Abstract
We have developed a strategy for isolating cry genes from Bacillus thuringiensis. The key steps are the construction of a DNA library in an acrystalliferous B. thuringiensis host strain and screening for the formation of crystal through optical microscopy observation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses. By this method, three cry genes—cry55Aa1, cry6Aa2, and cry5Ba2—were cloned from rice-shaped crystals, producing B. thuringiensis YBT-1518, which consists of 54- and 45-kDa crystal proteins. cry55Aa1 encoded a 45-kDa protein, cry6Aa2 encoded a 54-kDa protein, and cry5Ba2 remained cryptic in strain YBT-1518, as shown by SDS-PAGE or microscopic observation. Proteins encoded by these three genes are all toxic to the root knot nematode Meloidogyne hapla. The two genes cry55Aa1 and cry6Aa2 were found to be located on a plasmid with a rather small size of 17.7 kb, designated pBMB0228.
Bacillus thuringiensis is a gram-positive, spore-forming bacterium that produces crystal inclusions during the sporulation phase. The crystals comprise one or more Cry proteins (δ-endotoxins) that are specifically toxic to insect orders such as Lepidoptera, Diptera, and Coleoptera and also to some nematodes, mites, and protozoa (23). The insecticidal Cry proteins are encoded by cry genes. Since the cloning of the first cry gene by Schnepf and Whiteley (24), more than 300 cry genes have been isolated from B. thuringiensis (refer to Crickmore et al. [7] and the toxin nomenclature website http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/toxins2.html). These Cry proteins are classified into families Cry1 to Cry54 on the basis of their amino acid sequence homology. Among these 54 families, Cry5, Cry6, Cry12, Cry13, Cry14, and Cry21 have been shown to display nematicidal activity.
Agriculturally important nematodes usually live underground (29), which makes them difficult to control using traditional B. thuringiensis insecticides. One of the most effective approaches for controlling plant-parasitical nematodes has been constructing transgenic plants with nematicidal cry genes. However, most of the nematicidal cry genes have been described only in patents with sparse data (28); except for Cry6, most of the nematicidal Cry proteins are large proteins (90 to 140 kDa) that are difficult to use in transgenic manipulations. It is therefore imperative that more-detailed studies be carried out on the cloning of nematicidal cry genes, especially those whose products have small molecular weights.
Typically, cry genes have been cloned by constructing B. thuringiensis DNA libraries for Escherichia coli and then screening a great number of colonies by Western blotting (20, 24) or a Southern hybridization-based method (4, 13, 15, 19). Unfortunately, these methods require large amounts of Cry proteins in order to prepare the antibody or to facilitate amino acid sequencing, thus making it impossible to isolate cryptic or silent cry genes. More recently, cry genes have been screened with PCR-based methods (5, 6, 9, 14, 27), which are very rapid and effective in principle. However, these methods depend upon there being a high similarity between different cry genes; in using these methods, novel cry genes that have little or no identity to known genes would be lost. Furthermore, genes with little or no identity in the primer regions would not be amplified using this strategy. New strategies for identifying such novel cry genes have yet to be developed.
Most B. thuringiensis cry genes reside on large plasmids (23). The only exception to date was reported by Loeza-Lara et al. in 2005 (17): a cry-like gene, cry14-4, found in a small plasmid, pBMBt1, from a B. thuringiensis strain. The predicted protein sequence showed low identity with the proteins CryC53 (24.6%) and Cry15Aa (27.8%). Apart from the coding sequence, there is no detailed experimental evidence available regarding the Cry14-4 protein; whether other small plasmids from B. thuringiensis harbor cry genes or not remains uncertain.
