Abstract
The heterodimeric actin capping protein, referred to here as “CP,” is an essential element of the actin cytoskeleton, binding to the barbed ends of actin filaments and regulating their polymerization. In vitro, CP has a critical role in the dendritic nucleation process of actin assembly mediated by Arp2/3 complex, and in vivo, CP is important for actin assembly and actin-based process of morphogenesis and differentiation. Recent studies have provided new insight into the mechanism of CP binding the barbed end, which raises new possibilities for the dynamics of CP and actin in cells. In addition, a number of molecules that bind and regulate CP have been discovered, suggesting new ideas for how CP may integrate into diverse processes of cell physiology.
Introduction
CP was discovered, defined and named based on its ability to bind to the barbed ends of actin filaments, i.e. to “cap” them. The presence of CP at the barbed end inhibits the addition and loss of actin subunits at that end. In cells, CP is important for the dynamics of actin filament assembly, and this is important for the control of cell shape and movement. CP was called β-actin in when first characterized and purified from muscle by Maruyama and colleagues in the 1960’s and 70’s in a remarkably prescient series of studies (Maruyama and Obinata, 1965; Maruyama et al., 1977; Maruyama, 1966; Maruyama, 2002). Nonmuscle CP was purified to homogeneity from Acanthamoeba in 1980 and shown to cap barbed ends (Isenberg et al., 1980).
CP has continued to be an active subject of research, in part because it is found in essentially every eukaryotic organism and every metazoan cell type. Recent studies have produced new insights into the biochemistry of the interaction of CP with the actin filament, the mechanism of how this interaction can influence the architecture of actin filaments nucleated by Arp2/3 complex, the role of CP’s actin-binding activity in cells, and the identities and roles of molecules that bind and regulate CP. This review focuses on these recent discoveries. Other reviews of CP include the following: (Cooper et al., 1999; Schafer and Cooper, 1995; Wear and Cooper, 2004b; Wear et al., 2000).
Background
Physical and Chemical Properties
CP is an α / β heterodimer with each subunit having a mass of ~30 kD. Individual subunits are unstable, but the heterodimer is very stable. The heterodimer remains folded in 0.6 M KI or 1% non-ionic detergent (Wear and Cooper, 2004a), and it melts at 58 °C in a single irreversible transition (Sizonenko et al., 1996). Individual subunits expressed in bacteria are largely insoluble, but they can be renaturated as heterodimers from urea (Remmert et al., 2000). Simultaneous expression of both subunits in bacteria produces large quantities of soluble active protein; the development of this expression system was a major technological advance in the field (Soeno et al., 1998). CP remains soluble, folded and active for capping actin under a variety of physiological conditions, including the presence or absence of divalent cation, and in a variety of salt concentrations, osmolality and pH.
The CP molecule has the shape of a mushroom (Yamashita et al., 2003). The two subunits have very similar secondary structures, which is remarkable given their essentially complete lack of sequence similarity. The secondary structural elements of the subunits are arranged such that the molecule has a pseudo-two-fold axis of rotational symmetry down the center of the mushroom (Fig 1 A) (Yamashita et al., 2003). On the top surface of the mushroom, both subunits have C-terminal amphipathic α helixes, which appear to bind actin (Wear et al., 2003).
Figure 1.
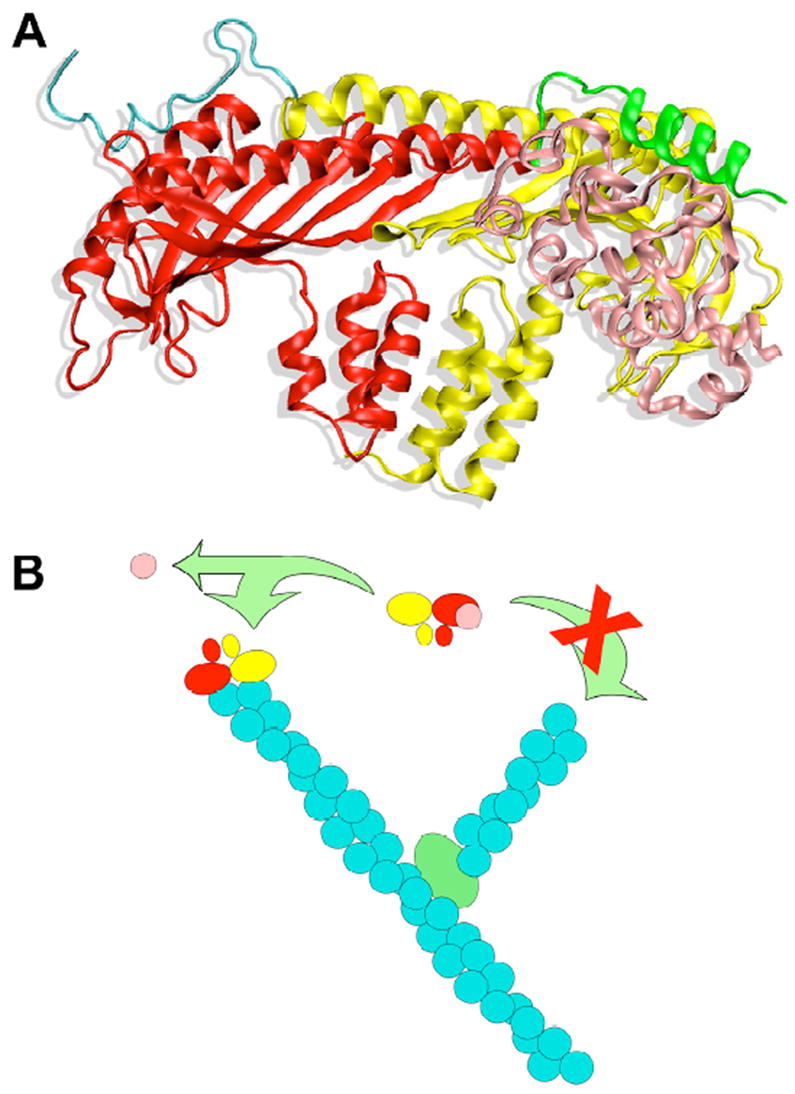
Illustrations of a model for the interaction of V-1 with CP and how their interaction inhibits actin capping activity. A) The structure of the proposed molecular interaction between capping protein and V-1/ myotrophin. The α subunit of capping protein is yellow, and its C-terminal actin-binding region is teal. The β subunit of capping protein is red, and its C-terminal actin-binding region is green. V-1 / myotrophin is in pink. B) The binding of V-1/myotrophin prevents capping protein from binding to actin filament barbed ends, i.e. “capping.” Growth of free barbed ends is an essential of the dendritic nucleation model for actin assembly, which involves Arp2/3 complex as the nucleating and branching agent. The color scheme is similar to the one in panel A, with Arp2/3 complex as the green oval at an end-to-side branch point for two actin filaments, whose subunits are teal.
Biochemical Activities
CP was named for its ability to inhibit growth of the actin filament at the barbed end, i.e. to “cap” that end (Isenberg et al., 1980). CP binds to barbed ends with sub-nanomolar affinity (Wear et al., 2003). The presence of CP at the barbed end prevents the loss of the terminal actin subunit at the end of the filament, thus preventing depolymerization of the filament from that end.
The critical concentration for actin polymerization is lower at the barbed end than at the pointed end, and the rate constants for actin elongation are higher at the barbed end than at the pointed end. These facts mean that capping of barbed ends by CP leads to an increase in the critical concentration, i.e. the actin monomer concentration at steady state. Cell cytoplasm has a high concentration of unpolymerized actin, for which capping of barbed ends is probably necessary.
One molecule of CP appears to be sufficient to bind and attach a filament barbed end to an object, based on direct observation of single actin filaments by light microscopy (Bearer, 1991), including recent TIRF microscopy (Pavlov et al., 2007). TIRF microscopy confirms that the presence of CP at the barbed end abrogates the addition and loss of actin subunits (Kim et al., 2007).
