Abstract
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium that differentiates heterocysts in response to deprivation of combined nitrogen. A hetF deletion strain lacked heterocysts and had aberrant cell morphology. Site-directed mutagenesis of the predicted active-site histidine and cysteine residues of this putative caspase-hemoglobinase fold protease abolished HetF function, supporting the hypothesis that HetF is a protease. Deletion of patA, which is necessary for the formation of most intercalary heterocysts, or hetF resulted in an increase in HetR protein, and extra copies of hetF on a plasmid functionally bypassed the deletion of patA. A hetR-gfp translational fusion expressed from an inducible promoter demonstrated that hetF-dependent downregulation of HetR levels occurs rapidly in vegetative cells, as well as developing heterocysts. “Mosaic” filaments in which only one cell of a filament had a copy of hetR or hetF indicated that hetF is required for differentiation only in cells that will become heterocysts. hetF was required for transcription from a hetR-dependent transcription start point of the hetR promoter and induction of transcription from the patS promoter. The inverse correlation between the level of HetR protein and transcription from hetR-dependent promoters suggests that the transcriptional activity of HetR is regulated by HetF and PatA.
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium capable of reducing atmospheric dinitrogen in specialized cells known as heterocysts, which provide the microaerophilic environment required for nitrogen fixation by the O2-labile nitrogenase enzyme complex. When sources of fixed nitrogen are scarce, approximately 10% of the cells in filaments differentiate into heterocysts to form a regular pattern (24). This simple, one-dimensional pattern of two interdependent cell types makes heterocyst formation in Anabaena sp. strain PCC 7120 one of the paradigm systems for the regulation of cellular differentiation and pattern formation in bacteria. Although many genes regulating the process of heterocyst formation have been discovered (11, 30), a clear model of the genetic network regulating this process is still emerging.
The master regulator of heterocyst formation is thought to be HetR. A hetR mutant is incapable of differentiating heterocysts, and ectopic overexpression leads to the formation of multiple contiguous heterocysts, even in the presence of nitrate or ammonia, conditions that suppress differentiation by the wild type (4, 5). Therefore, expression of hetR is both necessary and sufficient for heterocyst differentiation in a wild-type genetic background. Transcriptional regulation of hetR is complex. Four transcription start points (TSPs) have been defined within the hetR promoter region (5). The −728 and −696 TSPs are dependent on ntcA via nrrA, which binds directly to the region of DNA upstream of these two TSPs, while the −271 TSP is dependent on hetR itself, an example of positive autoregulation (7, 18). Transcription from the −184 TSP appears to be constitutive (18). Induction of hetR transcription occurs early in the developmental process and is enhanced in developing heterocysts (2).
HetR has been reported to possess DNA-binding and autoproteolytic, Ca2+-dependent serine-protease activities, both of which appear to be essential for heterocyst formation (13, 33). HetR has been shown to bind to the promoter regions of patS, hepA, and hetR in vitro, and hetR is required for transcriptional upregulation of these three genes in vivo (13). Serine residues at positions 152 and 179 of HetR have been reported to be required for protease activity, with the serine at residue 152 reported to be the active site (33). However, a recent study conflicts with the original reports, demonstrating that the serine at position 152 is not required to promote heterocyst formation, suggesting that this may not be the active site of HetR protease activity or that HetR protease activity may not be required for heterocyst differentiation (21). Although the role of HetR protease activity is unclear, proper turnover of HetR appears to correlate with the ability of HetR to promote heterocyst formation, since glycine, histidine, and serine residues at positions 36, 69, and 179, respectively, are required for both normal HetR protein turnover and heterocyst formation (21, 32).
The genes patS and hetN, which encode negative regulators of heterocyst formation, act at the level of HetR. The patS-derived pentapeptide RGSGR is capable of inhibiting HetR DNA-binding activity in vitro and heterocyst formation in vivo (13, 14, 27). Along with hetR, patA has been shown to positively influence heterocyst formation. However, unlike hetR, inactivation of patA results in heterocyst formation primarily at the filament termini. Inactivation of patA prevents the patterned upregulation of transcription of hetR found in the wild type, but ectopic overexpression of hetR is insufficient to bypass inactivation of patA, suggesting that patA affects positive autoregulation of hetR transcription and HetR activity (15). The function of patA may be to attenuate patS- or hetN-dependent inhibition of heterocyst differentiation because strains with patA and patS or patA, patS, and hetN inactivated have a partially restored ability to form intercalary heterocysts (20). The C terminus of PatA is similar in sequence to the response regulator CheY, and the N terminus contains a recently defined PATAN domain, which is predicted to be involved in protein-protein interaction (15, 16). To date, however, no biochemical characterization has elucidated the exact function of PatA.
