Abstract
G protein-coupled receptors comprise the largest family of eukaryotic signal transduction proteins that communicate across the membrane. We report the crystal structure of a human β2-adrenergic receptor—T4 lysozyme fusion protein bound to the partial inverse agonist carazolol at 2.4 Å resolution. The structure provides a high-resolution view of a human G protein-coupled receptor bound to a diffusible ligand. Ligand-binding site accessibility is enabled by the second extracellular loop which is held out of the binding cavity by a pair of closely spaced disulfide bridges and a short helical segment within the loop. Cholesterol, a necessary component for crystallization, mediates an intriguing parallel association of receptor molecules in the crystal lattice. Although the location of carazolol in the β2-adrenergic receptor is very similar to that of retinal in rhodopsin, structural differences in the ligand binding site and other regions highlight the challenges in using rhodopsin as a template model for this large receptor family.
Introduction
G protein-coupled receptors (GPCRs) comprise the largest integral membrane protein family in the human genome, with over one thousand members (1, 2). These receptors actively participate in the transduction of signals across cellular membranes in response to an astonishing variety of extracellular stimuli, including light, proteins, peptides, small molecules, hormones, protons and ions. Once activated, GPCRs trigger a cascade of intracellular responses, primarily through interactions with their cognate heterotrimeric G proteins, although G protein independent signaling pathways have also been described (3-5). GPCRs are major contributors to the information flow into cells and, as such, are associated with a multitude of diseases that make members of this family important pharmacological targets (6).
GPCRs have been grouped into five classes (2) based on sequence conservation, with class A being the largest and most studied. Class A receptors are further divided into groups associated with particular ligand specificity, such as the opsin, amine, peptide, cannabinoid, and olfactory receptors. Historically, the adrenergic receptors in the amine group are some of the most thoroughly investigated of the class A GPCRs (7-12), and are composed of two main subfamilies, α and β, which differ in tissue localization and ligand specificity, as well as in G protein coupling and downstream effector mechanisms (13). Genetic modifications of adrenergic receptors are associated with diseases as diverse as asthma, hypertension, and heart failure (14). β2-adrenergic receptors (β2ARs) reside predominantly in smooth muscle throughout the body, and β2AR agonists are used in the treatment of asthma and preterm labor (15-17).
Despite extensive efforts, structural information for only one member of the eukaryotic GPCR family, bovine rhodopsin, is available to date (18-21). Rhodopsin is unusual in that it is highly abundant from natural sources and structurally stabilized by the covalently bound ligand 11-cis-retinal, which maintains the receptor in a dark-adapted, non-signaling conformation. In contrast, all other GPCRs are activated by diffusible ligands and are expressed at relatively low levels in native tissues. These receptors are structurally more flexible and equilibrate among multiple conformational states, some of which are prone to instability (22). While the structure determination of rhodopsin was important, many questions remain on the conformational changes between different activation states for each receptor, as well as the structural differences amongst receptors that accommodate the very large diversity of ligands. Specifically: (i) What structural features enable GPCRs to recognize and bind diffusible ligands? (ii) How structurally conserved are the class A GPCRs, and what is the importance of both similarities and differences?
To address these questions, we modified the human β2AR to facilitate the growth of diffraction quality crystals by inserting T4-lysozyme (T4L) in place of the third intracellular loop (β2AR-T4L) and solved the three-dimensional crystal structure in the presence of a partial inverse agonist carazolol (2-propanol, 1-(9H-carbazol-4-yloxy)-3-[(1-methylethyl)amino]) at 2.4 Å resolution (23, 24). We provide a comprehensive analysis of the crystal packing and intramolecular contacts between the β2AR and T4L to identify potential receptor perturbing interactions. The overall receptor topology and the ligand binding pocket are described, as are the main similarities and differences between β2AR-T4L and rhodopsin, and the implications for modeling of other GPCR-ligand complexes.
Structure determination
The engineering, functional properties, expression and purification of crystallization grade β2AR-T4L protein are described fully in the companion paper (25, 26). Briefly, β2AR-T4L was expressed in Sf9 insect cells, solubilized in 1 % dodecylmaltoside, and purified by sequential antibody and ligand affinity chromatography. Following the reported success with microbial rhodopsins in lipidic cubic phase (LCP) (27), we were able to produce crystals of β2AR-T4L that diffract to a resolution of 2.2 Å with a modified LCP procedure, and to solve and refine the structure at 2.4 Å resolution (28). Compared to crystallization in detergents, LCP provides a more native, lipid environment for crystallization, as well as a confinement of protein molecules to two-dimensional membrane sheets that may facilitate the crystallization process through the formation of Type I packing interactions (29-31). In agreement with prior biological evidence that cholesterol improves β2AR stability (32) and may mediate receptor-receptor interactions, crystals were grown from a cholesterol-doped monoolein cubic phase. An automated, nanovolume LCP crystallization protocol (33) significantly reduced the time and amount of protein required for the exhaustive, multi-dimensional optimization trials needed to arrive at these conditions. Crystals of b2AR-T4L were also obtained in lipid bicelles, but they did not diffract as well as those obtained in LCP (28).
Diffraction data for β2AR-T4L were measured to a resolution of 2.4 Å from a total of 27 microcrystals (average size 30 × 15 × 5 μm) using a high intensity, highly parallel minibeam with a diameter of 10 microns at the GM/CA-CAT beamline of the Advanced Photon Source, Argonne National Laboratory (34). Phase information was obtained by molecular replacement using both T4 lysozyme (PDB ID Code 2LZM) and a polyalanine model of the transmembrane regions of rhodopsin (PDB ID Code 1U19) as search models. Additional crystallization, data collection, processing, and refinement statistics are reported in Table 1 and discussed in detail in (28).
Table 1.
