Abstract
A highly supported maximum-likelihood species phylogeny for the genus Bradyrhizobium was inferred from a supermatrix obtained from the concatenation of partial atpD, recA, glnII, and rpoB sequences corresponding to 33 reference strains and 76 bradyrhizobia isolated from the nodules of Glycine max (soybean) trap plants inoculated with soil samples from Myanmar, India, Nepal, and Vietnam. The power of the multigene approach using multiple strains per species was evaluated in terms of overall tree resolution and phylogenetic congruence, representing a practical and portable option for bacterial molecular systematics. Potential pitfalls of the approach are highlighted. Seventy-five of the isolates could be classified as B. japonicum type Ia (USDA110/USDA122-like), B. liaoningense, B. yuanmingense, or B. elkanii, whereas one represented a novel Bradyrhizobium lineage. Most Nepalese B. japonicum Ia isolates belong to a highly epidemic clone closely related to strain USDA110. Significant phylogenetic evidence against the monophyly of the of B. japonicum I and Ia lineages was found. Analysis of their DNA polymorphisms revealed high population distances, significant genetic differentiation, and contrasting population genetic structures, suggesting that the strains in the Ia lineage are misclassified as B. japonicum. The DNA polymorphism patterns of all species conformed to the expectations of the neutral mutation and population equilibrium models and, excluding the B. japonicum Ia lineage, were consistent with intermediate recombination levels. All species displayed epidemic clones and had broad geographic and environmental distribution ranges, as revealed by mapping climate types and geographic origins of the isolates on the species tree.
Soybean (Glycine max) is the most important grain legume in the world, with an annual production of around 180 million tons and a market value of more than 36 billion euros. This crop is planted on 5.7 million, 8.3 million, 29 million, and 30 million hectares in India, China, South America, and North America, respectively (66). It is a major cash crop for small farmers in Asia, in South America, and also in some African countries. The diversity of soybeans today is the result of more than 5,000 years of cultivation, which started in China, where more than 20,000 land races were selected and later globally distributed and further domesticated by modern breeding programs (4). Soybeans were introduced in India around 1000 CE via the silk route from China (26).
Bradyrhizobium japonicum, Bradyrhizobium elkanii, Bradyrhizobium liaoningense, Ensifer (Sinorhizobium) fredii, Ensifer xinjiangense, and Mesorhizobium tianshanense are the microsymbionts currently known to nodulate soybeans naturally under field conditions (19, 23, 36, 45, 46, 57, 68). Soybean-nodulating B. japonicum strains have been isolated from different continents and climatic zones. Recently, two B. japonicum biovars (symbiotic ecotypes) were described (61). The B. japonicum bv. glycinearum isolates nodulate soybeans, whereas the B. japonicum bv. genistearum strains nodulate genistoid legumes such as Adenocarpus, Lupinus, Spartocytisus, or Teline, but not soybeans, and vice versa. Bradyrhizobium elkanii-like isolates have been recovered from diverse legumes, including soybeans, growing in tropical soils (1, 34) and in subtropical and temperate regions (32, 58). Bradyrhizobium liaoningense isolates from soybeans and peanuts (Arachis hypogaea) have been isolated only in Chinese locations with cold or temperate humid climates (65, 68).
The other three validly published Bradyrhizobium species are B. yuanmingense, isolated from the root nodules of Lespedeza cuneata in China (69); B. betae, from tumor-like structures of sugar beet (Beta vulgaris) in northern Spain (42); and B. canariense bv. genistearum, recovered from the nodules of diverse legume genera in the tribes Genisteae and Loteae growing naturally in the Canary Islands, in Morocco, in Spain, along the Mediterranean Basin, and in the Americas (18, 61). B. canariense bv. genistearum has recently been reported to nodulate lupins and serradela plants in South Africa and western Australia (54), thus being a truly cosmopolitan species.
Multilocus sequence analysis (10) has been recently employed to infer highly resolved Bradyrhizobium species phylogenies, to elucidate microevolutionary processes of particular species, to determine their geographic distribution ranges, and to formulate initial phylogeographic hypotheses (54, 61, 65). The aim of this study was to assess the power and practical utility of the multilocus sequence analysis approach (10) for Bradyrhizobium molecular systematics. This bacterial genus is considered a “taxonomically difficult” group of organisms due to their highly conserved rrs sequences and poor correlation between the groupings formed on the basis of genotypic and phenotypic traits, raising questions about the suitability of the polyphasic taxonomic approach to Bradyrhizobium systematics (51, 60).
Here we present a multilocus sequence-based analysis of 80 soybean nodule isolates obtained from India, Myanmar, Nepal, and Vietnam. Thirty-three reference strains were included in the combined phylogenetic and population genetic approaches used for species demarcation in order to estimate the magnitude of evolutionary forces acting within lineages and to gain further insights into their geographic and environmental distribution ranges. We took advantage of recently developed fast maximum-likelihood (ML) phylogeny algorithms (2, 11) and the power of multiprocessor computing to make thorough searches of tree space, compute bipartition significance values, and evaluate competing phylogenetic hypotheses in order to ground our taxonomic classifications. We discuss the advantages and potential pitfalls of phylogenetic supermatrix analyses in the frameworks of bacterial molecular systematics and ecological inference.
MATERIALS AND METHODS
Isolation of symbiotic Bradyrhizobium strains from soybean root nodules.
Rhizospheric soil samples were taken from traditionally managed soybean fields in India, Myanmar, Nepal, and Vietnam without known inoculation records, except for two Indian locations that had been inoculated with B. japonicum (Table 1). The geographic coordinates, Köppen-Geiger climatic types, and agroecological land uses of the soil sampling sites are summarized in Table 1. The isolates were obtained from the nodules induced by bacteria present in the rhizosphere soil samples on Glycine max trap plants cultivated for 4 weeks using the Leonard jar setting, cultivation conditions, and isolation protocols described elsewhere (62). Five-gram aliquots of air-dried soil were mixed with the sterile perlite-vermiculite substrate used to fill each cultivation unit. Three jars containing two axenically germinated plantlets were used for each site, whereas jars without soil inoculum served as negative nodulation controls. Genomic DNAs from purified isolates and reference strains were isolated using a cetyltrimethylammonium bromide-based protocol, as described previously (62).
TABLE 1.
Geographic coordinates, climate types, and land uses of the sampling sites
| Isolate prefixa | Country/localityb | Geographic coordinates | Climatec | Land used | G. max cultivare |
|---|---|---|---|---|---|
| BuCeG | Bu/Heho | 97.04°E, 20.78°N | Aw | Diverse Phaseolus beans | Gm-JS335 |
| BuCeR | Bu/Heho | 97.04°E, 20.78°N | Aw | Diverse Phaseolus beans | Ra |
| BuMiN | Bu/Nan-daw-kyun | 96.03°E, 22.05°N | Cwa | Vigna cajan L. | MA |
| BuMiT | Bu/Tha-min-chan | 96.05°E, 22.03°N, 22.02°N | Cwa | G. max, Lablab niger | MA |
| BuNoG | Bu/Mandalay | 96.09°E, 21.98°N | Cwa | Diverse Phaseolus beans | Gm-JS335 |
| BuNoR | Bu/Mandalay | 96.09°E, 21.98°N | Cwa | Diverse Phaseolus beans | Ra |
| InBu | In/Rajasthan/Bundi | 75.6°E, 25.5°N | BSh | G. max L., previous inoculation | MA |
| InIn | In/Madhya Pradesh-Indore | 75.86°E, 22.72°N | Aw | G. max L., no inoculation | MA |
| InJa | In/Madhya Pradesh-Jabalpur | 79.94°E, 23.17°N | Aw | G. max L., no inoculation | MA |
| InKo | In/Rajasthan-Kota | 75.83°E, 25.18°N | Bsh | G. max L., previous inoculation | MA |
| InRo | In/Uttaranchal-Roorkee | 77.89°E, 29.87°N | Cwa | Alluvial soils, Ganges plains, rice fields | Ra |
| NeMa | Nep/Kathmandu | 85.26°E, 27.42°N | Cwa | Soybean/rice/maize/potatoes | MA |
| NeRa | Nep/Kathmandu | 85.26°E, 27.42°N | Cwa | Soybean/rice/maize/potatoes | Ra |
| ViHaG | Vi/Halong | 107°E, 21°N | Cwa | Diverse vegetables | Gm-JS71-05 |
| ViHaR | Vi/Halong | 107°E, 21°N | Cwa | Diverse vegetables | Ra |
These prefixes are the ones given to the isolates obtained from each of the indicated sites, followed by the isolate number.
