Abstract
Serine/threonine phosphorylation of the nonstructural protein 5 (NS5) is conserved feature of flaviviruses, but the kinase(s) responsible and function(s) remain unknown. Mass spectrometry was used to characterize phosphorylated residues of yellow fever virus (YFV) NS5 expressed in mammalian cells. Multiple different phosphopeptides were detected. Mutational and additional mass spectrometry data implicated serine 56 (S56), a conserved residue near the active site in the NS5 methyltransferase domain, as one of the phosphorylation sites. Methyltransferase activity is required to form a methylated RNA cap structure and for translation of the YFV polyprotein. We show the 2’-O- methylation reaction requires the hydroxyl side chain of S56, and replacement with a negative charge inhibits enzymatic activity. Furthermore mutational alteration of S56, S56A or S56D, prevents amplification in a viral replicon system. Collectively our data suggest phosphorylation of NS5 S56 may act to shut down capping in the viral life cycle.
Keywords: Phosphorylation, Methyltransferase, RNA cap, flavivirus
Introduction
Phosphorylation of amino acids is a common mechanism to regulate protein function in eukaryotes and is controlled by a wide variety of tightly regulated cellular kinases and phosphatases. Viral protein function and viral replication in several different viruses are dependent upon conserved phosphorylation sites. For example, Vaccinia virus virion assembly is dependent upon the phosphorylation state of the protein A13 (Unger and Traktman, 2004). Phosphorylation of serine clusters in the Ebola virus protein VP30 has been shown to directly affect whether VP30 localizes to inclusion bodies within the cell or activates transcription of viral RNA. This phosphorylation may serve as a switch between packaging and transcription in the Ebola lifecycle (Modrof et al., 2002). Despite these examples, relatively few phosphorylation sites have been mapped on the amino acid level in viruses.
The family Flaviviridae includes many diverse medical pathogens including hepatitis C virus (HCV) as well as arthropod-borne diseases such as dengue virus (DV), yellow fever virus (YFV) and west nile virus (WNV). More than two billion people are at risk from dengue alone, leading to an estimated 50 million cases annually, which may increase further as the range of the mosquito vector increases due to climate change (World Health Organization, 2002). Among Flaviviridae, the most studied example is phosphorylation of NS5A of HCV, which exists in both a basal (termed p56) and hyperphosphorylated (termed p58) state. CK1α mediates some basal phosphorylation and the hyperphosphorylation event. Silencing CK1α decreases HCV RNA replication, but selective blockade of p58 formation actually increases HCV RNA replication in the replicon model suggesting a complex interplay between RNA replication and host kinases (Neddermann et al., 2004; Quintavalle et al., 2007). Phosphorylation of NS5B, the RNA-dependent RNA polymerase (RdRp) has also been shown to affect replicon activity (Kim et al., 2004). Other Flaviviridae such as DV and YFV have phosphorylated NS5 proteins (Kapoor et al., 1995; Morozova et al., 1997; Reed, Gorbalenya, and Rice, 1998). DV phosphorylation of NS5 affects NS5 interactions with the viral helicase NS3. A hyperphosphorylated form of NS5 was found to localize to the nucleus away from the cytoplasmic sites of viral replication (Forwood et al., 1999; Kapoor et al., 1995). A nuclear localization sequence is present in DV NS5 and is phosphorylated in vitro by host CKII but the relationship between phosphorylation and nuclear localization has yet to be fully elucidated (Pryor et al., 2007).
NS5 has two separate activities encoded in distinct domains. The C-terminal 600 amino acids of NS5 contain RdRp activity and the N-terminal 300 amino acids represent the methyltransferase domain, which methylates the cap structure found on the 5’ end of viral RNA. Both RNA polymerase and methyltransferase activity are essential for the viral lifecycle (Khromykh, Kenney, and Westaway, 1998; Khromykh et al., 1999, Ray, 2006 #271). Viruses require host translational machinery to synthesize viral proteins and a methylated cap structure greatly increases translation of RNA (Furuichi and Shatkin, 2000). The flavivirus NS5 activity includes both 2’-O methylation and N7 methylation, which is rare since other methyltransferases typically carry out only one type of methylation reaction (Ray et al., 2006; Schubert, Blumenthal, and Cheng, 2003).
This study reports the first mapping of YFV NS5 phosphorylation sites on an amino acid level. Both mass spectrometry and site-directed mutagenesis were used to identify S56 of YFV NS5 as a phosphorylated amino acid. Our data show that not only that S56 phosphorylated, but also that the hydroxyl group is required for 2’-O methylation in vitro, yet is not essential for SAH binding. Finally, S56 is required for replication of viral RNA, but not for translation of exogenously capped viral RNA. While it is unclear the extent to which S56 phosphorylation occurs in during cellular infection, our results suggest phosphorylation of S56 could alter the methyltransferase activity and shut down viral translation.
Results
Multiple phosphorylation sites occur in YFV NS5, including serine 56
Phosphorylation is an important way to regulate signaling cascades and protein activity, and has been shown to play a role in several viral processes (Jakubiec and Jupin, 2007; Mohr, 2006). To determine phosphorylation sites in YFV NS5, the protein was expressed in a form amenable to purification from mammalian cells, and then characterized by mass spectrometry (MS). Amino acids 1–900 of YFV NS5 were amplified from YF-R.luc2A-RP and cloned into a pIRES eukaryotic expression vector in frame with a 6XHis nickel affinity tag. DNA was transfected into HEK293T cells, which support viral replication (data not shown). YFV NS5 was purified from cell lysates by metal affinity chromatography, excised from SDS-PAGE, and after tryptic digest, was analyzed by MS. For background corrections, an empty vector control was transfected and prepared in parallel for MALDI-TOF analysis even though the ~100kD YFV NS5 was absent from this preparation (Fig 1). The MS peaks obtained from this negative control spectra did not correspond to any peaks in the MS identified as potential phosphopeptides. Fig. 2 shows six peptides that were identified as 80 daltons larger than the predicted mass. Each contains at least one serine/threonine phosphoacceptor. Only masses from approximately 50% of the YFV NS5 protein were identified (with or without an extra phosphate) so some phosphorylation sites may not have been identified. The masses for each of these peptides were no longer detected upon treatment of the purified NS5 with Calf Intestinal Phosphotase consistent with them being phosphorylated peptides.