CP was one of the proteins found to be required for the reconstitution of motility based on actin assembly from pure proteins, in a landmark study (Loisel et al., 1999). Other molecules that cap barbed ends, not only CP, can serve this function (Revenu et al., 2007). One idea about the essential role of CP in the reconstitution system is that CP caps barbed ends that are older and thus located away from the surface of the object to be moved. By preventing actin subunits from adding in these undesired locations, the addition of actin subunits in the optimal locations is promoted, which can be considered as “funneling” of the subunits to these locations (Carlier and Pantaloni, 1997). Recent studies with synthetic systems show that CP can cause the shell of Arp2/3-nucleated actin filaments that assemble around a bead to break symmetry, which is necessary to produce polarity and movement (Orkun Akin and Dyche Mullins, personal communication).
In reconstituted systems, high concentrations of CP lead to decreased actin assembly, making the plot of actin assembly or motility vs CP concentration a bell-shaped curve (Loisel et al., 1999). This biphasic nature of the effect of CP makes it difficult to interpret results in complex systems, such as mixtures of multiple actin regulators or even the cell cytoplasm. Mathematical modeling can help make predictions in such cases. For example, the concentration dependence of actin assembly on CP in the presence of Arp2/3 complex provided information about the end vs side nature of Arp2/3-mediated branching, based on predictions from mathematical modeling of the alternatives (Carlsson et al., 2004).
Cellular Studies
The abilities of CP to cap barbed ends and to tether barbed ends to objects appear to be a physiologically relevant in cells. The concentration of CP in cells is in the μM range, comparable to the number of actin filament barbed ends, and the binding affinity is in the sub-nM range (Cooper et al., 1984; Wear et al., 2003). Analysis of a set of CP mutants in yeast showed a correlation of capping activity with the ability to rescue the null mutant phenotype (Kim et al., 2004). In cultured myotubes, injection of an anti-CP mAb that inhibited the actin-binding ability of CP caused a disruption in the early steps in myofibril-logenesis, as did expression of a mutant form of the CP β subunit that caps actin poorly (Schafer et al., 1995). In the mouse heart, expression of a capping-deficient CP β subunit during development caused disruption of myofibril architecture (Hart and Cooper, 1999). Other potential functions for CP are discussed below.
Sequence Conservation and Isoforms
One of the most interesting and surprising features of the CP crystal structure was the two-fold rotational similarity between the tertiary structures of the two subunits (Yamashita et al., 2003). In vertebrates, the sequence similarity between the α and β subunits is very low, and given the lack of symmetry at the end of the actin filament, there was little reason to expect such structural symmetry in the CP heterodimer. When comparing the individual subunits in different organisms, sequence similarity is much higher. BLAST searches readily reveal apparent homologs of both subunits in vertebrates, invertebrates, plants, fungi, insects and protozoa (Fig 2 A, B). The sequences of the β subunits appear to be more strongly conserved than those of α subunits (Fig 2 C).
Figure 2.
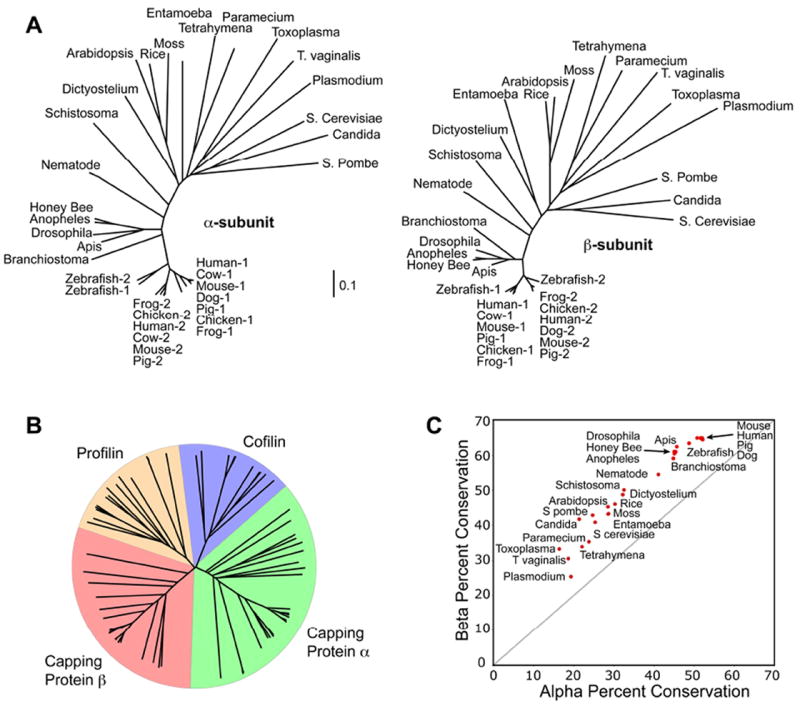
Phylogenetic analysis of CP subunits. Amino acid sequences were identified in a wide range of eukaryotes by BLAST. The sequences were aligned with CLUSTALW. A) Phylogenetic trees for the α and β subunits are remarkably similar. Vertebrates have up to three isoforms of each subunit, while invertebrates and lower organisms have single isoforms of each subunit. Vertebrate isoforms 1 and 2 represent nearly all the CP outside of germ cells, they cluster into distinct groups. B) Phylogenetic analysis of CP subunits compared with cofilin and profilin, other actin-binding proteins. The α and β subunits of CP are not more similar to each other than they are to the cofilin and profilin families, despite their similar secondary structures and interactions with actin. C) For each organism, the similarity of its β subunit to the β subunits of other organisms is plotted versus the similarity of its α subunit to the α subunits of other organisms. For vertebrates, only a single isoform of each subunit was included per species. The results show that the β subunit sequences are more similar to each other than are those of the α subunits.
The regions of conservation and variability are localized in a complementary manner on the two subunits. Within the β subunit, the actin-binding C-terminal region, the β-tentacle, shows the highest sequence variability. In contrast, the body of the α subunit is more weakly conserved than is the C-terminal region, giving rise to an inherent asymmetry in the molecule where the half of the molecule containing the β subunit and α tentacle is more conserved than the half with the α subunit and the β tentacle. This asymmetry may have implications for how CP interacts with the barbed end of the actin filament, and it appears to be consistent with structural studies discussed next.
Organisms other than vertebrates have single genes encoding each of the CP subunits. Vertebrates, in contrast, have two somatically-expressed isoforms of each subunit and one additional male germ-cell specific isoform (Hart et al., 1997b; Hurst et al., 1998; Schafer et al., 1994; von Bulow et al., 1997). For the α subunit, the somatic isoforms, termed α1 and α2, are encoded by different genes (Hart et al., 1997a), while the β subunit isoforms are produced from a single gene by alternative splicing (Schafer et al., 1994). The sequences of the α1 and α2 isoforms are conserved across vertebrates, as are those of the β1 and β2 isoforms, suggesting that they have distinct functions in vertebrates. Little evidence exists regarding specific functions of the α isoforms, but they are expressed at varying ratios in different cells and tissues (Hart et al., 1997b).
The β1 isoform is located specifically at the Z-disc of the sarcomere of striated muscle; β2 is also present in the same cells, but it localizes elsewhere (Schafer et al., 1994). The β1 and β2 isoforms were not able to substitute for each other in muscle cells, supporting the hypothesis of distinct functions (Hart and Cooper, 1999). The biochemical nature of the functional difference has not been discovered. One would suspect that the β1 isoform interacts specifically with one or more components of the Z-disc. CP isoforms appear to bind equally well to actin and nebulin, as purified proteins in vitro (Pappas et al., 2007; Schafer et al., 1994), and other components remain to be tested.
Mechanism of Binding Actin
Structural Studies
An X-ray crystal structure of CP shows that the molecule has the shape of a mushroom and that the two subunits are arranged with a pseudo-two-fold axis of rotational symmetry (Yamashita et al., 2003). The N-termini of the subunits are located at the base of the stalk of the mushroom, and the subunits are extensively intertwined, with a large β sheet at the core of the mushroom cap structure. On the top surface of the mushroom, each subunit has an extended α helix oriented perpendicular to the strands of the β sheet on which it lies. Each subunit ends with a C-terminal amphipathic α helix on the top surface of the mushroom.