This work presents a strategy for isolating new cry genes by screening a plasmid library in an acrystalliferous B. thuringiensis host strain. This method enables the isolation of the cry genes cry55Aa1, cry6Aa2, and cryptic cry5Ba2 from the YBT-1518 strain, which produces rice-shaped crystals and displays nematicidal activity. Of these three genes, the first two were found to be located on a 17.7-kb plasmid.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultural conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The nematicidal B. thuringiensis strain YBT-1518 was isolated in China (30). The E. coli mutant strain DH10B and the acrystalliferous “B. thuringiensis subsp. kurstaki” strain BMB171 from Li and Yu (16) were used as the intermediate and final hosts, respectively, in the plasmid library construction. The cloning vector was the E. coli-Bacillus shuttle vector pHT304 (4). B. thuringiensis was grown at 28°C and E. coli at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Organism | Plasmid | Characteristic | Cry protein production | Source |
|---|---|---|---|---|---|
| YBT-1518 | B. thuringiensis | Wild type | Cry55Aa1, Cry6Aa2 | Stored in our lab | |
| BMB171 | B. thuringiensis | None | Host strain | None | Li and Yu (16) |
| DH10B | E. coli | Host strain | Stored in our lab | ||
| BMB0222 | B. thuringiensis | pBMB0222 | Recombinant strain | Cry55Aa1 | This study |
| BMB0223 | B. thuringiensis | pBMB0223 | Recombinant strain | Cry55Aa1 | This study |
| BMB0224 | B. thuringiensis | pBMB0224 | Recombinant strain | Cry55Aa1 | This study |
| BMB0249 | B. thuringiensis | pBMB0249 | Recombinant strain | Cry6Aa2 | This study |
| BMB0250 | B. thuringiensis | pBMB0250 | Recombinant strain | Cry6Aa2 | This study |
| BMB0215 | B. thuringiensis | pBMB0215 | Recombinant strain | Cry5Ba2 | This study |
Plasmid isolation and DNA manipulation.
Plasmids were extracted from B. thuringiensis according to the procedure of Andrup et al. (2) and from E. coli according to the methods of Sambrook and Russell (22). The extraction of DNA from gel was performed using an AxyPrep DNA gel extraction kit (Axygen Scientific, Inc.). The DNA restriction and ligation operations were performed according to the methods of Sambrook and Russell (22). The preparation of the cloning vector pHT304 and determination of the insert size were carried out as described by Luo and Wing (18).
Southern hybridization.
DNA samples were separated in 0.8% agarose gel and transferred onto a nylon membrane, according to the procedure of Sambrook and Russell (22). The PCR-amplified probe labeling, hybridization, and detection of the result were all performed according to the insert of the DIG High Prime DNA labeling and detection starter kit I (Roche Applied Science, Germany).
Sequence analysis.
Open reading frame prediction was performed using the CLC Free Workbench 4 program (CLC bio, Denmark). DNA and protein sequence homology searches were performed using the BLAST algorithm (1) against the GenBank database.
Electroporation of E. coli and B. thuringiensis.
E. coli DH10B electroporation-competent cells were prepared according to the procedure of Sambrook and Russell (22); the transformation procedure was performed according to the method of Luo and Wing (18). B. thuringiensis BMB171 transformation was according to the modified method of Silo-Suh et al. (26). The transformants were selected on LB (22) agar plates with erythromycin (25 μg/ml).
Microscopy observation.
An optical microscope was used to observe crystal formation. Bacteria were cultivated on CCY agar plates containing casein hydrolysate and yeast for more than 72 h at 28°C (9); the cells were then stained with basic fuchsin by a simple staining procedure and observed with an oil immersion lens. Initially, 10 colonies per group were observed together, and then every colony in the crystal-producing groups was observed separately. For scanning electron microscopy observation, the bacteria were treated by following the methods of Shao et al. (25).
SDS-PAGE and crystal protein purification.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to examine the Cry proteins from the positive colonies. For crystal protein purification, bacteria were cultivated in liquid ICPM medium (25) for 36 h at 28°C with erythromycin (25 μg/ml). The crude spore lysate pellets were then treated using the method of Griffitts et al. (10). The purified proteins were treated and loaded as described by Shao et al. (25).
Nematode toxicity bioassay.
The purified Cry proteins were used for bioassay. Meloidogyne hapla eggs were harvested from the root knot of infected tomato, and the second-stage juveniles were reared at 18 to 25°C and used to test the toxicity of the Cry toxin. The bioassay procedure and 50% lethal concentration (LC50) evaluation were undertaken according to the method of Yu et al. (30).
Nucleotide sequence accession numbers.
The nucleotide sequences published in this paper have been submitted to GenBank and assigned accession numbers EU121521 (cry55Aa1), AF499736 (cry6Aa2), and EU121522 (cry5Ba2).
RESULTS
Construction of a library of plasmid DNA from strain YBT-1518 in BMB171.