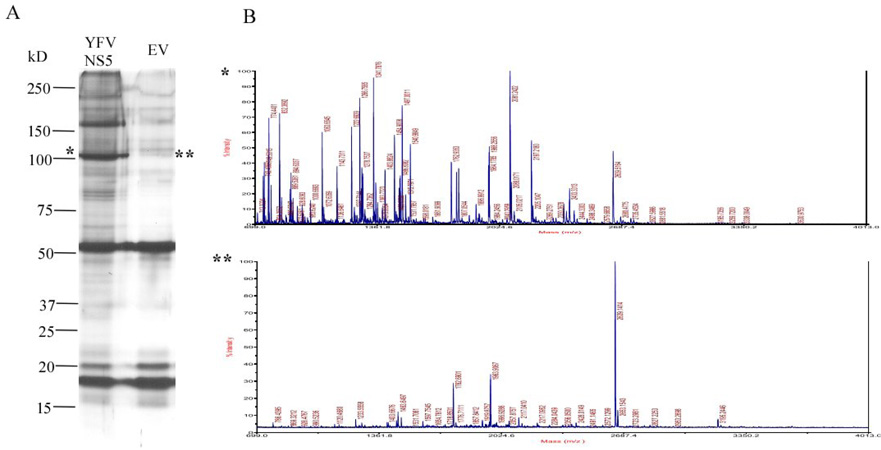
Fig. 1. Expression of wild type (WT) YFV NS5, empty vector control (EV) and their mass spectrometric (MS) analysis.
(A) HEK293T cells were transfected with WT YFV NS5 and EV separately. After 48 hours of transfection the proteins were pull down by 6X His and identified in 10% SDS-PAGE gel with colloidal staining method. 100 kD band is correspond to WT YFV NS5 which is absent EV control lane. (B) MALDI-TOF full scan spectra correspond to 100 kD size band of WT YFV NS5 (*) and EV control lane (**). The EV MS data was used as a background control in each peptide analysis.
Fig. 2. Map of identified phosphorylation sites.
A schematic diagram of the YFV Nonstructural Protein5 (NS5) showing the location and coordinate number of amino acids of identified phosphopeptides by mass spectrometry. The peptide 49–61 was further characterized because it contained a completely conserved serine and crystallographic data was available. Potential phosphorylated residues are shown in bold italics.
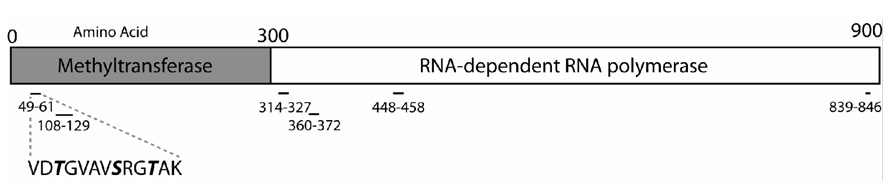
Further analysis was performed on peptide 49–61 because it had both structural data (Assenberg et al., 2007; Egloff et al., 2002; Mastrangelo et al., 2007; Zhou et al., 2007) and it contained a serine that was completely conserved for all viruses in the flavivirus genus. Fig. 3A is a MALDI-TOF plot showing the peptide (49–61, VDTGVAVSRGTAK) with a mass of 1341.7876, which is an increase of 80 daltons over the expected mass of this peptide, suggesting a phosphorylated amino acid. In order to confirm that Fig. 3A corresponds to a phosphopeptide, half of tryptic digested NS5 peptides was treated with calf intestinal alkaline phosphatase (CIAP). Fig. 3B shows the peptide present at 1341.7876 was lost after CIAP treatment suggesting the presence of a phosphate on one of three acceptors, T51, S56, or T59. Identification of the resultant dephosphosphorylated peptide is shown in Fig 3C with a mass of 1262.5821 correspond the same peptide as Fig 3A (49–61).
Fig. 3. Mass spectrometric analysis of tryptic digested peptides of wild type YFV NS5 expressed in HEK293T cells by MALDI-TOF.
(A) Spectra of singly charged monoisotopic peak of single phosphorylated peptide corresponding amino acid 49–61 with a mass of 1341.7876. The mass was calculated with an addition of 79.9 for posttranslational modification of one phosphorylated site. (B) YFV NS5 phosphopeptides treated with 20U of CIAP to remove phosphate showing the loss of 1341.7876 peak as present in A. (C) Recovery of the spectra of singly charged monoisotopic peak of YFV NS5 corresponding amino acid 49–61 with a mass of 1262.5821 indicated the loss of phosphate following CIAP treatment.
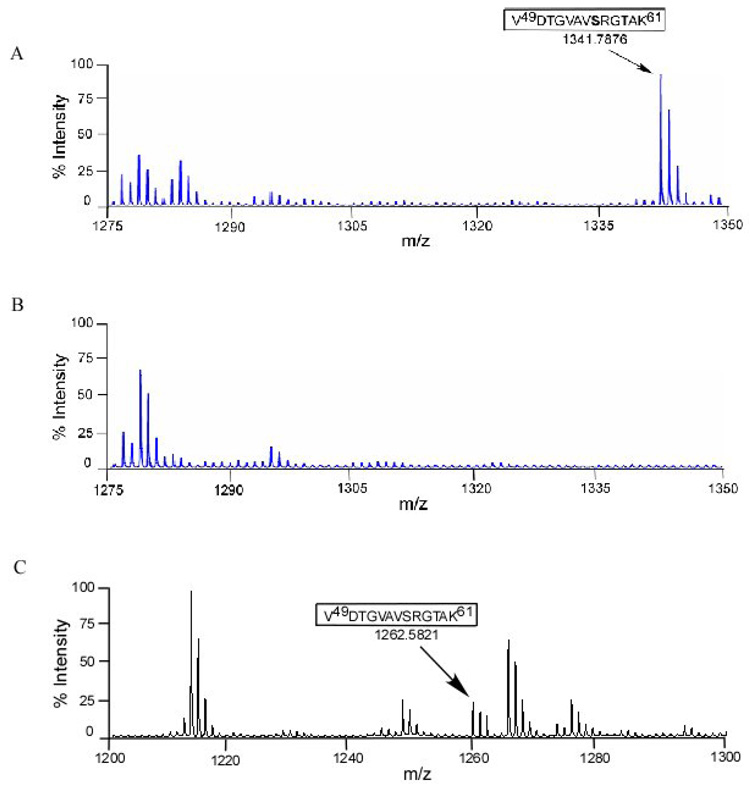
Both S56 and T59 were mutated to alanine in the NS5 expression vector and then analyzed for shifts in mass corresponding to a loss of phosphorylation (Fig. 4A, 4B). Fig. 4A shows the mass of 1522.7387 was detected for the S56A mutant, which is the expected mass for the alanine containing peptide in an unphosphorylated state. As T51 is also present in fragment 43–57, the lack of an increase in mass suggests that T51 is not phosphorylated, or that S56 is required for T51 phosphorylation. To determine whether T59 is phosphorylated the T59A mutant was analyzed by mass spectrometry in a similar fashion. We observed a peptide with a mass of 1309.6541, which is the predicted size for a VDTGVAVSRGAAK peptide with a single phosphorylation, suggesting that S56 (or possibly T51) is the most likely phosphorylated amino acid in the fragment originally observed in Fig. 3A. To identify the phosphorylated amino acid definitively, the wild-type expressed YFV NS5 was analyzed by ESI LC-MS/MS (Fig. 5). The peptide 49–57 (VDTGVAVSR) was found to have a single phosphorylation site and by measuring the masses of individual amino acids, S56 was determined to be phosphorylated.
