Abstract
The goal of this study was to follow ceftiofur-treated and untreated cattle in a normally functioning dairy to examine enteric Escherichia coli for changes in antibiotic resistance profiles and genetic diversity. Prior to treatment, all of the bacteria cultured from the cows were susceptible to ceftiofur. Ceftiofur-resistant E. coli was only isolated from treated cows during and immediately following the cessation of treatment, and the 12 blaCMY-2-positive isolates clustered into two genetic groups. E. coli bacterial counts dropped significantly in the treated animals (P < 0.027), reflecting a disappearance of the antibiotic-susceptible strains. The resistant bacterial population, however, did not increase in quantity within the treated cows; levels stayed low and were overtaken by a returning susceptible population. There was no difference in the genetic diversities of the E. coli between the treated and untreated cows prior to ceftiofur administration or after the susceptible population of E. coli returned in the treated cows. A cluster analysis of antibiotic susceptibility profiles resulted in six clusters, two of which were multidrug resistant and were comprised solely of isolates from the treated cows immediately following treatment. The antibiotic treatment provided a window to detect the presence of ceftiofur-resistant E. coli but did not appear to cause its emergence or result in its amplification. The finding of resistant isolates following antibiotic treatment is not sufficient to estimate the strength of selection pressure nor is it sufficient to demonstrate a causal link between antibiotic use and the emergence or amplification of resistance.
Concerns persist regarding the potential negative impacts of antimicrobial use in livestock and, in particular, the potential for the emergence of antimicrobial resistance in human and animal pathogens (2, 27, 31). Studies that assess the biological consequences of these antimicrobial administrations in treated and untreated hosts in their natural environments are needed (6, 17, 26). Specific antimicrobial uses in animal agriculture must be evaluated to determine their importance in maintaining a healthy animal population and whether they are capable of selecting for resistance over variable time periods. Topics that need to be addressed include the frequency at which novel resistances or resistance gene arrangements evolve following antimicrobial administration, the degree and duration of amplification of the resistant pool following antimicrobial use, and the frequency at which resistant bacteria are transmitted to untreated hosts (24, 25).
Some antibiotics that are used therapeutically in animal agriculture are also important therapeutic options in human medicine, and therefore, judicious use of these antibiotics is critical (10). For example, third-generation cephalosporins have important applications to both human and veterinary medicine due to their broad-spectrum, generally bactericidal effects (33). Ceftriaxone is a third-generation cephalosporin with many applications in human medicine, including the treatment of severe salmonellosis cases in humans (36). Ceftiofur, while similar to ceftriaxone, is the only third-generation cephalosporin approved for use in cattle in the United States and is currently labeled for the treatment of bovine pneumonia, interdigital necrobacillosis, acute metritis (in cows 0 to 14 days postpartum), and mastitis (http://www.ceftiofur.com). All administrations of ceftiofur are by prescription of a licensed veterinarian only. When used according to the label insert, ceftiofur products produce minimal impact on dairy farm production, as there is no milk withdrawal time (except for the 72-h withholding for intramammary preparations), and depending on the product administered, there is a minimal required slaughter withdrawal time of 0 to 72 h.
A number of different genes that code for proteins that confer reduced susceptibility to third-generation cephalosporins have been identified. AmpC β-lactamase genes, originally characterized as chromosomal genes of Citrobacter freundii and Enterobacter cloacae, have since been found on plasmids in many Enterobacteriaceae (1, 3, 19, 37). One of the first documented human cases of a ceftriaxone-resistant Salmonella infection in the United States was a strain of Salmonella enterica serovar Typhimurium that possessed a plasmid with the AmpC β-lactamase gene termed blaCMY-2 (11). The prevalence of ceftriaxone resistance in Salmonella isolated from humans has increased over time, with the blaCMY-2 gene commonly being responsible for the resistance in the United States (8). Although evidence of AmpC plasmid transfer between Salmonella and Escherichia coli has been documented (37, 38), the dynamics of this plasmid in enteric bacteria have yet to be fully characterized.
Because ceftiofur use in animal agriculture has the potential to select for resistance to third-generation cephalosporins, several studies have been conducted to assess the biological consequences of this administration. In general, these studies have focused on resistant bacteria in the treated hosts but not necessarily under natural field conditions. In one study of ceftiofur administration in dairy calves, all treated and untreated animals were housed separately, thus precluding the investigation of potential horizontal transmission of bacteria among treated and untreated hosts (17). In studies that did allow for bacterial transmission among hosts, such as a study conducted in feedlot cattle, evaluating the microbial dynamics during and after treatment was not possible because no genotyping was performed on the E. coli investigated (26). Therefore, the goal of the current study was to follow ceftiofur-treated and untreated cattle in a normally functioning dairy to examine enteric E. coli for changes in the prevalence of ceftiofur resistance, particularly resistance mediated by the plasmid-borne blaCMY-2 gene. We wanted to assess whether treated cattle would pose a risk to untreated cattle for the potential horizontal transmission of resistant bacteria or resistance genes. Furthermore, we wanted to evaluate the genetic diversity of E. coli isolates in the treated and untreated cattle before, during, and after the administration of ceftiofur. Based on previous studies, we hypothesized that the prevalence of blaCMY-2 genes in the E. coli strains of the treated cows would temporarily increase following treatment but that the impact on the untreated cohort would not be significant. If ceftiofur-resistant E. coli strains were detected in the untreated cattle, we expected these isolates to be identical to the strains present in the treated cattle. After the selection pressure due to ceftiofur administration was removed, we hypothesized that any ceftiofur-resistant E. coli isolates would disappear. We hypothesized that the E. coli population that would return in the treated cows after the effects of the antibiotic had disappeared would be genetically similar to that in the untreated cows.