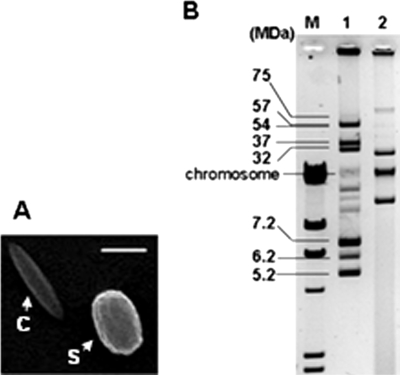
B. thuringiensis strain YBT-1518 produces rice-shaped crystals (Fig. 1A) and has at least three plasmids (Fig. 1B). The plasmid DNA from YBT-1518 was isolated and partially digested with HindIII. The resulting 5- to 12-kb fragments were extracted and ligated into pHT304. The ligation mix was transformed into E. coli DH10B. Plasmids present in the recombinant E. coli pool were then extracted and transferred into the acrystalliferous B. thuringiensis strain BMB171 by electroporation, resulting in a library of strain YBT-1518 plasmids.
FIG. 1.
Crystal morphology and plasmid profile of YBT-1518. (A) Crystal morphology of YBT-1518. Abbreviations: C, crystal; S, Spore. Spores and crystals from strain YBT-1518 were harvested and washed three times with 1 M NaCl and then three times with water. The spore and crystal mix was resuspended in water and spread on a microscope slide to be ion coated; it was then observed under a scanning electron microscope (JEOL JSM-6390LV). The bar is 1 μm. (B) Plasmid profile of YBT-1518 in 0.8% agarose gel. Lane M, DNA molecular mass λ DNA/HindIII markers; lane 1, plasmids from B. thuringiensis HD-2 as size references; lane 2, plasmids from YBT-1518.
The average size of the inserts in the library was about 8 kb. Approximately 1,000 recombinants were stored in the library, of which 300 were first randomly selected for the screening process described below.
Screening recombinants from the library harboring cry genes.
Each of the 300 recombinants selected in the previous step was observed under an optical microscope to determine whether it formed crystals. Seven recombinants were found to produce the same rice-shaped crystal as that in strain YBT-1518. SDS-PAGE analysis revealed that three of them could produce a 45-kDa crystal protein (for example BMB0222 and BMB0223) and that four of them could produce a 54-kDa protein (such as BMB0249). There was one recombinant, BMB0215, that produced a bipyramidal crystal (Fig. 2) comprising a 140-kDa protein (Fig. 3).
FIG. 2.
Optical microscope graphs of parasporal crystal proteins from the wild-type strain B. thuringiensis YBT-1518 and the recombinant strains BMB0224, BMB0250, and BMB0215. (A) YBT-1518; (B) BMB0224 (harboring the 45-kDa protein gene cry55Aa1); (C) BMB0250 (harboring the 54-kDa protein gene cry6Aa2); (D) BMB0215 (harboring the 140-kDa protein gene cry5Ba2). Magnification, ×1,000.
FIG. 3.
SDS-PAGE analysis of crystal proteins from the wild-type strain and recombinant strains of B. thuringiensis. Lane M, molecular mass standards; lane 1, YBT-1518; lane 2, BMB0224 (harboring the 45-kDa protein gene cry55Aa1); lane 3, BMB0250 (harboring the 54-kDa protein gene cry6Aa2; lane 4, BMB0215 (harboring the 140-kDa protein gene cry5Ba2).
Identification of cry55Aa1, cry6Aa2, and cry5Ba2.
The analysis of a 45-kDa protein-producing recombinant, BMB0223, showed that the recombinant plasmid it contained, pBMB0223, possessed a 6.7-kb HindIII insertion fragment. The fragment was predicted to bear six potential open reading frames (ORFs), among which orf2 encoded a hypothetical protein with a molecular mass of 40 kDa and a pI of 5.15. Significantly, the first 15 N-terminal sequence of this putative protein was identical to the 45-kDa crystal protein from strain YBT-1518, which was previously determined (data not shown). A 2.1-kb HindIII-HpaI fragment containing only orf2 was then subcloned into pHT304, resulting in pBMB0224, and transferred to BMB171, resulting in BMB0224. The strain BMB0224 formed rice-shaped crystals (Fig. 2) consisting of a 45-kDa protein (Fig. 3). A BLASTP homology search revealed that the putative Cry protein encoded by orf2 did not bear any homology to known crystal proteins in the GenBank database. The proposed gene was designated cry55Aa1 by the B. thuringiensis δ-endotoxin nomenclature committee.
