191-6. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in genetic analysis of population structure. Proc R Soc Lond B Biol Sci. 1996;263:1619–1626. [Google Scholar]
- Bossuyt F, Milinkovitch MC. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc Natl Acad Sci USA. 2000;97:6585–6590. doi: 10.1073/pnas.97.12.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey C, Cohen N, Rollins-Smith L. Amphibian declines: an immunological perspective. Dev Comp Immunol. 1999;23:459–472. doi: 10.1016/s0145-305x(99)00028-2. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:379–384. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchar VG, Bryan L, Silphadaung U, Noga E, Wade D, Rollins-Smith L. Inactivation of viruses infecting ectothermic animals by amphibian and piscine antimicrobial peptides. Virology. 2004;323:268–275. doi: 10.1016/j.virol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Clark AG, Wang L. Molecular population genetics of Drosophila immune system genes. Genetics. 1997;147:713–724. doi: 10.1093/genetics/147.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon JM, Kolodziejek J, Nowotny N. Antimicrobial peptides from ranid frogs: taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim Biophys Acta. 2004;1696:1–14. doi: 10.1016/j.bbapap.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Infectious disease and amphibian population declines. Divers Distrib. 2003;9:141–150. [Google Scholar]
- Di Candia MR, Routman EJ. Cytonuclear discordance across a leopard frog contact zone. Mol Phylogenet Evol. 2007;45:564–575. doi: 10.1016/j.ympev.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Frost DR, Grant T, Faivovich J, et al. (19 co-authors) The amphibian tree of life. Bull Am Mus Nat Hist. 2006;297:1–370. [Google Scholar]
- Garrigan D, Hedrick PW. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- Goraya J, Wang Y, Li Z, O'Flaherty M, Knoop FC, Platz JE, Conlon JM. Peptides with antimicrobial activity from four different families isolated from the skins of the North American frogs Rana luteiventris, Rana berlandieri, and Rana pipiens. Eur J Biochem. 2000;267:894–900. doi: 10.1046/j.1432-1327.2000.01074.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Green DE, Converse KA, Schrader AK. Epizootiology of sixty-four amphibian morbidity and mortality events in the USA, 1996–2001. Ann N Y Acad Sci. 2002;969:323–339. doi: 10.1111/j.1749-6632.2002.tb04400.x. [DOI] [PubMed] [Google Scholar]
- Greer AL, Berrill M, Wilson PJ. Five amphibian mortality events associated with ranavirus infection in south central Ontario, Canada. Dis Aquat Org. 2005;67:9–14. doi: 10.3354/dao067009. [DOI] [PubMed] [Google Scholar]
- Hancock REW. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- Hoffman EA, Blouin MS. Evolutionary history of the northern leopard frog: reconstruction of phylogeny, phylogeography, and historical changes in population demography from mitochondrial DNA. Evolution. 2004;58:145–159. doi: 10.1111/j.0014-3820.2004.tb01581.x. [DOI] [PubMed] [Google Scholar]
- Hoffman EA, Schueler FW, Jones AG, Blouin MS. An analysis of selection on a colour polymorphism in the northern leopard frog. Mol Ecol. 2006;15:2627–2641. doi: 10.1111/j.1365-294X.2006.02934.x. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JAL. Directional and balancing selection in human beta-defensins. BMC Evol Biol. 2008;8:113. doi: 10.1186/1471-2148-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Tajima F. The effect of selection on the amount of nucleotide variation within and between allelic classes. Genet Res [Camb] 1999;73:15–28. [Google Scholar]
- Jensen LF, Hansen MM, Mensberg K-LD, Loeschcke V. Spatially and temporally fluctuating selection at non-MHC immune genes: evidence from TAP polymorphism in populations of brown trout (Salmo trutta, L.) Heredity. 2008;100:79–91. doi: 10.1038/sj.hdy.6801067. [DOI] [PubMed] [Google Scholar]
- Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1179–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Clark AG. Molecular population genetics of inducible antibacterial peptide genes in Drosophila melanogaster. Mol Biol Evol. 2003;20:914–923. doi: 10.1093/molbev/msg109. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford: Oxford University Press; 2000. [Google Scholar]
- Pahdi A, Verghese B. Molecular diversity and evolution of myticin-C antimicrobial peptide variants in the Mediterranean mussel, Mytilus galloprovincialis. Peptides. 2008;29:1094–1101. doi: 10.1016/j.peptides.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Thorpe RS. Can microsatellites be used to infer phylogenies? Evidence from population affinities of the western Canary Island lizard (Gallotia galloti) Mol Phylogenet Evol. 2001;20:351–360. doi: 10.1006/mpev.2001.0981. [DOI] [PubMed] [Google Scholar]
- Rollins-Smith LA, Conlon JM. Antimicrobial peptide defenses against chytridiomycosis, an emerging infectious disease of amphibian populations. Dev Comp Immunol. 2005;29:589–598. doi: 10.1016/j.dci.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Seger J. Dynamics of some simple host-parasite models with more than two genotypes in each species. Philos Trans R Soc Lond B Biol Sci. 1988;319:541–555. doi: 10.1098/rstb.1988.0064. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, Hayes JP. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Nei M. Fifty-million-year-old polymorphism at an immunoglobulin variable region gene locus in the rabbit evolutionary lineage. Proc Natl Acad Sci USA. 1999;96:9710–9715. doi: 10.1073/pnas.96.17.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) version 4.0b10. Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA. Enhanced synonymous site divergence in positively selected vertebrate antimicrobial peptide genes. J Mol Evol. 2005a;61:445–455. doi: 10.1007/s00239-004-0330-2. [DOI] [PubMed] [Google Scholar]
- Tennessen JA. Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J Evol Biol. 2005b;18:1387–1394. doi: 10.1111/j.1420-9101.2005.00925.x. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Blouin MS. Selection for antimicrobial peptide diversity in frogs leads to gene duplication and low allelic variation. J Mol Evol. 2007;65:605–615. doi: 10.1007/s00239-007-9045-5. [DOI] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Tiffin P, Hacker R, Gaut BS. Population genetic evidence for rapid changes in intraspecific diversity and allelic cycling of a specialist defense gene in Zea. Genetics. 2004;168:425–434. doi: 10.1534/genetics.103.023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Moeller DA. Molecular evolution of plant immune system genes. Trends Genet. 2006;22:662–670. doi: 10.1016/j.tig.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Woodhams DC, Rollins-Smith LA, Carey C, Reinert L, Tyler MJ, Alford RA. Population trends associated with skin peptide defenses against chytridiomycosis in Australian frogs. Oecologia. 2006;146:531–540. doi: 10.1007/s00442-005-0228-8. [DOI] [PubMed] [Google Scholar]
- Woodhams DC, Voyles J, Lips KR, Carey C, Rollins-Smith LA. Predicted disease susceptibility in a Panamanian amphibian assemblage based on skin peptide defenses. J Wildl Dis. 2006;42:207–218. doi: 10.7589/0090-3558-42.2.207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.