Data collection and refinement statistics
| β2AR-T4L | |
|---|---|
| Data collection (APS GM/CA CAT 23ID-B, 10 μm beam) * | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 106.32, 169.24, 40.15 |
| β (°) | 105.62 |
| No. of reflections processed | 245,571 |
| No. unique reflections | 26,574 |
| Resolution (Å) | 50 - 2.4 (2.5 - 2.4) |
| Rsym | 12.7 (67.8) |
| Mean I/σ(I) | 9.6 (2.2) |
| Completeness (%) | 99.5 (99.1) |
| Redundancy | 9.4 (4.8) |
| Refinement* | |
| Resolution (Å) | 20 - 2.4 (2.46 - 2.4) |
| No. reflections (test set) | 25,247 (1,310) |
| Rwork / Rfree | 19.8(27.0) / 23.2(30.1) |
| No. atoms | 3,805 |
| Protein | 3,544 |
| Ions, lipids, ligand and other | 213 |
| Water | 48 |
| Overall B-values (Å2) | 82 |
| β2AR | 77 |
| T4-Lysozyme | 75 |
| Carazolol | 55 |
| Lipid | 100 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.5 |
| Ramachandran plot statistics (%) (excl. Gly, Pro): | |
| Most favored regions | 94.8 |
| Additionally allowed regions | 5.0 |
| Generously allowed regions | 0.2 |
| Disallowed regions | 0 |
Highest resolution shell is shown in parenthesis.
Rsym = Σhkl |I(hkl) - <I(hkl)>| / Σhkl(hkl), where <I(hkl)> is the mean of the symmetry equivalent reflections of I(hkl).
Overall receptor topology
The final model of β2AR-T4L includes 442 amino acids. The model also includes a palmitic acid covalently bound to Cys341 and an acetamide molecule bound to Cys2656.27 (throughout the text residues are designated by their position within the β2AR sequence and their Ballesteros-Weinstein designation as a superscript where applicable) (35, 36), as well as one carazolol molecule, three cholesterol molecules, two sulfate ions and two butanediol molecules that interact with β2AR. There are also four sulfate ions, a putative disaccharide (modeled as maltose) and a molecule of PEG 400 bound to T4L. For β2AR, excellent electron density is observed for residues 29-342, including the ligand carazolol and the two disulfide bonds Cys1063.25-Cys1915.30 and Cys1844.76-Cys1905.29. The palmitic acid at Cys341 is clearly visible in Fo-Fc omit maps; however, the quality of the electron density is lower than for the rest of the receptor. The N-terminus (residues 1 to 28) and the majority of the C-terminus (residues 343 to 365) are disordered and not visible in the structure.
The β2AR has a fold composed of seven transmembrane helices forming a helical bundle (Figure 1A). The residues that make up the helices (I to VII) in β2AR are as follows: helix I 291.28 to 601.59, helix II 672.38 to 962.67, helix III 1033.22 to 1363.55, helix IV 1474.39 to 1714.63, helix V 1975.36 to 2295.68, helix VI 2676.29 to 2986.60, and helix VII 3057.32 to 3287.55. The residues forming the intracellular loops (ICL) and extracellular loops (ECL) of β2AR are: ICL1 611.60 to 662.37, ECL1 972.68 to 1023.21, ICL2 1373.56 to 1464.38, ECL2 1724.64 to 1965.35, ICL3 2305.69 to 2666.28 (residues 231 to 262 are replaced by T4-lysozyme residues 2 to 161), and ECL3 2996.61 to 3047.31. Helices II, V, VI and VII each have a proline-induced kink at conserved positions along the span of the transmembrane segments. These kinks are thought to enable the structural rearrangements required for activation of G protein effectors (37). In addition to the seven membrane spanning helices, β2AR possesses two other helical segments: helix VIII, which is believed to be common to all rhodopsin-like GPCRs (38), and an unexpected, short helical segment in the middle of ECL2, which is not present in rhodopsin, and was not predicted by computational secondary structure analysis (Figure 1A).
Figure 1.
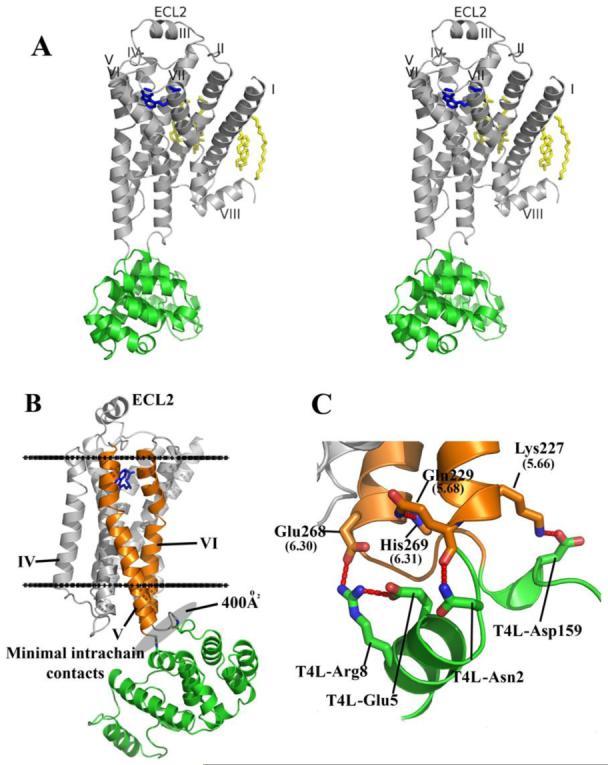
Overall fold of the β2AR-T4L fusion with its predicted orientation in the plasma membrane and key intramolecular interactions. A. Stereoview of the overall fold of β2AR-T4L. The receptor and T4L are colored gray and green, respectively. Carazolol is colored blue and the lipid molecules bound to the receptor are colored yellow. B. The receptor is aligned to a rhodopsin model that was positioned in a lipid membrane (boundaries indicated by horizontal black lines) as found in the orientations of proteins in membranes (OPM) database (74). T4L is fused internally into the third intracellular loop of β2AR and maintains minimal intramolecular packing interactions by tilting away from the receptor. C. Specific intramolecular interactions between β2AR and T4L are represented.