MATERIALS AND METHODS
Study design.
This observational study was conducted on a dairy farm in central Illinois that milks approximately 150 cows. Calves are raised on-site, but after weaning, the calves are sent to another facility until they are confirmed pregnant. They are then returned to the dairy just prior to calving, where they remain for the rest of their lives. The dairy has a single large barn in which all the cows live. All the cows can commingle except when they are walked to the milking facility twice a day. During these milking times, groups of cows (milking strings) are brought to the milking facility together, and these groups have a consistent composition over time.
Five cows on this dairy were diagnosed with infertility due to Leptospira borgpetersenii serovar Hardjo-bovis, and following the veterinarian's recommendation, these five cattle were to be treated with ceftiofur according to label instructions (2.2 mg/kg, intramuscularly, once daily for 5 days). A cohort of five untreated cows in the same milking string was randomly selected. These cows were individually matched to the treated cows based on the number of times that the cow had had a calf, also known as the lactation number.
All five treated cows started therapy at the same time. Animals were sampled prior to (days −1 and 0), during (days 2 and 4), and after (days 5 to 11, 14, 18, 25, and 32) ceftiofur therapy. On day 0, sample collection occurred approximately 1 hour before the first ceftiofur injection was administered. Fecal samples were collected from the rectum of each cow, stored in sterile Whirl-pak bags, and transported on ice until processing (within 4 h of collection).
Microbiological analysis.
E. coli was cultured from the bovine fecal samples using standard techniques. One gram of fecal material was suspended in 9 ml Luria-Bertani broth. Tenfold dilutions were prepared from this initial dilution. For quantification, 100 μl of each dilution was spread onto individual MacConkey plates by using sterile glass beads and the plates were incubated at 37°C for 18 to 24 h. For isolation of individual E. coli colonies, dilution plates with well-isolated lactose-positive colonies were used. Twenty-four presumptive E. coli colonies from each animal were picked from these plates and transferred to MacConkey agar plates. A subset of up to 10 colonies was randomly selected from the 24 transferred colonies from each animal, and each colony was positively identified as E. coli if it yielded a typical reaction on Simmons citrate agar and triple sugar iron agar. These isolates were then grown in skim milk and frozen at −80°C for storage until further analysis.
Three confirmed E. coli isolates that were randomly collected from each cow on each day were assayed for susceptibility to 16 antimicrobials by using broth microdilution. The antibiotics amikacin, amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole were tested with a commercially available panel (CMV1AGNF, Sensititre gram-negative NARMS plate; Trek Diagnostic Systems, Westlake, OH). The antibiotic florfenicol was tested using custom plates. All antimicrobial preparations, laboratory techniques, MIC breakpoints, and control strains were in accordance with the Clinical Laboratory Standards Institute (CLSI) guidelines (28) except for the breakpoint for florfenicol. There is no CLSI florfenicol breakpoint for E. coli, but in our previous work, we determined that E. coli isolates possessing the plasmid-borne florfenicol resistance flo gene have MICs in excess of 16 μg/ml, and therefore, we used 16 μg/ml as the breakpoint (32). Each isolate tested was inoculated to nonselective semisolid media and grown overnight. Well-isolated colonies were picked and suspended in 0.85% saline and were visually adjusted to a turbidity equivalent to the 0.5 McFarland standard. Ten microliters of this suspension was added to 5 ml of Mueller-Hinton broth, and this bacterial suspension was then added to the commercial 96-well plates containing 16 antibiotics and to the 96-well plates containing florfenicol. Each plate was covered and incubated at 37°C for 18 to 24 h. The plates were visually inspected to determine the MIC for each isolate, and the MIC, defined as the lowest concentration of antibiotic giving complete inhibition of visible growth, was recorded manually. E. coli ATCC 25922, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains.
DNA extraction from feces.
DNA was extracted from each fecal sample by using the QIAamp DNA stool mini kit (Qiagen Inc., Valencia, CA) and a modified protocol as previously described (29). Briefly, the following modifications were made to the manufacturer's recommended protocol: 0.1 g of sample was used instead of 0.2 g; all centrifugation steps were increased by 30 s; samples were incubated at 95°C instead of 70°C during the lysis step (no. 3); the volume of proteinase K was increased to 20 μl (14); the incubation step (no. 12) was changed from 70°C for 10 min to 55°C for 30 min (14); and the two wash steps (no. 15 and 16) were repeated.
PCR amplification of blaCMY-2.
We designed, optimized, and validated with nucleotide sequencing a multiplex PCR assay for the detection of the blaCMY-2 gene in the E. coli isolates that included 16S rRNA primers as an internal amplification control. The primer set for the blaCMY-2 gene (cmyT-F, 5′-ACA GCC TCT TTC TCC ACA TTT G-3′ [forward]; cmyT-R, 5′-CTG GTC ATT GCC TCT TCG TAA C-3′ [reverse]) yielded a predicted amplicon of 551 bp. The 16S rRNA primers (16S-1, 5′-CTT GCT CTT TGC TGA GTG-3′ [forward]; 16S-2, 5′-GGG TAT CTA ATC CTG TTT GCT CC-3′ [reverse]) yielded a predicted amplicon of 714 bp. PCR primers were designed with Mac Vector (Accelrys, San Diego, CA) by using the sequences of the plasmid-borne blaCMY-2 gene in E. coli and the 16S sequence for E. coli.