Truncation and point mutations of the C-terminal regions reveal that both are important for high-affinity capping (Wear et al., 2003), with the C-terminal region of the α subunit being more important than that of the β. CP binds to the barbed end of the actin filament with high affinity, generally less than 1 nM. The second-order association rate constant is high, approaching the range of the diffusion limit, and the first-order dissociation rate constant is accordingly low. CP containing only one (either one) of the C-terminal regions is able to cap, and the C-terminal region of the β subunit alone is sufficient to cap, with decreased affinity. The binding affinity and rate constants have been inferred largely from actin polymerization experiments, and physical binding studies would provide a valuable confirmation of those results.
Recent cryoEM work from the Maéda lab has resulted in a low-resolution (23 Å) structure of CP on the barbed end of an actin filament (Narita and Maeda, 2007; Narita et al., 2006). CryoEM analysis of actin filament binding proteins that decorate the sides of the filament can benefit from helical averaging. Here, only one molecule of CP was present on the barbed end of each filament, so this analysis depended on the collection and averaging of single-particle images, a challenging task. A novel method for combining the images that were collected produced a new model for the structure of the CP-capped filament. This structure was able to unambiguously identify the α and β subunits, and their positions with respect to the actin protomers at the barbed end suggest that the body of the β subunit, along with the α subunit C-terminal region, make the primary contacts with the last two protomers of the filament. This finding is supported by the sequence conservation data discussed above. Mutational analysis confirmed the importance of residues in the C-terminal region of the α subunit and on the top surface. Computational modeling analysis showed that the β subunit C-terminus, a tentacle-like amphipathic helix, can bind to a hydrophobic cleft on the actin subunit, in a manner comparable to that of a WH2 domain (Dominguez, 2004; Hertzog et al., 2004). Based on these results, the authors proposed a model, shown in Figure 3, in which CP binds to the barbed end in two steps, first by the α subunit C-terminus and surrounding residues and second by the flexible β subunit C-terminus (Narita et al., 2006).
Figure 3.
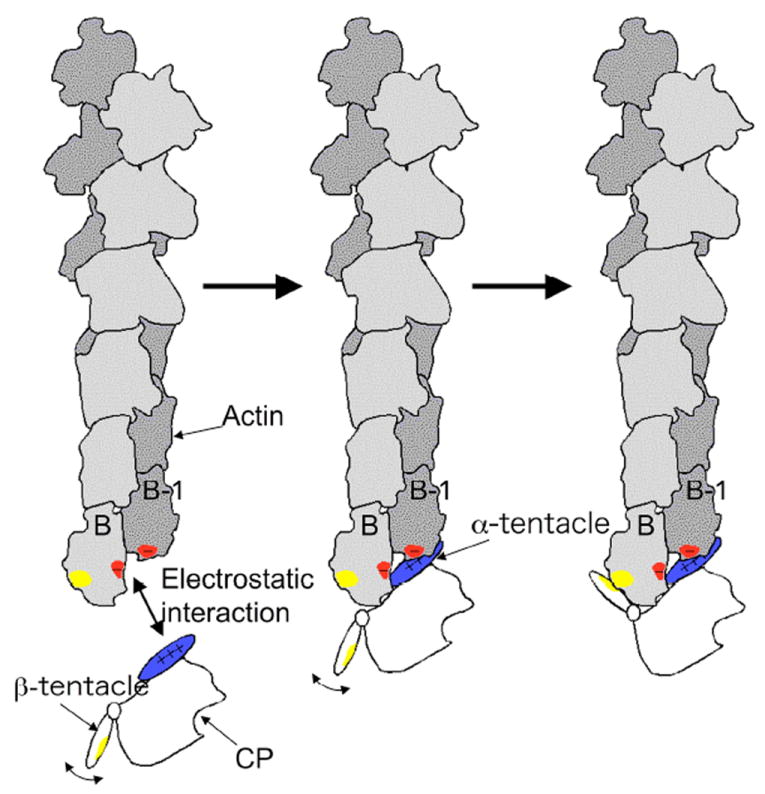
A model for the binding of CP to the actin filament barbed-end proposed by Narita and colleagues (Narita et al., 2006), kindly provided by the authors and reproduced with their permission. Basic residues on the CP α C-terminal region /(blue) are attracted to acidic residues on the barbed end of the actin filament (red). These acidic residues include ones from the terminal and the penultimate protomers of the filament, labeled B and B-1. Next, the mobile β tentacle searches for its binding position on the filament. Finally, the hydrophobic surface (yellow) of the amphipathic β tentacle binds to the hydrophobic cleft (yellow) on the terminal protomer, B.
This model raises the possibility that CP bound to the barbed end of the actin filament might dissociate from the α subunit site and thus be attached only by the β subunit tentacle. If this were to occur, the mobility of the β tentacle might allow the body of CP to move, or “wobble”, in place. If another molecule would bind to CP in this wobble state, then the presence of that molecule might inhibit rebinding and thus favor complete detachment, or “uncapping.”
Uncapping will be an important subject for further study, because of its potential relevance in cells. In vitro, the dissociation rate for CP to leave the barbed end is quite long relative to the time scale on which actin filaments assemble and disassemble in cells and to the time scale on which CP appears to dissociate from the actin cytoskeleton in cells (Iwasa and Mullins, 2007; Miyoshi et al., 2006). Inducing uncapping might be a mechanism for cells to induce assembly or disassembly of the filament network, depending on other conditions in the local environment.
Mobility of the C-terminal Regions
As described above, the C-terminal region of the CP α subunit is an amphipathic helix that lies on the top surface of the protein in the crystal structure. Its hydrophobic side is oriented toward the body of the protein (i.e. the top of the mushroom) (Yamashita et al., 2003), and in molecular dynamics simulations, this region remains in that position on the mushroom surface (Bhattacharya et al., 2006). Residue Trp271 of the amphipathic helix occupies a hydrophobic pocket on the surface of the body (Yamashita et al., 2003). A short peptide corresponding to part of this same region of the CP α subunit was found to bind to the protein S100 (Ivanenkov et al., 1995; Ivanenkov et al., 1996), and an NMR structure of the S100 - peptide complex showed that the Trp residue corresponding to position 271 occupies a hydrophobic pocket in S100 (Inman et al., 2002).
This peptide from CP α can bind S100, and full-length unfolded CP α can bind S100, but native CP was found not to bind S100 (Schafer et al., 1996; Wear and Cooper, 2004a). Treatment of CP with high concentrations of non-ionic detergent enabled S100 to bind weakly (Wear and Cooper, 2004a). Thus, the C-terminal region of the α subunit, which is necessary for binding actin, appears to be immobile in the native solution structure, as implied by the crystal structure and molecular dynamics results. Mutating the Trp271-analogous residue of yeast CP to Ala caused a large loss of capping activity, which may be due to alteration of the structure of the amphipathic helix (Kim et al., 2004).
In the crystal structure, the C-terminal region of the CP β subunit is also an amphipathic helix, but the helix extends out from the body of the protein, surrounded by solvent (Yamashita et al., 2003). In molecular dynamics simulations, this region is highly mobile, as expected (Bhattacharya et al., 2006).
The mobilities of the C-terminal regions of the subunits are incorporated into the current model for CP binding to the barbed end of the actin filament proposed by Maéda and colleagues (Narita and Maeda, 2007; Narita et al., 2006). In terms of the wobble hypothesis, the mobilities of the C-terminal actin-binding regions helps to predict that CP will not wobble when it is bound to a barbed end only by the C-terminal region of the α subunit, i.e. when the β subunit’s C-terminal tentacle is not bound to actin. In contrast, CP will wobble if the α subunit C-terminal region dissociates, leaving only the β tentacle attached (Bhattacharya et al., 2006).