In the β2AR-T4L construct, T4L is fused to the truncated cytoplasmic ends of helices V and VI. In the crystal structure, the T4L moiety is tilted slightly away from the center axis of β2AR drawn normal to the membrane (Figure 1B). As a result, interactions between T4L and β2AR are minimal, with only 400Å2 of surface area buried between them. The intramolecular contacts between T4L and β2AR include salt bridges between the side chains of T4L-Asp159 and the side-chain amine of β2AR-Lys2275.66 (distance 3.4 Å) and between the guanidinium group of T4L-Arg8 with the side-chain carboxyl of β2AR-Glu2686.30 on helix VI (distance 3.2 Å) (Figure 1C, Table S2). The latter interaction is noteworthy, as in rhodopsin, Glu6.30 forms an ionic bond with Arg3.50 of the conserved D(E)RY motif (18). This interaction is postulated to be important for maintaining rhodopsin in the inactive state, but the charged groups of the two residues [Arg1313.50 (NH1) and Glu2686.30 (OE1)] are 10 Å apart in the β2AR-T4L structure. Possible functional implications of this disruption are discussed in the companion manuscript (26). The remainder of the lysozyme molecule provides important crystal packing interactions, but does not appear to influence significantly the receptor structure.
Crystal packing interactions
The β2AR-T4L protein is packed in a C-centered monoclinic lattice with one molecule per asymmetric unit (Figure 2A). As observed in all previous lipidic mesophase grown crystals (39), the β2AR-T4L crystals adopt Type I packing (40), featuring a multilayered arrangement in accordance with proposed crystallization mechanism (29, 41). Within each layer, protein molecules form arrays of parallel, symmetry-related dimers. There are four distinct crystal-packing interactions within each layer, three of which are mediated by T4L. The fourth interaction in the array is between two receptor molecules related by a crystallographic two-fold rotation axis. This is the sole interaction between symmetry-related receptors, and is mediated primarily by ordered lipids consisting of six cholesterol and two palmitic acid molecules, the latter being covalently attached to Cys341 in the C-terminal portion of the receptor (42) (Figure 2B). These eight lipid molecules form a two-fold symmetric sheet between receptors. The only direct receptor-receptor contact involves a 2.7 Å pair of ionic interactions between the charged amine group of Lys601.59 in helix I and the carboxylate of Glu338 in helix VIII from the symmetry-related receptor. Remarkably, of the 515 Å2 buried at the receptor symmetry interface, 73% of the crystal contact surface area is mediated by ordered lipid, while only 27% is contributed by protein-protein contacts. The stacking interactions between layers are formed between T4L and extracellular loops ECL2 and ECL3 of the receptor (Figure 2A). It is unlikely that these contacts affect the orientation of these loops due to the small size of ECL3 and the rigid architecture of ECL2.
Figure 2.
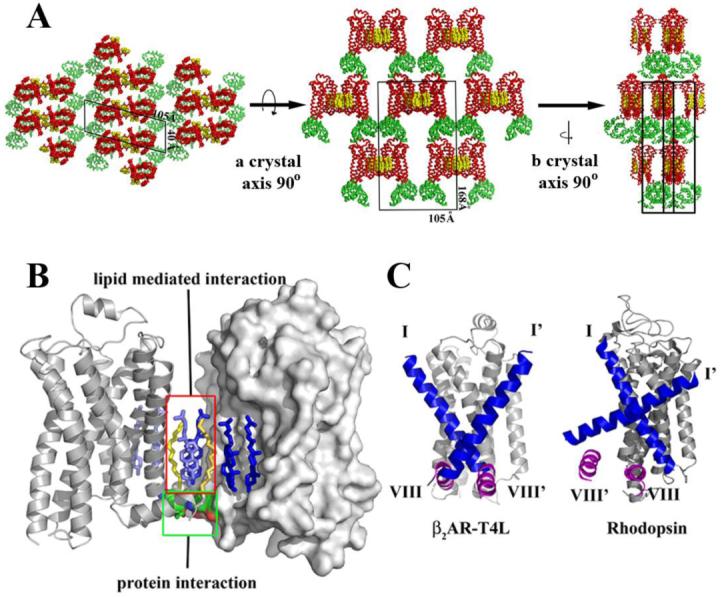
Crystal packing interactions in the lipidic mesophase crystallized β2AR-T4L. A. There are four main contact areas, two of which are mediated by T4L in the plane of the membrane with itself through a two-fold symmetry axis and translation. The third interaction is normal to the membrane plane between T4L and lumen exposed loops of β2AR. The fourth interaction is generated by the two-fold symmetry axis, packing one receptor to receptor in the plane of the membrane. B. The receptor crystal packing interface is composed mainly of lipids with two cholesterol molecules and two palmitic acid molecules forming the majority of the interactions. A network of ionic charge interactions exists on the cytoplasmic end of the interface forming the only inter-receptor protein contacts. C. Comparison between β2AR-T4L and rhodopsin (PDB ID Code 2I35) parallel receptor association interface. Helices I (blue) and VIII (magenta) are highlighted in both structures. Only one monomer is shown for each receptor representation along with helices I’ and VIII’ only from the opposing symmetry related molecule. The rhodopsin interface is twisted significantly relative to β2AR-T4L resulting in a significant offset from the parallel orientation required for a physiological dimer interface. β2AR-T4L associated monomers are in a highly parallel orientation.
Lipid mediated receptor association
Many GPCRs including β2AR are thought to exist as dimers in the plasma membrane, although the location of the dimer interface and the functional significance of dimerization is not clear (43). The observation of ordered lipids in the helix I and VIII interface between two symmetry related molecules makes it tempting to speculate on the physiological relevance of this association (44-46). Associations between the equivalent regions of rhodopsin have been found in crystal structures (21, 47) (Figure 2C). On the other hand, studies in native membranes suggest that helix VI may form the dimer interface for the β2AR (48), and helix IV may form the dimer interface for the closely related D2 dopamine receptor (49).