Bu, Myanmar (formerly Burma); In, India; Ne, Nepal; Vi, Vietnam. The region or geographic location from which the soil samples were obtained is indicated after the slash.
Köppen-Geiger climate classification: Aw, humid equatorial climate with dry winters; Cwa, humid temperate climate with dry winters and hot summers; BSh, dry climate, semiarid, hot.
Land use on the soybean plantations from which the soil samples were taken for the trapping experiments.
G. max (soybean) cultivars used in the trapping experiments: MA, Mapple Arrow; Ra, Ramson.
Amplification and sequencing of atpD, glnII, recA, and rpoB gene fragments.
Partial atpD, glnII, and recA gene fragments were amplified with the primers and conditions reported previously (65). Here we developed and validated an additional molecular marker for the genus Bradyrhizobium, tagging the RNA polymerase beta subunit (rpoB) locus, which had been previously used to study phylogenetic relationships between Afipia and Bosea species, two close relatives of bradyrhizobia (20). A partial rpoB fragment of 910 bp was amplified with primers rpoB-454F (ATCGTCTCGCAGATGCACCG) and rpoB-1364R (TCGATGTCGTCGATYTCGCC) using the protocol developed for the recA locus. The digits in the primer designations correspond to their binding coordinates on the B. japonicum USDA110 rpoB gene.
All amplifications were performed with Taq polymerase (USB-Amersham). Amplification products were purified using the PCR product purification system of Roche. Both strands were commercially sequenced by Macrogen, Korea.
Evolutionary analyses of nucleotide sequence alignments.
Diverse data parsing and transformation tasks were automated using ad hoc Perl scripts. Nucleotide sequences were translated and aligned using Muscle 3.52 (6). The resulting multiple-sequence alignments of proteins were used as masks to generate the corresponding codon alignments using custom Perl scripts.
Models of nucleotide substitution were selected by the Akaike information criterion, using MODELTEST3.7 (38). Among-site rate variation was modeled by a gamma distribution, approximated with four rate categories (7), with each category being represented by its mean. ML trees were inferred for each data set under the models of nucleotide substitution selected by the Akaike information criterion (37), using PhyML v2.4.5 (2, 11). In order to make a more thorough search of tree space, 100 random stepwise-addition parsimony trees were generated for each locus with PAUP*4b10 (55) and used to initiate a corresponding number of ML searches on a cluster of 27 dual core Pentium IV processors under Linux Rocks 3.3.0. A default search using a starting BioNJ tree was also run for all loci. The tree yielding the highest ln L value was selected among the 101 independent searches. The robustness of the ML topologies was evaluated using a recently developed Shimodaira-Hasegawa (SH)-like test (47) for branches implemented in PhyML v2.4.5 (2). In brief, the test assesses whether the branch being studied provides a significant likelihood gain, in comparison with the null hypothesis that involves collapsing that branch, but leaving the rest of the tree topology identical. We chose the SH-like procedure for assessing bipartition significance because the test is nonparametric and much less liberal than the diverse (parametric) approximate-likelihood ratio tests that are also implemented in that program. The resulting SH-like P values therefore indicate the probability that the corresponding split is significant. SH tests (47) were used to evaluate the global phylogenetic congruence of trees inferred from single gene partitions, as well as those inferred from all possible combinations of partitions, as implemented in PAUP* (55), using 10 random sequential-addition starting trees and TBR branch swapping. The statistical significance of conflicting phylogenetic hypotheses for different partitions and specific clades was also determined by SH tests using the phylograms resulting from constrained versus unconstrained tree searches under best-approximating substitution models.
Population genetic analyses of sequence polymorphisms were performed with DnaSP4.5 (44) in order to test the neutral mutation and population equilibrium hypotheses, to infer the population mutation (θ = 2Neμ) and recombination (C = 2Neρ) parameters (13), and to obtain estimates of population differentiation (15) and gene flow (17), as detailed in the relevant sections. Coalescent simulations based on 104 genealogy replications were performed with DnaSP to estimate the 95% confidence interval of the RM (minimal number of recombination events) and R2 (population growth) test statistics (16, 41). Permutation analyses with 104 replicates were run to test the significance of the population subdivision test statistics (15).
Nucleotide sequence accession numbers.
The GenBank accession numbers for all of the sequences generated in this study are listed in Table S1 in the supplemental material. These include 80 atpD, glnII, recA, and rpoB sequences for a corresponding number of Asiatic soybean nodule isolates and 37 rpoB sequences for selected reference strains, for which the other three loci had been sequenced in previous studies (61, 65).
RESULTS
Isolation of novel soybean root nodule rhizobial strains from India, Myanmar, Nepal, and Vietnam.
A total of 112 soybean microsymbionts were isolated. All soils used as inocula contained soybean-compatible bradyrhizobia. In this study, we present the analyses performed on 80 of these isolates. A list linking the sampling sites with the origin of the isolate is provided in Table 1.
Phylogenetic classification of the new Asiatic soybean root nodule isolates based on total evidence.
Sequence data for the atpD, glnII, recA, and rpoB loci were obtained for all 80 isolates. In addition, we sequenced the rpoB fragment for 37 reference Bradyrhizobium strains for which the former three loci had been previously sequenced and analyzed (61, 65). Therefore, a total of 357 new sequences were deposited in GenBank (see Table S1 in the supplemental material). These loci are unlinked and therefore provide independent genealogies from which to infer a species tree (43).
ML tree searches under best-approximating models were individually performed for the four sequence partitions containing 115 aligned sequences (comprising 80 Asiatic isolates and 35 reference strains). The resulting gene trees are provided in Fig. S1 to S4 in the supplemental material. These analyses identified several xenologous sequences. The Nepalese B. japonicum type Ia NeRa14 isolate had a B. japonicum type I rpoB locus, while the B. yuanmingense BuCeR1, BuCeR2, and BuMiT10 isolates were recipients of B. elkanii-like rpoB loci (highlighted in Fig. S4 in the supplemental material). These isolates were excluded from further analyses.
