Abstract
DLC-1 encodes a Rho GTPase-activating protein (RhoGAP) and negative regulator of specific Rho family proteins (RhoA-C and Cdc42). DLC-1 is a multi-domain protein, with the RhoGAP catalytic domain flanked by an amino-terminal sterile α motif (SAM) and a carboxyl-terminal START domain. The roles of these domains in the regulation of DLC-1 function remain to be determined. We undertook a structure-function analysis involving truncation and missense mutants of DLC-1. We determined that the amino-terminal SAM domain functions as an autoinhibitory domain of intrinsic RhoGAP activity. Additionally, we determined that the SAM and START domains are dispensable for DLC-1 association with focal adhesions. We then characterized several mutants for their ability to regulate cell migration and identified constitutively activated and dominant negative mutants of DLC-1. We report that DLC-1 activation profoundly alters cell morphology, enhances protrusive activity, and can increase the velocity but reduce directionality of cell migration. Conversely, the expression of the amino-terminal domain of DLC-1 acts as a dominant negative and profoundly inhibits cell migration by displacing endogenous DLC-1 from focal adhesions.
Members of the Rho family of small GTPases are intimately involved in many aspects of cell function including cell cycle progression, intracellular trafficking, and control of cell division (1). The well studied Rho GTPases RhoA, Cdc42, and Rac1 are best known for their key roles in regulation of the actin cytoskeleton and cell migration. Thus RhoA is essential to actin stress fiber and focal contact formation, Rac1 is vital to formation of actin networks at the leading edge of migrating cells, and Cdc42 triggers actin filament extension and bundling in filopodia (2).
The activation state of Rho GTPases is regulated primarily by the conjoint effects of Rho-specific guanine nucleotide exchange factors (GEFs)4 and GTPase-activating domains (GAPs). RhoGEFs promote GDP/GTP exchange to favor formation of the active GTP-bound protein, whereas RhoGAPs stimulate the intrinsic weak GTP hydrolysis activity of Rho GTPases and promote formation of the inactive GDP-bound GTPase (1). Although there are 20 members of the Rho family, there is far greater complexity with regards to the regulatory proteins that control GDP/GTP cycling. There are ∼90 human RhoGEFs and 80 RhoGAPs based on genome analysis (1, 3, 4). This diversity reflects the ability of each Rho GTPase to be regulated by a diverse spectrum of extracellular stimuli and in precise temporal and spatial patterns to dictate their divergent roles in cell physiology. In particular, the role and regulation of specific RhoGEFs in controlling Rho GTPase activity has been the subject of intense research evaluation. In contrast, far less attention has been focused on the role and mechanisms by which RhoGAPs may regulate the spatiotemporal activation and function of Rho GTPases.
The human RhoGAP DLC-1 and its rat homolog p122RhoGAP have elicited substantial interest of late. DLC-1 exhibits characteristics of a tumor suppressor gene with a demonstrated role in growth inhibition in many types of carcinomas (5-8). The highly related proteins DLC-2 and DLC-3 have been linked to similar inhibitory effects on cell growth and function (9, 10). Absence of the DLC-1 protein in tumors may be due either to deletion of the gene or to suppression of its expression caused by methylation of the promoter (11). Because activation of Rho GTPases has been associated with human oncogenesis (12), the tumor suppressor function has been attributed to its ability to negatively regulate Rho GTPase activity, although RhoGAP-independent mechanisms of DLC-1 growth regulation may also exist.
DLC-1/p122RhoGAP and related isoforms are comprised of three readily recognizable functional domains (13). There is a sterile α motif (SAM) domain at the amino terminus, a central RhoGAP catalytic domain found in all RhoGAPs, and a carboxyl-terminal steroidogenic acute regulatory related lipid transfer (START) domain. The ∼70-amino acid SAM domains are found in over 200 human proteins and are known to serve as protein-protein interaction domains (14). However, the recent structural determinations of the DLC-2 SAM domain suggest that this SAM domain is structurally distinct and hence may be functionally distinct from canonical SAM domains (15, 16). The RhoGAP domain of DLC-1 has been shown to stimulate RhoA inactivation in vitro and in vivo (5, 17). GAP activity in vitro has also been described for Cdc42 but not Rac. Consistent with RhoA GAP activity, overexpression of p122RhoGAP in fibroblasts led to a loss of focal contacts and stress fibers (18); this was also true of DLC-1 when overexpressed in carcinoma cells (6). The ∼210-amino acid START domains are found in 15 distinct proteins, either alone (e.g. STARD4) or as with DLC-1, associated with other protein domains (19). Some have been shown to bind lipids or sterols, and START domain-containing proteins exhibit very distinct subcellular locations. A recent study evaluated the role of the SAM and START domains in DLC-1 function and found that the START domain was critical for DLC-1 inhibition of actin stress fiber formation and growth (6). However, as described in the present study, these analyses may have utilized an inactive RhoGAP domain. Therefore, the precise functions of the SAM and START domains in the regulation of DLC-1 subcellular localization and RhoGAP catalytic activity remain to be determined.
Studies of the subcellular localization of DLC-1 present a complex picture. Several reports suggest that DLC-1 is localized at focal adhesions (20-22), whereas another study described localization to caveolae (23), and a third study found a diffuse distribution in the cytosol (7). The related DLC-2 and DLC-3 proteins have also been reported to localize to focal adhesions (10, 24). These reports may not be incompatible; a recent study has DLC-1 linked to focal adhesions via binding of the DLC-1 amino terminus to the focal contact protein tensin, as well a binding of the tensin-DLC-1 complex to caveolin (21). In three recent studies the interactions between DLC family members and the tensin family of focal adhesion proteins (tensins 1-3, c-ten) were determined. Thus the interaction between DLC-1 and c-ten has been mapped to a nonphosphorylation dependent binding of the Src homology 2 domain of c-ten to a short peptide motif (440SIYDNV) in the amino terminus of DLC-1; mutation of the critical Tyr442 residue abolishes the interaction (22). Another study (25) showed that DLC-1 and DLC-3 bound tensin1 via their Src homology 2 and phosphotyrosine-binding domains, with the former predominating.
Although substantial work has been done on the role of DLC-1 in cell growth regulation, there is less information available concerning its effects on cytoskeletal function. To further understand DLC-1 structure-function relationships in that context, we developed a series of deletion and point mutants of DLC-1 and characterized them in terms of GAP activity, subcellular localization, and effects on cell morphology and motility. Our findings identified both gain-of-function constitutively activated and dominant inhibitory mutants of DLC-1. Utilizing these mutants, we determined that activation of DLC-1 can contribute to increased velocity of migration but decreased directionality. The amino-terminal domain of DLC-1 can block the effects of DLC-1 on migration, likely by displacing active DLC-1 from focal adhesions.
EXPERIMENTAL PROCEDURES
Cell Culture—HEK293 cells were maintained in Dulbecco's modification of Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Sigma). MDA-MB-231 and MDA-MB-468 breast cancer cells were obtained from the ATCC (Rockville, MD) and grown in DMEM/Ham's F-12 medium supplemented with 10% fetal bovine serum.