Fig. 4. Mass spectrometric analysis of tryptic digested peptides of mutated YFV NS5 expressed in HEK293T cells by MALDI-TOF.
(A) Spectra of singly charged S56A mutated peptide at the mass of 1522.7387 corresponding to amino acid 43–57 showing YFV NS5 T51 was not phosphorylated. (D) Recovery of single charged monoisotopic peak of phosphorylated T59A mutated peptide with a mass of 1309.6541 corresponding amino acid 49–61 showing S56 could be the only phosphosite.
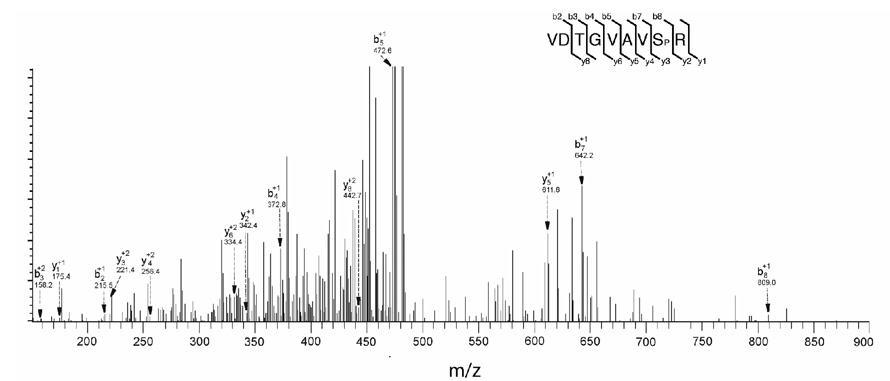
Fig. 5. LC-MS/MS ionizations of the singly phosphorylated peptide corresponding to amino acid 49–57 after in gel tryptic digest of YFV NS5 protein.
The peptide was passed through a C-18 HPLC column before fragmentations. Major fragment ions are labeled with their corresponding b (C-terminal) and y (N-terminal) ion and their charge state. The peak at m/z 809.00 represents the singly charged phosphorylated Serine (Sp) at the position of 56 in YFV NS5.
S56 may interact with S-adenosyl homocysteine in the YFV NS5 methyltransferase
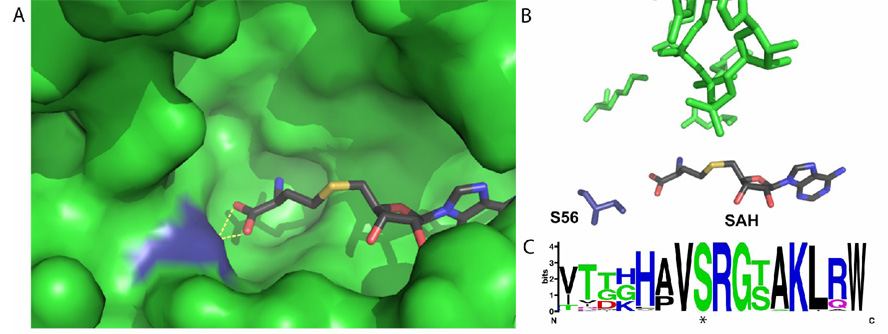
The methyltransferase domains of WNV and DV have recently been crystallized (Egloff et al., 2002; Zhou et al., 2007). Using biochemical and crystallographic analysis, several amino acids required for the methyltransferase reaction were identified as well as the binding sites for the 5’ cap or GTP and SAM, which are all highly conserved among flaviviruses (Dong et al., 2008). As shown in Fig. 6A, S56 lies very near the KDKE active site tetrad that catalyzes the transfer of a methyl group from SAM to the viral RNA. The hydroxyl of S56 directly interacts with S-adenosyl homocysteine, a product of SAM-dependent methylation (Egloff et al., 2002; Zhou et al., 2007). The addition of a phosphate to S56 would most likely alter the interaction between S56 and SAM, changing the orientation of the methyl donor of SAM required for methylation and possibly interfering with the binding of SAM (Fig. 6B). A sequence alignment and WebLogo analysis of more than 700 flavivirus NS5s shows a high level of conservation of not only S56, but also surrounding residues that typically constitute a kinase recognition sequence (Fig 6C) suggesting this region or at least S56 has evolutionary constraints on it.
Fig. 6. Crystallographic and bioinformatics analysis of the flavivirus methyltransferase active site.
(A) Pocket of Dengue Virus methyltransferase, with SAH in atomic coloring and S56 in blue. Hydrogen bonds between SAH and S56 are shown by yellow dotted lines (B) Direct interactions between S56 (dark blue) and SAH in the active site of Dengue Virus methyltransferase. The atoms shown in green are the canonical KDKE active site of all methyltransferases. The orientation of (A) and (B) are the same. (C) Logo analysis of the region around S56 from 715 distinct flaviviral genomes. The phosphorylated serine (S56) is starred. (alignment: http://athena.bioc.uvic.ca/workbench.php?tool=vocs&db=, for Logo: http://weblogo.berkeley.edu/).