PCR conditions in a final reaction volume of 25 μl using 2 μl template DNA were as follows: 1.25 U of Taq polymerase (Invitrogen, Carlsbad, CA) with 0.2 mM each of adenine, guanine, and cytosine and 0.175 mM thymine (Invitrogen); 2.5 mM MgCl2; 20 mM Tris; 50 mM KCl; 2.0 μM of each 16S primer; and 3.0 μM of each blaCMY-2 primer. The PCR template was prepared by adding one well-isolated colony from a freshly grown agar plate to 100 μl of autoclaved 5% Chelex-100 solution (Bio-Rad Laboratories, Hercules, CA) and 1 μl of 20 mg/ml proteinase K and incubating at 56°C for 45 min. This was followed by 8 min of boiling at 100°C. PCR templates were then stored at 4°C until used in PCR amplification. Amplification was performed on a PTC-200 thermocycler (MJ Research, Waltham, MA). PCR amplification was performed with the following program: 94°C for 2 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. An E. coli isolate with the blaCMY-2 gene was included in every PCR amplification as a positive control. Amplicons were separated on 1.2% agarose gels for 1 h at 5 V/cm, stained with ethidium bromide, and visualized under UV light.
For detecting the plasmid-borne blaCMY-2 gene in the extracted fecal community DNA, we used a previously published protocol for which specificity had already been established (29). Briefly, a nested PCR design was used to ensure the most sensitive results, and this increased sensitivity was evaluated previously (29). Samples were amplified by one set of primers, and the product of that reaction was diluted and used as the template for a second PCR amplification. The second primer pair was designed to amplify an internal region of the first PCR product and to amplify at a higher annealing temperature. The first primer pair is unable to amplify at the higher annealing temperature. Primers used in the first PCR were cmy-F, 5′GAC AGC CTC TTT CTC CAC A-3′, and cmy-R, 5′TGG AAC GAA GGC TAC GTA-3′ (39). This primer pair amplifies a 1,100 bp product. PCR conditions in a final reaction volume of 25 μl using 2 μl template DNA were as follows: 0.625 U of Taq polymerase (Invitrogen) in 1× buffer as supplied by the manufacturer with 0.25 mM deoxynucleoside triphosphates (Invitrogen); 2.0 mM MgCl2; 0.1 mg/ml bovine serum albumin (Promega); and 25 pmol of each primer. Acetamide (Sigma, St. Louis, MO) was added to a final concentration of 2.5%. Cycling conditions were as follows: 94°C for 2 min, followed by 29 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min. A final extension of 72°C was carried out for 7 min. Samples were then held at 4°C until processed further. The second nested PCR used 2 μl of 1:10 diluted template from the first PCR amplification. Primers used in the nested PCR were cmy F2, 5′CTC AGG AAT GAG TTA CGA AGA GG-3′, and cmy R2, 5′AAT CCA CCA GTG GAG CCC 3′. These primers amplify a product of 550 bp. Reaction conditions were the same as for the first PCR. Cycling conditions were as follows: 94°C for 2 min, followed by 29 cycles of 94°C for 30 s, 65.6°C for 45 s, and 72°C for 1 min. A final extension of 72°C was carried out for 7 min. Amplicons were analyzed on agarose gels as outlined above.
Rep-PCR.
E. coli isolates from each animal from each day were genotyped by repetitive element PCR (Rep-PCR) (13). For days −1, 0, and 2, up to 10 colonies per animal per day were analyzed. For days 4, 5, 6, and 8, up to five colonies per animal per day were analyzed. For days 9 and 10, up to three colonies per animal per day were analyzed. The Rep-PCR was carried out in 25 μl volumes containing 3.5 mM MgCl2, a 0.3 mM concentration of each deoxynucleoside triphosphate, 2.0 mM box AIR primer (5′-CTACGGCAAGGCGACGCTGACG-3′), and 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), with 1× buffer II (without MgCl2) and 2 μl of Chelex-treated template prepared as for gene-specific amplification above. Amplification was performed on a PTC-200 thermocycler (MJ Research) by using a touchdown program similar to that described by Johnson et al. (18). The preliminary denaturation step was for 10 min at 95°C. The 10-cycle touchdown included denaturation at 94°C for 30 s, ramping at 1.5°C/s to the annealing temperature (which starts at 70°C then decreases by 0.5°C per cycle until the ultimate annealing temperature of 65°C is reached), annealing for 1 min, ramping at 0.1°C/s to 72°C, and an extension for 4.5 min at 72°C. This touchdown program was followed by 25 amplification cycles of denaturation at 94°C for 30 s, ramping at 1.5°C/s to 65°C, annealing for 1 min at 65°C, ramping at 0.1°C/s to 72°C, and extension for 4.5 min at 72°C. A final extension of 72°C was carried out for 7 min. For quality control, a standard lab strain was amplified in each set of reactions to demonstrate repeatability and consistency across all experiments. Immediately following completion of the PCR, a 6× electrophoresis loading dye (Amresco) containing Ficoll, EDTA, and sodium dodecyl sulfate was added to a 1× final concentration to preserve the integrity and conformation of the single-stranded amplicons. Samples were stored at 4°C. Amplicons were separated on a 1.5% agarose gel for 5.5 h at 3.7 V/cm at 4°C. For normalization, 1 kb DNA ladder (Invitrogen) was loaded onto four equally distant points in the gel. Gels were stained with ethidium bromide and visualized under UV light. Digital images of each gel were then imported into BioNumerics 4.0 (BioSystematica, United Kingdom) for analysis.