CP Inhibitors and Uncapping
Contrasting Results with CARMIL and V-1
CARMIL and V-1/myotrophin are two different proteins that can bind to CP and inhibit its ability to bind to the barbed end of the actin filament, i.e. to cap. When CP is already present on the barbed end, CARMIL appears to be able to remove it. This conclusion is based on physical and functional assays. First, CP is found in the supernatant after sedimentation of actin filaments following the addition of CARMIL (Uruno et al., 2006). Second, the concentration of free barbed ends increases rapidly on addition of CARMIL (Uruno et al., 2006; Yang et al., 2005). This increase occurs on the time scale of ~ 10 sec (Uruno et al., 2006; Yang et al., 2005), while the spontaneous dissociation rate of CP from the barbed end appears to be on the time scale of ~ 10 min (Schafer et al., 1996). This difference suggests that CARMIL can bind to the CP / barbed-end complex in some manner, and we hypothesize that this interaction occurs in the wobble state. In support of this hypothesis, titration of CP with increasing concentrations of CARMIL in an actin-capping assay does not lead to complete inhibition of CP (Yang et al., 2005). Less than complete inhibition can be explained by the CARMIL / CP complex having a very low level of capping activity, suggesting that CARMIL and CP can co-exist in a ternary complex with the barbed end. In the future, identification of the CARMIL-binding site on CP should provide an important test of the wobble hypothesis, and imaging of single CP molecules on actin filaments should further our understanding of uncapping.
V-1 / myotrophin provides an important and interesting contrast with CARMIL. V-1 also binds to CP and inhibits its ability to cap the barbed end (Fig 1) (Bhattacharya et al., 2006; Taoka et al., 2003). However, V-1 has little or no uncapping activity in functional assays. That is, addition of a high concentration of V-1 to CP-capped actin filaments, at a level sufficient to inhibit all the CP in the reaction, produces little increase in the number of free barbed ends (Bhattacharya et al., 2006). Another difference between CARMIL and V-1 is that high concentrations of V-1 completely inhibit the capping activity of CP (Bhattacharya et al., 2006). Thus, both lines of evidence fail to indicate that V-1 can bind to CP that is bound to the barbed end.
In the hypothetical wobble state, CP is attached to the barbed end only by the β subunit’s C-terminal region, i.e. the tentacle. V-1 requires the C-terminal region of the β subunit but not that of the α subunit for optimal binding to CP (Bhattacharya et al., 2006), suggesting that V-1 may interact with the β tentacle, among other parts of CP. Thus, V-1 would not be predicted to bind to the hypothetical wobble state, and this predicts that V-1 should not be able to uncap, which is the case.
The binding site on CP for V-1 appears to include an area at the base of the β tentacle, as indicated in Figure 1 A. This conclusion is based on several results. First, truncation of the CP α subunit C-terminal region was found to weaken the interaction of CP with V-1 by a small amount (Bhattacharya et al., 2006). The simplest interpretation of this observation alone would be a direct interaction between V-1 and the α subunit C-terminus. However, the dynamics of the α C-terminus are coupled to the rest of the protein, in contrast to the situation for the highly mobile “tentacle” of the β subunit. Molecular dynamics simulations of a CP molecule from which the C-terminal region of the α subunit was truncated revealed decreased dynamics of the α subunit at locations distant from the C-terminus, near the base of the β tentacle (Bhattacharya et al., 2006). The surface residues at the affected location were also part of a V-1 binding site identified on CP by computational docking analysis. In addition, these residues are among the relatively few that differ between the α 1 and α 2 isoforms, and V-1 binds slightly differently to the α isoforms. To account for all these observations, we suggest that V-1 binds to CP as depicted in Figure 1 A. Structural data about the CP / V-1 complex should provide valuable information to test this model.
Interaction of Polyphosphoinositides with CP
Polyphosphoinositides, including PIP2, can bind to CP and inhibit its capping activity (Heiss and Cooper, 1991). PIP2 can also cause rapid uncapping, demonstrated recently by observations of the polymerization of single actin filaments by TIRF microscopy (Kim et al., 2007). In those studies, addition of PIP2 to a flow chamber with actin filaments that were capped by CP and thus not able to polymerize resulted in the rapid and complete conversion of ends from the non-growing to the growing state. Computational docking analysis suggested that PIP2 binds to three conserved basic residues on the surface of CP near the α subunit C-terminus, and mutations of those residues weakened the affinity of PIP2 for CP, measured with functional and physical assays (Kim et al., 2007). Some of these residues were predicted to sit at the interface between CP and actin based on the cryoEM CP / actin filament structure (Narita et al., 2006), and mutations of these residues affected the ability of CP to cap actin (Kim et al., 2007; Narita et al., 2006). Thus, the PIP2 and actin binding sites on CP may overlap. In addition, these observations are consistent with the wobble model in that they suggest that the region of the α C-terminus is available for PIP2 binding when CP is on the barbed end, i.e. in the wobble state.
In older work, studies of the actin assembly that accompanies platelet activation suggested that an early step was uncapping of CP-capped actin filaments by polyphosphoinositides(Barkalow et al., 1996). In many other cells systems, CP appears to terminate actin assembly by capping free barbed ends that are created by other mechanisms. The actin assembly that results from treatment of Dictyostelium cells with chemoattractant appears to be such a case (Eddy et al., 1997). Since polymerization of free barbed ends at membranes appears to drive the movement of those membranes, one attractive hypothesis is that PIP2 generated in the membrane helps to inhibit capping by CP near the membrane.
CARMIL, CKIP-1 and CD2AP - A motif for inhibition of CP
Motif for Inhibition of CP
The existence of a motif for binding and inhibiting CP has been suggested by comparative analysis of the sequences and biochemical properties of the proteins CARMIL, CKIP-1 and CD2AP (Bruck et al., 2006; Canton et al., 2005; Canton et al., 2006; Uruno et al., 2006). Each of the three proteins was found to bind directly to CP and inhibit the actin capping activity of CP. Structure / function analysis of each protein revealed an essential region with a common set of essential amino-acid residues. The potential CP-binding motif appears to be LXHXTXXRPK(6X)P (Bruck et al., 2006)
CARMIL
Acan125 was the original name for CARMIL when the protein was discovered in amoeba as a binding partner for the SH3 domain of certain class-I myosins (Xu et al., 1995). Later, this protein was found to bind CP and Arp2/3 complex as well, leading to the acronym CARMIL (Jung et al., 2001). The protein is relatively large, with a long leucine-rich repeat (LRR) region of unknown function (Xu et al., 1997). The LRR region may participate in autoinhibition of the CP-binding activity of CARMIL (Uruno et al., 2006). CARMIL binds tightly to CP, with a Kd in the nM range (Yang et al., 2005). CARMIL purified from Acanthamoeba contains CP in near-stoichiometric amounts (Remmert et al., 2004), but the large majority of Acanthamoeba CP in cell extracts is free and able to cap actin (Cooper et al., 1984).
CARMIL is important for actin-based motility, based on knockout and knockdown studies in Dictyostelium and cultured vertebrate cells (Jung et al., 2001; Yang et al., 2005). The multiple biochemical functions associated with CARMIL raise many possibilities for its mechanism of action in cells. Loss of the CP-binding site, by internal deletion of ~100 aa residues, produced a mutant form of CARMIL unable to rescue the knockdown phenotype in cultured vertebrate cells(Yang et al., 2005).
CKIP-1
CKIP-1 was discovered as an interaction partner for casein kinase 2, helping to recruit CK2 to the plasma membrane (Olsten et al., 2004). CKIP-1 was also found to interact biochemically with CP in cultured cells (Canton et al., 2005). The binding of CKIP-1 and CK2 to CP inhibits capping activity (Canton et al., 2005), and CKIP-1 expression in cultured cells causes changes in cell morphology and the actin cytoskeleton that depend on its interaction with CP (Canton et al., 2006). CK2 can phosphorylate CP by CK2 (Canton et al., 2005), which may be a novel regulatory mechanism.