Plasmids—For mammalian cell expression of green fluorescent protein tagged DLC-1 proteins, the full-length (1-1091 amino acids) and truncated sequences of the DLC-1 cDNA (GenBank™ accession number NM_006094) were generated by PCR and subcloned into the BamHI site of pEGFP-N1 (BD Biosciences). The truncated fragments of DLC-1 are as follows: DLC-1 ΔSAM (77-1091 amino acids), DLC-1 ΔN (609-1091 amino acids), DLC-1 START (875-1091 amino acids), DLC-1 ΔSTART (1-878 amino acids), DLC-1 N1 (1-638 amino acids), DLC-1 SAM (1-83 amino acids), and DLC-1 RhoGAP (609-878 amino acids). Serial amino-terminal truncations of DLC-1 ending with residues 252, 439, 609, and 629 were generated by PCR from full-length pEGFP-DLC-1, and serial carboxyl-terminal truncations of DLC-1 ending with residues 850, 828, and 878 were generated by PCR from pEGFP-DLC-1 ΔN (609-1091) and subcloned into the BamHI site of pEGFP-N1. Additional mutant constructs are explained in the figure legends and text. For expression of glutathione S-transferase (GST) fusion recombinant protein, cDNA sequences for full-length DLC-1, DLC-1 ΔSAM, and DLC-1 RhoGAP domain were subcloned into the pGEX-5X-3 (GE Healthcare) bacterial expression vector. All of the plasmid cDNA coding sequences were sequence-verified before use. pGEX plasmids for expression of GST fusions of wild type human RhoA have been described previously (26). The pAX142-RhoA (63L) and pAX142-Cdc42(12V) mammalian expression vectors for activated Rho GTPases have been described previously (27).
Confocal Microscopy—MDA-MB-468 cells on coverslips were transiently transfected with various EGFP-tagged DLC-1 constructs using Lipofectamine™ 2000 (Invitrogen). After 24 h of transfection, the cells were fixed with 4% paraformaldehyde for 5 min, permeabilized with 0.2% (v/v) Triton X-100 for 5 min at room temperature, and stained with either Alexa Fluo 568-phalloidin (Molecular Probes) for F-actin or anti-vinculin antibody (Sigma) or monoclonal anti-DLC-1 antibody (BD Biosciences) followed by incubation with fluorescently conjugated secondary antibody. The anti-DLC-1 antibody was shown to be specific versus other members of the DLC family (data not shown). For cotransfection experiments, HEK293 cells plated in 24-well plates were transiently transfected with 0.15 μg of pEGFP-DLC-1 ΔSAM and 0.45 μg of pAX142-RhoA(63L) or pAX142-Cdc42(12V) using Lipofectamine™ 2000. Twenty-four hours after transfection, the cells were trypsinized and replated on fibronectin-coated coverslips (10 μg/ml) in DMEM supplemented with 10% fetal bovine serum for 1 h. The cells were fixed, stained as described above, and observed on an Olympus confocal FV300 fluorescent microscope with a 60× oil immersion objective; the images were acquired by using Olympus Fluoview software.
In Vitro RhoGAP Assay—The in vitro GAP activity of DLC-1 was measured with a fluorescence-based technique as we described previously (5). Briefly, plasmids encoding GST fusion proteins of DLC-1 and Rho GTPases were transformed into the BL-21 Escherichia coli strain, and expression of the GST fusion proteins of DLC-1 and RhoA was induced with 100 mm and 250 mm isopropyl β-d-1-thiogalactopyranoside for 16 h at room temperature, respectively. Bacterially expressed Rho GTPases were purified by glutathione-Sepharose 4B chromatography and were preloaded with GTP in an exchange buffer for 1 min at 37 °C. Hydrolysis assays were initiated with adding 0.30 mm DLC-1 in an assay buffer containing 15 mm MDCC-phosphate-binding protein and 2 mm GTP-bound GTPase. The increases in Pi production from GTP hydrolysis were measured with a SpectraMAX Gemini (Molecular Devices) spectrofluorimeter by checking increases in fluorescence (λex = 425 nm and λem = 465 nm).
RhoA Activation Assay—A GST fusion of the Rho-GTP-binding domain of Rhotekin (amino acids 7-89), an effector of RhoA, was used in pulldown assays to detect expression of activated RhoA-GTP as described previously (28). In brief, HEK293 cells plated in 100 mm of culture were transfected with DLC-1 plasmids for 20 h, treated with lysophosphatidic acid for 30 min, and lysed in 300 μl of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 500 mm sodium chloride, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100, 0.5 mm magnesium chloride, 1 mm sodium orthovanadate, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mm phenylmethanesulfonyl fluoride) and clarified by centrifugation at 14,000 rpm for 20 min at 4 °C. A 5% aliquot was removed for determination of total quantities of the GTPase being analyzed. Clarified lysates were then incubated with 5 μg of GST-Rhotekin Rho-GTP-binding domain fusion protein for 1 h at 4 °C that had been precoupled to glutathione-Sepharose 4B beads (Amersham Biosciences) and washed three times with the lysis buffer. The samples were analyzed by SDS-PAGE and Western blotting using anti-RhoA antibody (BD Biosciences) to detect bound activated GTPases. Whole cell lysates were also analyzed for the presence of expressed RhoA and tubulin for normalization.
Migration Analysis—Cells plated overnight were transiently transfected with various pEGFP-DLC-1 plasmids using Lipofectamine™ 2000. After 12-18 h of transfection, the cells were trypsinized, replated sparsely on fibronetin-coated (10 μg/ml) glass-bottomed dishes in DMEM supplemented with 10% fetal bovine serum and placed in a temperature- and CO2-controlled chamber of a microscope equipped with 40× objective lenses. Time lapse recording was started 30 min or 1 h after plating, and differential interference contrast images were collected at 5-min intervals over 6 h using a CoolSnap HQ cooled charge-coupled device (Roper Scientific) linked to a Zeiss Axiovert 200M microscope controlled by SLIDEBOOK software (Intelligent Imaging, Denver, CO), or alternatively brightfield images were collected with a high content screening instrument, BD Pathway 855 Bioimager (BD Biosciences). To explore the effects of Rho kinase (ROCK) inhibition on cell migration, HEK293 cells on fibronectin-coated (10 μg/ml) glass-bottomed dishes in DMEM supplemented with 10% fetal bovine serum were treated for 3 h with 50 μm of the ROCK inhibitor Y27632 (Calbiochem Inc.) before starting time lapse recording. The velocity and persistence of migratory directionality (D/T) were measured by manually tracking of the location of cell centroids at each frame using Image J or Metamorph software (Molecular Devices). Each figure shown is representative of a minimum of two to three independent experiments analyzing 10-25 cells each. The velocity was calculated as [total length of migration paths (μm)/time (min)] and the persistence of migration was calculated as [net displacement (μm)/total length of migration paths (μm)].