Data analysis.
To determine if ceftiofur treatment had a significant effect on the total E. coli count in the fecal samples over time, we modeled the change in E. coli count (expressed as log CFU/g) with a repeated-measures analysis of variance. The repeated-measures design was used because the same animals were sampled on each day of the study. There were 12 days of sampling included in the model for each cow, and difference contrasts were used to assess the daily change in E. coli count. A variable for the treatment group was included in the model. Significance of the effect of treatment and the day x treatment contrasts were considered statistically significant if the P value was <0.05. Standard statistical software was used for all analyses (SPSS version 14.0; SPSS Inc., IL).
To measure the overall resistance levels in the E. coli populations of the treated and untreated cows over time, an antibiotic resistance index (ARI) was calculated for each group on each day (15, 23). The ARI is a quantitative measure of the level of antibiotic resistance in a bacterial population. It is useful for making comparisons among populations within a single study (35). The ARI of a population is expressed as:
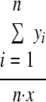 |
where n is the number of isolates in the population, x is the number of antibiotics in the panel against which an isolate is tested, and yi is the number of antibiotics to which isolate i is resistant. The ARI, which has a range from 0 to 1, therefore, expresses the total amount of resistance in the population of isolates, calculated as the proportion of total possible resistances present in the population. With five animals per treatment group and up to three E. coli isolates collected per animal per day, each ARI calculation uses antibiogram data for up to 15 isolates. Rep-PCR profiles were not considered in this analysis, so it is possible that multiple identical colonies from the same animals were used in this analysis.
A cluster analysis was performed to describe the E. coli antibiograms generated during the study. The goal of the cluster analysis was to determine if isolates from both treatment groups would have similar antibiograms pre- and posttreatment and if the treatment group, following treatment, would experience shifts in antibiograms, evidenced by isolates forming distinct clusters. To perform the cluster analysis, the MIC of each isolate to each antibiotic was first log2 transformed. Clusters were then determined using the squared Euclidean distance, which creates a dissimilarity matrix between isolates, and Ward's minimum variance hierarchical method (4). Analyses that generated 5 through 15 clusters were performed (11 total analyses). The order of isolates was randomized prior to the cluster analysis, and the analyses were repeated for three separate random orderings. For each cluster in each analysis, the MIC50 of the isolates in the cluster to each antibiotic was calculated. The optimal analysis should minimize the number of clusters, maximize the intercluster variability, and result in the fewest number of isolates misclassified. Criteria for selecting the analysis with the optimal number of clusters included (i) no two clusters having the same antibiogram (based on the MIC50 profile), (ii) a preference for fewer total clusters, (iii) clusters containing ≥1% of the total number of isolates, and (iv) repeatability of cluster assignments across the three random orderings of isolates. As stated previously, the analysis was performed without consideration of the Rep-PCR profiles.
The relationships among the E. coli isolates collected during the study were analyzed in two ways. First, a cluster analysis of the Rep-PCR fingerprints was conducted using the BioNumerics software program. All isolates from days −1 through 10 were included in this analysis (n = 468). Patterns were analyzed using curve-based methods with an optimization value of 8.0, and Pearson's product-moment algorithm was used to correlate the densitometric curves (13). Second, analysis of molecular variance (AMOVA) (9) was used to partition the genetic variation observed in the E. coli isolates to quantify the population genetic substructure. The AMOVA was conducted using the GENALX add-in to Microsoft Excel (30), and the significance of the variance components were calculated by 9,999 random permutations of the data. Several different AMOVA analyses were performed. First, the genetic variations within cows, among cows within treatment groups, and between treatment groups were assessed for the two sampling days prior to ceftiofur treatment (days −1 and 0 in Fig. 1A). Second, the genetic variations within cows, among cows within treatment groups, and between treatment groups were assessed for the three sampling days after the effects of the ceftiofur treatment had disappeared (days 8, 9, and 10 in Fig. 1A). Finally, to determine the relationship between the E. coli populations in ceftiofur-treated cows before and after treatment, genetic variations were assessed within cows, among cows within a time period, and between time periods (days −1 and 0 versus days 8, 9, and 10 in Fig. 1A).
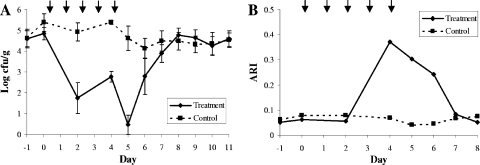
FIG. 1.
Total E. coli load (A) and ARI of the E. coli (B) in the treatment and control groups over time. ↓ indicates that a ceftiofur treatment was administered to the treated animals on that day, immediately after a sample was collected.
RESULTS
Using the repeated-measures analysis of variance, we found a significant overall difference between the treatment groups with respect to the total E. coli counts over time. The control group had significantly higher E. coli counts than the treatment group (P < 0.027) (Fig. 1A), but given the significant contrasts between treatment group and day, the two treatment groups were not significantly different from each other on all days of the study. Based on the contrasts of the interaction between treatment group and day in the repeated-measures analysis, there were significant daily changes between the groups for the two sampling days during treatment (day 0 to 2 and day 2 to 4 in Fig. 1A; P < 0.01) and the two days immediately following the cessation of treatment (day 4 to 5, P < 0.01, and day 5 to 6, P < 0.05). Three days posttreatment, the treatment group had lower E. coli counts than the control group, although this daily change from day 6 was not significant (day 7 in Fig. 1A; P = 0.4), and for the remainder of the study, the daily changes in E. coli counts were not significantly different between the treatment and control groups.