CD2AP
CD2-associated protein (CD2AP) and its relative Cin85 (Cbl-interacting protein) have been found to bind to CP and inhibit its capping activity (Bruck et al., 2006; Hutchings et al., 2003). CD2AP and Cin85 appear to be adaptor proteins that provide signaling pathway connections from membrane receptors to the actin cytoskeleton (Dustin et al., 1998; Lynch et al., 2003; Shih et al., 1999). CD2AP and Cin85 also interact with cortactin (Lynch et al., 2003; Nam et al., 2007), which promotes actin assembly via Arp2/3 complex, so these adaptors have multiple potential connections to actin assembly.
Indirect Regulators of CP
Formin proteins act as competitors of CP at the actin filament barbed end. Formins are a large family of proteins, with a diversity of structures and functions that have yet to be understood fully (Goode and Eck, 2007; Staiger and Blanchoin, 2006). A formin dimer can bind to an actin filament barbed end, which can inhibit the binding of CP (Fig 4) (Zigmond et al., 2003). However, the presence of formin at the barbed end still allows an actin subunit to add to that barbed end. Most remarkably, one formin dimer can remain bound to the barbed end while more and more actin subunits add over time; the formin essentially “surfs” with the growing barbed end of the actin filament. Thus, formins are very effective anti-cappers, promoting the growth of barbed ends. A number of cellular studies of formins support the relevance of this mechanism in vivo (Goode and Eck, 2007; Staiger and Blanchoin, 2006).
Figure 4.
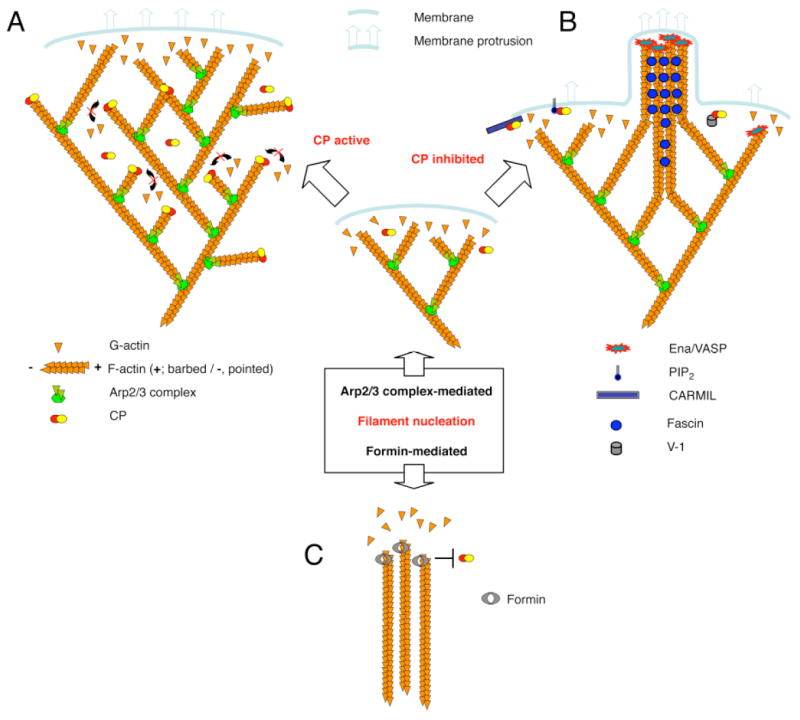
Illustration of potential modes of actin assembly in cells with respect to CP, based on one created and provided by Dr. Martin Wear (Wear and Cooper, 2004b). A) When CP is active and actin nucleation is Arp2/3-mediated, lamellipodial assembly predominates. Newly created free barbed ends are near the membrane. They elongate to push the membrane forward and / or the actin filament network backward. After some time, CP caps those barbed ends, which would seem to be efficacious because the ends are no longer near the membrane and their further growth would not produce useful work. B) In this setting, when CP is inactivated in one location, by any of several potential inhibitors, then the filaments in that small region may continue to grow, producing a thin protrusion that contains a bundle of actin filaments. C) An alternative mechanism that may produce actin filament bundles, perhaps not associated with a plasma membrane, is the nucleation of actin polymerization by a formin. Formins allow actin subunits to add and do not allow CP to add. Thus, the filaments continue to grow.
VASP also functions as an antagonist of CP, via interactions with the actin filament. VASP is a member of the Ena/VASP family of proteins, which have been implicated in actin-based motility and morphogenesis. VASP antagonizes the capping activity of CP in vitro (Barzik et al., 2005). The anti-capping effect of VASP is not specific for CP in that other barbed-end cappers, such as gelsolin, are also antagonized by VASP. Ena/VASP proteins are found at the tips of filopodia, where they may prevent capping, which would allow barbed ends of actin filaments to grow and filopodia to elongate (Fig 4) (Applewhite et al., 2007; Mejillano et al., 2004).
Other Interactors
Twinfilin was characterized as a protein that binds and sequesters actin monomers, thus inhibiting actin polymerization (Goode et al., 1998; Lappalainen et al., 1998; Palmgren et al., 2002). Twinfilin binds directly to CP, and that interaction does not affect the interaction of either protein with actin (Falck et al., 2004). In yeast, twinfilin’s ability to bind CP and its ability to bind actin are both necessary for its function in actin dynamics (Falck et al., 2004). Twinfilin alone can also cap barbed ends, with a preference for ADP-actin (Helfer et al., 2006; Paavilainen et al., 2007), raising important questions about the relative contributions of these various biochemical activities to cell physiology.
Extracts of neutrophils were found to contain a low-molecular-weight inhibitor of CP (Huang et al., 2005), which has not yet been identified. This inhibitor was able to inhibit and reverse capping of barbed ends by purified CP in functional assays. The biochemical properties of the inhibitor indicated that it was not PIP2, V-1, CARMIL or VASP. In an earlier study with neutrophil extracts, Cdc42-induced actin polymerization was found to be insensitive to CP, relative to polymerization induced by actin seeds (Huang et al., 1999). In retrospect, this Cdc42-induced anti-capper may have been a formin.
Role of CP in Complex Cellular Processes
Actin-based Motility at the Plasma Membrane
In metazoan cells in culture, the ability to form lamellipodial type protrusions was found to depend on CP in siRNA knockdown studies (Iwasa and Mullins, 2007; Mejillano et al., 2004). In mouse melanoma cells, inhibition of lamellipodial assembly was accompanied by an increase in filopodia formation (Mejillano et al., 2004), consistent with older results in Dictyostelium with antisense (Hug et al., 1995). In contrast, filopodia were not increased on CP knockdown in Drosophila cultured S2 cells (Iwasa and Mullins, 2007). In the mouse melanoma cells, the increased filopodia formation depended on VASP (Mejillano et al., 2004), so the Drosophila S2 cells may have lacked sufficient activity of VASP or some other filopodial component (Iwasa and Mullins, 2007).
The need for CP in lamellipodial assembly supports the relevance of a key element of the dendritic nucleation model proposed to account for the assembly of branched networks of actin filaments associated with membranes and Arp2/3 complex (Nicholson-Dykstra et al., 2005). In that model, the reason why capping of barbed ends by CP is important has been proposed to be to “funnel” actin assembly to the new filament ends at the membrane, as described above, or to keep the actin filaments short and highly branched, to strengthen the network (Fig 4). Testing these ideas will likely require mathematical modeling and measurements of physical parameters on a microscopic time scale with high time resolution.
Speckle and single-molecule fluorescence imaging of lamellipodial regions of cultured cells reveals that CP binds to the actin filament network very near the membrane and that it dissociates from the network after a short time and distance (Iwasa and Mullins, 2007; Miyoshi et al., 2006), also consistent with the proposed role for CP in the dendritic nucleation model. CP dissociation may result from severing-induced depolymerization of filaments or it may be the direct effect of an uncapper. Distinguishing these possibilities will require observing the behavior of CP in cells carrying a specific defect in severing or uncapping, which will require careful biochemical characterization of mutant proteins. A challenge for these studies will be identifying which of several potential uncappers or severing agents are the relevant actors in a given cell system.