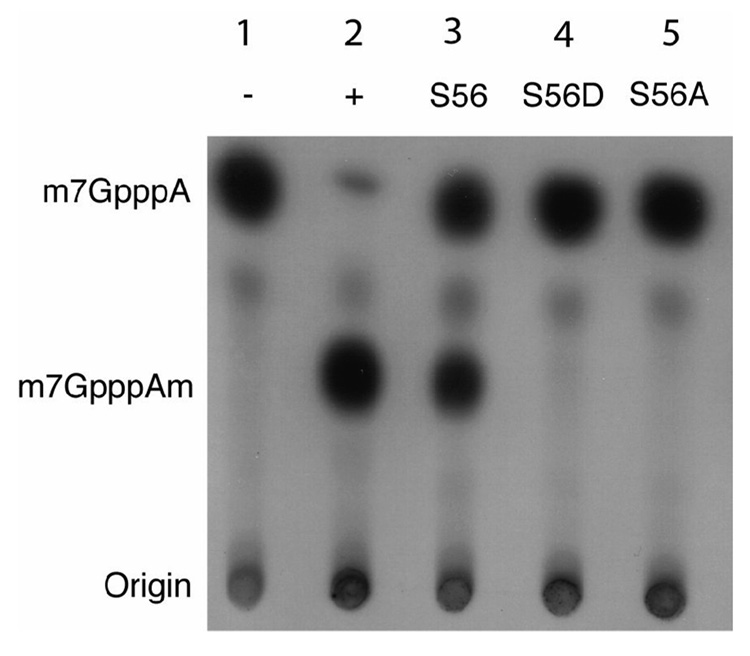
S56 is required for in vitro 2’-O methylation of viral RNA
S56 may serve a critical function in methyltransferase activity. In vitro methylation studies were performed to measure the effects of mutations on YFV NS5 methyltransferase activity. Wild type YFV NS5 methyltransferase, as well as two mutants, S56A and S56D, were cloned into an E. coli expression vector. After purification the 2’-O methyltransferase activity of each mutant was compared to both vaccinia 2’-O methyltransferase and wild-type YFV 2’-O methyltransferase activity and the products analyzed by thin layer chromatography (Dong et al., 2007). As shown in Fig. 7, WT NS5 MT was able to 2’-O methylate the N-7 methylated 5’ 250 bp fragment of the YFV 5’ RNA. S56A mutant NS5 was unable to carry out 2’-O methylation, indicating that this residue is essential for methyltransferase activity. Similarly, the S56D mutant, that mimics a phosphorylated serine, lacked 2’-O methyltransferase activity as well. These data suggest that phosphorylation of S56 may inhibit the methyltransferase activity of YFV NS5. When the same experiment was carried out with purified DV NS5 methyltransferase, similar results were observed (data not shown).
Fig. 7. In vitro methylating activity of YFV NS5s.
Authentic YFV RNA corresponding to the first 250bp that had been capped by VP39 of Vaccinia virus as well as S-adenosyl methionine were incubated with either a mock (1), vaccinia 2-0 methyltransferase (2), the wild type YFV methyl transferase (3), and mutant YFV NS5s S56D (4) and S56A (5). This reaction was then run out by TLC and the ability to perform 2’-O methylation was compared (middle spot, m7GpppA).
S56 hydroxyl is not required for SAH binding in the active site of YFV methyltransferase
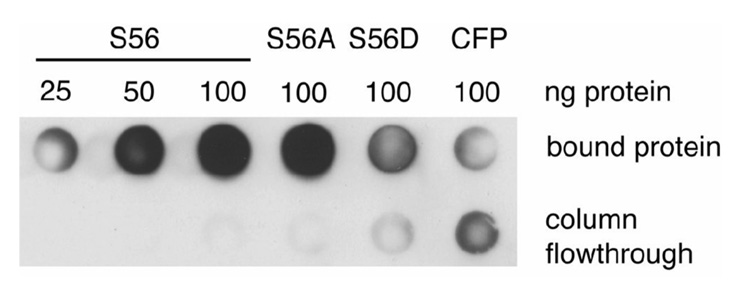
Since structural studies have shown that S56 interacts with SAH, it is possible that S56 is involved in the binding of SAH/SAM to the active site of the YFV methyltransferase (Zhou et al., 2007). In order to measure the effects of mutations of S56 on SAH binding, SAH was anchored to Sepharose 4B beads via an N-hydroxysuccinimidyl linkage. Purified protein was incubated with the SAH-Sepharose beads overnight and after being washed with 25 column volumes of wash buffer, bound protein was eluted and spotted on a dot blotter. The amount of protein was measured via western blot and there was no difference detected between the ability of the wild-type YFV methyltransferase and the S56A mutant to bind to SAH (Fig. 8). While some S56D binding to SAH-Sepharose beads could be detected above the level of a nonSAH binding control protein (CFP) it was clearly impaired relative to either S56A or wild-type.
Fig. 8. SAH binding of YFV methyltransferases.
SAH was covalently bound via the free amino group of the homocysteine moiety to Sepharose using an N-hydroxysuccinimidyl linkage. Purified 6X His tagged methyltransferases (amino acid number G1 to D296) of YFV wild type (WT), S56A, S56D and CFP were incubated overnight with SAH bound to Sepharose and after being washed with 50 column volumes of PBS+0.5% NP40, bound protein was eluted, spotted on a PVDF membrane via a dot blot apparatus and the amount of bound protein was measured by western blot with anti-His antibody.