In the analysis of the ARI over time, there was no conspicuous difference between the treatment and control groups except for on days 4, 5, and 6 (Fig. 1B). On these days, the treatment group had a much higher ARI than the control group due to the multidrug resistance in the isolates collected on these days. The return of the ARI in the treatment group to pretreatment levels mirrored the return of the total E. coli count in this group to pretreatment levels (day 7).
The blaCMY-2 gene was detected by PCR in 12 of the 203 E. coli isolates evaluated from cows in the treatment group but was never found in the E. coli isolates evaluated from the control group (0 out of 265). On day 4, two of the treated animals had E. coli isolated from their feces that were positive for blaCMY-2, although the total E. coli counts for these animals were 1,700 CFU/g and 2,700 CFU/g. On day 5, four of the cows in the treatment group had blaCMY-2-positive E. coli, but E. coli counts among the treatment group animals on this day ranged from 0 to 400 CFU/g. By day 6, only one cow still had blaCMY-2-positive E. coli, and this animal had an E. coli count of 6,700 CFU/g. The gene was not detected in any of the cows after day 6. The blaCMY-2-positive isolates were clustered into two groups with the isolates in each group having similarities of >95% when the Rep-PCR data were analyzed (see the supplemental material). The blaCMY-2-negative E. coli isolates that were most closely related to the blaCMY-2-positive isolates had similarities of <90%. The blaCMY-2 gene was detected in the community DNA from cows in both the treatment and control groups on all days of the study. At least two of the cows in each group were blaCMY-2 positive on each day of the study.
The optimal cluster analysis contained six different E. coli clusters (Table 1). One of the clusters (cluster D) had a pan-susceptible profile based on the MIC50 values of the 83 isolates in the cluster. Clusters A to D had resistances between 0 and 2 and were comprised of isolates from both the treatment and control groups. The 14 isolates in clusters E and F all came from cows in the treatment group on the days immediately following treatment, and all isolates in these clusters were multidrug resistant. The blaCMY-2-positive isolates all clustered within cluster E.
TABLE 1.
Antibiotic susceptibility patterns for the six clusters derived by Ward's minimum variance method describing the patterns of resistance for the 209 E. coli isolates
| Cluster | Treatment group(s)b | MIC50 with indicated antibiotica
|
Total no. of isolates | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMI | AMP | AUG | AXO | CHL | CIP | COT | FLO | FOX | GEN | KAN | NAL | SOX | STR | TET | TIO | |||
| A | 1, 2 | 2 | 64 | 4 | 0.25 | 8 | 0.015 | 0.12 | 4 | 4 | 0.5 | 8 | 2 | 16 | 32 | 64 | 0.25 | 21 |
| B | 1, 2 | 2 | 4 | 4 | 0.25 | 8 | 0.015 | 0.12 | 4 | 4 | 0.5 | 8 | 4 | 512 | 32 | 4 | 0.25 | 66 |
| C | 1, 2 | 2 | 2 | 4 | 0.25 | 8 | 0.015 | 0.12 | 4 | 4 | 0.5 | 8 | 2 | 512 | 32 | 64 | 0.25 | 25 |
| D | 1, 2 | 2 | 2 | 4 | 0.25 | 8 | 0.015 | 0.12 | 4 | 2 | 0.5 | 8 | 2 | 16 | 32 | 4 | 0.25 | 83 |
| E | 2 | 2 | 64 | 64 | 4 | 64 | 0.015 | 0.25 | 32 | 64 | 32 | 128 | 2 | 512 | 128 | 64 | 16 | 11 |
| F | 2 | 2 | 64 | 64 | 0.5 | 8 | 0.015 | 0.12 | 8 | 16 | 0.5 | 8 | 4 | 512 | 128 | 64 | 1 | 3 |
Boldface type represent MIC50s for the cluster that are classified as resistant according to the CLSI guidelines for E. coli, except for florfenicol, for which a value of >16 μg/ml was used. AMI, amikacin; AMP, ampicillin; AUG, amoxicillin-clavulanic acid; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; COT, trimethoprim-sulfamethoxazole; FLO, florfenicol; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SOX, sulfisoxazole; TET, tetracycline; TIO, ceftiofur.
The treatment group that is represented in each cluster. 1, the nontreated group; 2, the treated group.
There was high genetic diversity among the 468 E. coli isolates collected and analyzed during this study. As shown by the AMOVA, the majority of the genetic variation existed within the individual animal (Table 2). For example, within-animal variation explained more than 84% and 81% of the genetic variation in the E. coli population before antibiotic treatment and after the ceftiofur effects had disappeared, respectively (P < 0.001). Very little genetic variation existed between the treatment and control groups pretreatment (1.86%; P < 0.001) or after the ceftiofur effects had disappeared (1.79%; P < 0.001). No analysis was performed for the period between days 2 and 6 because at least one animal had no cultivable E. coli on each of these days (Fig. 1A). When the pretreatment (days −1 and 0) genetic diversity of the E. coli isolates in the treated animals was compared to the diversity posttreatment (days 8, 9, and 10), the majority of the variation still existed within the individual animal (81%; P < 0.001). In this analysis, the difference among time periods (pretreatment versus posttreatment) explained approximately 6% of the total genetic variation.
TABLE 2.