The plasmid pBMB0249 harbored in the 54-kDa crystal protein-producing recombinant strain BMB0249 contained an insertion with three HindIII fragments (4.2, 2.4, and 2.9 kb). When the 2.9-kb HindIII fragment was subcloned into pHT304, the resulting recombinant strain, BMB0250, formed rice-shaped crystals (Fig. 2) consisting of 54-kDa proteins (Fig. 3). The cry gene in this fragment was identical to the cry6Aa2 crystal gene that we have described previously (30).
One recombinant strain, BMB0215, was found to produce a bipyramidal crystal. The plasmid pBMB0215 harbored a 4.6-kb HindIII fragment including an ORF that encoded a 140-kDa protein displaying 99.99% identity to the known protein Cry5Ba1. The gene was designated cry5Ba2. The presence of the cry5Ba2 gene in wild-type strain YBT-1518 was confirmed by PCR amplification (data not shown). Since the production of the bipyramidal crystal and the 140-kDa crystal protein could not be detected in YBT-1518, the cry5Ba2 gene may be cryptic in this strain.
Localization of the 45- and 54-kDa protein genes.
Alignment of the relevant DNA sequences showed that there was a 4.2-kb overlap between the insert in pBMB0222 carrying the cry55Aa1 gene and the insert in pBMB0249 carrying the cry6Aa2 gene, suggesting that both cry genes may be located in close proximity within the genome. In support of this, an E. coli recombinant strain isolated from a routine DNA library of strain YBT-1518, EMB0228, was found to harbor a plasmid, designated pEMB0228, with a 17.7-kb insert fragment that contained the complete inserts from pBMB0222 and pBMB0249 within its sequence.
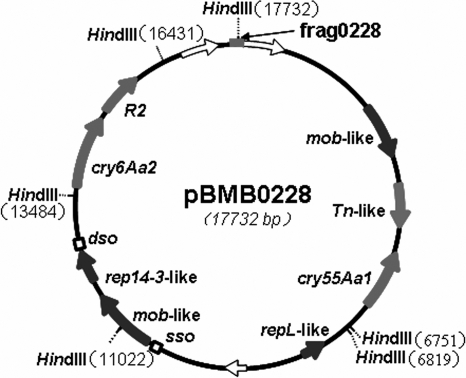
Sequence alignments revealed the presence in pBMB0249 of features typical of rolling circle plasmids like pTX14-3 (3), such as replication-associated protein, dso (double-stranded origin), and sso (single-stranded origin) sequences, suggesting that the plasmid harboring the two cry genes may not be very large. To obtain the possible remaining sequence of this plasmid not represented in the insertion fragment in plasmid pEMB0228, PCR was performed using primers designed according to the end sequence of the insertion fragment borne by pEMB0228. The sequence alignment of the PCR product frag0228 (Fig. 4) showed that, in YBT-1518, there was no gap between the two HindIII sites at the end of pEMB0228.
FIG. 4.
Physical map of plasmid pBMB0228, harboring both cry55Aa2 and cry6Aa2. Genes are indicated by arrows. The filled arrows indicate the genes with known functions, and the open arrows indicate genes having unknown functions. The filled box indicates the position of the PCR fragment frag0228, while the open boxes indicate the predicted positions of dso (double-stranded origin) and sso (single-stranded origin). The rep14-3-like and repL-like genes are the putative replication initiator protein genes, while two mob-like genes contribute to the mobilization of this plasmid. R2 is a negative-regulator gene of cry6Aa2, referred to as orf2 by Yu et al. (30).
The above data suggested that the 17.7-kb insert in pEMB0228 contained the entire plasmid bearing the cry55Aa1 and cry6Aa2 genes in strain YBT-1518. The native plasmid harboring the two genes was designated pBMB0228. Southern hybridization was performed against the plasmids from strain YBT-1518 with a digoxigenin-labeled probe corresponding to nucleotides 482 to 1032 of the cry55Aa1 gene to check whether the plasmid pBMB0228 was indeed present and whether it corresponded in size with that in the native strain. The result showed that this probe hybridized to the smallest detectable plasmid, with an estimated size of 18 kb, which is consistent with the predicted size of 17.7 kb in strain YBT-1518 (Fig. 5).
FIG. 5.