For CP, the ability to alter protein activity protein locally and rapidly should provide powerful information for testing predictions of models. Acute inactivation of CP has been achieved in fibroblasts by laser inactivation of GFP-CP. The result was a local increase in the concentration of free barbed ends, the polymerization of actin and the formation of actin-based protrusive structures (Vitriol et al., 2007). These results support the notion that the CP caps barbed ends and that barbed-end capping prevents actin polymerization. Note that intuitive reasoning from the dendritic nucleation model seems capable of explaining the increase in actin-based protrusions in this experiment but also the decrease in lamellipodial protrusions in the set of knockdown experiments discussed above. One can rationalize the opposing predictions for the effect of the loss of CP activity in these two experiments on the basis of the laser inactivation effect being local and acute, while the knockdown inhibition effect is global and chronic. The rigor and certainty of the conclusions would be greatly enhanced by the application of mathematical modeling so that predictions are based on more than intuition and rationalization.
Other membrane movements, in addition to lamellipodial protrusions of the plasma membrane, appear to be based on actin assembly. Endocytosis is another good example, based on recent studies in yeast and vertebrate systems (Engqvist-Goldstein and Drubin, 2003; Kaksonen et al., 2006). The endocytic process is composed of multiple steps of actin assembly and actin-based movement. Membrane receptors, endocytic adaptors, and actin-binding proteins, including Arp2/3 complex, are involved, with distinct roles at various steps in the process. Yeast CP null mutants showed a decrease in the initial movement of the cortical actin patch, the site of endocytosis, away from the plasma membrane, and the actin filaments of the patch still assembled (Kim et al., 2006), all of which appears to be consistent with the dendritic nucleation model. In contrast, other steps of endocytic traffic showed little to no effect from the loss of CP, so the model may not apply in these cases.
The Z-line of the Sarcomere in Striated Muscle
CP purified from skeletal muscle was called “CapZ” because of its presence at the Z-disc of the sarcomere (Casella et al., 1987). The barbed ends of the actin-based thin filaments are also located at the Z-disc, and one molecule of CapZ appears to cap each barbed end (Schafer et al., 1993). The reason for capping the barbed end may be to help anchor the thin filament to the Z-disc, or it may be to prevent the growth of the thin filament into the adjacent sarcomere. CapZ and its actin-binding activity appear to be important for assembly of the sarcomere, as noted above.
Recent studies have uncovered a biochemical interaction between nebulin, a giant protein of the sarcomere, and CapZ (Pappas et al., 2007; Witt et al., 2006). Nebulin knockdown in developing muscle cells leads to decreased accumulation of CapZ at the Z-disc and poor alignment of thin filament barbed ends (Pappas et al., 2007), consistent with a role for nebulin as a “ruler” specifying thin filament length by interacting with CapZ as the capper of the barbed end. The location of the CapZ binding site in nebulin suggests a model for the Z-disc in which nebulin connects one thin filament with an adjacent one, thus serving as a structural cross bridge to impart strength to the disc (Fig. 5).
Figure 5.
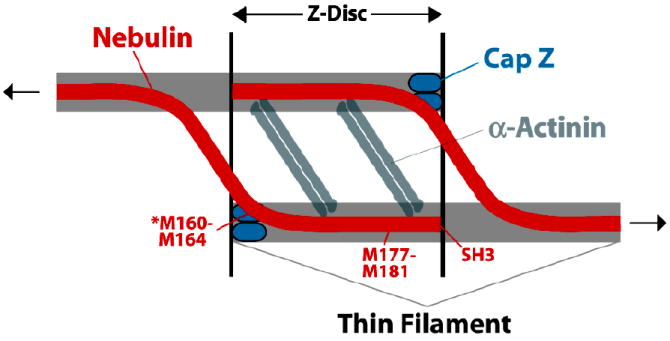
An illustration of a new model for the structure of the Z-disc, provided by Dr. Carol Gregorio. Based on a structure / function analysis of the interaction of nebulin with CapZ (Pappas et al., 2007), the model proposes that the nebulin molecule crosses from one actin-based thin filament to another one, within the Z-disc.
Drosophila development
In Drosophila, CP is essential for viability of the organism, and loss-of-function mutants die as embryos (Hopmann et al., 1996). In the bristles of the adult fly, actin bundles underlie and define the surface structure of the bristle, and the assembly of these actin bundles depends on CP and other actin regulators, including profilin and Arp2/3 complex (Frank et al., 2006; Hopmann and Miller, 2003). The effects of CP and the other proteins on the actin filament bundles appears to be an indirect one, mediated by their effects on a separate dynamic pool of actin filaments termed snarls (Frank et al., 2006). Studies of eye and wing development have also revealed an important role for CP, most likely through effects on actin assembly and morphogenesis (Iwasa and Mullins, 2007; Janody and Treisman, 2006).
Dynactin
CP is a biochemical component of dynactin, a multi-subunit complex necessary for the function of dynein (Schroer, 2004). Dynactin contains an actin-like filament composed largely of Arp1 (actin-related protein 1), and one mol per mol of CP. Single-particle EM image averaging of purified dynactin reveals lobes at the barbed end of the actin-like filament, which are likely to correspond to the subunits of CP (Hodgkinson et al., 2005; Imai et al., 2006). The presence of CP may be important to control the number of Arp1 subunits in the filament, which is remarkably constant among dynactin molecules.
Whether the presence of CP affects the function of dynactin in cells has not been thoroughly tested. In yeast, CP null mutations produce no measurable effect on dynein function (Moore et al., 2007). Null mutations of some other dynactin subunits, including Arp1, produce a complete loss of dynein function. Biochemical approaches have not revealed CP to be a component of dynactin. Therefore, CP is either not an important component of dynactin in yeast or not a component at all. Yeast dynactin may lack other subunits of dynactin as well (Moore et al., 2007). To our knowledge, no tests of the functional role of CP have been done in other systems, which would be useful.
Summary and Future Directions
The emerging multiplicity of molecules that interact with CP and the diversity of their biochemical actions raises many new questions about how CP functions and is regulated in cells. The potential complexity is amplified by the cases where the interactors are proteins with multiple domains and may serve as adaptors with other molecules. Dissecting the individual roles of these interactions in biochemical and physiological terms will be an important challenge.
New insight into the dynamic nature of the interaction of CP with the actin filament barbed end raises interesting possible mechanisms for the action of CP in the rapid assembly and disassembly of actin in cells. The potential existence of a wobble state needs to be established with direct physical methods, which will allow one to test whether the wobble state is part of the mechanism of uncapping.
CP is present in essentially all cells and tissues of vertebrates, and actin filaments are proving to have multiple distinct roles in various cell settings. The roles that CP may play in these settings, especially the possibility of different roles for the conserved vertebrate isoforms of CP, will be an important avenue for the future.
Acknowledgments
The authors are grateful to Drs. Carol Gregorio, Martin Wear, Yuichiro Maéda, and Akihiro Narita for providing illustrations. Research in this area in the authors’ laboratories is supported by NIH GM 38542 to J.A.C. and GM 67426 to D.S.