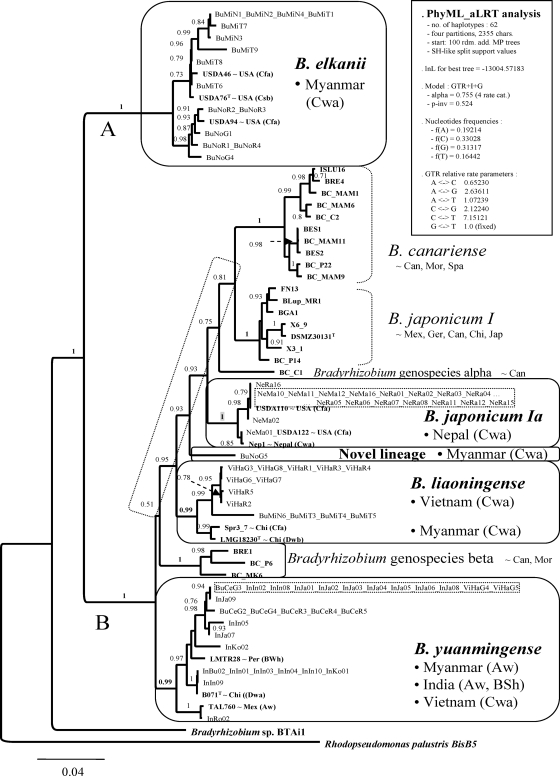
Figure 1 shows the ML phylogram obtained under the GTR+I+G model for 62 unique haplotypes recorded among concatenated atpD-glnII-recA-rpoB sequences from the 110 Bradyrhizobium isolates and reference strains. This was the best-scoring tree found among 100 independent PhyML searches that started from a corresponding number of random sequential-addition seed trees. Their ln L scores ranged from −13,004.57183 to −13,038.67491 (best to worst). These values correspond to 86 tree islands with unique trees and 7 islands with two trees. A default PhyML search starting with a BioNJ tree found a slightly worse tree (ln L = −13,007.21494) than the best one indicated above. Better-scoring ML trees could be found in all the single-locus analyses performed in this study when multiple distinct seed trees were used to initiate ML searches, compared to the score of the tree found by default PhyML searches starting from a BioNJ tree (data not shown). The fact that most of the tree islands, including the highest-scoring one, were hit only once reveals the complexity of the likelihood surface and strongly suggests that better trees remain to be found.
FIG. 1.
ML species tree estimated under the GTR-I-G model, showing the relationships among 62 atpD-glnII-recA-rpoB haplotypes found among 76 Asiatic Bradyrhizobium isolates and 34 reference strains (bold). This was the best tree found among 101 independent PhyML searches started from 100 random sequential addition trees and 1 NJ seed tree. The support values on the bipartitions correspond to SH-like P values, which denote the probability of the particular branch being correct. Ten Bradyrhizobium sp. lineages were resolved. The Asiatic isolates grouped in five of them, which are enclosed in rounded boxes. Dotted boxes highlight epidemic clones. The vertical rectangular box shows the parameterization of the PhyML tree search resulting in the best ln L score. The scale indicates the expected number of substitutions per site under the specified substitution model. The following country or regional abbreviations were used to indicate the geographic origins of the reference strains: Can, Canary Islands; Chi, China; Ger, Germany; Jap, Japan; Mex, Mexico; Mor, Morocco; Per, Peru; Spa, Spain; USA, United States. The following abbreviations were used to indicate the Köppen-Geiger world climate classes: BWh, dry, arid, hot; Cfa, humid, temperate, without dry season and hot summer; Csb, humid, temperate, with dry cool summer; Dwa, humid, cold, with dry winter and hot summer; Dwb, humid, cold, with dry winter and cool summer. The remaining abbreviations are explained in Table 1.
The tree is rooted with the homologous sequences from Rhodopseudomonas palustris BisB5. The phylogeny resolves two major and deeply branching clades with maximal SH-like support (Fig. 1A and B) and a total of 10 Bradyrhizobium lineages (Fig. 1). The Asiatic isolates were recovered in five of them. Clade A groups B. elkanii strains, whereas clade B groups strains from B. japonicum types I and Ia, B. canariense, B. yuanmingense and B. liaoningense, Bradyrhizobium genospecies alpha and beta, and a novel lineage represented by isolate BuNoG5.
Ten multilocus haplotypes from 15 Myanmarese isolates were recovered in clade A, forming two subclades related to the B. elkanii strains USDA76T and USDA94, respectively. All BuMi* isolates grouped in the former, while all BuNo* isolates clustered in the latter (the asterisk denotes any alphanumerical character). Eight very significantly supported subclades (SH-like P values of ≥0.99) were resolved within clade B (Fig. 1). The remaining Asiatic isolates were recovered in four of these subclades. The largest number of the isolates, (21 Indian, 6 Myanmarese, and 2 Vietnamese), comprising a total of nine haplotypes, clustered with the B. yuanmingense reference strains CCBAU10071T, LMTR28, and TAL760. No obvious correlation was found between the internal subdivisions of this clade and the geographic origin of the isolates, with the most abundant haplotype being shared by Myanmarese, Indian, and Vietnamese isolates (Fig. 1). Four Myanmarese isolates (BuMi*) and 9 Vietnamese isolates (ViHa*) grouped with B. liaoningense LMG18230T and Spr3-7 from China. The three lineages resolved within the B. liaoningense clade correlate perfectly with the geographic origin of the strains (China, Myanmar, and Vietnam). All 19 Nepalese NeMa* and NeRa* isolates clustered tightly with B. japonicum type Ia strains, such as USDA110 and USDA122, representing six haplotypes. One of them was shared by 15 Nepalese isolates, corresponding to a highly epidemic clone (Fig. 1). The isolate NeRa14 was found to harbor a xenologous rpoB allele from a B. japonicum type I donor (see Fig. S4 in the supplemental material), but none of the Asiatic isolates studied here was recovered within the clade grouping those strains (Fig. 1). The Myanmarese isolate BuNoG5 most likely represents a novel Bradyrhizobium species within clade B.
The splits separating the highly significant subclades (B. canariense, B. japonicum I and Ia, B. liaoningense, and Bradyrhizobium genospecies beta) resolved within clade B are significant (≥0.95) in only one case (Fig. 1). Interestingly, species clades could be recognized as the most inclusive clades, with a long subtending branch (≥26 expected substitutions) with SH-like P-values of ≥0.99, separated from other such clades by short branches (≤22 expected substitutions) with P values of ≤0.95. One of these deep internal branches is particularly short and not supported at all (P = 0.51), indicating that the phylogenetic relationships between some species may not be properly determined, particularly for sister clades having very short and poorly supported (P ≪ 0.90) subtending branches.
Global phylogenetic congruence among single-gene partitions and their combinations.
Figure 2 shows the result of pairwise SH tests (47) performed between all pairs of single-gene partitions and all possible combinations of them. All single-gene partitions were significantly incongruent between them. However, as shown in Fig. 2 and in Table S2 in the supplemental material, the mean and median congruence levels of trees increases with the number of concatenated partitions used to infer them. The species tree shown in Fig. 1 has the highest mean and median P values (0.40 and 0.38, respectively) for all pairwise comparisons (see Table S2 in the supplemental material).
FIG. 2.
Matrix showing P values of SH phylogenetic congruence tests among all pairs of single and combined sequence partitions, as indicated. White corresponds to P = 0 and black to P = 1, meaning completely incongruent and congruent trees, respectively.
Taxonomic implications of significant phylogenetic incongruences found among sequence partitions for specific clades.