Detection of plasmid pBMB0228. (A) Plasmid profile by 0.8% agarose gel electrophoresis. (B) Southern hybridization, detecting the existence of pBMB0228 in the native strain YBT-1518. The PCR fragment corresponding to nucleotides 482 to 1032 of the cry55Aa1 gene was used as a probe. Lane M, DNA molecular mass λ DNA/HindIII markers; lane 1, total plasmids from YBT-1518; lane 2, plasmid pBMB0223 digested with HindIII, as a positive control; lane 3, total plasmids from YBT-1518 digested with HindIII; lane 4, cloning vector pHT304 digested with HindIII as a negative control. The arrows point to the position of plasmid pBMB0228.
Sequence alignment showed that, besides having the two cry genes and pTX14-3-like rolling circle plasmid features, pBMB0228 encoded two Mob proteins (Fig. 4) and a RepL-like protein having the conserved domain of the RepL family of plasmid replication proteins characterized for the Firmicutes. This unusual collection of features suggested that pBMB0228 was not an ordinary plasmid, by B. thuringiensis standards.
Toxicity to root knot nematode Meloidogyne hapla.
Since the three cry genes identified in this study all originated from a nematicidal strain of B. thuringiensis (30) and cry5B is a known nematicidal protein gene (28), we examined the toxicity of their protein products against the root knot nematode Meloidogyne hapla. Three kinds of Cry proteins—Cry55Aa1 protein from strain BMB0224, Cry6Aa2 from strain BMB0250, and Cry5Ba2 from strain BMB0215—were prepared and assayed in parallel against a second-stage juvenile of Meloidogyne hapla. As a result, all three proteins were found by the bioassay to be toxic to Meloidogyne hapla, with LC50s of 23.2 μg/ml, 23.9 μg/ml, and 18.1 μg/ml, respectively (Table 2).
TABLE 2.
Activities of Cry55Aa1, Cry6Aa2, and Cry5Ba2 against Meloidogyne haplab
| Crystal protein | Regression equation | Regression coefficient | LC50 (μg/ml)a |
|---|---|---|---|
| Cry55Aa1 | y = 2.6911x + 1.4288 | 0.9789 | 23.2 |
| Cry6Aa2 | y = 1.8736x + 2.4415 | 0.9711 | 23.9 |
| Cry5Ba2 | y = 1.6544x + 2.9475 | 0.9692 | 18.1 |
The second-stage juvenile of Meloidogyne hapla was tested, and LC50s were determined on the fifth day.
Bovine serum albumin (BSA) was used as the negative control; it has no toxicity to Meloidogyne hapla.
DISCUSSION
In this study, we screened cry genes for the formation of a crystal from a B. thuringiensis plasmid library. Compared to previous methods for screening E. coli libraries (4, 8, 13, 15, 19, 20, 24) or PCR amplification (5, 6, 9, 27), this strategy is advantageous in terms of isolating cry genes that have little identity to known genes or to some cryptic genes with functional promoters. In this process, the high electroporation efficiency of B. thuringiensis BMB171 played an important role. By modifying the method of Silo-Suh et al. (26), the electroporation efficiency reached 8 × 104 CFU μg−1 ml−1, while 29.1-kb plasmids were transferred to BMB171; for the small 6.5-kb plasmids, the efficiency was 5 × 109 CFU μg−1 ml−1 (21). In addition, the shuttle vector pHT304 was used for transfer to the intermediate host, E. coli DH10B, for converting the ligation mix to covalently closed circular plasmids, making the recombinant plasmids much more likely to be transferred (22); use of the method of Luo and Wing (18) made it possible to produce a highly efficient vector. All these crucial technical points of the protocol made the DNA library construction in B. thuringiensis possible.
By applying the method described above, we cloned the cry genes cry55Aa1 and cry6Aa2 and the cryptic cry5Ba2 gene from the B. thuringiensis rice-shaped crystal strain YBT-1518. Bioassay data showed that all three proteins displayed effective toxicity to the nematode Meloidogyne hapla, which causes tremendous crop damage throughout the world (29). In this context, the 45-kDa protein gene cry55Aa1, which showed no identity to other known crystal protein genes, is particularly interesting, because the small molecular weight of its protein makes it a prime candidate for use in the creation of nematode-resistant transgenic plants.