A third gene that positively regulates heterocyst formation, hetF, has been characterized in the closely related species Nostoc punctiforme (26). Like hetR, hetF is essential to heterocyst formation, and the presence of hetF on a multicopy plasmid results in the differentiation of clusters of cells to give a multiple contiguous heterocyst phenotype. Induction of hetR transcription was delayed and failed to localize to specific cells in a hetF-null mutant. Extra copies of hetR on a plasmid were insufficient to bypass inactivation of hetF, suggesting that, like patA, hetF affects positive autoregulation of hetR transcription and HetR activity. A hetF deletion strain carrying extra copies of hetR on a plasmid also had an irregular cell morphology, and a functional hetR-gfp translational fusion suggested that levels of HetR are higher in the deletion strain than in the wild type (26). It has been suggested that the mechanism of HetF in cellular differentiation may be promotion of cell-specific accumulation of HetR. Since the report of its discovery, HetF has been predicted to contain a caspase-hemoglobinase fold (CHF), the proteolytic domain in a wide range of cysteine-dependent proteases, and to be most closely related to the eukaryotic separases (1). The hetF homologue in Anabaena sp. strain PCC 7120 has been shown to be necessary for nitrogen fixation (25), but its role in cellular differentiation has not been characterized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The growth of Escherichia coli and Anabaena sp. strain PCC 7120 and its derivatives; concentrations of antibiotics; the induction of heterocyst formation in BG-110 medium, which lacks a combined-nitrogen source; the regulation of PpetE and Pnir expression; and conditions for photomicroscopy were as previously described (21). Plasmids were conjugated from E. coli to Anabaena sp. strain PCC 7120 and its derivatives as previously described (8).
Construction of plasmids created in this study.
The plasmids and strains used in this study are described in Table 1. The primers used in PCR are described in Table 2. All PCR-generated constructs were sequenced to verify their integrity. Plasmid pDR256 was used to delete the hetF coding region. A 673-bp region upstream of hetF containing the first 9 bp of the hetF coding region was amplified from the chromosome (with primers hetF-Up-F and hetF-Up-R) and cloned into pBluescript SK+ (Stratagene) as a BamHI-KspI fragment by using restriction sites introduced on the primers. An 808-bp region downstream of hetF including the last 9 bp of the hetF coding region was amplified from the chromosome (with primers hetF-Down-F and hetF-Down-R) and cloned into this plasmid as a KspI-SacI fragment by using restriction sites introduced on the primers. A BamHI-SacI fragment containing the regions both upstream and downstream of hetF was moved into pRL277 (2) at the BglII and SacI sites to create pDR256.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Anabaena sp. strainsa | ||
| PCC 7120 | Wild type | Pasteur Culture Collection |
| 216 | hetR(S179N) | 4 |
| UHM100 | PpetE-hetN ΔpatS | 3 |
| UHM101 | ΔpatA | 20 |