RESULTS
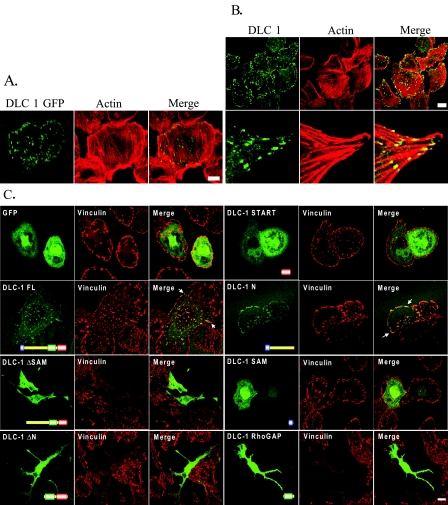
Distinct Roles of Amino- and Carboxyl-terminal Sequences in Regulation of DLC-1 Subcellular Localization—Recent studies suggest that amino-terminal sequences between the SAM and RhoGAP domains regulate DLC-1 subcellular localization (22, 25). To further evaluate the contribution of amino- and carboxyl-terminal sequences in regulation of DLC-1, we generated expression vectors that encoded more precise deletion and truncation mutants that were coupled at the carboxyl terminus to enhanced green fluorescent protein (EGFP) to allow us to monitor DLC-1 expression and localization in live cells (Fig. 1).
FIGURE 1.
Truncation and missense mutants of DLC-1 for DLC-1 structure-function analyses. A schematic representation of the DLC-1 truncation constructs is shown. The full-length and various fragments of DLC-1 cDNA were subcloned into the EGFP-tagged expression vector, pEGFP-N1, resulting in the expression of carboxyl-terminal GFP-tagged fusion proteins.
First, we wanted to verify that our ectopic expression of a tagged DLC-1 protein showed physiologic subcellular localization. We utilized Western blot analyses of a series of breast carcinoma cell lines for expression of DLC-1 and determined that MDA-MB-468 and MDA-MB-231 cells are negative and positive for endogenous DLC-1 protein expression, respectively (data not shown).
In agreement with previous studies using amino-terminally tagged DLC-1 fusion proteins (20-22), we found that full-length DLC-1 with a carboxyl-terminally fused GFP partially localized at the tips of actin filaments in MDA-MB-468 cells (Fig. 2A) as well as showing some diffuse cytosolic distribution. Because proteins that are ectopically expressed can be mislocalized, we also examined the distribution of endogenous DLC-1 in MDA-MB-231 cells, and we found that a significant portion of endogenous DLC-1 also seems to be localized at actin tips within focal adhesion-like structures (Fig. 2B). These results confirm that the ectopically expressed GFP-tagged DLC-1 has a similar subcellular localization as endogenous DLC-1 and may thus be an appropriate surrogate for the endogenous protein.
FIGURE 2.
Morphology of cells transfected with DLC-1 constructs and DLC-1 subcellular localization. A, DLC-1 negative MDA-MB-468 cells grown on glass coverslips were transiently transfected with EGFP-tagged full-length DLC-1, stained with rhodamine-phalloidin, and imaged with a confocal fluorescence microscope to observe subcellular localization and the effects on cell morphology. Scale bar, 10 μm. B, DLC-1 positive MDA-MB-231 cells were fixed and stained with anti-DLC-1 antibody followed by anti-mouse secondary antibodies conjugated to Alexa Fluor 488 and rhodamine-phalloidin (upper panel). A higher magnification of the analysis is shown in the lower panels. Scale bar, 20 μm. C, the various EGFP-tagged DLC-1 constructs indicated were transiently transfected into MDA-MB-468 cells, and the cells were stained with an anti-vinculin antibody followed by a Alexa Fluor 594 labeled secondary antibody. Selected overlap regions of vinculin and DLC-1 are indicated with white arrows. Scale bar, 10 μm.
To assess the role of amino- and carboxyl-terminal sequences in regulation of DLC-1 function, we next evaluated the subcellular location of several GFP-tagged DLC-1 truncation mutants when transiently expressed in MDA-MB-468 cells (Fig. 2C). EGFP alone exhibited a diffuse cytoplasmic and nuclear distribution. Full-length DLC-1 largely colocalized with vinculin-containing focal contacts. The isolated SAM domain (DLC-1 SAM) and the isolated START domain (DLC-1 START) did not show localization to focal adhesions but showed the same localization as EGFP alone. Hence, the SAM or START domains alone are not sufficient for focal adhesion targeting. However, inclusion of amino-terminal sequences upstream of the RhoGAP domain (DLC-1 N1) showed an apparent focal adhesion localization similar to that of full-length DLC-1. These observations suggested that the aminoterminal region between the SAM and GAP domains was necessary for the focal adhesion localization of DLC-1. These results are consistent with recent studies determining that DLC-1 association with focal adhesions is mediated through tensin family protein binding to a nonphosphorylated Tyr442 residue that serves as a binding site for the tensin Src homology 2 domains (20-22). Unexpectedly, we found that expression of any truncation mutant lacking the amino-terminal SAM domain caused drastic cell rounding and formation of protrusions that complicated our ability to accurately visualize the subcellular localization abilities of these truncation mutants. However, as we demonstrate in a section below, this problem can be overcome. Supplemental Table S1 provides a semiquantitative analysis of the morphological effects and subcellular localization of the various DLC-1 constructs.
The SAM Domain Functions as an Autoinhibitory Regulator of RhoGAP Activity in Vitro and in Vivo—We further evaluated the consequences of deletion of the SAM domain for DLC-1 function. As seen in Fig. 2C truncation mutants of DLC-1 lacking the SAM domain alone (DLC-1 ΔSAM) or with the entire amino-terminal region deleted (DLC-1 ΔN) produced dramatic changes in morphology including the development of long “neurite-like” protrusions that often terminated in lamellipodia, as well as the loss of mature vinculin staining focal adhesions. A similar result was produced by expression of DLC-1 RhoGAP comprised of only the functional RhoGAP domain (amino acids 609-878) linked to GFP. However, these morphological changes and the loss of vinculin staining did not occur upon expression of GFP alone or GFP-tagged full-length and other mutants of DLC-1. These observations suggest that amino-terminal truncations may activate DLC-1, thus leading to the changes in cell shape. Similar morphological alterations were also produced in other cell types including MDA-MB-231, MDA-MB-361, and MCF-7 cells (data not shown).
The morphological changes caused by SAM domain-deleted DLC-1 are similar to those that we have seen with inhibition of endogenous RhoA function by overexpression of the p190 RhoGAP and the Rnd3 antagonist of RhoA (29). Therefore, we speculated that deletion of the SAM domain rendered DLC-1 RhoGAP constitutively activated, causing inactivation of endogenous RhoA function. Consistent with this possibility, we found that any of the three disabling mutations in the DLC-1 RhoGAP domain (R677E, K714E, or K718E) resulted in DLC-1 ΔSAM mutants that were unable to induce morphological changes in cells (see supplemental Fig. S1).