References
- Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkalow K, Witke W, Kwiatkowski DJ, Hartwig JH. Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J Cell Biol. 1996;2:389–399. doi: 10.1083/jcb.134.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL. Direct observation of actin filament severing by gelsolin and binding by gCap39 and CapZ. J Cell Biol. 1991;115:1629–1638. doi: 10.1083/jcb.115.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya N, Ghosh S, Sept D, Cooper JA. Binding of Myotrophin/V-1 to Actin-capping Protein: Implications for How Capping Protein binds to the Filament Barbed End. J Biol Chem. 2006;281:31021–31030. doi: 10.1074/jbc.M606278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck S, Huber TB, Ingham RJ, Kim K, Niederstrasser H, Allen PM, Pawson T, Cooper JA, Shaw AS. Identification of a Novel Inhibitory Actin-capping Protein Binding Motif in CD2-associated Protein. J Biol Chem. 2006;281:19196–19203. doi: 10.1074/jbc.M600166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton DA, Olsten ME, Kim K, Doherty-Kirby A, Lajoie G, Cooper JA, Litchfield DW. The pleckstrin homology domain-containing protein CKIP-1 is involved in regulation of cell morphology and the actin cytoskeleton and interaction with actin capping protein. Mol Cell Biol. 2005;9:3519–3534. doi: 10.1128/MCB.25.9.3519-3534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton DA, Olsten ME, Niederstrasser H, Cooper JA, Litchfield DW. The Role of CKIP-1 in Cell Morphology Depends on Its Interaction with Actin-capping Protein. J Biol Chem. 2006;281:36347–36359. doi: 10.1074/jbc.M607595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Control of actin dynamics in cell motility. J Mol Biol. 1997;4:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- Carlsson AE, Wear MA, Cooper JA. End versus Side Branching by Arp2/3 Complex. Biophys J. 2004;2:1074–1081. doi: 10.1016/S0006-3495(04)74182-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella JF, Craig SW, Maack DJ, Brown AE. Cap Z(36/32), a barbed end actin-capping protein, is a component of the Z-line of skeletal muscle. J Cell Biol. 1987;1:371–379. doi: 10.1083/jcb.105.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Blum JD, Pollard TD. Acanthamoeba castellanii capping protein: Properties, mechanism of action, immunologic cross-reactivity, and localization. J Cell Biol. 1984;99:217–225. doi: 10.1083/jcb.99.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Hart MC, Karpova TS, Schafer DA. In: Capping Protein. Kreis T, Vale R, editors. Oxford University Press; New York: 1999. pp. 62–64. [Google Scholar]
- Cooper JA, Blum JD, Pollard TD. Acanthamoeba castellanii capping protein: properties, mechanism of action, immunologic cross-reactivity, and localization. J Cell Biol. 1984;99:217–225. doi: 10.1083/jcb.99.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, Widder P, Rosenberger F, van der Merwe PA, Allen PM, Shaw AS. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;5:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- Eddy RJ, Han J, Condeelis JS. Capping protein terminates but does not initiate chemoattractant-induced actin assembly in Dictyostelium. J Cell Biol. 1997;139:1243–1253. doi: 10.1083/jcb.139.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Falck S, Paavilainen VO, Wear MA, Grossmann JG, Cooper JA, Lappalainen P. Biological role and structural mechanism of twinfilin-capping protein interaction. EMBO J. 2004;15:3010–3019. doi: 10.1038/sj.emboj.7600310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DJ, Hopmann R, Lenartowska M, Miller KG. Capping protein and the Arp2/3 complex regulate nonbundle actin filament assembly to indirectly control actin bundle positioning during Drosophila melanogaster bristle development. Mol Biol Cell. 2006;17:3930–3939. doi: 10.1091/mbc.E06-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Drubin DG, Lappalainen P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J Cell Biol. 1998;3:723–733. doi: 10.1083/jcb.142.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and Function of Formins in the Control of Actin Assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Hart MC, Cooper JA. Vertebrate isoforms of actin capping protein beta have distinct functions In vivo. J Cell Biol. 1999;6:1287–1298. doi: 10.1083/jcb.147.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MC, Korshunova YO, Cooper JA. Mapping of the Mouse Actin Capping Protein Alpha Subunit Genes and Pseudogenes. Genomics. 1997a;3:264–270. doi: 10.1006/geno.1996.4506. [DOI] [PubMed] [Google Scholar]
- Hart MC, Korshunova YO, Cooper JA. Vertebrates have conserved capping protein alpha isoforms with specific expression patterns. Cell Motil Cytoskeleton. 1997b;2:120–132. doi: 10.1002/(SICI)1097-0169(1997)38:2<120::AID-CM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Heiss SG, Cooper JA. Regulation of CapZ, an actin capping protein of chicken muscle, by anionic phospholipids. Biochemistry. 1991;30:8753–8758. doi: 10.1021/bi00100a006. [DOI] [PubMed] [Google Scholar]
- Helfer E, Nevalainen EM, Naumanen P, Romero S, Didry D, Pantaloni D, Lappalainen P, Carlier MF. Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J. 2006;25:1184–1195. doi: 10.1038/sj.emboj.7601019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- Hodgkinson JL, Peters C, Kuznetsov SA, Steffen W. Three-dimensional reconstruction of the dynactin complex by single-particle image analysis. Proc Natl Acad Sci U S A. 2005;10:3667–3672. doi: 10.1073/pnas.0409506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Cooper JA, Miller KG. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J Cell Biol. 1996;133:1293–1305. doi: 10.1083/jcb.133.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Miller KG. A balance of capping protein and profilin functions is required to regulate actin polymerization in Drosophila bristle. Mol Biol Cell. 2003;14:118–128. doi: 10.1091/mbc.E02-05-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Pring M, Yang C, Taoka M, Zigmond SH. Presence of a novel inhibitor of capping protein in neutrophil extract. Cell Motil Cytoskeleton. 2005;62:232–243. doi: 10.1002/cm.20097. [DOI] [PubMed] [Google Scholar]
- Huang M, Yang C, Schafer DA, Cooper JA, Higgs HN, Zigmond SH. Cdc42-induced actin filaments are protected from capping protein. Curr Biol. 1999;9:979–982. doi: 10.1016/s0960-9822(99)80428-x. [DOI] [PubMed] [Google Scholar]
- Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, Cooper JA. Capping protein levels influence actin assembly and cell motility in Dictyostelium. Cell. 1995;81:591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Hurst S, Howes EA, Coadwell J, Jones R. Expression of a testis-specific putative actin-capping protein associated with the developing acrosome during rat spermiogenesis. Molecular Reproduction & Development. 1998;1:81–91. doi: 10.1002/(SICI)1098-2795(199801)49:1<81::AID-MRD9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hutchings NJ, Clarkson N, Chalkley R, Barclay AN, Brown MH. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J Biol Chem. 2003;25:22396–22403. doi: 10.1074/jbc.M302540200. [DOI] [PubMed] [Google Scholar]
- Imai H, Narita A, Schroer TA, Maeda Y. Two-dimensional averaged images of the dynactin complex revealed by single particle analysis. J Mol Biol. 2006;359:833–839. doi: 10.1016/j.jmb.2006.03.071. [DOI] [PubMed] [Google Scholar]
- Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;5:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- Isenberg G, Aebi U, Pollard TD. A novel actin binding protein from Acanthamoeba which regulates actin filament polymerization and interactions. Nature. 1980;288:455–459. doi: 10.1038/288455a0. [DOI] [PubMed] [Google Scholar]
- Ivanenkov VV, Dimlich RVW, Jamieson GA. Interaction of S100a(0) protein with the actin capping protein, CapZ - Characterization of a putative S100a(0) binding site in CapZ-Alpha-Subunit. Biochemical & Biophysical Research Communications. 1996;1:46–50. doi: 10.1006/bbrc.1996.0542. [DOI] [PubMed] [Google Scholar]