Analysis of the Rep-PCR fingerprint data for E. coli isolates collected during the study using a hierarchical AMOVA
| Variance component | Observed partition value
|
φ statistic | P valuea | |
|---|---|---|---|---|
| Variance (σ2) | % Total | |||
| Pretreatment (days −1 and 0) | ||||
| Between treatment groups | 0.136 | 1.86 | 0.019 | <0.001 |
| Among individuals within treatment groups | 1.030 | 14.02 | 0.143 | <0.001 |
| Within individuals | 6.181 | 84.13 | 0.159 | <0.001 |
| Posttreatment (days 8, 9, and 10) | ||||
| Between treatment groups | 0.155 | 1.79 | 0.018 | 0.004 |
| Among individuals within treatment groups | 0.825 | 9.52 | 0.097 | <0.001 |
| Within individuals | 7.69 | 88.69 | 0.113 | <0.001 |
| Pretreatment vs posttreatment in treated animals | ||||
| Between time periods | 0.535 | 6.20 | 0.062 | <0.001 |
| Among individuals within time periods | 1.077 | 12.48 | 0.133 | <0.001 |
| Within individuals | 7.014 | 81.32 | 0.187 | <0.001 |
P values represent the probability of obtaining a more extreme variance component and φ statistic than the observed values by chance, calculated through 9,999 permutations of the data.
DISCUSSION
The selection pressure conferred by the use of antibiotics is generally considered to result in the creation, persistence, amplification, and dissemination of resistant strains. Because ceftiofur-resistant E. coli was detected in the ceftiofur-treated cows but not in the control cows, this study would appear to demonstrate the emergence of resistance following the use of a therapeutic antibiotic in dairy production. There are several pieces of information, however, which lead to an alternate conclusion. First, the E. coli bacterial counts dropped significantly in the treated animals, reflecting a disappearance of the antibiotic-susceptible strains. The reduction (Fig. 1A) implies that the susceptible population could have outnumbered the resistant population by 4 to 5 logs prior to treatment. Consequently, the probability of isolating a resistant colony without antibiotic selection would have been low. Second, the incorporation of community DNA PCR enabled us to detect the resistance gene in the feces of many of the cows in the study over multiple time points. This detection probability was independent of treatment status. It must be emphasized that a blaCMY-2-positive result with the community DNA approach indicates that a plasmid-borne blaCMY-2 gene exists in the sample, but it is impossible to know if this gene came from an E. coli isolate. Finally, the resistant bacterial population did not increase in abundance within the treated cows. The levels stayed low and were overtaken by a returning susceptible population when the effects of the antibiotic diminished. Based on these findings, it appears that the antibiotic treatment provided a window to detect the presence of this specific resistance phenotype and genotype but did not cause its emergence or result in its amplification. Interestingly, the resistant strain did not have a genotype resembling that of any of the susceptible E. coli in the population, so it does not appear that the plasmid possessing the blaCMY-2 gene was transferred to other E. coli hosts as a result of the selection pressure. Additional studies should be performed to aid in the interpretation of these findings and to determine if there is a critical window posttreatment during which resistant strains are detected at a higher frequency.
During our longitudinal investigation of this herd, the ceftiofur-resistant strains that were isolated from the treated cows had previously been isolated from the calves on the farm (data not shown). No antibiotic selection was needed to detect the ceftiofur-resistant strains in the calves. Occasionally, we detected these isolates in the cows on the farm, including cows that had not received antibiotics recently. Based on the results of this study, it would appear that these isolates were circulating throughout the herd over an extended period of time, but the probability of detecting the strains in adults was low unless the use of an antibiotic enhanced our ability to detect the strains.
Prior to treatment, the E. coli population in the cows was diverse, with considerable overlap of E. coli strains between those cows that were to be treated and those cows that were not. These isolates had little antibiotic resistance, as reflected in the ARI values and in the assignments to clusters A to D. After treatment, when the susceptible bacterial population began to return in the treated cows, the bacteria again were largely indistinguishable between the treated and untreated animals. The return of this population is most likely due to the cattle being repopulated with an environmental source of the susceptible bacterial population present in the herd or with susceptible strains that survived in the gut at low levels during the antibiotic treatment. Once the pressure of the antibiotic was removed, the bacteria were able to recolonize. The lack of detectable horizontal transmission of the resistant strain between the treated and untreated animals is noteworthy. Because the cows in this herd are likely sharing E. coli strains on a continual basis, the reduction of the susceptible population in the treated animals should have increased the probability that these animals would transmit the ceftiofur-resistant strains (25). The inability to detect this strain in the untreated animals suggests that the strain might be less competitive than the susceptible E. coli in adult cows. Alternatively, the lack of detectable horizontal transmission could be due to a low sensitivity and a small sample size. The inclusion of selective culture media, such as an agar with ceftiofur, could have enhanced the ability to detect ceftiofur-resistant strains. The analysis of additional isolates per animal per day may have also provided more power to detect a transmission event.
Specific bacterial strains are more successful at colonizing individual animals under different circumstances. This competitive advantage of specific strains appears to change as the host immune system and gastrointestinal microbiota change. For example, calves often shed resistant bacterial strains with high frequency, and as the cow ages, the population shifts to a more susceptible set of strains (5, 7, 16, 21). Because calves are often fed a milk replacer that contains antibiotics, part of this increased shedding probability in calves might be attributed to this feeding practice. However, in studies of antibiotic resistance in animals, age is typically the strongest predictor of resistance, with younger animals shedding a higher prevalence of resistant bacteria. This finding has been observed even in the absence of antibiotic administrations (5, 7, 16, 21). In competition experiments, it has been demonstrated that a resistant E. coli strain isolated from dairy calves was more competitive in calves than the susceptible strain, even in the absence of antibiotics (21, 22). In heifers, there was no competitive advantage for the resistant strain. When the wild-type strain had its resistance removed, the new strain was equally as competitive as the original resistant strain in calves, implying that the resistance is not the factor responsible for the competitive advantage (22). Furthermore, an additional study found that a dietary supplement fed to dairy calves in milk selected for E. coli of the SSuT phenotype (resistance to streptomycin, sulfadiazine, and tetracycline), regardless of whether the milk contained oxytetracycline (20).