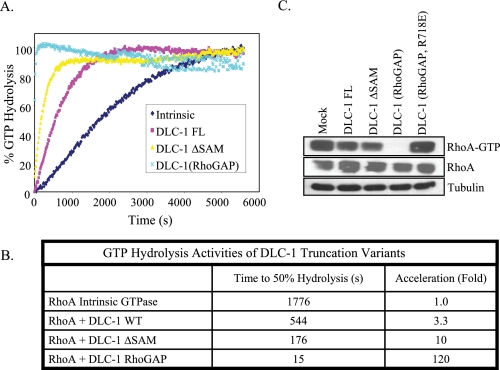
The increased biological effects of DLC-1 ΔSAM, suggested that the SAM may serve as an autoinhibitor domain and regulate intrinsic DLC-1 activity. To determine whether this was based on an increase in intrinsic GAP catalytic activity we undertook biochemical studies of full-length or truncated DLC-1 proteins expressed in bacteria and analyzed via an in vitro assay for GAP activity. We recently reported that the isolated RhoGAP domain displays substantially greater GTP hydrolysis than full-length DLC-1 in vitro (5), indicating that the amino-terminal and/or carboxyl-terminal regions of DLC-1 negatively regulate GAP activity. We found that full-length DLC-1 accelerated the intrinsic GTP hydrolysis activity of RhoA 3.3-fold and that DLC-1 ΔSAM stimulated GTP hydrolysis to a 3-fold greater degree than full-length DLC-1, whereas the isolated RhoGAP domain stimulated to an even greater degree (36-fold greater than the full length) (Fig. 3, A and B). Thus we suggest that the SAM domain functions as a negative intramolecular regulator of intrinsic DLC-1 RhoGAP catalytic activity. This may account for the greater in vivo effects of DLC-1 ΔSAM versus full-length DLC-1 in causing morphological changes. However, the greater activity of DLC RhoGAP suggests that additional amino- and/or carboxyl-terminal sequences may also function as autoinhibitory sequences.
FIGURE 3.
SAM domain deleted DLC-1 shows enhanced catalytic activity for RhoA. A, bacterially expressed full-length, SAM domain deleted (amino acids 77-1091), and RhoGAP domain fragment (amino acids 609-878) of DLC-1 were purified for analysis of in vitro GAP activity. Purified GST-RhoA fusion proteins were preloaded with GTP, and GTP hydrolysis was monitored by incubation with a phosphate-binding protein that undergoes a major increase in fluorescence upon binding inorganic phosphate. B, GTP hydrolysis activities of DLC-1 constructs. C, GTP loading of RhoA in cells was monitored by a Rhotekin pull-down assay as described under “Experimental Procedures.”
Because in vitro assays may not precisely reflect spatially regulated events in living cells, we also evaluated the GAP activity of DLC-1 mutants using a pulldown assay with a GST fusion protein containing the Rhotekin Rho GTPase-binding domain, which associates preferentially with RhoA-GTP. As seen in Fig. 3C, transient expression of full-length DLC-1 in HEK293 cells partially reduced the level of RhoA-GTP, whereas DLC-1 ΔSAM caused a further reduction. As we observed in vitro (Fig. 3A), the isolated RhoGAP domain exhibited greater activity in vivo, but expression of the RhoGAP-dead version of DLC-1 RhoGAP (R718E) was without effect. Thus the cellular assay results closely parallel the in vitro biochemical studies and illustrate the activating effect of the SAM domain truncation. These results also support the possibility that DLC-1 ΔSAM functions as a constitutively activated version of DLC-1 and may be a useful reagent for evaluating the role and mechanism of DLC-1 tumor suppression.
The striking change in morphology produced by activated DLC-1 gave us the chance to more precisely define the boundaries of the RhoGAP domain. A previous report had indicated that the START domain was indispensable for RhoGAP catalytic activity in vivo (6). However, our deletion analyses more precisely defining the minimal sequences required for a functional RhoGAP domain (609-878; see supplemental Fig. S2 for a detailed analysis) suggest that the previous study may have utilized a nonfunctional RhoGAP domain for their analyses. Note that our definition of the RhoGAP domain (609-878) differs from the conventional definition depicted in green in Fig. 1 (655-801) that is based on homologies to other RhoGAPs.
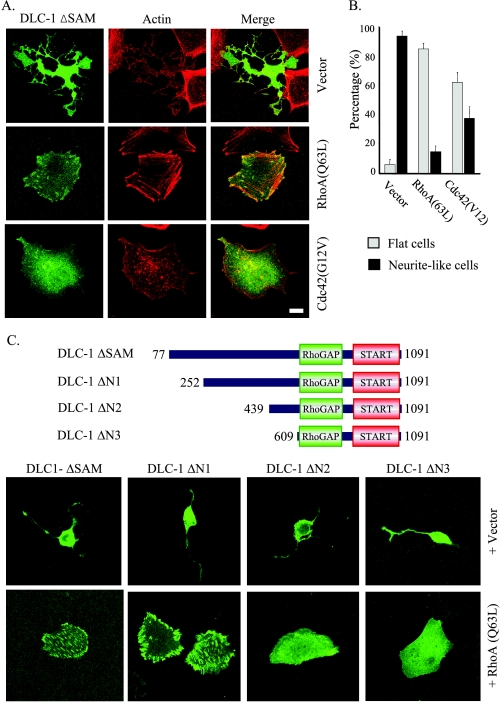
Reversal of Effects of Activated DLC-1 by Rho and CDC42—DLC-1 has been reported to have robust GAP activity for RhoA and the closely related RhoB and RhoC proteins and to a lesser degree for Cdc42, based on in vitro biochemical studies. Thus we tested whether transient cotransfection of GAP-insensitive, constitutively activated versions of RhoA and Cdc42 could reverse the dramatic morphological changes produced by expression of DLC-1 ΔSAM or DLC-1 ΔN in HEK293 cells. As seen in Fig. 4A, cotransfection with GAP-insensitive RhoA(Q63L) fully reversed the morphological effects and the loss of focal adhesions caused by DLC-1 ΔSAM, whereas cotransfection of GAP-insensitive, active Cdc42(G12V) largely restored the overall shape but failed to restore the presence of mature focal contacts or actin stress fibers. The fraction of cells with approximately normal morphology versus those with extensive neurite-like protrusions was quantitated and summarized in Fig. 4B.
FIGURE 4.