All ML gene trees support the monophyly of B. canariense, B. japonicum type I and Ia, B. yuanmingense, and B. elkanii (see Fig. S1 to S4 in the supplemental material). However, the B. japonicum I and Ia lineages are not grouped in a clade on the atpD, glnII, and rpoB phylograms, whereas the BuMiN*, BuMiT*, ViHaG*, and ViHaR* strains are significantly associated with the bona fide B. liaoningense strains LMG18230T and Spr3-7 in the atpD and rpoB phylogenies but not in the recA or glnII trees. SH tests (47) were performed on constrained and unconstrained ML tree searches in order to test the strengths of the alternative phylogenetic hypotheses suggested by the individual gene trees. As shown in Table 2, the constrained glnII and recA tree searches forcing the monophyly of BuMiN*, BuMiT*, ViHaG*, and ViHaR* isolates (excluding ViHAG1 and ViHaG5, recovered in the B. yuanmingense clade) with the bona fide B. liaoningense strains LMG18230T and Spr3-7 did not result in significantly worse trees (P > 0.2 in both cases). Therefore, we classified the former isolates as B. liaoningense, which is consistent with their highly significant monophyletic grouping in the ML phylogeny shown in Fig. 1. However, tree searches imposing the monophyly constraint on the B. japonicum I and Ia clades resulted in significantly worse trees for both the atpD and rpoB loci, as well as for the concatenated data set. The monophyly hypothesis was therefore rejected in this case.
TABLE 2.
Evaluation of constrained versus unconstrained tree searches for selected clades and lineages in an ML framework
| Locusa | Constraint (monophyly)b | −ln Lc | Difference in −ln Ld | Pe |
|---|---|---|---|---|
| atpD | B. japonicum I and Ia | 2,489.38670 | 56.16313 | 0.000*** |
| rpoB | B. japonicum I and Ia | 3,479.73551 | 22.05898 | 0.017* |
| Concat | B. japonicum I and Ia | 13,360.14170 | 359.42271 | 0.000*** |
| glnII | B. liaoningense | 3,094.23147 | 8.68990 | 0.208 (NS) |
| recA | B. liaoningense | 2,968.76600 | 5.18924 | 0.245 (NS) |
Concat refers to the concatenated data set (atpD-glnII-recA-rpoB) used to infer the phylogeny shown in Fig. 1.
Constraints used in ML tree searches under best-fitting substitution models for the indicated sequence partition.
The values correspond to those for the constrained topology.
Score differences between the nonconstrained and constrained trees.
Significance of the difference in −ln L scores achieved by the constrained and unconstrained trees, as assessed by the SH test. *, 0.05 ≥ P > 0.01: ***, 0.001 ≥ P; NS, not significant.
Quantification of the phylogenetic signal contents of individual sequence partitions and their concatenations.
The relative phylogenetic information contents of the individual sequence partitions and their concatenations were evaluated by computing diverse descriptive statistics of the SH-like P values parsed from the corresponding trees using ad hoc Perl scripts. Table 3 shows the means, medians, and standard deviations of SH-like P values for each tree, along with the percentages of bipartitions having particular P cutoff values. This analysis indicates that the atpD and rpoB partitions have the lowest median P values, whereas recA has the highest mean and median P values of the single-locus partitions. However, based on the cutoff value of P ≥ 0.95, the partitions are ranked in decreasing order of significant split percentages as follows: glnII > rpoB > recA > atpD. The additive nature of the phylogenetic signals contained in the individual partitions is evident when these values are compared with those achieved by the concatenated glnII-recA and atpD-glnII-recA-rpoB data sets. The latter one has the lowest percentage of nonsignificant (P < 0.95) and the highest proportion of significantly supported (P ≥ 0.95) bipartitions (Table 3).
TABLE 3.
Relative performances of individual molecular markers and some of their combinations assessed using SH-like P values of branch significance under the ML criterion
| Partition | No. of:
|
P valuea
|
% of bipartitions withb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sites | Haplotypes | Bipartitionsc | Mean | Median | SD | Variance | P < 0.95 | P ≥ 0.95 | 0.95 ≤ P < 0.99 | P ≥ 0.99 | |
| atpD | 483 | 44 | 41 | 0.76 | 0.78 | 0.17 | 0.0293 | 85.37 | 14.64 | 7.32 | 7.32 |
| glnII | 591 | 46 | 43 | 0.75 | 0.84 | 0.27 | 0.0727 | 65.11 | 34.88 | 20.93 | 13.95 |
| recA | 510 | 46 | 43 | 0.79 | 0.87 | 0.25 | 0.0613 | 83.72 | 16.28 | 11.63 | 4.65 |
| rpoB | 771 | 51 | 48 | 0.63 | 0.77 | 0.36 | 0.1299 | 77.09 | 22.91 | 14.58 | 8.33 |
| glnII-recA | 1,101 | 56 | 53 | 0.86 | 0.92 | 0.17 | 0.0287 | 64.15 | 35.85 | 18.87 | 16.98 |
| atpD-glnII-recA-rpoB | 2,355 | 62 | 59 | 0.78 | 0.94 | 0.31 | 0.0980 | 55.93 | 44.07 | 16.95 | 27.12 |
Values for the best tree found for each partition among the 101 ML searches initiated from 100 random sequential addition parsimony trees and a neighbor-joining tree, using the sequences from the organisms included in the analysis shown in Fig. 1.
Percentage of bipartitions having SH-like P values with the indicated P value cutoffs.
Number of bipartitions (splits) on the tree.
Population genetic analysis of the DNA sequence polymorphisms found in selected lineages.
Table 4 summarizes the results of basic descriptive statistics of DNA polymorphisms, neutrality, and population growth tests based on the concatenated data set used to infer the phylogeny shown in Fig. 1 for B. japonicum I and Ia, B. liaoningense, B. yuanmingense, and B. elkanii. The analyses were based on the segregating sites, excluding those that violate the infinite-sites model (i.e., those segregating more than one base). Bradyrhizobium elkanii was the lineage with the highest level of DNA polymorphism, both in terms of haplotype (Hd) and nucleotide (π) diversity, whereas the B. japonicum Ia lineage displayed the lowest diversity. The observed patterns of nucleotide substitution were compatible with those expected under the neutral equilibrium model, as revealed by Tajima's D (56) and Fu and Li's D* and F* (9) statistics, which were all nonsignificant. They are all based on intraspecifc data of DNA polymorphisms and are designed to test the hypothesis that all mutations are selectively neutral (21). The small negative D values could be the result of population bottlenecks (8, 56). However, the powerful R2 test statistic (41), which is particularly suited for small sample sizes with recombination, also failed to reject the population equilibrium model, as revealed by coalescent simulations (14) run under the assumption of intermediate levels of recombination. Therefore, all evidence indicates that the observed polymorphisms in the concatenated data sets conform to the neutral equilibrium model (8, 41, 56).
TABLE 4.
Descriptive statistics of nucleotide polymorphisms, along with neutrality and growth tests for the concatenated atpD-glnII-recA-rpoB partitions (2,355 sites), based on segregating sites
| Type or species (no. of sequences) | No. of sites:
|
ka/ksa | kb | h/Hdc | θd | πe | Tajima's Df | Fu and Li's D*f | Fu and Li's F*f | R2g | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Segregating | Parsimony informative | ||||||||||
| B. japonicum I (7) | 83 | 39 | 80/6 | 33.667 | 7/1 | 0.1475 | 0.01430 | −0.03628 | −0.23415 | −0.21017 | 0.1293 |
| B. japonicum Ia (21) | 25 | 22 | 24/1 | 6.067 | 6/0.495 | 0.00297 | 0.00258 | −0.48593 | 0.96108 | 0.61592 | 0.1171 |
| B. elkanii (18) | 151 | 89 | 135/22 | 45.84 | 13/0.948 | 0.01864 | 0.01947 | 0.18707 | −0.77994 | −0.57540 | 0.1357 |
| B. liaoningense (15) | 112 | 93 | 101/14 | 38.01 | 7/0.838 | 0.01463 | 0.01614 | 0.45443 | 0.83018 | 0.83579 | 0.1601 |
| B. yuanmingense (32) | 129 | 93 | 122/8 | 28.81 | 12/0.817 | 0.01407 | 0.01225 | −0.51068 | −0.26557 | −0.41353 | 0.1061 |
Total number of synonymous/nonsynonymous changes.