S56 mutant YFV replicons are unable to replicate in HEK293T cells
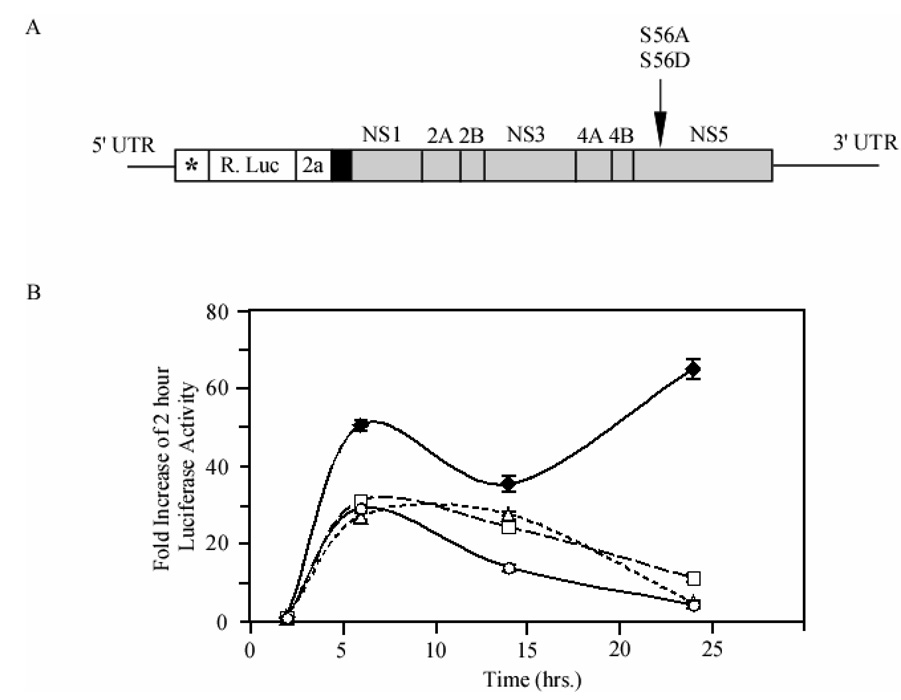
Individual YFV replicons were constructed containing the S56A or the S56D mutant (Fig. 9A). In vitro synthesized capped replicon RNA was transfected into HEK293T cells to measure the ability of various replicons to replicate over 24 hours. Methylation is essential for flavivirus replicons to replicate (Dong et al., 2008; Khromykh et al., 1999). Using a luciferase-reporter replication assay we monitored the production of protein from the input RNA, and the later replication of the RNA by viral proteins via luciferase that correlates with RNA levels (Jones, Patkar, and Kuhn, 2005). At 6 hours post-transfection, both wild type and mutant replicons showed increased luciferase readings compared to the initial 2-hour time point (Fig. 9B). The initial increase represents translation of transfected RNA prior to RNA replication, similar to the GAA mutant replicon, which is unable to copy RNA due to mutations in the active site of the polymerase. There was a decrease in signal at fourteen hours for all replicons. The wild-type replicon, however, was able to replicate the input RNA replicate and had the highest luciferase readings 24 hours post-transfection. S56A and S56D both showed little translation after initial transfection, a trend that continued 72 hours post-transfection (data not shown). Wild-type replicon showed maximum growth after 24 hours, in accordance with the length of YFV replication cycle and the wild-type growth curves looked similar to those seen for other flavivirus replicons (Holden et al., 2006). These data indicate S56 is required for replication of viral RNA and that methylation is an important part of this process.
Fig. 9. YFV replicon constructs and comparison of the effects of S56 mutations of YFV replicon growth.
(A) Schematic representation of YFV replicon construct (Jones et.al., 2005) where the NS5 S56A and S56D mutations were done and used to measured the luciferase activity. * indicates first 22 amino acids of capsid protein for RNA cyclization sequences; R. Luc, Renilla luciferase; 2a, 17 amino acid residue auto proteolytic peptide from foot and mouth disease virus; black box, NS1 signal sequence; gray box, nonstructural protein sequences; arrow indicates in NS5 where S56A and S56D mutations were done. (B) YFV replicon RNA was transfected into HEK293T cells and incubated over 24 hours. At various time points, cells were lysed and luciferase activity corresponding to translation of viral RNA was measured. Wild type virus (filled diamonds), a GAA polymerase-dead mutant (circles) were used as controls for S56A (triangles) and S56D (squares). Values were normalized to readings obtained at 2 hours post transfection to control for transfection efficiency.
Discussion
In this work we performed the most detailed analysis of flaviviral phosphorylation to date and identify six phosphorylated peptides from YFV NS5 all containing at least one serine or threonine. The putative serine/ threonine phosphoacceptor sites were largely conserved between different YFV genomes, but the conservation weakened when comparisons were made with more distant relatives including Tick borne flaviviruses. At least one peptide (peptide 839 see logos in supplemental data) contained a tyrosine highly conserved throughout the genus, while the threonine (position 844) was specific to YFV genomes. Previous studies detected only phosphothreonine/ serines in flavivirus (Kapoor et al., 1995; Morozova et al., 1997; Reed, Gorbalenya, and Rice, 1998). Additional experiments will be required distinguish between a YFV specific phosphothreonine, and a phosphotyrosine that is now detectable due to advances in methodology. One peptide corresponding to a portion of the active site the methyltransferase portion of NS5 with three potential phosphoacceptor sites was examined in greater detail. Both mutational and additional mass spectrometry data identified the phosphoacceptor site as serine 56, which is completely conserved in the genus flavivirus. Mutational and biochemical analyses indicated this residue is essential for YFV RNA 2’-O methylation and replication.
RNA capping and methylation are essential processes for viruses that depend upon the full host translational apparatus for translation of viral proteins. RNA capping methyltransferases are hypothesized to function by aligning the methyl donor group of SAM with the RNA such that methyl transfer may occur via an SN2 reaction (Fabrega et al., 2004). S56 interacts with the carbonyl group of SAM in order to position the donor methyl group and the addition of a phosphate at S56, from both a charge and steric standpoint, would almost certainly reposition the methyl donor group of SAM. In the crystal structure of Ecm1, a eukaryotic 2’-O RNA methyltransferase, Lysine 54 showed similar bonding geometry as S56 in relation to the carbonyl group of SAM. K54 mutants were found to be catalytically inactive (Fabrega et al., 2004). Recently, a serine to alanine mutant at S56 in WNV was also shown to be defective for methylation and viral replication (Dong et al., 2008). Dong et al also found that R57 may play a role in RNA binding and is required for methyltransferase activity in WNV. The addition of a phosphate at S56 may inhibit methyltransferase activity in two ways. First, the phosphate blocks direct access to S56 and secondly may reposition another essential amino acid, R57. Both S56 and R57 are conserved among all 715 flavivirus examined (Fig. 6C). While both S56A and S56D defective in 2’-O methylation the hydroxyl group of S56 does not appear to be essential for SAM binding as long as it is not replaced with a negative charge (Fig. 8).
Our work is consistent with and extends prior work that multiple viral serines and threonines in NS5 can be phosphorylated (Kapoor et al., 1995; Reed, Gorbalenya, and Rice, 1998) and these modifications may be reversible. Neither the phosphomimetic D, nor the unphosphorylatable A at position 56 were functional in a replicon (Fig 9). This may imply a transient phosphorylation at this site. At least in the case of dengue virus, one event occurs late in the lifecycle and coincides with a “hyperphosphorylated” form of NS5 (Kapoor et al., 1995). Molecular mapping of phosphorylation sites gives clues toward the identities of kinases involved. For example the S56 site is predicted by neural net bioinformatic program (NetPhosK (Blom et al., 2004)) to be a CK1 recognition site, the same kinase responsible for hyperphosphorylation in HCV (Quintavalle et al., 2007). Our data do not directly show that S56 is phosphorylated in the course of a normal infection. Our data does suggest that the hydroxyl sidechain of S56 is required for methylation to occur. We do not predict that S56 would be phosphorylated early in the course of infection if at all. One attractive possibility though, is that at least some NS5 molecules are phosphorylated at S56 late in infection. Phosphorylation at S56 of even a portion of NS5 molecules would serve to decrease capping and protein synthesis viral protein and allow more viral RNA to be used for replication. At present we have no data that argues phosphorylation regulates the conversion from translation to replication, but the shunting of some RNA molecules for replication is an essential step for the virus. Further investigation is required to determine the role of phosphorylation in this step. The identification of these phosphopeptides may facilitate the identification of specific kinases that interact with flaviviruses.