During this study, the ARI increased in the treated animals (Fig. 1B), and this increase was entirely due to the blaCMY-2-positive isolates (cluster E in Table 1) as well as the three additional multidrug-resistant isolates that had ampicillin resistance and decreased susceptibility to ceftiofur (cluster F in Table 1). The blaCMY-2-positive isolates have the ACSSuT phenotype and are resistant to 11 of the 16 antibiotics for which we tested. Many of the resistance genes in these isolates are located on a large multidrug-resistant plasmid, approximately 165 kb in size (data not shown). A similar plasmid has been observed in Salmonella and E. coli in other studies (1, 12, 19, 37). Even though much of the resistance, including blaCMY-2, is localized on a plasmid, we did not observe the horizontal transfer of this plasmid among other E. coli isolates during the study. It is possible, however, that the blaCMY-2 gene or the plasmid possessing this gene could have been transferred to other bacterial genera that were not studied, thus allowing this ceftiofur-resistant E. coli strain to serve as a reservoir for the blaCMY-2 gene.
The results of this study corroborate those of other studies, in which ceftiofur-resistant E. coli appeared for a short duration, following the use of ceftiofur in cattle, and that overall E. coli populations declined immediately after treatment. In one investigation of dairy farms, those dairies that used ceftiofur were significantly more likely to have cows shedding E. coli with reduced susceptibility to cephalosporins (34). However, there was no significant relationship between individual animals that had received ceftiofur treatment and the odds of isolating E. coli with reduced susceptibility from the treated animals. Given the results of our study, the enhanced probability of detecting ceftiofur-resistant E. coli lasted only 2 to 3 days after the cessation of antibiotic therapy, and therefore, the probability of finding a statistically significant relationship between ceftiofur administration and ceftiofur-resistant E. coli in individual cows by using a random sampling scheme would be low. Other studies that followed animals that had been treated with ceftiofur documented a reduction in the E. coli population during and after treatment, along with an increase in the prevalence of ceftiofur-resistant E. coli (17, 26). In one study, the animals continued to shed ceftiofur-resistant E. coli for at least 17 days after the initial treatment, a duration much longer than that observed in our study (17). Of note is that this latter study used dairy calves. As described previously, young animals can shed high levels of antibiotic-resistant bacteria for reasons that have nothing to do with antibiotic administration, and therefore, the results from the calf study do not necessarily reflect the risk associated with the adult animal. In the other study, beef cattle were followed over time (26). The animals that were treated exhibited a decline in the total E. coli population, but the drop was not as extreme as that which we observed in this study or that which was observed in the dairy calf study (17).
This present study demonstrated an apparent emergence of resistance following treatment. However, the interpretation of this finding was revised when additional data, such as colony counts, community DNA PCR, and genotyping of isolates, were included in the analysis. Treatment with ceftiofur resulted in a significant drop in the gram-negative enteric bacterial population, which allowed for the detection of E. coli with the blaCMY-2 resistance gene. With the conclusion of the treatment regimen, the selection pressure of ceftiofur declined, and fecal E. coli counts rapidly returned to pretreatment levels and pretreatment diversity. The prevalence of E. coli with the blaCMY-2 gene returned to a low frequency not detected by our sampling methods. Importantly, this study emphasizes that the finding of resistant isolates following antibiotic treatment is not sufficient to estimate the strength of a selection pressure, nor is it sufficient to demonstrate a causal link between antibiotic use and the emergence or amplification of resistance. There are background populations of resistant bacteria and resistance genes that must be understood in any studied population so that accurate conclusions about the relationship between antibiotic use and antibiotic resistance can be made.
Supplementary Material
Acknowledgments
We thank Rich Clem, Jessica Gibson, Josh Kammerer, Ann Kremer, Kevin Lang, Hannah Lee, Carol Maddox, Anne Meier, and Marie Sienkewicz for technical assistance. We also thank the three anonymous reviewers, whose comments helped to improve the manuscript.
The research in this project has complied with all relevant animal use federal guidelines and institutional policies.
This project was supported by National Research Initiative competitive grants 2000-35212-9398 and 2003-35212-13853 (R.S.S.) from the USDA Cooperative State Research, Education, and Extension Service Epidemiological Approaches to Food Safety program.