While the role of cholesterol in promoting β2AR association is speculative, its role in the physiologic function of β2AR is well documented. Depletion of cholesterol from the membranes of neonatal cardiac myocytes alters the signaling behavior of endogenous β2AR (50). In untreated cells, activation of β2AR results in sequential coupling to the G proteins Gs and Gi, producing a biphasic effect on myocyte contraction rate. Upon depletion of cholesterol, the β2AR couples more strongly to Gs. This effect may be due to a role of cholesterol in regulating interactions between the β2AR and G proteins, or possibly to the effect of cholesterol on β2AR dimerization. The β2AR couples efficiently to Gs as a monomer (51), so it is possible that cholesterol mediated association (dimerization) reduces the efficiency of β2AR coupling to Gs. The effects of cholesterol depletion on β2AR signaling may also be a secondary effect of altering subcellular signaling compartments. There is evidence that cells may concentrate signaling molecules, such as GPCRs and their cognate G proteins, by way of membrane microdomains or compartments, such as caveolae (52). This compartmentalization may be a major regulator of receptor-effector coupling. Thus, the importance of cholesterol in forming the observed crystallographic association is consistent with its role in β2AR signaling. Additional experiments will be required to determine how relevant the association of monomers observed in the crystal is to β2AR packing within membrane microdomains.
Electrostatic charge distribution
Electrostatic charge distribution was calculated using APBS (53) and mapped onto a molecular surface representation of β2AR. The analysis reveals three polarized areas within the molecule (Figure 3A). First, the cytoplasmic face of the receptor is involved in G protein interaction and carries a net positive charge even in the absence of ICL3, which also has a predicted overall positive charge (Figure 3B). The second site is an electrostatically negative region located within the membrane between helices III, IV and V potentially exposed to the lipid alkyl chains, which is unexpected as the burial of charge within the plasma membrane is thermodynamically unfavorable. A glutamate residue at position 1223.41 may partially account for the observed charge distribution. Finally, the binding site cleft is negatively charged and exposed to solvent by an unusual ECL2 architecture and lack of N-terminal interactions. This negative charge may facilitate ligand binding through electrostatic funneling of positively charged catecholamines (Figure 3B).
Figure 3.
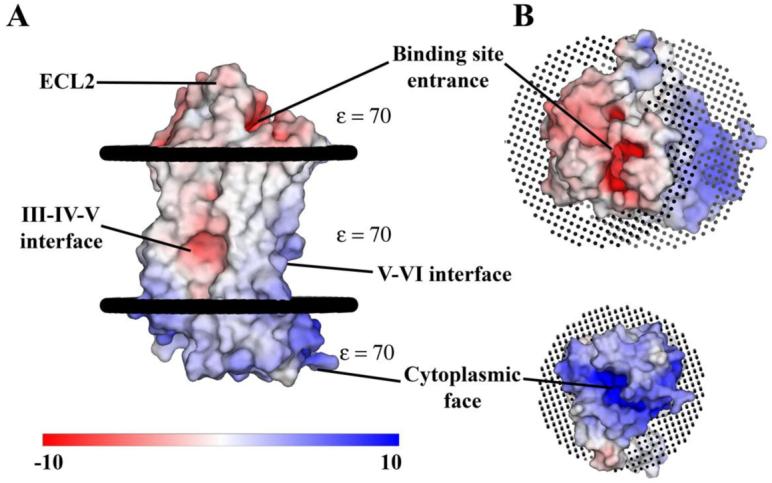
Surface representation of β2AR colored by calculated charge from red (-10 kbT/ec) to blue (+10 kbT/ec) using a dielectric constant of 70. A. Three main areas of interest are indicated. The binding site cleft is negatively charged as is a groove between helices III, IV and V. The third region is an overall positive charge in the region of the ionic lock and DRY motif on the cytoplasmic face. The overall result is a highly polarized molecule that may utilize its negative charge to facilitate binding of catecholamine ligands. The presence of a negative charge in the groove between helices III, IV and V is unexpected as it is in the middle of the lipid membrane. This charge may be partially derived from the presence of an unpaired glutamate at position 1223.41. The effective charge in this region is likely greater than shown here due to its location in the low dielectric environment of the lipid membrane. B. View rotated 90° from A. Showing both the negatively charged binding site cleft (top) and positively charged cytoplasmic face (bottom). Poisson-Boltzmann electrostatics were calculated using the program APBS (53) as implemented in Pymol (75). Pymol was used exclusively in the preparation of all figures.
Extracellular region
The ECLs and amino termini of GPCRs, together with the extracellular halves of the transmembrane helices, are believed to define the ligand-binding site of each receptor (44). Therefore, the ECLs play an important role in the overall pharmacology of any particular receptor. In general, small molecule ligands are thought to bind deeper within the space created by the transmembrane domain helices, whereas larger ligands such as peptides bind closer to the membrane surface near the ECLs (54, 55). Mutagenesis studies suggest that the β2AR binds its ligand deep within the transmembrane helix bundle, which may be related to the observation that the extracellular regions have a rather simple structure with short loops connecting transmembrane helices II and III, and VI and VII (Figure 4A). ECL2, which links helices IV and V, has a somewhat more extensive architecture that is unanticipated. In contrast to the buried, β-sheet structure of this loop in rhodopsin (Figure 4B), ECL2 in β2AR is more exposed to the solvent and contains an extra helical segment. Additionally, there is an intra-loop disulfide bond between Cys1844.76 and Cys1905.29 that may help stabilize the more exposed ECL2. A second disulfide bond between Cys1915.30 and Cys1063.25 in helix III effectively ties ECL2 to the transmembrane core (56). The distal portion of ECL2 makes close contacts with ECL1 and contains a glycosylation site at Asn1875.26 (57), which may serve to mask a grouping of aromatic residues on ECL1; in this construct, Asn1875.26 has been mutated to glutamate to aid in crystallization.
Figure 4.