Effects of active RhoA and Cdc42 on the morphology of cells expressing DLC-1. A, HEK293 cells were cotransfected with pEGFP-DLC-1 ΔSAM and vector alone (pAX142) or constructs expressing active forms of RhoA(Q63L) or Cdc42(12V) and replated on fibronectin-coated coverslips (10 μg/ml) for 1 h. The overall cell shapes as well as the localization of DLC-1 and of actin filaments were imaged by confocal fluorescence microscopy. Scale bar, 10 μm. Unusual, multi-branched shapes were frequently seen in cells transfected with pEGFP-DLC-1 ΔSAM as illustrated in the top row. B, the bar graph shows the fraction of cells displaying long, branched, neurite-like projections (black bars) and cells displaying flat morphology similar to untransfected HEK293 cells (gray bars) in the experiment described in A. The means ± S.D. of ∼100 cells analyzed in three independent assays are shown. C, HEK293 cells were cotransfected with various amino-terminal truncations of pEGFP-DLC-1 and with RhoA(Q63L) or control vector. The overall cell shapes as well as the localization of DLC-1 were imaged as above.
The ability of coexpressed RhoA(Q63L) to block the morphologic effects of SAM-deleted truncations mutants provided us with an approach to better evaluate the role of amino-terminal sequences in regulation of DLC-1 subcellular localization. Therefore, we evaluated the focal adhesion localization of additional amino-terminal deletion mutants of DLC-1 when coexpressed with RhoA(Q63L) (Fig. 4C). Deletion of the SAM domain alone did not disrupt DLC-1 association with focal adhesions. Although deletion of residues 1-252 (DLC-1 ΔN1) did not perturb DLC-1 association with focal adhesions, as expected, deletion of residues 1-609 (DLC-1 ΔN3) that includes the tensin Src homology 2 domain binding motif (SIYDNV; amino acids 440-445) was sufficient to impair DLC-1 association with focal adhesions. Deletion of 1-439 (DLC-1 ΔN2) also dramatically abolished focal adhesion localization; this may be due to disruption of the adjacent tensinbinding site.
Active DLC-1 Increases Cell Migration Velocity but Reduces Directionality—Spatially directed RhoA activation at the rear of migrating cells is thought to facilitate directional cell migration (1). We thus examined DLC-1 effects in wound type migration assays and observed that, as reported by others (6), expression of DLC-1 results in a reduction in directional migration in this assay (data not shown). To determine the role of specific domains in regulating the role of DLC-1 in cell migration, we utilized a dynamic live cell imaging approach.
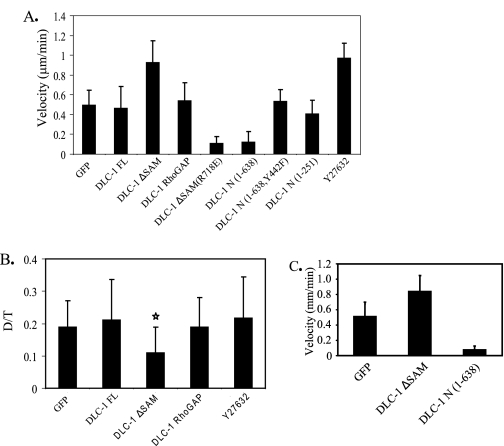
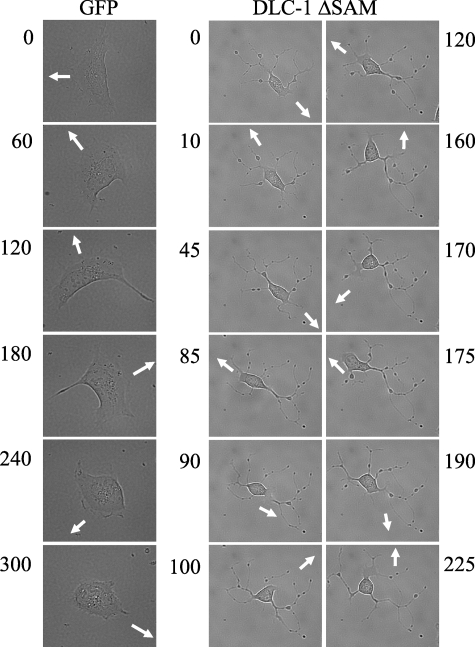
For these analyses, we additionally utilized HEK 293 cells, because they have been used extensively to study the involvement of RhoA in cell migration. HEK 293 or breast tumor cells were transiently transfected with various DLC-1 constructs, and the effects on the velocity and directionality of cell migration were observed. Digital images were taken every 5 min for a total of 6 h/experiment. Blot analyses verified comparable expression of wild type and truncated DLC-1 proteins, except that DLC-1 N (1-638) was expressed at slightly lower levels than the other constructs (see supplemental Fig. S3). Our analyses revealed profound differences in migration behavior between controls and cells expressing wild type and mutant DLC-1 proteins. Quantitation of results drawn from observations of multiple cells in terms of cell velocity and directionality (Fig. 5, A and B) led to the following observations. Whereas ectopic expression of wild type DLC-1 did not increase cell velocity, when compared with GFP-expressing controls, cells expressing constitutively activated DLC-1 ΔSAM showed a marked increase in velocity (Fig. 5A) but reduced directionality (persistence of motion) (Fig. 5B). In contrast, cells expressing constitutively activated DLC-1 (DLC-1 RhoGAP) displayed similar velocity and directionality as controls. These observations suggest that both activation of the RhoGAP domain and as yet unknown functions of the amino-terminal domain of DLC-1 are involved in effects on motility. Interestingly, the ROCK inhibitor Y27632 produced an increase in the velocity of migration similar to that produced by transfection of DLC-1 ΔSAM (Fig. 5A); however, it did not affect directionality (Fig. 5B). This suggests that effects of active DLC-1ΔSAM on velocity are mediated primarily through Rho/ROCK, whereas effects on directionality may be mediated through Cdc42, which would not be affected by Y27632 or by other Rho effectors not sensitive to this inhibitor. The dramatic effect of DLC-1 ΔSAM on directionality is visually illustrated in Fig. 6. Whereas a control cell expressing GFP changed direction only five times during a 300-min time period, a cell expressing DLC-1 ΔSAM changed direction 10 times in less than 200 min.
FIGURE 5.
Migration of cells transfected with various truncations or mutations of DLC-1. For migration experiments HEK293 cells transfected with various truncation constructs or mutants of DLC-1 were plated on fibronectin-coated (10 μg/ml) 35-mm glass-bottomed culture dishes for 30 min to 1 h. Digital images were taken every 5 min for a total of 6 h/experiment. A, the migration speeds of the cells (15-25 cells/condition) were determined by dividing the total length of migration path by the total time elapsed. B, directional persistence (D/T) was determined by dividing the net displacement (D) by total length of the migration path (T). A and B illustrate results for HEK293 cells transfected with various pEGFP-DLC-1 constructs or treated with the Rho kinase inhibitor Y27632. In B only the DLC-1 ΔSAM transfected cells (star) displayed a statistically significant difference from EGFP transfected controls. This observation was repeated in several independent experiments. C, MDA-MB-231 cells were transfected with various DLC-1 truncation constructs and migration speeds of the cells (15-20 cells/condition) were determined as described above.
FIGURE 6.