Footnotes
Published ahead of print on 26 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alcaine, S. D., S. S. Sukhnanand, L. D. Warnick, W. L. Su, P. McGann, P. McDonough, and M. Wiedmann. 2005. Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob. Agents Chemother. 49:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo, F. J., V. N. Nargund, and T. C. Chiller. 2004. Evidence of an association between use of antimicrobial agents in food animals and antimicrobial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:374-379. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, M., and B. G. Hall. 2002. Origin and evolution of the AmpC β-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge, A. C., E. R. Atwill, and W. M. Sischo. 2003. Assessing antibiotic resistance in fecal Escherichia coli in young calves using cluster analysis techniques. Prev. Vet. Med. 61:91-102. [DOI] [PubMed] [Google Scholar]
- 5.Berge, A. C., E. R. Atwill, and W. M. Sischo. 2005. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69:25-38. [DOI] [PubMed] [Google Scholar]
- 6.Berge, A. C., W. B. Epperson, and R. H. Pritchard. 2005. Assessing the effect of a single dose florfenicol treatment in feedlot cattle on the antimicrobial resistance patterns in faecal Escherichia coli. Vet. Res. 36:723-734. [DOI] [PubMed] [Google Scholar]
- 7.Donaldson, S. C., B. A. Straley, N. V. Hegde, A. A. Sawant, C. DebRoy, and B. M. Jayarao. 2006. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72:3940-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 9.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes-application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.FAO/WHO/OIE. 2008. Joint FAO/WHO/OIE expert meeting on critically important antimicrobials. Report of a meeting held in FAO headquarters, Rome, 26-30 November 2007. FAO, Rome, Italy.
- 11.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 12.Giles, W. P., A. K. Benson, M. E. Olson, R. W. Hutkins, J. M. Whichard, P. L. Winokur, and P. D. Fey. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg, T. L., T. R. Gillespie, and R. S. Singer. 2006. Optimization of analytical parameters for inferring relationships among Escherichia coli isolates from repetitive-element PCR by maximizing correspondence with multilocus sequence typing data. Appl. Environ. Microbiol. 72:6049-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gookin, J. L., A. J. Birkenheuer, E. B. Breitschwerdt, and M. G. Levy. 2002. Single-tube nested PCR for detection of Tritrichomonas foetus in feline feces. J. Clin. Microbiol. 40:4126-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinton, M., A. J. Hedges, and A. H. Linton. 1985. The ecology of Escherichia coli in market calves fed a milk-substitute diet. J. Appl. Bacteriol. 58:27-35. [DOI] [PubMed] [Google Scholar]
- 16.Hinton, M., A. H. Linton, and A. J. Hedges. 1985. The ecology of Escherichia coli in calves reared as dairy-cow replacements. J. Appl. Bacteriol. 58:131-138. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, X., H. Yang, B. Dettman, and M. P. Doyle. 2006. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3:355-365. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. R., and T. T. O'Bryan. 2000. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin. Diagn. Lab. Immunol. 7:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang, M. S., T. E. Besser, and D. R. Call. 2006. Variability in the region downstream of the blaCMY-2 β-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50:1590-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khachatryan, A. R., T. E. Besser, D. D. Hancock, and D. R. Call. 2006. Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl. Environ. Microbiol. 72:4583-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2006. Antimicrobial drug resistance genes do not convey a secondary fitness advantage to calf-adapted Escherichia coli. Appl. Environ. Microbiol. 72:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumperman, P. H. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipsitch, M. 2001. The rise and fall of antimicrobial resistance. Trends Microbiol. 9:438-444. [DOI] [PubMed] [Google Scholar]
- 25.Lipsitch, M., and M. H. Samore. 2002. Antimicrobial use and antimicrobial resistance: a population perspective. Emerg. Infect. Dis. 8:347-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowrance, T. C., G. H. Loneragan, D. J. Kunze, T. M. Platt, S. E. Ives, H. M. Scott, B. Norby, A. Echeverry, and M. M. Brashears. 2007. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 68:501-507. [DOI] [PubMed] [Google Scholar]
- 27.Mølbak, K. 2004. Spread of resistant bacteria and resistance genes from animals to humans—the public health consequences. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:364-369. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard, 2nd edition. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 29.Patterson, S. K., and R. S. Singer. 2006. Development of a polymerase chain reaction assay for the detection of antibiotic resistance genes in community DNA. J. Vet. Diagn. Investig. 18:172-181. [DOI] [PubMed] [Google Scholar]
- 30.Peakall, R., and P. E. Smouse. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips, I. 2007. Withdrawal of growth-promoting antibiotics in Europe and its effects in relation to human health. Int. J. Antimicrob. Agents 30:101-107. [DOI] [PubMed] [Google Scholar]
- 32.Singer, R. S., S. K. Patterson, A. E. Meier, J. K. Gibson, H. L. Lee, and C. W. Maddox. 2004. Relationship between phenotypic and genotypic florfenicol resistance in Escherichia coli. Antimicrob. Agents Chemother. 48:4047-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenover, F. C. 2006. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control 34:S3-10. [DOI] [PubMed] [Google Scholar]
- 34.Tragesser, L. A., T. E. Wittum, J. A. Funk, P. L. Winokur, and P. J. Rajala-Schultz. 2006. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67:1696-1700. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, B. A., D. A. Dargatz, P. S. Morley, T. J. Keefe, and M. D. Salman. 2003. Analysis methods for evaluating bacterial antimicrobial resistance outcomes. Am. J. Vet. Res. 64:1570-1579. [DOI] [PubMed] [Google Scholar]
- 36.Whichard, J. M., K. Joyce, P. D. Fey, J. M. Nelson, F. J. Angulo, and T. J. Barrett. 2005. Beta-lactam resistance and Enterobacteriaceae, United States. Emerg. Infect. Dis. 11:1464-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, S., S. Qaiyumi, S. Friedman, R. Singh, S. L. Foley, D. G. White, P. F. McDermott, T. Donkar, C. Bolin, S. Munro, E. J. Baron, and R. D. Walker. 2003. Characterization of Salmonella enterica serotype Newport isolated from humans and food animals. J. Clin. Microbiol. 41:5366-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.