Materials and methods
Genetic constructs
The YF-R.luc2A-RP, as well as YF-R.luc2A-RP GAA, replicons were provided by Dr. Richard Kuhn, Purdue University, West Lafayette, IN (Jones, Patkar, and Kuhn, 2005). YF-R.luc2A-RP contains an additional G to facilitate T7 transcription yet is competent for both replication and expresses Renilla luciferase as a reporter. The YF-R.luc2A-RP GAA replicon is deficient in RNA replication due to mutations in the active site of the polymerase.
YFV NS5/IRES/GFP construc
Full length YFV NS5 (PFLYF17D, GenBank Accession No. X03700) was amplified from a DNA template by PCR with primers that introduce a XhoI site, Kozak sequences and an in frame start codon ATG 5’ to the NS5 sequence and a PacI site, 6XHis in frame and a stop codon at 3’ end. ~2.8kb PCR product was digested and cloned to pIRES/GFP bicistronic mammalian expression vector (provided by Dr Kouacou V. Konan, Penn State University, University Park, PA) in XhoI-PacI sites and sequenced.
YFV NS5 mutant/IRES/GFP constructs
YFV NS5 S56A and YFV NS5 T59A were generated by an overlap PCR strategy (Ho et al., 1989) with introduction of individual mutations in overlapping primer pairs. Second step PCR products were directly cloned into WT pYFV NS5 6XHis IRES/GFP construct by replacing XhoI-SacII fragment and sequenced. YFV NS5 methyltransferase constructs: 888 nt long (YFV NS5 amino acid coordinate number G1 to D296) fragment was amplified from YFV NS5/IRES/GFP construct by PCR with introduction of BamHI site at the 5’ and HindIII site at 3’ ends. The PCR products were then cloned directly to pQE-30 N-terminus 6XHis prokaryotic expression vector (Qiagen, Valencia, CA) in frame at BamHI-HindIII sites and sequenced. YFV NS5 S56A and YFV NS5 S56D were generated by overlap PCR strategy (Ho et al., 1989) with introduction of individual mutation in overlap primer pairs. Second step PCR products were directly cloned to pQE-30 N-terminus 6XHis WT YFV NS5 MTase construct by replacing XhoI-SacII fragment and sequenced. For negative control construct full length (717nt long) of enhanced cyan fluorescent protein (CFP) was cloned in same pQE-30 N-term 6X His vector and sequenced.
YF-R.luc2A-RP NS5 S56A and S56D replicons
Quick-change method was performed to generate YF-R.luc2A-RP NS5 S56A and YF-R.luc2A-RP NS5 S56D replicons. The PCR reactions consisted of 200ng YF-R.luc2A-RP replicon template DNA, 40pmol of each mutated primer, 25mM dNTPs and 2.5U Pfu Ultra II Fusion HS DNA polymerase (Stratagene, La Jolla, CA). Amplifications were carried out for 30 cycles at 92°C for 10s followed by both annealing and extension at 68°C for 30s/kb of plasmid. 40µl of the PCR reactions were directly treated with 20U of DpnI (New England Biolabs, Ipswitch, MA) at 37°C for 2 hours and inactivated DpnI enzyme at 80°C for 20min. 2 µl of these DpnI treated linear PCR products were electroporated in to E. coli Top10 electrocompetent cells. Desalted primers were synthesized (IDT, Coralville, IA) and were as follows: for NS5 S56A the forward primer was 5’-GGGGTGGCGGTCGCCAGGGGGACCGCAAAGTTAAGG – 3’ and for NS5 S56D the forward primer was 5’-GGGTGGCGGTCGACAGGGGGACCGCAAAGTTAAGG – 3’ (bold letters are where the mutation introduced). The reverse primers were exactly the complementary sequences of the respective forward primers.
Expression and purification of YFV NS5 proteins in HEK293T cells
GFP in the IRES vector was used as transfection/expression marker. Transfection of the empty vector was performed in parallel in each case as a negative control. 15µg of either YFV NS5 DNA or empty vector DNA were transfected in 70–80% confluent HEK293T cells in a T75 flask. HEK 293T cells were maintained at 37°C in 5% CO2 incubator in DMEM (Invitrogen, La Jolla, CA) supplemented with 10% FBS and 1X Penicillin-Streptomycin solution. TransIT-LT1 (MirusBio, Madison, WI) was used as a transient transfection reagent and the transfection was carried out in presence of Opti-MEM1 (Invitrogen, CA). After 48 hours of transfection the cells were collected, washed several times in 10mM phosphate buffer and homogenized in ice cold lysis buffer (50mM NaH2PO4, 2H2O; 300mM NaCl; 10mM Imidazole; 0.05% Tween-20; 1% NP-40, pH 8.0) in presence of EDTA-free 1X halt protease inhibitor cocktail and 1X halt phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA). After collecting the lysate by centrifugation, the 6XHis recombinant proteins were bound with lysis buffer equilibrated Ni-NTA magnetic agarose beads (Qiagen, CA) at 4°C for 2hrs 30min. The beads were washed thoroughly in wash buffer (50mM NaH2PO4, 2H2O; 300mM NaCl; 10mM Imidazole; 0.05% Tween-20; ph 8.0) and the 6XHis proteins were eluted from magnetic beads in elution buffer (50mM NaH2PO4, 2H2O; 300mM NaCl; 250mM Imidazole; 0.05% Tween-20; ph 8.0).
Sample preparations for MS
6XHis purified proteins (both experimental and empty vector control) from HEK293T cells were run in 10% SDS-PAGE gels. The gels were stained in GelCode Blue stain (Thermo Fisher Scientific, MA). An ~100kD band was excised from the gel. Gel pieces containing protein were destained in 25mM NH4HCO3 in 50% ACN at 37°C, reduced in 50mM TCEP in 25mM NH4HCO3 at 60°C, alkylated in 100mM IAA in 25mM NH4HCO3 in the dark at room temperature, dehydrated in 100% ACN at room temperature and digested with trypsin (Trypsin Gold, Mass Spectrometry Grade - Promega, Madison, WI) at 30°C overnight. Peptides were collected and divided in two. Half of these peptides were enzymatically dephosphorylated with 20U of CIAP (New England Biolabs, Beverly, MA) at 37°C for 2hours and the reaction was stopped by adding 50mM EDTA. Both phosphorylated and dephosphorylated peptides were cleaned up with OMIX C18 Zip-Tip (Varian, Harbor City, CA) and eluted either in 0.1% TFA in 75% ACN for MALDI-TOF or in 0.1% formic acid in 50% ACN for ESI analysis.