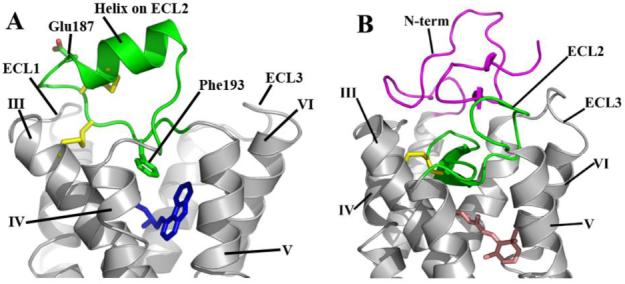
Comparison of the extracellular sides of β2AR-T4L and rhodopsin. A. The N-terminus is missing from the experimental density in the β2AR-T4L structure and is not shown. ECL2 is shown in green and contains a short α-helix and two disulfide bonds (yellow). The intraloop disulfide bond constrains the tip of ECL2 which interacts with ECL1. The second disulfide bond links ECL2 with helix III. There is one interaction between ECL2 and carazolol (blue) through Phe1935.32. The entire loop is held out of the ligand binding site by a combination of the rigid helical segment and the two disulfide bonds. B. In contrast, ECL2 (green) in rhodopsin assumes a lower position in the structure that occludes direct access to the retinal-binding site and forms a small β-sheet in combination with the N-terminal region (magenta) directly above the bound retinal (pink).
Electron density corresponding to the N-terminus was not apparent in the maps and, therefore, residues 1-28 are not included in the model. This disorder contrasts with rhodopsin, in which the N-terminus interacts extensively with the ECLs, forming a small four-strand β-sheet in conjunction with ECL2. This sheet structure forms a cap that effectively isolates the retinal binding site in a hydrophobic pocket (Figure 5B). The lack of interactions between the N-terminus of β2AR and ECL2 further enables diffusible ligand access to the binding site. However a completely disordered N-terminus may be an artifact induced by the presence of the N-terminal Flag tag which carries an overall positive charge and may disrupt N-terminal interactions.
Figure 5.
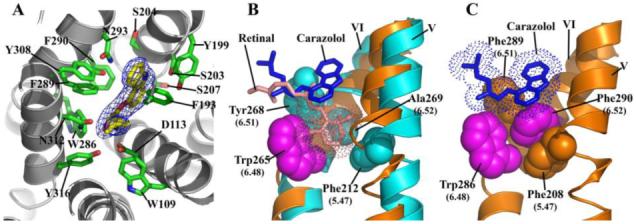
Ligand binding characterization and comparison to rhodopsin. A. A view looking down on the plane of the membrane from the extracellular surface showing a detailed representation of the carazolol binding site in β2AR-T4L. Carazolol is shown as sticks with carbon atoms colored yellow. β2AR-T4L residues contributing to carazolol binding are shown in green and labeled. Electron density is contoured at 5σ from an Fo-Fc omit map calculated without the contribution of carazolol. B. Binding orientation comparison between 11-cis-retinal in rhodopsin and carazolol in β2AR-T4L. Van der Waals’ surfaces for carazolol and retinal are represented as dots to accentuate the close packing interactions. Retinal in the all-cis conformation (pink), binds deep in the active site of rhodopsin as compared to carazolol (blue), packing its β-ionone ring between Tyr2686.51 and Phe2125.47 (cyan), blocking movement of Trp2656.48 (magenta) into the space. The β-ionone ring of trans-retinal in activated rhodopsin would not block Trp2656.48 from rotating into the space allowing a rotameric shift into its proposed active form. C. There are four residues involved in the toggle switch mechanism of β2AR-T4L as shown. Phe2906.52 (magenta) is sandwiched between Phe2085.47 (tan) and Phe2896.51 (tan) forming a ring-face aromatic interaction. Like rhodopsin, an activation step is thought to occur by a rotameric change of Trp2866.48 (magenta) which would displace Phe2906.52. Carazolol is shown to interact extensively with the sandwich motif as shown: however, few interactions are seen with Trp2866.48. The 6.52 position in β2AR-T4L is occupied by Phe2906.52 as opposed to Ala2696.52 in rhodopsin where the β-ionone ring replaces an aromatic protein side chain in forming the sandwich interactions. The aromatic character of the sandwich is otherwise maintained by Phe2896.51 and Phe2085.47 in β2AR-T4L.
The short helical region on ECL2 adds a rigid structural element that, along with the two disulfide bonds, constrains the loop to a small range of conformations and helps stabilize the receptor by linking three transmembrane helices (Figure 5A). This rigid conformation may help to stabilize the core of the receptor and lock ECL2 in a conformation that does not hinder access to the binding pocket.
Ligand binding site and comparison to rhodopsin
Carazolol is a partial inverse agonist that binds with picomolar affinity to β2AR-T4L producing a reduction of the basal activity of the receptor (24). The crystal structure reveals extensive interactions between the receptor and carazolol that position the carbazole moiety adjacent to Phe2896.51, Phe2906.52, and Trp2866.48 (Figure 5A, S1 and Table S3). In contrast, cis-retinal is a full inverse agonist covalently bound to rhodopsin, which suppresses all activity towards transducin (58). Carazolol and retinal occupy similar spaces in their respective receptors, with significant overlap of the non-aromatic regions of carazolol. However, the β-ionone ring of retinal extends deep into the binding pocket of rhodopsin and contacts residues on helix V and VI, where it is sandwiched between Phe2125.47 and Tyr2686.51, and interacts with the highly conserved Trp2656.48 (Figure 5B). It has been proposed that changes in the rotamer of Trp2656.48 occur upon activation of rhodopsin and related family members, and constitutes the “toggle switch” for receptor activation (59). Accordingly, the interactions between cis-retinal and Trp2656.48 are likely to contribute to the absence of basal activity in rhodopsin. Carazolol does not interact directly with the toggle switch on helix VI, however it lowers the basal activity of the receptor, and may do so by interacting with Phe2896.51 and Phe2906.52, which form an extended aromatic network surrounding the highly conserved Trp2866.48. As a result, Trp2866.48 adopts the rotamer associated with the inactive state. Thus, the steric constraints imposed by Phe2906.52 appear to structurally mimic the interaction of the β-ionone ring of retinal with the conserved Trp2656.48 and Phe2125.47 on rhodopsin (60) (Figure 5C).