Active DLC-1 reduces persistence of cell movement. The images were taken at the indicated time (min) and illustrate the rapid changes in directionality (lack of persistence) in cells transiently transfected with pEGFP-DLC-1 ΔSAM as compared with control cells transfected with the pEGFP empty vector. The arrows indicate the direction of cell movement. The images presented are representative of multiple observations.
The Amino-terminal Domain of DLC-1 Acts as a Dominant Negative to Block Cell Migration—We had evaluated a GAP-deficient variant of activated DLC-1 (DLC-1 ΔSAM (R718E)) as a control to verify that the enhancement in velocity was due to inactivation of Rho GTPase function. Unexpectedly, we observed that DLC-1 ΔSAM (R718E), which we anticipated to be an inactive protein, rendered cells almost completely immobile (Fig. 5A). We then addressed the possibility that DLC-1 ΔSAM (R718E) may function as a dominant inhibitory mutant by antagonizing endogenous DLC-1 association with focal adhesions. If so, then the isolated amino-terminal region of DLC-1, which localizes at focal adhesions, should also inhibit cell motility. To probe this further, we compared the effects of expressing the isolated amino-terminal domain (DLC-1 N1 (1-638)) versus the same amino-terminal fragment that contains a missense mutation (Y442F) at the site reported (22, 25) to be involved in binding to tensins and required for focal adhesion association (Fig. 1). We also examined an amino-terminal fragment of DLC-1, DLC-1 N2 (1-251), which is truncated prior to the tensin-binding site. As seen in Fig. 5A, a striking difference was observed, with the wild type DLC-1 N1 causing a marked inhibition of cell motility, whereas the two tensin binding-deficient amino-terminal fragments were without effect. These results suggest that the isolated wild type amino-terminal domain of DLC-1 can act as a dominant inhibitor of cell migration that is dependent on association with tensin and focal adhesions.
We also performed a similar series of cell migration experiments in MDA-MB-231 cells (Fig. 5C). These results were similar to those seen in HEK293 cells, even though the expression levels of the various DLC-1 constructs were substantially lower (data not shown). Thus constitutively activated DLC-1 ΔSAM enhanced cell migration velocity, whereas dominant negative DLC-1 ΔSAM (R718E) or DLC-1 N1 strongly inhibited cell movement.
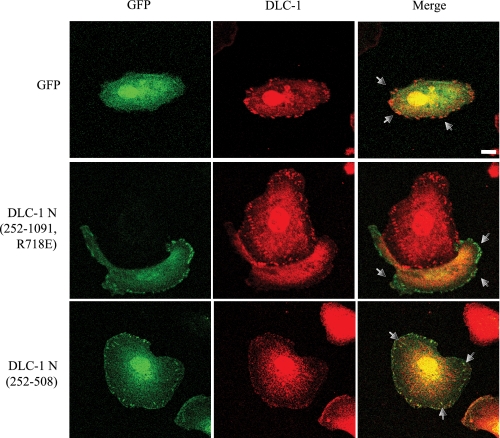
The Amino-terminal Domain Can Displace DLC-1 from Focal Adhesion Sites—Our observations with HEK293 and MDA-MB-231 cell lines (Fig. 5) suggested that DLC-1 activity at focal adhesion sites may be critical for cell motility and that expression of the native amino-terminal domain construct may displace endogenous DLC-1 and thus block motility. We examined this by determining whether amino-terminal fragments of DLC-1 (ΔN 252-1091 or ΔN 252-508) that strongly associate with focal adhesions but are not recognized by the monoclonal antibody that reacts with the region amino acids 47-249 can disrupt endogenous DLC-1 subcellular localization. As seen in Fig. 7, expression of these constructs in MDA-MB-231 cells resulted in reduced levels of endogenous DLC-1 at focal adhesion sites. This suggests that the dominant negative effect of the amino-terminal domain could be due to displacement of endogenous DLC-1 from focal adhesions, possibly by blocking its interaction with tensin.
FIGURE 7.
The amino terminus of DLC-1 displaces endogenous DLC-1 from focal contacts. MDA-MB-231 cells were transiently transfected with pEGFP, pEGFP-DLC-1 ΔN (252-508), or pEGFP-DLC-1 ΔN (252-1091, R718E). These proteins retain the tensin-binding sequence that leads to focal adhesion localization but do not have Rho GAP activity, nor are they recognized by the antibody that binds full-length DLC-1. Transfected cells were then were replated on fibronectin-coated coverslips (10 μg/ml) for 4-5 h and stained with anti-DLC-1 monoclonal antibodies (to visualize endogenous DLC-1) followed by anti-mouse secondary antibodies conjugated to Alexa Fluor 594 and imaged with a confocal fluorescence microscope. The arrows indicate the focal adhesions. Scale bar, 10 μm. The images presented are representative of multiple observations.
DLC-1 Affects Cell Protrusions—Another aspect of DLC-1 function was also revealed from our live cell imaging analyses. Initially we interpreted the long projections seen in cells expressing highly active DLC-1 as tail retraction fibers caused by a reduction in RhoA activity (2). However, it is clear that cells expressing active DLC-1 also showed a high degree of protrusive activity. Thus, as illustrated in Fig. 6, cells that express activated DLC-1 ΔSAM displayed extensive ruffled membranes that rapidly switch direction, as well as displaying multiple branched projections. Some of these projections were due to defects in tail retraction. However, in addition to retraction fibers, the cells expressing constitutively activated DLC-1 also clearly displayed enhanced protrusive activity, with many cells with rapidly extending long, branched, neurite-like projections (see supplemental Fig. S4).
DISCUSSION
Restoration of DLC-1 activity in DLC-1-deficient tumor cells causes reduced growth rates, restores a more normal phenotype (7, 8, 17), and impairs invasion and metastasis (30). Because a key aspect of DLC-1 involves inhibition of Rho function and Rho GTPases are implicated in cell motility, in the present study we focused on the role of DLC-1 function in regulation of actin organization, focal contact organization, and cell migration (note: additional images of the subcellular localization of various DLC-1 constructs and their relation to focal adhesions are presented in supplemental Figs. S5-S10). We undertook a structure-function analysis to evaluate the contributions of the various domains of DLC-1 to its overall effects on the RhoGAP activity and its consequences on actin cytoskeletal organization, cell morphology, and cell motility. First, we determined that the amino-terminal SAM domain is a negative regulator of the intrinsic RhoGAP activity in vitro and in vivo. Second, we established that a DLC-1 variant lacking the SAM domain had strong effects on the velocity and directionality of cell migration. Further, the isolated amino terminus (positions 1-638) can function as a dominant inhibitory variant of DLC-1 and dramatically reduce migration.