Unlike with the previous report (23), in our study, the two typical cry genes, cry55Aa1 and cry6Aa2, were found to be located on a plasmid of uncharacteristically modest size (17.7 kb) designated pBMB0228 (Fig. 4). The plasmid harbors two Rep-like protein genes—rep14-3-like and repL-like, which play critical roles in its replication (12)—and two Mob protein genes, both of which have significant similarity with Mob14-4, which is necessary for plasmid pBMbt1 mobilization (17). The apparent functional redundancies of the replication and mobilization elements could be explained as a result of a recombination process that fuses two plasmids into one in the evolutionary history of this plasmid. In any case, these features, combined with the fact that pBMB0228 harbors cry genes, make this unusual plasmid a very interesting subject for further research.
It was interesting to observe that the cry5Ba2 gene was expressed in the BMB171 host strain, as evidenced by the formation of a bipyramidal crystal, but not in the original strain, YBT-1518. According to previous reports, cryptic or silent cry genes have been found to lack functional promoters, resulting in their display of little or no expression (8, 11, 27). In the case of cry5Ba2, with this strategy, it has been shown that the cry5Ba2 gene possesses a functional promoter that allows it to be expressed in the acrystalliferous B. thuringiensis strain BMB171. The reason for the gene's cryptic behavior in its original strain therefore seems to be different from those mentioned above. What has been proved is that the high level of expression of cry55Aa1 and cry6Aa2 should not affect the issue of cry5Ba2 expression in the wild-type strain. The expression of gene cry5Ba2 still cannot be detected (data not shown) after pBMB0228 is cured by deletion of the two genes but not cry5Ba2 from YBT-1518. In this strain, a negative regulator of cry6Aa2 had been isolated downstream of the gene by Yu et al. (30). There was likely another negative regulator for the cry5Ba2 gene in the strain. The substantive mechanism responsible for the lack of expression in strain YBT-1518 is still under investigation in our laboratory.
In conclusion, in this study, we proposed a strategy for isolating cry genes from B. thuringiensis. This process was applied to a strain that produces rice-shaped crystals—YBT-1518—resulting in the cloning of three nematicidal cry genes. One of those genes was genuinely novel, one was cryptic, and one was a traditional cry gene, and we also found that a plasmid smaller than the usual Cry plasmids (17.7 kb) could encode typical cry genes.
Acknowledgments
We thank Geraldine A. Van der Auwera (Laboratory of Food and Environmental Microbiology in Belgium) for her critical review of the manuscript.
This research was supported by grants from the Key Project of China National Programs for Fundamental Research and Development (grant 2003CB114201) and the National Programs for High Technology Research and Development of China (grants 2006AA02Z174 and 2006AA10A212).
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andrup, L., J. Damgaard, and K. Wassermann. 1993. Mobilization of small plasmids in Bacillus thuringiensis subsp. israelensis is accompanied by specific aggregation. J. Bacteriol. 175:6530-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrup, L., G. B. Jensen, A. Wilcks, L. Smidt, L. Hoflack, and J. Mahillon. 2003. The patchwork nature of rolling-circle plasmids: comparison of six plasmids from two distinct Bacillus thuringiensis serotypes. Plasmid 49:205-232. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian, P., R. Jayakumar, P. Shambharkar, N. Unnamalai, S. K. Pandian, N. S. Kumaraswami, R. Ilangovan, and V. Sekar. 2002. Cloning and characterization of the crystal protein-encoding gene of Bacillus thuringiensis subsp. yunnanensis. Appl. Environ. Microbiol. 68:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beron, C. M., L. Curatti, and G. L. Salerno. 2005. New strategy for identification of novel Cry-type genes from Bacillus thuringiensis strains. Appl. Environ. Microbiol. 71:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceron, J., A. Ortiz, R. Quintero, L. Guereca, and A. Bravo. 1995. Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 61:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore, N., D. R. Zeigler, J. Feitelson, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, and D. H. Dean. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delecluse, A., M. L. Rosso, and A. Ragni. 1995. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl. Environ. Microbiol. 61:4230-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gleave, A. P., R. Williams, and R. J. Hedges. 1993. Screening by polymerase chain reaction of Bacillus thuringiensis serotypes for the presence of cryV-like insecticidal protein genes and characterization of a cryV gene cloned from B. thuringiensis subsp. kurstaki. Appl. Environ. Microbiol. 59:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffitts, J. S., J. L. Whitacre, D. E. Stevens, and R. V. Aroian. 2001. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science 293:860-864. [DOI] [PubMed] [Google Scholar]