Structural alignment and helix bundle reorganization
It has long been thought that class A GPCRs share a similar architecture due to their predicted seven transmembrane helical bundles and sequence conservation within the membrane spanning regions (61). Nonetheless, given the common ability to activate G proteins, yet the astonishing variety of ligand specificities among the class A receptor family, the similarities and differences in ligand binding modes remains an open question. To this end, we aligned the structure of β2AR-T4L to highest resolution structure of rhodopsin (PDB ID Code 1U19). We used difference distance matrices to select non-divergent areas between the two structures that align to reveal the differences in helix orientation between β2AR-T4L and rhodopsin (62).
Relative to rhodopsin, the following helical shifts are seen in β2AR-T4L: the extracellular portions of helices I and III angle away from the center of the receptor, helix IV is translated away from the center of the receptor, helix V is translated closer to the center of the receptor and helix VI angles away from the receptor on the cytoplasmic end (Figure 6). The largest difference is in helix I, which lacks a proline-induced kink found in rhodopsin and is comparatively straight. The angle between the rhodopsin and β2AR positions of helix I is approximately 18° with a shift of 7 Å at the apex on the extracellular face. This structural difference may arise from the need for an accessible binding site in β2AR, which is provided in part by a lack of interactions between the N-terminus and extracellular loop segments. In contrast the N-terminal region in rhodopsin occludes the retinal-binding site through extensive interactions with the extracellular loops (Figure 4B). Helix V of β2AR is closer to the binding pocket by approximately 3.5 Å on average and its lumenal end is angled more towards helix VI. Helix IV of β2AR is further from the binding site, possibly to remove steric clashes resulting from the modified position of helix V (Figure 6B, C). Helix III pivots further from the binding site about a fulcrum located close to the cytoplasmic end (Figure 6C). The angle formed between rhodopsin helix III and the β2AR helix III is approximately 7°, yielding a 4 Å displacement out of the binding pocket at the cytoplasmic end of the helix. Helix VI is positioned further from the center of the receptor at the cytoplasmic end as compared to rhodopsin, which is caused by a slight difference in the angle about the proline-induced kink in the helix (Figure 6C).
Figure 6.
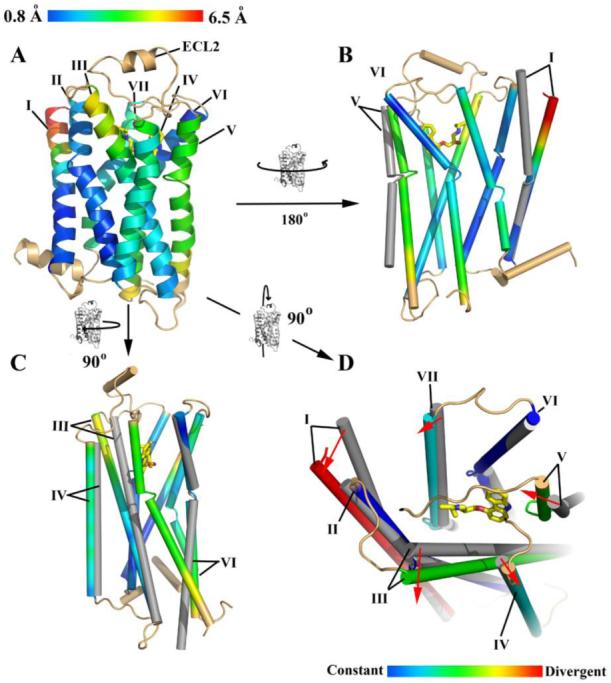
Comparison of β2AR-T4L helical orientations with rhodopsin (PDB ID Code 1U19). A. β2AR-T4L is rendered as a ribbon trace colored with a blue to red spectrum corresponding to observed distances between Cα positions in the two structures (RMSD 2.7 Å between all residues in the transmembrane region). Helix II shows very little movement, whereas the entire lengths of helices III, IV, V shift significantly. Helix VIII and loops were not included in the comparison and are colored in tan. B. Movements of helices I and V of rhodopsin (grey) are shown relative to β2AR-T4L. C. Movements of helices III, IV and VI. D. Ligand binding site representation. Carazolol is shown with yellow carbons. Entire helices are assigned a single designation based on their divergence from the rhodopsin position in the area of the ligand binding site as shown. Helix I is highly divergent, Helices II and VI are similar to rhodopsin. Helices IV and VII are moderately constant. Helices III and V are moderately divergent.
The ligand-binding pocket is formed by both structurally conserved and divergent helices as compared to rhodopsin (Figure 6D). Helices III and V are two of the most conformationally shifted helices and contain the canonical catecholamine binding residues associated with activation of adrenergic family of receptors (63-65). The comparison with rhodopsin suggests that the structurally conserved helices provide a common core present throughout the class A GPCRs, whereas the variable helices confer binding site plasticity with a resulting architecture capable of binding a large spectrum of ligands.