Average number of nucleotide differences.
Number of haplotypes/haplotype (gene) diversity.
Theta per bp (65a), assuming the infinite-sites model.
Nucleotide diversity.
Calculations using the total number of segregating sites; all the values are nonsignificant.
Population growth test statistic of Ramos-Onsis and Rozas (41); all the values were nonsignificant as determined by neutral coalescence simulations considering recombination if necessary.
Table 5 shows the estimates obtained for the population recombination parameter C using the methods of Hudson and Kaplan (16) and Hudson (13). The first method is based on RM, or minimum number of recombination events, observed in the sample. Estimates of the observed RM were used to compute average C and 95% credibility intervals by neutral coalescent simulations (14). The Hudson (13) method is based on the variance of the number of differences between pairs of sequences; in this case, the estimate of C can be obtained numerically. Both estimates of C are consistent with intermediate levels of recombination in all but the B. japonicum Ia lineage, which appears to have a clonal and highly epidemic population structure. Interestingly, the B. japonicum I lineage has the highest level of average RM values (estimated under the neutral coalescent), which is almost 4 orders of magnitude higher than that estimated for the Ia lineage.
TABLE 5.
Recombination estimates based on the segregating sites from the concatenated atpD-glnII-recA-rpoB partitions (2,355 sites) of selected Bradyrhizobium populations
| Type or species (no. of sequences) | Ra | RMb | Coalescence simulationsc
|
||
|---|---|---|---|---|---|
| Confidence intervald | P (RM ≤ observed RM)e | Avg RMf | |||
| B. japonicum I (7) | 125 | 13 | 5.0, 15.0 | 0.922 | 9.98 |
| B. japonicum Ia (21) | 0.001 | 1 | 0.0, 0.0 | 1.0 | 0.002 |
| B. elkanii (18) | 9.3 | 15 | 2.0, 9.0 | 1.0 | 5.0 |
| B. liaoningense (15) | 0.99 | 2.0 | 0.0, 2.0 | 0.98 | 0.683 |
| B. yuanmingense (32) | 2.6 | 20 | 0.0, 5.0 | 1.0 | 2.372 |
Estimate of the population recombination parameter R (13) corrected for haploid organisms.
Observed minimum number of recombination events (16).
Neutral coalescence simulations (104) given the number of segregating sites, with an intermediate level of recombination.
Confidence interval (lower limit, upper limit) for RM under the neutral coalescent process.
Probability that RM is less than or equal to the observed RM under the neutral coalescent process.
Average value of RM under the neutral coalescent process.
Further evidence for the distinctness of the B. japonicum I and Ia lineages was gained from genetic differentiation and gene flow analyses (Table 6). The highest average nucleotide substitutions per site between lineages (Dxy) was found precisely for this pair. Both the haplotype (χ2) and sequence-based (KST*) genetic differentiation statistics for this comparison were also the most significant ones found (Table 6). This differentiation cannot be explained solely by disjunct geographic origins of the isolates, since both groups contain isolates from different continents. High fixation indices (FST) and low effective numbers of migrants (Nm) between these lineages reveal a high level of genetic isolation.
TABLE 6.
Genetic differentiation and gene flow estimates
| Populations (no. of isolates per species or lineage) | Fixed differencesa | Dxyb | Genetic differentiation
|
Gene flow
|
||||
|---|---|---|---|---|---|---|---|---|
| χ2 (df)c | Pd | KST*e | Pf | FSTg | Nmh | |||
| B. japonicum I vs Ia (7, 21) | 49 | 0.03928 | 28.0 (12) | 0.0055** | 0.38907 | 0.0000*** | 0.78523 | 0.14 |
| B. elkanii USDA76 vs USDA94 (11, 7) | 29 | 0.02930 | 18.0 (12) | 0.1157 (NS) | 0.51744 | 0.0001*** | 0.68310 | 0.23 |
| B. liaoningense | ||||||||
| Vietnam vs Burma (9, 4) | 60 | 0.02647 | 13.0 (4) | 0.0113* | 0.94092 | 0.0007*** | 0.97950 | 0.01 |
| Vietnam vs China (9, 2) | 42 | 0.02222 | 11.0 (5) | 0.0514 (NS) | 0.49644 | 0.0055** | 0.81316 | 0.11 |
| Burma vs China (4, 2) | 79 | 0.03694 | 6.0 (2) | 0.0498* | 1.0 | 0.0677 (NS) | 0.90230 | 0.05 |
| B. yuanmingense Burma vs India (6, 21) | 0 | 0.01201 | 21.8 (8) | 0.0053** | 0.15049 | 0.0006*** | 0.43216 | 0.66 |
Number of fixed differences between populations.
Average number of nucleotide substitutions per site between populations or lineages.
Haplotype-based statistic (15); degrees of freedom are indicated in parenthesis.
Probability of rejecting the null hypothesis that the two populations are not genetically differentiated, based on the critical values from the χ2 distribution. *, 0.05 ≥ P > 0.01; **, 0.01 ≥ P > 0.001; NS, not significant.
Sequence-based statistic described by Hudson et al. (15).
Probability obtained by the permutation test (15) with 1,000 replicates. **, 0.01 ≥ P > 0.001; ***, 0.001 ≥ P; NS, not significant.
Sequence-based estimate described by Hudson et al. (15).
Effective number of migrants.
Highly significant KST* values were also found for the pairwise comparisons between the B. elkanii USDA76 and USDA94 lineages and for the B. liaoningense isolates originating from Burma and Vietnam. However, their genetic differentiation is not as marked, as judged from their higher KST* values, lower or nonsignificant haplotype (χ2) differentiation levels, and ∼25% lower Dxy values, compared with those obtained for the first pairwise comparison.
The populations from all lineages presented epidemic clones, that is, multilocus haplotypes that appear in high frequency in the collection (28, 48). The two most prevalent atpD-glnII-recA-rpoB haplotypes found in our collection belong to the B. yuanmingense and B. japonicum Ia clades, with 12 and 15 isolates, respectively.
Broad geographic and environmental distribution of four Bradyrhizobium species nodulating soybean.
A preliminary definition of the environmental distribution ranges of four Bradyrhizobium species could be defined when the Köppen-Geiger climate types of the sites sampled in this study were mapped on the species phylogeny shown in Fig. 1. B. yuanmingense was recovered from sites with humid equatorial climates (Aw) or dry, hot, semiarid climates (BSh) with marked seasonal fluctuations in water availability. The Bradyrhizobium japonicum Ia, B. liaoningense, and B. elkanii isolates in our collection were preferentially recovered from areas with humid, temperate climates with dry winters and hot summers (Cwa).
Taking also the reference strains into account indicates that at least B. japonicum I and Ia, B. yuanmingense, and B. elkanii have a very broad geographic distribution across the Northern hemisphere. B. liaoningense seems to be broadly distributed across East and Southeast Asia. The distribution range of B. yuanmingense reaches the Southern hemisphere, since strain LMTR28 was isolated in Peru from lima beans (31). Therefore, the environmental range for this species also includes dry arid and hot environments (BWh), as well as humid cold climates with dry winters and hot summers (Dwa). Larger samples of taxonomically well-characterized strains are obviously needed to better define the environmental and geographic distribution ranges of these species. Hence, those provided here represent only minimal ranges.