- 11.Jain, D., V. Udayasuriyan, P. I. Arulselvi, S. S. Dev, and P. Sangeetha. 2006. Cloning, characterization, and expression of a new cry2Ab gene from Bacillus thuringiensis strain 14-1. Appl. Biochem. Biotechnol. 128:185-194. [DOI] [PubMed] [Google Scholar]
- 12.Khan, S. A. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126-136. [DOI] [PubMed] [Google Scholar]
- 13.Kongsuwan, K., J. Gough, D. Kemp, A. McDevitt, and R. Akhurst. 2005. Characterization of a new Bacillus thuringiensis endotoxin, Cry47Aa, from strains that are toxic to the Australian sheep blowfly, Lucilia cuprina. FEMS Microbiol. Lett. 252:127-136. [DOI] [PubMed] [Google Scholar]
- 14.Kuo, W. S., and K. F. Chak. 1996. Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl. Environ. Microbiol. 62:1369-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, H. K., and S. S. Gill. 1997. Molecular cloning and characterization of a novel mosquitocidal protein gene from Bacillus thuringiensis subsp. fukuokaensis. Appl. Environ. Microbiol. 63:4664-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, L., and Z. Yu. 1999. Transformation and expression properties of a Bacillus thuringiensis plasmid-free derivative strain BMB171. Chin. J. Appl. Environ. Biol. 5:395-399. [Google Scholar]
- 17.Loeza-Lara, P. D., G. Benintende, J. Cozzi, A. Ochoa-Zarzosa, V. M. Baizabal-Aguirre, J. J. Valdez-Alarcon, and J. E. Lopez-Meza. 2005. The plasmid pBMBt1 from Bacillus thuringiensis subsp. darmstadiensis (INTA Mo14-4) replicates by the rolling-circle mechanism and encodes a novel insecticidal crystal protein-like gene. Plasmid 54:229-240. [DOI] [PubMed] [Google Scholar]
- 18.Luo, M., and R. A. Wing. 2003. An improved method for plant BAC library construction. Methods Mol. Biol. 236:3-20. [DOI] [PubMed] [Google Scholar]
- 19.Masson, L., W. J. Moar, K. van Frankenhuyzen, M. Bosse, and R. Brousseau. 1992. Insecticidal properties of a crystal protein gene product isolated from Bacillus thuringiensis subsp. kenyae. Appl. Environ. Microbiol. 58:642-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLinden, J. H., J. R. Sabourin, B. D. Clark, D. R. Gensler, W. E. Workman, and D. H. Dean. 1985. Cloning and expression of an insecticidal k-73 type crystal protein gene from Bacillus thuringiensis var. kurstaki into Escherichia coli. Appl. Environ. Microbiol. 50:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng, D., Y. Luo, S. Guo, H. Zeng, S. Ju, Z. Yu, and M. Sun. Elaboration of an electroporation protocol for large plasmids and wild-type strains of Bacillus thuringiensis. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnepf, H. E., and H. R. Whiteley. 1981. Cloning and expression of the Bacillus thuringiensis crystal protein gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:2893-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao, Z., Z. Liu, and Z. Yu. 2001. Effects of the 20-kilodalton helper protein on Cry1Ac production and spore formation in Bacillus thuringiensis. Appl. Environ. Microbiol. 67:5362-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silo-Suh, L. A., B. J. Lethbridge, S. J. Raffel, H. He, J. Clardy, and J. Handelsman. 1994. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol. 60:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, F., J. Zhang, A. Gu, Y. Wu, L. Han, K. He, Z. Chen, J. Yao, Y. Hu, G. Li, and D. Huang. 2003. Identification of cry1I-type genes from Bacillus thuringiensis strains and characterization of a novel cry1I-type gene. Appl. Environ. Microbiol. 69:5207-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, J. Z., K. Hale, L. Carta, E. Platzer, C. Wong, S. C. Fang, and R. V. Aroian. 2003. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl. Acad. Sci. USA 100:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widmer, T. L., and G. S. Abawi. 2000. Mechanism of suppression of Meloidogyne hapla and its damage by a green manure of Sudan grass. Plant Dis. 85:562-568. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Z., P. Bai, W. Ye, F. Zhang, L. Ruan, Z. Yu, and M. Sun. 2008. A novel negative regulatory factor for nematicidal Cry protein gene expression in Bacillus thuringiensis. J. Microbiol. Biotechnol. 18:1033-1039. [PubMed] [Google Scholar]