Sample loading and analysis
Both Matrix-Assisted Laser Desorption/Ionization–Time of Flight (MALDI-TOF) and Electrospray Ionzation (ESI) were done at the University of Wisconsin mass spectrometry facilities. 10mg/ml α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, St. Louis, MO) was used as matrix and added in the samples in a ratio of 1:1. Samples were spotted in a MALDI plate and dried at room temperature. MALDI data were obtained using a MDS SCIEX 4800 MALDI TOF/TOF (Applied Biosystem, Foster City, CA) instrument. The mass spectra were collected from m/z 700 to 4000 and preliminary analysis was performed in GPS Explorer software (Applied Biosystem, Foster City, CA). After Mascot database (Matrix Science, London, UK) search and the data comparison before and after CIAP treatment (wild-type only), detailed matching of mass with peptide identification was performed in MS-Digest tools (a web based software in ProteinProspector, v 4.27.1). ESI mass spectrometry was performed for LC-MS/MS analysis. ESI precursor ion scan mass spectra were obtained using Finnigan LTQ Linear Ion Trap mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source. Zorbax 300SB-C18 nanoflow HPLC column (150mm × 75µm, Agilent Technologies, Santa Clara, CA) was eluted with a linear gradient of water-acetonitrile in the presence of 0.1% formic acid at a flow rate of 0.2µl/min. Rest of the protocol was followed as described elsewhere (Black et al.,2007) and full scan spectra were recorded from m/z 200–2000 followed by MS/MS spectra in Xcalibur software (Thermo Fisher Scientific). MS/MS spectra of detected phosphopeptide were searched against the FASTA database using Sequest search program within BioWorks Rev3.3 software (Thermo Fisher Scientific).
Expression and purification of YF methyltransferase domain and CFP in bacterial cells
The pQE-30 6XHis tag methyltransferase constructs and pQE-30 6X His tag CFP construct were transformed in M15[pREP4] E.coli (Qiagen, CA). The cells were grown at 37°C until an OD600 of 0.7 and expression was induced by adding 1mM isopropylthio-β-D-galactoside (IPTG) at 28°C for 5 hours. The recombinant proteins were then bound to Ni-NTA agarose (Qiagen) and purified according to manufacturer protocol.
SAH affinity chromatography
Five ml of SAH-labeled Sepharose was made as follows: NHS-Sepharose (Sigma, St. Louis, MO) was spun at 1000g for one min and the supernatant was removed. 40ml of 1mM HCl was added to the Sepharose and then removed by centrifugation. 75mmol of SAH was dissolved in coupling buffer (0.2M NaHCO3, 0.5M NaCl, pH 8.3) and incubated with the activated NHS-Sepharose overnight in a rotary shaker at 4°C. Supernatant was removed by centrifugation and the beads were blocked for two hours in ten ml of 0.1M Tris HCl, pH 8.5. After blocking, beads were washed with five cycles of alternating low pH (0.1M Acetate buffer, 0.5M NaCl, pH 4.5) and high pH (0.1M Tris HCL pH 8.5) and stored in 20% ethanol at 4°C. To measure the ability of each YFV methyltransferase to bind SAH, 100µl of beads were sedimented by centrifugation to remove ethanol and washed twice with 500µl of binding buffer (0.2M KCl, 50mM Tris HCl pH 7.0, 1mM β-mercaptoethanol, 1mM dithiothreitol). Beads were then incubated with 100ng of purified recombinant 6XHis-NS5 in 500µl of binding buffer overnight at 4°C. Unbound protein was collected by centrifugation at 1000g for one min and beads were washed with 50 column volumes of PBS + 0.5% NP40 and collected. Bound protein was eluted in 4M guanidium HCl and spotted via a Minifold I microsample filtration manifold dot blotter (Schleicher & Schuell, Keene, NH) onto Immobilon-P PVDF membrane (Millipore, Billerica, MA). After drying, protein was visualized via Western blot using a 6XHis antibody (Qiagen) and developed via ECL.
RNA substrates
RNA substrates for methyltransferase assays were generated similar way described elsewhere (Dong et al., 2007). YFV RNA representing the first 250bp of the genome was in vitro transcribed from PCR products generated with primers 1 (TAA TAC GAC TCA CTA TTA GTA AAT CCT GTG TGC TAA TTG AG) and 2 (AAC ACC TCT TGA AGG TCC AG). Primer 1 contains a bacteriophage T7 class II Φ2.5 promoter (italicized) that allows for ATP-initiated transcript using T7 RNA polymerase (Coleman, Wang, and Huang, 2004). In vitro YFV transcripts were capped with (α-32P) G*TP and Vaccinia virus guanylyltansferase (Applied Biosystems) in the absence or presence of SAM to generate G*pppA-RNA or m7-G*pppA-RNA. To enhance the generation of G*pppA-RNA, (0.1 units) inorganic pyrophosphatase (New England Biolabs, MA) was added to the capping reaction. The resulting capped transcripts were purified through a G-50 column (Amersham, Piscataway, NJ) and used for N7 and 2–O methylations.
Cap methylation assays
The N-7 methylation reaction was performed at 22°C for 30 min in N7 buffer (50mM Tris, pH 7.0, 50mM NaCl, 2mM DTT, 1mM MgCl2, and 6mM KCl) containing 5000 CPM G*pppA-YFV RNA, 1mg purified Methyltransferase and 100 mM SAM. Control m7GpppA-RNA was prepared using Vaccinia virus guanylyltransferase as described by the manufacturer. The 2’-O methylation reaction was performed at 22°C for two hours in 2’-O buffer (50mM glycine, pH 10, 2mM DTT, 1mM MgCl2, and 6mM KCl) containing 5000 CPM m7G*pppA-YFV RNA, 1mg purified Methyltransferase and 100 mM SAM. Control m7GpppAm-RNA was prepared using Vaccinia virus Cap 1 methyltransferase (Epicentre, Madison, WI) following the manufacturers protocol. Methylation reactions were digested for one hour at 68°C with Nuclease P1 (US Biological, Swampscott, MA) and analyzed by polyethyleneimine cellulose thin layer chromatography in 0.2M ammonium sulfate.