DISCUSSION
Many studies have reported on the high diversity of native Bradyrhizobium strains found in contrasting ecosystems, on different continents, and associated with diverse agricultural and wild legumes (1, 18, 24, 29, 31, 35, 70, 71). However, in most of these and similar studies, no clear assertions were made about the number of Bradyrhizobium species that nodulate a particular host. Generally the strains are classified as a Bradyrhizobium sp. “related to” the B. japonicum or B. elkanii lineages, which is the equivalent of classifying the strains as belonging to clade A or B in Fig. 1 of this study. This level of taxonomic resolution is clearly insufficient to disclose geographic or environmental distribution ranges of particular species (40), or to make inferences about evolutionary forces and historic contingencies acting on them (53, 54, 63, 65). This situation is largely due to the predominant use of 16S rRNA gene sequences or PCR-restriction fragment length polymorphisms as the only molecular marker for diversity assessment and lineage classification. Several publications have shown that this marker has only limited utility in Bradyrhizobium diversity studies due to very low levels of polymorphism and frequent intragenic mosaicism, which yields a poor and often misleading signal (33, 58, 59, 61, 64, 67).
This work provides further empirical evidence showing the adequacy of multilocus sequence analyses (10) of protein-coding genes for Bradyrhizobium species demarcation (52, 61, 65) and their suitability for making refined ecological and evolutionary inferences. Because of the stochastic way in which lineages sort during speciation, gene trees generally differ in topology from each other and from the species tree, and therefore no single gene tree is likely to be a good approximation of a species phylogeny (30, 43, 50, 63-65), as clearly illustrated in this study with our phylogenetic congruence analyses. It has also been shown that the inference of a multispecies tree can be problematic when single individuals are analyzed per species, due to the presence of anomalous gene trees (5). A practical and powerful strategy to diminish the impact of this potential problem is to sample multiple individuals per species, as shown in this and other studies (5, 43). However, the strong phylogenetic incongruence detected between sequence partitions and the presence of at least one very short and nonsupported bipartition located deeply within the species tree indicate that, although the overall tree support is high, its accuracy is not definite (22). The short internal branches may reflect incomplete lineage sorting, but as the number of individuals per species increases, the corresponding species clades become more robust, because each individual from a species provides an independent opportunity to observe coalescence with an individual from the sister species (25). Therefore, based on these theoretical considerations and the very strong support of the relatively long branches subtending the species clades, we conclude that the species demarcation suggested by our species tree is robust. The use of multiple strains per species is also very useful to identify individuals harboring xenologous loci. The inference of accurate bacterial species trees from concatenated alignments demands the identification and removal of such individuals, which can strongly distort species phylogenies inferred using standard tree reconstruction methods that assume a single underlying evolutionary history (65). Moreover, the estimation of a multispecies tree with many multilocus haplotypes and several concatenated sequence partitions demands the use of complex substitution models (37, 39, 65) and, even more importantly, a thorough search of tree space. There are (2s − 5)!/2(s − 3)(s − 3)! unrooted and bifurcating trees for s sequences (7). Thus, for the inference problem with 62 multilocus haplotypes presented in this study, there are 1.945514 × 10181 possible topologies of this kind. Only heuristic tree searching algorithms are suitable to solve such a formidable computational task, implying that there is no guarantee to find the best global ML tree, since the search may easily “get trapped” in a local maximum (7). This explains why starting multiple heuristic searches from distinct random trees allowed, in all cases, better ML trees to be found than with a BioNJ starting tree, which is the default search option in PhyML (11).
Despite these computational limitations, a well-resolved species phylogeny could be inferred from the concatenated data set after exclusion of the individuals showing xenologous sequences. This tree had both the highest overall tree resolution level and the highest mean and median phylogenetic congruence levels compared with all possible single and combined partition combinations. These values underline the convenience of the supermatrix approach used here for species demarcation, although some uncertainty concerning the phylogenetic relationships between species was evident, based on the low support values (P ≪ 0.90) of some of the shorter branches found deep within the tree.
Five lineages of soybean-nodulating bradyrhizobia were found among the Asiatic isolates. These lineages could be classified with great statistical confidence as B. japonicum type Ia (12), B. elkanii (23), B. yuanmingense (69), B. liaoningense (68), and a novel lineage. We show that B. yuanmingense contains isolates capable of nodulating soybeans in diverse soils and countries (India, Myanmar, and Vietnam), confirming and extending the results of a report (3) that appeared during the review of this paper. Taking into account previous publications, we conclude that this species has very broad geographic and host ranges, nodulating not only Lespedeza spp. in northern China (65), but also lima beans in Peru (31, 59), Indigofera hirsuta in Mexico (59), soybeans in southern and southeastern Asia, and different Vigna species in southern Africa (52) and subtropical China (72). The fact that the B. yuanmingense isolates from Chinese Lespedeza cuneata plants do not nodulate soybeans (69) strongly suggests the existence of several symbiotic ecotypes (50, 61) within this cosmopolitan species. This species also has a very broad environmental distribution. It has been isolated from warm, semiarid regions such as the Indian Rajasthan or the arid coastal strip of Peru, from humid temperate and equatorial climates such as those found in Myanmar and Vietnam, but also from regions with a humid, cold climate with dry winters such as the Beijing province of China. It is noteworthy that the most abundant B. yuanmingense composite atpD-glnII-recA-rpoB haplotype was recovered from all three tropical and subtropical Asiatic countries sampled, revealing that some of its clones or clonal complexes also have a broad geographic and environmental distribution. Therefore, no clear geographic or ecological patterning of haplotypes was found for this species.
Very striking was the finding that 16 Nepalese isolates (NeMa* and NeRa*) had the same composite glnII-recA-rpoB haplotype as that of B. japonicum USDA110. These strains do not represent a contamination of the cultivation systems used for the trapping experiments with USDA110, because the Nepalese strains have a different atpD sequence. The B. japonicum Ia (12) lineage displayed the lowest DNA polymorphism level of all lineages analyzed. It essentially has a clonal (and highly epidemic) population structure (28), which contrasts with that of the B. japonicum I lineage, for which the highest haplotype diversity and RM values (16) were recorded. Kuykendall et al. (23) also found low genetic diversity among B. japonicum strains of the DNA homology group Ia based on DNA hybridization experiments with cosmid clones. Strong phylogenetic evidence against the monophyly of these two lineages was found, as reported by van Berkum and Fuhrmann based on bootstrap analysis of a neighbor-joining phylogeny reconstructed from ribosomal internal transcribed spacer sequences (58). Our results present compelling evidence that these two groups represent significantly differentiated and genetically isolated evolutionary lineages, therefore supporting the previously published opinion that strains USDA110 and USDA122 “need not necessarily be representative of B. japonicum” (58). In other words, current evidence suggests that homology group Ia (12) strains are misclassified as B. japonicum and probably represent a novel species.
A comparative evolutionary genetic analysis of multiple Bradyrhizobium species revealed that all have epidemic clones, as found in other population genetic studies of diverse rhizobia (28, 49, 50, 65), and that intermediate levels of recombination shape their population genetic structures (28, 49, 50, 65), with the notable exception of the B. japonicum Ia lineage. The recombination parameter values are most likely underestimated because they are based on data for all individuals and not on haplotypes (28, 50). Significant genetic structuring of haplotypes was found within the two B. elkanii lineages represented by the North American reference strains USDA76T and USDA94. Kuykendall and colleagues also reported significant genetic differentiation between these two groups of strains (B. elkanii homology groups II and IIa) (23). This issue requires further investigation, not only because of potential taxonomic implications but especially because it would be desirable that future reports on the diversity of strains described as “related to the B. elkanii clade” consider the evident genetic structuring that exists within this species, as currently defined.