DLC-1 is a multi-functional protein with additional domains and sequences beyond its RhoGAP catalytic sequences. These additional sequences may regulate RhoGAP activity as well as dictate RhoGAP-independent functions of DLC-1. In previous work we had shown that deletion of both amino- and carboxyl-terminal domain sequences resulted in a very active core RhoGAP domain (5). Here we show that deletion of the SAM domain alone substantially increased DLC-1 RhoGAP activity both in in vitro assays and within cells. The underlying mechanism is unclear at this point. One possibility is that the aminoterminal SAM domain regulates the catalytic domain via an intramolecular interaction. This type of regulation has been seen commonly in RhoGEFs and to a lesser extent in other RhoGAPs (1, 3, 4). Alternatively intermolecular interactions or the recruitment of accessory regulatory proteins may be involved. In any case, we suggest that the SAM domain may facilitate stimulus-mediated post-transcriptional mechanisms for the regulation of the intrinsic RhoGAP activity of DLC-1. Furthermore, because the signaling mechanisms that activated DLC-1 function remain to be identified, similar to activated mutants of Rho GTPases, this constitutively activated DLC-1 variant will be a very useful reagent for further delineation of DLC-1 activity in normal and neoplastic cell biology.
Expression of the active DLC-1 ΔSAM results in profound changes in cytoskeletal organization with the loss of actin stress fibers and reduction of focal adhesions. However, by reverting the effects of DLC-1 ΔSAM with a constitutively active Rho, we show that DLC-1 ΔSAM retains the ability to localize to focal contacts and thus can be a properly localized activated GAP. Expression of DLC-1 ΔSAM also profoundly affects cell migration, leading to an increase in velocity but a reduction in directionality. The dual effect of DLC-1 ΔSAM on cell velocity and directionality may be due to the fact that this protein has GAP activity for both Rho and for Cdc42, a key mediator of cell polarity (2, 5). Interestingly expression of the highly active construct DLC-1 RhoGAP fails to mimic the effect of DLC-1 ΔSAM, suggesting that both GAP activity and as yet undefined functions of the amino-terminal domain are essential for DLC-1 effects on cell motility. By contrast, the change in cell morphology resulting from expression of an activated DLC-1 is not dependent on the presence of amino-terminal sequences.
As expected, expression of the RhoGAP domain containing the inactivating R718E mutation had little effect on migration, indicating the critical role of GAP activity in motility. Surprisingly, however, expression of full-length DLC-1 containing the GAP-inactive R718E mutation caused a profound reduction in cell movement. Similarly, the isolated amino-terminal fragment had the same inhibitory action. This effect was not seen with a mutated version (Y442F) of the amino-terminal domain that fails to bind to the focal adhesion protein tensin, suggesting that focal contact localization is a key to these effects. The wild type version, but not the Y442F version, of the amino-terminal domain was able to displace endogenous DLC-1 from focal adhesions. Because DLC-2 and DLC-3 share similar tensin-binding sequences, it seems likely that the isolated DLC-1 amino-terminal can also displace endogenous DLC-2 and DLC-3 as well. Thus these observations argue that proper placement of appropriate (endogenous) levels of DLC family members at focal adhesions is critical for the maintenance of normal cell motility. Finally, we have found that interfering RNA suppression of endogenous DLC-1 in lung and breast cancer cells typically does not result in a robust biological consequences (data not shown), most likely because of the continued expression of the functionally related DLC-2 and DLC-3 isoforms. Therefore, this dominant inhibitory mutant of DLC-1, similar to the dominant inhibitory Rho GTPase mutants that block the activity of multiple Rho activators, will be an important reagent for further dissection of role of DLC loss of function in tumor progression and growth.
The mechanism(s) underlying the key role of DLC-1 in cell movement is not clearly understood. One likely possibility is that it may be critical to facilitate a precise spatial inactivation of RhoA activity to support migration. Consistent with this possibility, we recently found that ectopic expression of wild type DLC-1 caused inactivation of RhoA preferentially at the leading edge of migrating cells (5). Our observations on DLC-1 and cell migration seem consistent with a recently published model of the mechanisms relating lamellipodial activity and adhesion site formation (31). In this model, forward motion of the leading edge involves localized actin extension, adhesion formation, contraction, and backward flow of actin, all in balance. When DLC-1 is active, Arp 2/3 complex-mediated actin protrusion continues (even if Rac activity is not increased), but contractility and adhesion site formation will be reduced as Rho/ROCK activity goes down; this tilts the balance toward rapid leading edge protrusion and rapid migration. When DLC-1 is displaced from adhesion sites, for example by the amino-terminal fragment, local Rho is greatly activated, leading to increased contractility, to enhanced strength of focal adhesions, and to enhanced backward retraction of actin and thus paralysis of motion.
An interesting observation is that expression of activated DLC-1 stimulated extensive protrusive activity, but the mechanistic basis for these events is unclear. There is a well established antagonism between Rho and Rac in regulation of the cytoskeleton and protrusive activity (32). This antagonism may be based in part on FilGAP, a filamin-binding RacGAP that is negatively regulated by Rho and ROCK and reciprocally on down-regulation of Rho via Rac-generated reactive oxygen species (33, 34). Thus a reduction in Rho activity by DLC-1 could lead to an increase in Rac activity that would enhance cell protrusions. However, we have not been able to detect an overall increase in Rac-GTP loading in cells expressing activated DLC-1 (data not shown); possibly the effects are localized and difficult to detect at the biochemical level. In neuronal cells p190RhoGAP has been shown to play a positive role in axon extension and branching morphogenesis through suppression of an axon retraction pathway mediated by Rho and ROCK (35, 36). Our observations suggest that RhoGAPs, particularly DLC-1, may also play a key role in supporting protrusive activity.
In summary, our structure-function analyses established two critical roles for two distinct amino-terminal sequence elements in regulating DLC-1. Whereas the SAM domain functions as an autoinhibitory element of intrinsic RhoGAP catalytic activity, the tensin-binding sequences are critical for DLC-1 localization to focal adhesions, which is critical for regulation of cell migration. Our preliminary analyses of additional amino-terminal deletion mutants have identified a second autoinhibitory sequence that coincides with sequences important for tensin binding. Hence, our future studies will focus on determining whether tensin binding may also serve as a mechanism to regulate both the spatial and intrinsic activity of DLC-1. Finally, our studies additionally identified both gain-of-function and dominant negative variants of DLC-1 that will be very useful reagents for further delineation of the role of DLC-1 loss in human oncogenesis.
Supplementary Material
Acknowledgments
We thank Gary L. Johnson and Klaus M. Hahn for use of the microscope systems and Bryan Roth for use of the BD pathway instrument located in the National Institute of Mental Health Psychoactive Drug Screening Program at the University of North Carolina. We also thank Betsy Clarke for expert editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1 HL4500 (to R. L. J.) and CA063071, CA129610, and CA67771 (to C. J. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1-S10.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: GEF, guanine nucleotide exchange factor; GAP, GTPase-activating domain; SAM, sterile α motif; START, steroidogenic acute regulatory related lipid transfer; DMEM, Dulbecco's modified Eagle's medium; GST, glutathione S-transferase; GFP, green fluorescent protein; EGFP, enhanced GFP; ROCK, Rho kinase.