- Ivanenkov VV, Jamieson GA, Jr, Gruenstein E, Dimlich RV. Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J Biol Chem. 1995;24:14651–14658. doi: 10.1074/jbc.270.24.14651. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janody F, Treisman JE. Actin capping protein alpha maintains vestigial-expressing cells within the Drosophila wing disc epithelium. Development. 2006;133:3349–3357. doi: 10.1242/dev.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., 3rd The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;7:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kim K, Galletta BJ, Schmidt KO, Chang FS, Blumer KJ, Cooper JA. Actin-based Motility during Endocytosis in Budding Yeast. Mol Biol Cell. 2006;17:1354–1363. doi: 10.1091/mbc.E05-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, McCully ME, Bhattacharya N, Butler B, Sept D, Cooper JA. Structure/Function Analysis of the Interaction of Phosphatidylinositol 4,5-Bisphosphate with Actin-capping Protein: Implications for How Capping Protein Binds the Actin Filament. J Biol Chem. 2007;282:5871–5879. doi: 10.1074/jbc.M609850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Yamashita A, Wear MA, Maeda Y, Cooper JA. Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J Cell Biol. 2004;4:567–580. doi: 10.1083/jcb.200308061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Kessels MM, Cope MJ, Drubin DG. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;6753:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, Corthals G, Cordwell S, Daly RJ. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem. 2003;24:21805–21813. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- Maruyama K. Effect of beta-actinin on the particle length of F-actin. Biochim Biophys Acta. 1966;126:389–398. doi: 10.1016/0926-6585(66)90076-8. [DOI] [PubMed] [Google Scholar]
- Maruyama K. beta-Actinin, Cap Z, connectin and titin: what’s in a name? Trends Biochem Sci. 2002;27:264–266. doi: 10.1016/s0968-0004(02)02068-6. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Kimura S, Ishi T, Kuroda M, Ohashi K. beta-actinin, a regulatory protein of muscle. Purification, characterization and function. J Biochem (Tokyo) 1977;81:215–232. doi: 10.1093/oxfordjournals.jbchem.a131438. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Obinata T. Presence of beta-actinin in the soluble fraction of the muscle cells of the chick embryo. J Biochem (Tokyo) 1965;57:575–577. doi: 10.1093/oxfordjournals.jbchem.a128118. [DOI] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery; pivotal role of the filament barbed end. Cell. 2004;3:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JM, Onodera Y, Mazaki Y, Miyoshi H, Hashimoto S, Sabe H. CIN85, a Cbl-interacting protein, is a component of AMAP1-mediated breast cancer invasion machinery. EMBO J. 2007;26:647–656. doi: 10.1038/sj.emboj.7601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita A, Maeda Y. Molecular determination by electron microscopy of the actin filament end structure. J Mol Biol. 2007;365:480–501. doi: 10.1016/j.jmb.2006.06.056. [DOI] [PubMed] [Google Scholar]
- Narita A, Takeda S, Yamashita A, Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 2006;25:5626–5633. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson-Dykstra S, Higgs HN, Harris ES. Actin dynamics: growth from dendritic branches. Curr Biol. 2005;15:R346–57. doi: 10.1016/j.cub.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Olsten ME, Canton DA, Zhang C, Walton PA, Litchfield DW. The Pleckstrin homology domain of CK2 interacting protein-1 is required for interactions and recruitment of protein kinase CK2 to the plasma membrane. J Biol Chem. 2004;40:42114–42127. doi: 10.1074/jbc.M407628200. [DOI] [PubMed] [Google Scholar]
- Paavilainen VO, Hellman M, Helfer E, Bovellan M, Annila A, Carlier MF, Permi P, Lappalainen P. Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc Natl Acad Sci U S A. 2007;104:3113–3118. doi: 10.1073/pnas.0608725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren S, Vartiainen M, Lappalainen P. Twinfilin, a molecular mailman for actin monomers. J Cell Sci. 2002;Pt 5:881–886. doi: 10.1242/jcs.115.5.881. [DOI] [PubMed] [Google Scholar]
- Pappas CT, Bhattacharya N, Cooper JA, Gregorio CC. Nebulin Interacts with CapZ and Regulates Thin Filament Architecture within the Z-disc. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-07-0690. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert K, Olszewski TE, Bowers MB, Dimitrova M, Ginsburg A, Hammer JA., 3rd CARMIL is a bona fide capping protein interactant. J Biol Chem. 2004;4:3068–3077. doi: 10.1074/jbc.M308829200. [DOI] [PubMed] [Google Scholar]
- Remmert K, Vullhorst D, Hinssen H. In vitro refolding of heterodimeric CapZ expressed in E. coli as inclusion body protein. Protein Expr Purif. 2000;1:11–19. doi: 10.1006/prep.1999.1132. [DOI] [PubMed] [Google Scholar]
- Revenu C, Courtois M, Michelot A, Sykes C, Louvard D, Robine S. Villin severing activity enhances actin-based motility in vivo. Mol Biol Cell. 2007;18:827–838. doi: 10.1091/mbc.E06-05-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Cooper JA. Control of actin assembly at filament ends. Annu Rev Cell Dev Biol. 1995;11:497–518. doi: 10.1146/annurev.cb.11.110195.002433. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Hug C, Cooper JA. Inhibition of CapZ during myofibrillogenesis alters assembly of actin filaments. J Cell Biol. 1995;1:61–70. doi: 10.1083/jcb.128.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Korshunova YO, Schroer TA, Cooper JA. Differential localization and sequence analysis of capping protein b-subunit isoforms of vertebrates. J Cell Biol. 1994;2:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Waddle JA, Cooper JA. Localization of CapZ during myofibrillogenesis in cultured chicken muscle. Cell Motil Cytoskeleton. 1993;4:317–335. doi: 10.1002/cm.970250403. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Shih NY, Li J, Karpitskii V, Nguyen A, Dustin ML, Kanagawa O, Miner JH, Shaw AS. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 1999;5438:312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- Sizonenko GI, Karpova TS, Gattermeir DJ, Cooper JA. Mutational analysis of capping protein function in Saccharomyces cerevisiae. Mol Biol Cell. 1996;1:1–15. doi: 10.1091/mbc.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno Y, Abe H, Kimura S, Maruyama K, Obinata T. Generation of functional beta-actinin (CapZ) in an E coli expression system. J Muscle Res Cell Motil. 1998;6:639–646. doi: 10.1023/a:1005329114263. [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Blanchoin L. Actin dynamics: old friends with new stories. Curr Opin Plant Biol. 2006;9:554–562. doi: 10.1016/j.pbi.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Taoka M, Ichimura T, Wakamiya-Tsuruta A, Kubota Y, Araki T, Obinata T, Isobe T. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J Biol Chem. 2003;278:5864–5870. doi: 10.1074/jbc.M211509200. [DOI] [PubMed] [Google Scholar]
- Uruno T, Remmert K, Hammer JA. CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J Biol Chem. 2006;281:10635–10650. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Uetrecht AC, Shen F, Jacobson K, Bear JE. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc Natl Acad Sci U S A. 2007;104:6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bulow M, Rackwitz HR, Zimbelmann R, Franke WW. CP b-3, a Novel Isoform of an Actin-Binding Protein, is a Component of the Cytoskeletal Calyx of the Mammalian Sperm Head. Experimental Cell Research. 1997;1:216–224. doi: 10.1006/excr.1997.3564. [DOI] [PubMed] [Google Scholar]
- Wear MA, Cooper JA. Capping protein binding to S100B: implications for the tentacle model for capping the actin filament barbed end. J Biol Chem. 2004a;279:14382–14390. doi: 10.1074/jbc.M313412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci. 2004b;8:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wear MA, Schafer DA, Cooper JA. Actin dynamics: Assembly and disassembly of actin networks. Curr Biol. 2000;24:R891–R895. doi: 10.1016/s0960-9822(00)00845-9. [DOI] [PubMed] [Google Scholar]
- Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA. How capping protein binds the barbed end of the actin filament. Curr Biol. 2003;13:1531–1537. doi: 10.1016/s0960-9822(03)00559-1. [DOI] [PubMed] [Google Scholar]
- Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, Labeit S. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Mitchelhill KI, Kobe B, Kemp BE, Zot HG. The myosin-I-binding protein Acan125 binds the SH3 domain and belongs to the superfamily of leucine-rich repeat proteins. Proc Natl Acad Sci U S A. 1997;8:3685–3690. doi: 10.1073/pnas.94.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zot AS, Zot HG. Identification of Acan125 as a myosin-I-binding protein present with myosin-I on cellular organelles of Acanthamoeba. J Biol Chem. 1995;43:25316–25319. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Maéda K, Maéda Y. Crystal structure of CapZ: Structural basis for actin filament barbed end capping. EMBO J. 2003;7:1529–1538. doi: 10.1093/emboj/cdg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;2:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;20:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]