In conclusion, more studies using careful multilocus sequence analyses coupled with detailed descriptions of the habitats from which new strains are isolated are needed to build the databases required to make robust inferences about the biogeography and environmental distribution of rhizobial species (63). Current evidence clearly demonstrates that rhizobia belong to the class of bacteria with very broad geographic and environmental distribution ranges at the genus and species levels of taxonomic resolution (50, 54, 64). Much remains to be learned, however, about the relative contributions of history and environment to the distribution patterns of particular rhizobial species, as well as about the processes that shape their biogeography (27, 40).
Supplementary Material
Acknowledgments
We thank the European Union for long-term support in INCO DEV project ICA4-CT-2001-10057 and the DEGAPA-PAPIIT from the National Autonomous University of Mexico for grant DGAPA IN201806-2 to P.V.
We gratefully acknowledge Claudia Silva, Susana Brom, and Paul V. Dunlap for their critical reading of the manuscript.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ando, S., and T. Yokoyama. 1999. Phylogenetic analyses of Bradyrhizobium strains nodulating soybean (Glycine max) in Thailand with reference to the USDA strains of Bradyrhizobium. Can. J. Microbiol. 45:639-645. [DOI] [PubMed] [Google Scholar]
- 2.Anisimova, M., and O. Gascuel. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539-552. [DOI] [PubMed] [Google Scholar]
- 3.Appunu, C., A. N′Zoue, and G. Laguerre. 2008. Genetic diversity of native bradyrhizobia isolated from soybean (Glycine max L.) in different agro-eco-climatic regions of India. Appl. Environ. Microbiol. 74:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, T. E., T. Hymowitz, and R. L. Nelson. 2004. Biogeography, local adaptation, vavilov, and genetic diversity in soybean, p. 47-59. In D. Werner (ed.), Biological resources and migration. Springer Verlag, Heidelberg, Germany.
- 5.Degnan, J. H., and N. A. Rosenberg. 2006. Discordance of species trees with their most likely gene trees. PLoS Genet. 2:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 2004. Inferring phylogenies. Sinauer Associates, Inc., Sunderland, MA.
- 8.Fu, Y.-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu, Y.-X., and W.-H. Li. 1993. Statistical tests of neutrality of mutations. Genetics 133:693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrandt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739. [DOI] [PubMed] [Google Scholar]
- 11.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 12.Hollis, A. B., W. E. Kloos, and G. E. Elkan. 1981. DNA:DNA hybridization studies of Rhizobium japonicum and related Rhizobiaceae. J. Gen. Microbiol. 123:215-222. [Google Scholar]
- 13.Hudson, R. R. 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50:245-250. [DOI] [PubMed] [Google Scholar]
- 14.Hudson, R. R. 1990. Gene genealogies and the coalescent process. Oxf. Surv. Evol. Biol. 7:1-44. [Google Scholar]
- 15.Hudson, R. R., D. D. Boos, and N. L. Kaplan. 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9:138-151. [DOI] [PubMed] [Google Scholar]
- 16.Hudson, R. R., and N. L. Kaplan. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson, R. R., M. Slatkin, and W. P. Maddison. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarabo-Lorenzo, A., R. Pérez-Galdona, J. Donate-Correa, R. Rivas, E. Velázquez, M. Hernández, F. Temprano, E. Martínez-Molina, T. Ruiz-Argüeso, and M. León-Barrios. 2003. Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst. Appl. Microbiol. 26:611-623. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, D. C. 1982. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 32:136-139. [Google Scholar]
- 20.Khamis, A., P. Colson, D. Raoult, and B. L. Scola. 2003. Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences. Appl. Environ. Microbiol. 69:6740-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, MA.
- 22.Kubatko, L. S., and J. H. Degnan. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 56:17-24. [DOI] [PubMed] [Google Scholar]
- 23.Kuykendall, L. D., B. Saxena, T. E. Devine, and S. E. Udell. 1992. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 38:501-505. [Google Scholar]
- 24.Lafay, B., and J. J. Burdon. 2007. Molecular diversity of legume root-nodule bacteria in Kakadu National Park, Northern Territory, Australia. PLoS ONE. 2:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddison, W. P., and L. L. Knowles. 2006. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55:21-30. [DOI] [PubMed] [Google Scholar]
- 26.Mahna, S. K. 2005. Production, regional distribution of cultivars, and agricultural aspects of soybean in India, p. 43-66. In D. Werner and W. E. Newton (ed.), Nitrogen fixation in agriculture, forestry and the environment. Springer Verlag, Dordrecht, The Netherlands.
- 27.Martiny, J. B., B. J. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A. L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 28.Maynard-Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreira, F. M., K. Haukka, and J. P. Young. 1998. Biodiversity of rhizobia isolated from a wide range of forest legumes in Brazil. Mol. Ecol. 7:889-895. [DOI] [PubMed] [Google Scholar]
- 30.Nichols, R. 2001. Gene trees and species trees are not the same. Trends Ecol. Evol. 16:358-364. [DOI] [PubMed] [Google Scholar]
- 31.Ormeño-Orrillo, E., P. Vinuesa, D. Zúñiga-Dávila, and E. Martínez-Romero. 2006. Molecular diversity of native bradyrhizobia isolated from lima bean (Phaseolus lunatus L.) in Peru. Syst. Appl. Microbiol. 29:253-262. [DOI] [PubMed] [Google Scholar]
- 32.Parker, M. A. 2002. Bradyrhizobia from wild Phaseolus, Desmodium, and Macroptilium species in northern Mexico. Appl. Environ. Microbiol. 68:2044-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker, M. A. 2001. Case of localized recombination in 23S rRNA genes from divergent Bradyrhizobium lineages associated with neotropical legumes. Appl. Environ. Microbiol. 67:2076-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker, M. A. 2004. rRNA and dnaK relationships of Bradyrhizobium sp. nodule bacteria from four papilionoid legume trees in Costa Rica. Syst. Appl. Microbiol. 27:334-342. [DOI] [PubMed] [Google Scholar]
- 35.Parker, M. A. 2008. Symbiotic relationships of legumes and nodule bacteria on Barro Colorado Island, Panama: a review. Microb. Ecol. 55:662-672. [DOI] [PubMed] [Google Scholar]
- 36.Peng, G. X., Z. Y. Tan, E. T. Wang, B. Reinhold-Hurek, W. F. Chen, and W. X. Chen. 2002. Identification of isolates from soybean nodules in Xinjiang Region as Sinorhizobium xinjiangense and genetic differentiation of S. xinjiangense from Sinorhizobium fredii. Int. J. Syst. Evol. Microbiol. 52:457-462. [DOI] [PubMed] [Google Scholar]
- 37.Posada, D., and T. R. Buckley. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53:793-808. [DOI] [PubMed] [Google Scholar]
- 38.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 39.Posada, D., and K. A. Crandall. 2001. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50:580-601. [PubMed] [Google Scholar]
- 40.Ramette, A., and J. M. Tiedje. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53:197-207. [DOI] [PubMed] [Google Scholar]
- 41.Ramos-Onsins, S. E., and J. Rozas. 2002. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19:2092-2100. [DOI] [PubMed] [Google Scholar]
- 42.Rivas, R., A. Willems, J. L. Palomo, P. García-Benavides, P. F. Mateos, E. Martinez-Molina, M. Gillis, and E. Velázquez. 2004. Bradryrhizobium betae sp. nov. isolated from roots of Beta vulgaris affected by tumor-like deformations. Int. J. Syst. Evol. Microbiol. 54:1271-1275. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg, N. A. 2002. The probability of topological concordance of gene trees and species trees. Theor. Pop. Biol. 61:225-247. [DOI] [PubMed] [Google Scholar]
- 44.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz Sainz, J. E., J. C. Zhou, D.-N. Rodriguez-Navarro, J. M. Vinardell, and J. E. Thomas-Oates. 2005. Soybean cultivation and BNF in China, p. 67-87. In D. Werner and W. E. Newton (ed.), Nitrogen fixation in agriculture, forestry and the environment. Springer Verlag, Dordrecht, The Netherlands.