Comparison to rhodopsin-based GPCR models
Since the determination of the inactive dark-state rhodopsin structure (18), a number of homology models of other class A GPCRs have been reported (66-70). Typically, homology models start by alignment of so-called fingerprint motifs that are common among the family. These fingerprint motifs are extrapolated to assign coordinates for the entire helical bundle. Loop regions are either ignored or modeled based on databases of loop conformations depending on the application (66). A number of models exist for β2AR, some of which have been improved upon with supporting biochemical data (66, 70-73). When compared to the β2AR structure reported here, however, all of these models were more similar to rhodopsin, as were models for other receptors (e.g. dopamine, muscarinic, and chemokine) (28). This is not entirely surprising but highlights a general shortcoming in homology models generated from a single structural template. The structural divergence between β2AR and rhodopsin would be quite difficult to predict accurately using only rhodopsin as a template. The addition of a second class A GPCR structure should make it possible to correlate the sequence differences between rhodopsin and β2AR with the observed structural differences and extrapolate to other class A GPCRs. Highlighting interactions that constrain class A receptors into each of the two observed states will allow a more comprehensive analysis of structural divergence and should result in more accurate models. Furthermore, evidence provided in the companion publication (26) indicates that β2AR-T4L may not be in a completely inactive conformation like rhodopsin, providing an alternative signaling state on which to base homology models that will be more relevant for virtual ligand screening and structure-based drug design (66, 73). The addition of further structural templates and conformational states to the pool of information on GPCRs should pave the way to a new generation of more potent therapeutics targeting this expansive receptor family and enhance our understanding of the signaling properties within their associated pathways.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH Roadmap Initiative grant P50 GM073197 and Protein Structure Initiative P50 GM62411 (to R.C.S.) and NIH Roadmap Initiative grant R21 GM075811 and NINDS Grant NS028471 (to B.K.K.). The authors acknowledge the support of Janet Smith, Robert Fischetti and Nukri Sanishvili at the GM/CA-CAT beamline at the Advanced Photon Source, for assistance in development and use of the minibeam and beamtime. The GM/CA-CAT beamline (23-ID) is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). D.M.R. was supported in part by NIH grant F32 GM082028; S.G.F.R. was supported in part by the Lundbeck Foundation; H.-J.C. and W.I.W. were supported in part by NIH grant R01 GM056169. We thank Gebhard Schertler for help with the initial diffraction experiments on LCP crystals performed at ID-13 at the ESRF. The authors acknowledge Chris Roth, Veli-Pekka Jaakola, Alexander Alexandrov, Ellen Chien, Michael Bracey, Vsevolod Katrich, Ian Wilson and Mark Yeager for careful review of the manuscript, Yuan Zheng and Martin Caffrey from The Ohio State University for the generous loan of the in meso robot, and Angela Walker for assistance with manuscript preparation. Coordinates and structure factors have been deposited in the Protein Data Bank with identification code 2RH1.
Footnotes
One sentence summary: A 2.4 Å resolution structure of the human β2-adrenergic receptor reveals a distinct architecture and helical orientation suggestive of a structural plasticity associated with class A GPCRs.
References and Notes
- 1.Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. FEBS Lett. 2002;520:97. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. Mol Pharmacol. 2003;63:1256. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 3.Pierce KL, Premont RT, Lefkowitz RJ. Nat Rev Mol Cell Biol. 2002;3:639. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 4.Lefkowitz RJ, Shenoy SK. Science. 2005;308:512. doi: 10.1126/stke.2005/308/cm10. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y, et al. EMBO J. 2007;26:53. doi: 10.1038/sj.emboj.7601502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drews J. Science. 2000;287:1960. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 7.Kobilka B. Annu Rev Neurosci. 1992;15:87. doi: 10.1146/annurev.ne.15.030192.000511. [DOI] [PubMed] [Google Scholar]
- 8.Caron MG, Lefkowitz RJ. Recent Prog Horm Res. 1993;48:277. doi: 10.1016/b978-0-12-571148-7.50014-2. [DOI] [PubMed] [Google Scholar]
- 9.Strosberg AD. Protein Sci. 1993;2:1198. doi: 10.1002/pro.5560020802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hein L, Kobilka BK. Trends Cardiovasc Med. 1997;7:137. doi: 10.1016/S1050-1738(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 11.Rohrer DK. J Mol Med. 1998;76:764. doi: 10.1007/s001090050278. [DOI] [PubMed] [Google Scholar]
- 12.Xiang Y, Kobilka B. Adrenergic Receptors. 2006:267. [Google Scholar]
- 13.Milligan G, Svoboda P, Brown CM. Biochem Pharmacol. 1994;48:1059. doi: 10.1016/0006-2952(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 14.Taylor MR. Pharmacogenomics J. 2007;7:29. doi: 10.1038/sj.tpj.6500393. [DOI] [PubMed] [Google Scholar]
- 15.Bai TR. Lung. 1992;170:125. doi: 10.1007/BF00174316. [DOI] [PubMed] [Google Scholar]
- 16.Barnes PJ. Life Sci. 1993;52:2101. doi: 10.1016/0024-3205(93)90725-i. [DOI] [PubMed] [Google Scholar]
- 17.Smiley RM, Finster M. J Matern Fetal Med. 1996;5:106. doi: 10.1002/(SICI)1520-6661(199605/06)5:3<106::AID-MFM2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 18.Palczewski K, et al. Science. 2000;289:739. [Google Scholar]
- 19.Okada T, et al. J Mol Biol. 2004;342:571. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. J Mol Biol. 2004;343:1409. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 21.Salom D, et al. Proc Natl Acad Sci U S A. 2006;103:16123. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobilka BK, Deupi X. Trends Pharmacol Sci. 2007;28:397. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Inverse agonists act to reduce the basal activity of a receptor through interactions that shift the equilibrium to more of an inactive state. In contrast, antagonists bind to and block the active site, but do not affect the equilibrium between inactive and active states, and agonists shift the equilibrium to an active receptor state.
- 24.Rasmussen SGF, et al. Submitted to Nature [Google Scholar]
- 25.β2AR-T4L was generated by three distinct modifications to β2AR: (1) a fusion protein was created by replacement of the third intracellular loop with T4L, (2) the carboxyl terminal 48 amino acids were deleted, and (3) a glycosylation site at Asn187 was eliminated through a glutamate substitution. This modified version was created to assist in improved crystal formation.
- 26.Rosenbaum DM, et al. companion paper [Google Scholar]
- 27.Landau EM, Pebay-Peyroula E, Neutze R. FEBS Lett. 2003;555:51. doi: 10.1016/s0014-5793(03)01082-2. [DOI] [PubMed] [Google Scholar]
- 28.Methods are available as supporting material on Science Online.
- 29.Caffrey M. Curr Opin Struct Biol. 2000;10:486. doi: 10.1016/s0959-440x(00)00119-6. [DOI] [PubMed] [Google Scholar]
- 30.Deisenhofer J, Michel H. EMBO J. 1989;8:2149. doi: 10.1002/j.1460-2075.1989.tb08338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau EM, Rosenbusch JP. Proc Natl Acad Sci U S A. 1996;93:14532. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Z, Kobilka B. Anal Biochem. 2005;343:344. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. Acta Crystallogr D Biol Crystallogr. 2004;60:1795. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 34.The successful diffraction screening and data collection that led to the structure determination of β2AR-T4L required overcoming a number of technological barriers that encompassed the growth and harvest of microcrystals, crystal imaging, and the collection of diffraction data. Due to their transparency, crystals were often visually obstructed by the frozen lipidic mesophase material and, therefore, could not be confidently imaged by traditional beamline cameras, while their extremely small size made them susceptible to rapid radiation damage. Further details are provided in (28).