Analysis of the effects of mutations on replicon growth
YFV replicon DNA was linearized by treating with Xhol for four hours and ethanol precipitated. Linearized DNA was then used to generate full-length replicon RNA using the mMessage mMachine in vitro RNA transcription kit (Ambion, Austin, TX), and cleaned up via LiCl precipitation. HEK293T cells were transfected with purified RNA by plating 50,000 cells per well of a 48-well plate and incubating overnight. Media was removed and replaced with 25µL of new media before transfection. Transfection mixture contained 0.25µg of RNA per well in 25µL of OptiMEM-1 growth media, along with 0.25µl of mRNA Boost and 0.5µl of Trans-IT mRNA transfection mixture (MirusBio, Madison, WI). Transfection mixture was incubated for three minutes before being added to each well. Cells were then incubated for 2, 6, 12, or 24 hours at which time media was removed and cells were lysed with 50µl of Renilla Lysis buffer from the Renilla Luciferase Assay system (Promega, Madison, WI). 10µl of cell lysate was added to 50µl of Renilla Luciferase Assay buffer and read in triplicate on a Turner Designs TD20/20 Luminometer.
Crystallographic and bioinformatics analysis
All crystallographic analysis was performed on a crystal structure of the dengue methyltransferase including 7MeGpppA and SAH (pdb id 2P3O, (Egloff et al., 2007)) using MacPyMol (DeLano). Logo analysis was performed with WebLogo after aligning all 715 flaviviral methyltransferases available in the Viral Bioinformatics Resource Center using the Viral Orthologous Cluster program (Crooks et al., 2004; Hiscock and Upton, 2000; Schneider and Stephens, 1990).
Supplementary Material
Acknowledgements
This work was sponsored by the NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program. The authors wish to acknowledge membership within and support from the Region V 'Great Lakes' RCE (NIH award 1-U54-AI-057153). We thank the UW Madison Biotechnology mass spectrometry facility and the Human Proteomics Mass Spectrometry Facility supported by the "The Wisconsin Partnership Fund for a Healthy Future” for technical assistance. We also thank Janet Smith and Richard Kuhn for helpful discussions, and Deane Mosher for comments on drafts of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Assenberg R, Ren J, Verma A, Walter TS, Alderton D, Hurrelbrink RJ, Fuller SD, Bressanelli S, Owens RJ, Stuart DI, Grimes JM. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J Gen Virol. 2007;88(Pt 8):2228–2236. doi: 10.1099/vir.0.82757-0. [DOI] [PubMed] [Google Scholar]
- Black TM, Andrews CL, Kilili G, Ivan M, Tsichlis PN, Vouros P. Characterization of phosphorylation sites on Tpl2 using IMAC enrichment and a linear ion trap mass spectrometer. J Proteome Res. 2007;6(6):2269–2276. doi: 10.1021/pr0700293. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Coleman TM, Wang G, Huang F. Superior 5' homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 2004;32(1):e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System. 2002 on World Wide Web http://www.pymol.org.
- Dong H, Ray D, Ren S, Zhang B, Puig-Basagoiti F, Takagi Y, Ho CK, Li H, Shi PY. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J Virol. 2007;81(9):4412–4421. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, Shi PY. West Nile Virus Methyltransferase Catalyzes Two Methylations of the Viral RNA Cap through a Substrate-Repositioning Mechanism. J Virol. 2008;82(9):4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B. Structural and functional analysis of methylation and 5'-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol. 2007;372(3):723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13(1):77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, Vasudevan SG. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem Biophys Res Commun. 1999;257(3):731–737. doi: 10.1006/bbrc.1999.0370. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscock D, Upton C. Viral Genome DataBase: storing and analyzing genes and proteins from complete viral genomes. Bioinformatics. 2000;16(5):484–485. doi: 10.1093/bioinformatics/16.5.484. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3' stem-loop structure. Virology. 2006;344(2):439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Jakubiec A, Jupin I. Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation. Virus Res. 2007;129(2):73–79. doi: 10.1016/j.virusres.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Patkar CG, Kuhn RJ. Construction and applications of yellow fever virus replicons. Virology. 2005;331(2):247–259. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Kenney MT, Westaway EG. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72(9):7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol. 1999;73(12):10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kim JH, Kim YG, Lim HS, Oh JW. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J Biol Chem. 2004;279(48):50031–50041. doi: 10.1074/jbc.M408617200. [DOI] [PubMed] [Google Scholar]
- Mastrangelo E, Bollati M, Milani M, Selisko B, Peyrane F, Canard B, Grard G, de Lamballerie X, Bolognesi M. Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase. Protein Sci. 2007;16(6):1133–1145. doi: 10.1110/ps.072758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrof J, Muhlberger E, Klenk HD, Becker S. Phosphorylation of VP30 impairs ebola virus transcription. J Biol Chem. 2002;277(36):33099–33104. doi: 10.1074/jbc.M203775200. [DOI] [PubMed] [Google Scholar]
- Mohr I. Phosphorylation and dephosphorylation events that regulate viral mRNA translation. Virus Res. 2006;119(1):89–99. doi: 10.1016/j.virusres.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Morozova OV, Tsekhanovskaya NA, Maksimova TG, Bachvalova VN, Matveeva VA, Kit Y. Phosphorylation of tick-borne encephalitis virus NS5 protein. Virus Res. 1997;49(1):9–15. doi: 10.1016/s0168-1702(96)01433-5. [DOI] [PubMed] [Google Scholar]
- Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, Bartholomew L, De Francesco R. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J Virol. 2004;78(23):13306–13314. doi: 10.1128/JVI.78.23.13306-13314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8(7):795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- Quintavalle M, Sambucini S, Summa V, Orsatti L, Talamo F, De Francesco R, Neddermann P. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J Biol Chem. 2007;282(8):5536–5544. doi: 10.1074/jbc.M610486200. [DOI] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5'-cap structure is formed by sequential guanine N-7 and ribose 2'-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KE, Gorbalenya AE, Rice CM. The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol. 1998;72(7):6199–6206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28(6):329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger B, Traktman P. Vaccinia virus morphogenesis: a13 phosphoprotein is required for assembly of mature virions. J Virol. 2004;78(16):8885–8901. doi: 10.1128/JVI.78.16.8885-8901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WorldHealthOrganization. Dengue Factsheet. WHO; 2002
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.