- 46.Scholla, M. H., and G. H. Elkan. 1984. Rhizobium fredii sp. nov., a fast-growing species that effectively nodulates soybeans. Int. J. Syst. Bacteriol. 34:484-486. [Google Scholar]
- 47.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 48.Silva, C., L. E. Eguiarte, and V. Souza. 1999. Reticulated and epidemic population genetic structure of Rhizobium etli biovar phaseoli in a traditionally managed locality in Mexico. Mol. Ecol. 8:277-287. [Google Scholar]
- 49.Silva, C., P. Vinuesa, L. E. Eguiarte, E. Martínez-Romero, and V. Souza. 2003. Rhizobium etli and Rhizobium gallicum nodulate common bean (Phaseolus vulgaris) in a traditionally managed milpa plot in Mexico: population genetics and biogeographic implications. Appl. Environ. Microbiol. 69:884-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva, C., P. Vinuesa, L. E. Eguiarte, V. Souza, and E. Martínez-Romero. 2005. Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol. Ecol. 14:4033-4050. [DOI] [PubMed] [Google Scholar]
- 51.So, R. B., J. K. Ladha, and J. P. Young. 1994. Photosynthetic symbionts of Aeschynomene spp. form a cluster with bradyrhizobia on the basis of fatty acid and rRNA analyses. Int. J. Syst. Bacteriol. 44:392-403. [DOI] [PubMed] [Google Scholar]
- 52.Steenkamp, E. T., T. Stepkowski, A. Przymusiak, W. J. Botha, and I. J. Law. 2008. Cowpea and peanut in southern Africa are nodulated by diverse Bradyrhizobium strains harboring nodulation genes that belong to the large pantropical clade common in Africa. Mol. Phylogenet. Evol. 48:1131-1144. [DOI] [PubMed] [Google Scholar]
- 53.Stepkowski, T., C. E. Hughes, I. J. Law, L. Markiewicz, D. Gurda, A. Chlebicka, and L. Moulin. 2007. Diversification of lupine Bradyrhizobium strains: evidence from nodulation gene trees. Appl. Environ. Microbiol. 73:3254-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stepkowski, T., L. Moulin, A. Krzyzanska, A. McInnes, I. J. Law, and J. Howieson. 2005. European origin of Bradyrhizobium populations infecting lupins and serradella in soils of Western Australia and South Africa. Appl. Environ. Microbiol. 71:7041-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swofford, D. L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony and Other Methods. Sinuauer Associates, Sunderland, MA.
- 56.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan, Z. Y., X. D. Xu, E. T. Wang, J. L. Gao, E. Martinez-Romero, and W. X. Chen. 1997. Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int. J. Syst. Bacteriol. 47:874-879. [DOI] [PubMed] [Google Scholar]
- 58.van Berkum, P., and J. J. Fuhrmann. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 50:2165-2172. [DOI] [PubMed] [Google Scholar]
- 59.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalainen, K. Lindström, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 185:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Rossum, D., F. P. Schuurmans, M. Gillis, A. Muyotcha, H. W. Van Verseveld, A. H. Stouthamer, and F. C. Boogerd. 1995. Genetic and phenetic analyses of Bradyrhizobium strains nodulating peanut (Arachis hypogaea L.) roots. Appl. Environ. Microbiol. 61:1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinuesa, P., M. León-Barrios, C. Silva, A. Willems, A. Jarabo-Lorenzo, R. Pérez-Galdona, D. Werner, and E. Martínez-Romero. 2005. Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae:Genisteae) growing in the Canary Islands, along with B. japonicum bv. genistearum, Bradyrhizobium genospecies α and Bradyrhizobium genospecies β. Int. J. Syst. Evol. Microbiol. 55:569-575. [DOI] [PubMed] [Google Scholar]
- 62.Vinuesa, P., J. L. W. Rademaker, F. J. de Bruijn, and D. Werner. 1998. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl. Environ. Microbiol. 64:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinuesa, P., and C. Silva. 2004. Species delineation and biogeography of symbiotic bacteria associated with cultivated and wild legumes, p. 143-161. In D. Werner (ed.), Biological resources and migration. Springer Verlag, Berlin, Germany.
- 64.Vinuesa, P., C. Silva, M. J. Lorite, M. L. Izaguirre-Mayoral, E. J. Bedmar, and E. Martínez-Romero. 2005. Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst. Appl. Microbiol. 28:702-716. [DOI] [PubMed] [Google Scholar]
- 65.Vinuesa, P., C. Silva, D. Werner, and E. Martínez-Romero. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34:29-54. [DOI] [PubMed] [Google Scholar]
- 65a.Waterson, G. A. 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7:256-276. [DOI] [PubMed] [Google Scholar]
- 66.Werner, D., and W. E. Newton (ed.). 2005. Nitrogen fixation in agriculture, forestry and the environment. Springer Verlag, Dordrecht, The Netherlands.
- 67.Willems, A., R. Coopman, and M. Gillis. 2001. Phylogenetic and DNA-DNA hybridization analyses of Bradyrhizobium species. Int. J. Syst. Evol. Microbiol. 51:111-117. [DOI] [PubMed] [Google Scholar]
- 68.Xu, L. M., C. Ge, Z. Cui, J. Li, and H. Fan. 1995. Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int. J. Syst. Bacteriol. 45:706-711. [DOI] [PubMed] [Google Scholar]
- 69.Yao, Z. Y., F. L. Kan, E. T. Wang, G. H. Wei, and W. X. Chen. 2002. Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int. J. Syst. Evol. Microbiol. 52:2219-2230. [DOI] [PubMed] [Google Scholar]
- 70.Yokoyama, T., N. Tomooka, M. Okabayashi, A. Kaga, N. Boonkerd, and D. A. Vaughan. 2006. Variation in the nod gene RFLPs, nucleotide sequences of 16S rRNA genes, Nod factors, and nodulation abilities of Bradyrhizobium strains isolated from Thai Vigna plants. Can. J. Microbiol. 52:31-46. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, X., G. Nick, S. Kaijalainen, Z. Terefework, L. Paulin, S. W. Tighe, P. H. Graham, and K. Lindström. 1999. Phylogeny and diversity of Bradyrhizobium strains isolated from the root nodules of peanut (Arachis hypogaea) in Sichuan, China. Syst. Appl. Microbiol. 22:378-386. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Y. F., E. T. Wang, C. F. Tian, F. Q. Wang, L. L. Han, W. F. Chen, and W. X. Chen. 2008. Bradyrhizobium elkanii, Bradyrhizobium yuanmingense and Bradyrhizobium japonicum are the main rhizobia associated with Vigna unguiculata and Vigna radiata in the subtropical region of China. FEMS Microbiol. Lett. 285:146-154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.