References
- 1.Jaffe, A., and Hall, A. (2005) Annu. Rev. Cell Dev. Biol. 21 247-269 [DOI] [PubMed] [Google Scholar]
- 2.Raftopoulou, M., and Hall, A. (2004) Dev. Biol. 265 23-32 [DOI] [PubMed] [Google Scholar]
- 3.Rossman, K. L., Der, C. J., and Sondek, J. (2005) Nat. Rev. Mol. Cell Biol. 6 167-180 [DOI] [PubMed] [Google Scholar]
- 4.Moon, S. Y., and Zheng, Y. (2003) Trends Cell Biol. 13 13-22 [DOI] [PubMed] [Google Scholar]
- 5.Healey, K. D., Shutes, A., Kim, T. H., Juliano, R. L., Bang, Y.-J., and Der, C. J. (2008) Mol. Carcinog. 47 326-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong, C. M., Yam, J. W., Ching, Y. P., Yau, T. O., Leung, T. H., Jin, D. Y., and Ng, I. O. (2005) Cancer Res. 65 8861-8868 [DOI] [PubMed] [Google Scholar]
- 7.Zhou, X., Thorgeirsson, S. S., and Popescu, N. C. (2004) Oncogene 23 1308-1313 [DOI] [PubMed] [Google Scholar]
- 8.Ng, I. O., Liang, Z. D., Cao, L., and Lee, T. K. (2000) Cancer Res. 60 6581-6584 [PubMed] [Google Scholar]
- 9.Durkin, M. E., Ullmannova, V., Guan, M., and Popescu, N. C. (2007) Oncogene 26 4580-4589 [DOI] [PubMed] [Google Scholar]
- 10.Leung, T. H., Ching, Y. P., Yam, J. W., Wong, C. M., Yau, T. O., Jin, D. Y., and Ng, I. O. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15207-15212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, T. Y., Jong, H. S., Song, S. H., Dimtchev, A., Jeong, S. J., Lee, J. W., Kim, T. Y., Kim, N. K., Jung, M., and Bang, Y. J. (2003) Oncogene 22 3943-3951 [DOI] [PubMed] [Google Scholar]
- 12.Sahai, E., and Marshall, C. J. (2002) Nat. Rev. Cancer 2 133-142 [DOI] [PubMed] [Google Scholar]
- 13.Durkin, M. E., Yuan, B. Z., Zhou, X., Zimonjic, D. B., Lowy, D. R., Thorgeirsson, S. S., and Popescu, N. C. (2007) J. Cell Mol. Med. 11 1185-1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao, F., and Bowie, J. U. (2005) Sci. STKE 2005 re7. [DOI] [PubMed] [Google Scholar]
- 15.Kwan, J. J., and Donaldson, L. W. (2007) BMC Struct. Biol. 7 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, H., Fung, K. L., Jin, D. Y., Chung, S. S., Ching, Y. P., Ng, I. O., Sze, K. H., Ko, B. C., and Sun, H. (2007) Proteins 67 1154-1166 [DOI] [PubMed] [Google Scholar]
- 17.Wong, C. M., Lee, J. M., Ching, Y. P., Jin, D. Y., and Ng, I. O. (2003) Cancer Res. 63 7646-7651 [PubMed] [Google Scholar]
- 18.Sekimata, M., Kabuyama, Y., Emori, Y., and Homma, Y. (1999) J. Biol. Chem. 274 17757-17762 [DOI] [PubMed] [Google Scholar]
- 19.Alpy, F., and Tomasetto, C. (2005) J. Cell Sci. 118 2791-2801 [DOI] [PubMed] [Google Scholar]
- 20.Kawai, K., Yamaga, M., Iwamae, Y., Kiyota, M., Kamata, H., Hirata, H., Homma, Y., and Yagisawa, H. (2004) Biochem. Soc. Trans. 32 1107-1109 [DOI] [PubMed] [Google Scholar]
- 21.Yam, J. W., Ko, F. C., Chan, C. Y., Jin, D. Y., and Ng, I. O. (2006) Cancer Res. 66 8367-8372 [DOI] [PubMed] [Google Scholar]
- 22.Liao, Y. C., Si, L., DeVere White, R. W., and Lo, S. H. (2007) J. Cell Biol. 176 43-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaga, M., Sekimata, M., Fujii, M., Kawai, K., Kamata, H., Hirata, H., Homma, Y., and Yagisawa, H. (2004) Genes Cells 9 25-37 [DOI] [PubMed] [Google Scholar]
- 24.Kawai, K., Kiyota, M., Seike, J., Deki, Y., and Yagisawa, H. (2007) Biochem. Biophys. Res. Commun. 364 783-789 [DOI] [PubMed] [Google Scholar]
- 25.Qian, X., Li, G., Asmussen, H. K., Asnaghi, L., Vass, W. C., Braverman, R., Yamada, K. M., Popescu, N. C., Papageorge, A. G., and Lowy, D. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9012-9017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arthur, W. T., Ellerbroek, S. M., Der, C. J., Burridge, K., and Wennerberg, K. (2002) J. Biol. Chem. 277 42964-42972 [DOI] [PubMed] [Google Scholar]
- 27.Whitehead, I. P., Lambert, Q. T., Glaven, J. A., Abe, K., Rossman, K. L., Mahon, G. M., Trzaskos, J. M., Kay, R., Campbell, S. L., and Der, C. J. (1999) Mol. Cell Biol. 19 7759-7770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren, X. D., Kiosses, W. B., and Schwartz, M. A. (1999) EMBO J. 18 578-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wennerberg, K., Forget, M. A., Ellerbroek, S. M., Arthur, W. T., Burridge, K., Settleman, J., Der, C. J., and Hansen, S. H. (2003) Curr. Biol. 13 1106-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodison, S., Yuan, J., Sloan, D., Kim, R., Li, C., Popescu, N. C., and Urquidi, V. (2005) Cancer Res. 65 6042-6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giannone, G., Dubin-Thaler, B. J., Rossier, O., Cai, Y., Chaga, O., Jiang, G., Beaver, W., Dobereiner, H. G., Freund, Y., Borisy, G., and Sheetz, M. P. (2007) Cell 128 561-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burridge, K., and Wennerberg, K. (2004) Cell 116 167-179 [DOI] [PubMed] [Google Scholar]
- 33.Ohta, Y., Hartwig, J. H., and Stossel, T. P. (2006) Nat. Cell Biol. 8 803-814 [DOI] [PubMed] [Google Scholar]
- 34.Nimnual, A. S., Taylor, L. J., and Bar-Sagi, D. (2003) Nat. Cell Biol. 5 236-241 [DOI] [PubMed] [Google Scholar]
- 35.Billuart, P., Winter, C. G., Maresh, A., Zhao, X., and Luo, L. (2001) Cell 107 195-207 [DOI] [PubMed] [Google Scholar]
- 36.Brouns, M. R., Matheson, S. F., and Settleman, J. (2001) Nat. Cell Biol. 3 361-367 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.