- 35.GPCRs are frequently post-translationally modified with palmitoylate on cysteine residues at the C-terminal tail. β2AR-T4L was treated with iodoacetamide during purification to eliminate free thiols.
- 36.Ballesteros-Weinstein numbering is used throughout the text as superscripts to the protein numbering. Within each helix is a single most conserved residue among the class A GPCRs. This residue is designated x.50 where x is the number of the transmembrane helix. All other residues on that helix are numbered relative to this conserved position.
- 37.Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. Proc Natl Acad Sci U S A. 2004;101:959. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katragadda M, Maciejewski MW, Yeagle PL. Biochim Biophys Acta. 2004;1663:74. doi: 10.1016/j.bbamem.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Cherezov V, Clogston J, Papiz MZ, Caffrey M. J Mol Biol. 2006;357:1605. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 40.Membrane protein generally can form two types of crystal packing: Type I represents stacks of two dimensional crystals ordered in the third dimension via interactions of hydrophilic parts of membrane proteins. Type II crystals are composed of membrane proteins whose hydrophobic part is shielded by a detergent micelle and all crystal contacts are formed through hydrophilic, solvent exposed parts of protein molecules.
- 41.Nollert P, Qiu H, Caffrey M, Rosenbusch JP, Landau EM. FEBS Lett. 2001;504:179. doi: 10.1016/s0014-5793(01)02747-8. [DOI] [PubMed] [Google Scholar]
- 42.O’Dowd BF, Hnatowich M, Caron MG, Lefkowitz RJ, Bouvier M. J Biol Chem. 1989;264:7564. [PubMed] [Google Scholar]
- 43.Milligan G. Mol Pharmacol. 2004;66:1. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 44.Angers S, et al. Proc Natl Acad Sci U S A. 2000;97:3684. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Javitch JA. Mol Pharmacol. 2004;66:1077. doi: 10.1124/mol.104.006320. [DOI] [PubMed] [Google Scholar]
- 46.Mercier JF, Salahpour A, Angers S, Breit A, Bouvier M. J Biol Chem. 2002;277:44925. doi: 10.1074/jbc.M205767200. [DOI] [PubMed] [Google Scholar]
- 47.Schertler GF. Curr Opin Struct Biol. 2005;15:408. doi: 10.1016/j.sbi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Hebert TE, et al. J Biol Chem. 1996;271:16384. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 49.Guo W, Shi L, Javitch JA. J Biol Chem. 2003;278:4385. doi: 10.1074/jbc.C200679200. [DOI] [PubMed] [Google Scholar]
- 50.Xiang Y, Rybin VO, Steinberg SF, Kobilka B. J Biol Chem. 2002;277:34280. doi: 10.1074/jbc.M201644200. [DOI] [PubMed] [Google Scholar]
- 51.Whorton MR, et al. Proc Natl Acad Sci U S A. 2007;104:7682. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ostrom RS, Insel PA. Br J Pharmacol. 2004 Sep;143:235. doi: 10.1038/sj.bjp.0705930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci U S A. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji TH, Grossmann M, Ji I. J Biol Chem. 1998;273:17299. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 55.Gether U. Endocr Rev. 2000;21:90. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 56.Noda K, Saad Y, Graham RM, Karnik SS. J Biol Chem. 1994;269:6743. [PubMed] [Google Scholar]
- 57.Mialet-Perez J, Green SA, Miller WE, Liggett SB. J Biol Chem. 2004;279:38603. doi: 10.1074/jbc.M403708200. [DOI] [PubMed] [Google Scholar]
- 58.Palczewski K. Annu Rev Biochem. 2006;75:743. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Annu Rev Pharmacol Toxicol. 2006;46:481. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 60.Shi L, et al. J Biol Chem. 2002;277:40989. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 61.Lefkowitz RJ. Nat Cell Biol. 2000;2:E133. doi: 10.1038/35017152. [DOI] [PubMed] [Google Scholar]
- 62.For the alignment, residues on β2AR were aligned to equivalent residues on Rhodopsin, respectively: 43-59 to 47-63; 67-95 to 71-99; 122-135 to 126-139; 285-296 to 264-275.
- 63.Strader CD, et al. J Biol Chem. 1988;263:10267. [PubMed] [Google Scholar]
- 64.Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RA. J Biol Chem. 1989;264:13572. [PubMed] [Google Scholar]
- 65.Liapakis G, et al. J Biol Chem. 2000;275:37779. doi: 10.1074/jbc.M002092200. [DOI] [PubMed] [Google Scholar]
- 66.Bissantz C, Bernard P, Hibert M, Rognan D. Proteins. 2003;50:5. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 67.Fano A, Ritchie DW, Carrieri A. J Chem Inf Model. 2006;46:1223. doi: 10.1021/ci050490k. [DOI] [PubMed] [Google Scholar]
- 68.Hobrath JV, Wang S. J Med Chem. 2006;49:4470. doi: 10.1021/jm0501634. [DOI] [PubMed] [Google Scholar]
- 69.Nowak M, Kolaczkowski M, Pawlowski M, Bojarski AJ. J Med Chem. 2006;49:205. doi: 10.1021/jm050826h. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Devries ME, Skolnick J. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freddolino PL, et al. Proc Natl Acad Sci U S A. 2004;101:2736. doi: 10.1073/pnas.0308751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furse KE, Lybrand TP. J Med Chem. 2003;46:4450. doi: 10.1021/jm0301437. [DOI] [PubMed] [Google Scholar]
- 73.Gouldson PR, et al. Proteins. 2004;56:67. doi: 10.1002/prot.20108. [DOI] [PubMed] [Google Scholar]
- 74.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. Bioinformatics. 2006;22:623. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 75.DeLano WL. The PyMOL Molecular Graphics System. 2002 on World Wide Web http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