| UHM103 | ΔhetR | 3 |
| UHM109 | ΔhetR ΔpatA | 20 |
| UHM130 | ΔhetF | This study |
| UHM131 | PpetE-hetN ΔpatS ΔhetF | This study |
| UHM132 | ΔhetR ΔhetF | This study |
| UHM134 | ΔhetR ΔpatA ΔhetF | This study |
| UHM135 | ΔpatA ΔhetF | |
| Plasmids | ||
| pAM504 | Shuttle vector for replication in E. coli and Anabaena; Kmr Nmr | 23 |
| pAM505 | Same as pAM504 with multiple cloning site inverted | 23 |
| pAM1951 | pAM505 with PpatS-gfp | 29 |
| pAM1956 | pAM505 with promoterless gfp | 29 |
| pRL277 | Suicide vector; Smr Spr | 2 |
| pET21a | Expression vector; Apr | Novagen |
| pPROEX-1 | Expression vector for making polyhistidine epitope-tagged proteins; Apr | Life Technologies |
| pSMC127 | pAM504 carrying PhetR-gfp | 6 |
| pSMC138 | pAM505 carrying PpetE-gfp | This study |
| pSMC148 | pPROEX-1 carrying hetR | This study |
| pAM1951 | Shuttle vector carrying PpatS-gfp | 28 |
| pDR204 | pAM505 carrying Pnir-hetRH6 | 21 |
| pDR216 | pAM505 Pnir-hetR | 21 |
| pDR226 | pAM505 Pnir-hetRH6(S179N) | 21 |
| pDR255 | pET21a carrying hetFH6 | This study |
| pDR256 | Suicide plasmid based on pRL277 used to delete hetF | This study |
| pDR266 | pAM1956 with PhetF fused to gfp | This study |
| pDR267 | pAM505 carrying PhetF-hetF | This study |
| pDR281 | pAM505 carrying PhetF-hetFH6 | This study |
| pDR282 | pAM505 carrying PhetF-hetF (H201Y) | This study |
| pDR284 | pAM505 carrying PhetF-hetF (C246A) | This study |
| pDR293 | pAM504 PpetE-hetR-gfp | This study |
| pDR302 | pAM505 carrying PhetF-hetF and Pnir-hetRH6 | This study |
| pDR309 | pAM505 carrying Pnir-hetRH6 and PpetE-gfp | This study |
| pDR310 | pAM505 carrying PhetF-hetF and PpetE-gfp | This study |
| pDR311 | pAM504 carrying Pnir-lacZ | This study |
| pDR312 | pAM504 carrying PhetF-lacZ | This study |
| pDR324 | pAM1956 with Pnir fused to gfp | This study |
| pDR350 | pAM504 carrying PpetE-lacZ | This study |
| pPetHetR | Shuttle vector carrying PpetE-hetR | 5 |
For the Anabaena strains constructed in this study, mutations were introduced in the order indicated.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotidea | Sequence |
|---|---|
| hetF-NdeI-F | CATATGAATGACCCATTGAAATCAG |
| hetF-H6-BamHI-R | ATATAGGATCCCTAGTGATGGTGATGGTGATGCTTGGGGCTTTTTTGTTG |
| hetF-Up-F | ATATAGGATCCAGTCGGTGTTAGCCAGCTAG |
| hetF-Up-R | GATTTCCGCGGGTCATTCATCAAATGCCCAG |
| hetF-Down-F | AACAACCGCGGCCCAAGTAGAGTACGGGAAG |
| hetF-Down-R | TTGCAGAGCTCTGACCTAAAGCCGCCATTTTC |
| hetF-BamHI-R | ATATAGGATCCCTACTTGGGGCTTTTTTGTTG |
| hetFH201Y-F | CTACTCCGGTTATAGCAATTTAGGCGCGAACG |
| hetFH201Y-R | CTAAATTGCTATAACCGGAGTAGTGTAAAACATG |
| hetFC246A-F | GTTTAACTCCGCGTGGGGTACCTACACCGCGAG |
| hetFC246A-R | GGTACCCCACGCGGAGTTAAACACCGCCATTTG |
| TNL-GFP-F | TTTGGATCCAATCCCGGGGATCGGCGTCAGCTAGTAAAGGAGAAGAACTTTTCACTGGA |
| TNL-GFP-R | ACAGAGCTCTTATTTGTATAGTTCATCCATGCCATG |
| PhetF-SacI-F | ATATAGAGCTCTGTAGCTCTGTGTCTCTTGTTG |
| PhetF-SmaI-R | ATATACCCGGGCCGCCAACCAATCAGCTAC |
| PpetE-BamHI-F | ATATAGGATCCCTGAGGTACTGAGTACACAG |
| PpetE-SmaI-R | ATATACCCGGGACCTGTAGTTTTATTTTTCTTATTTC |
| Pnir-SmaI-R | ATATACCCGGGAAGTTTTTTTGCTCAAGATCAATCC |
| Pnir-BamHI-F | GCGCGCGGATCCAGCTACTCATTAGTTAAGTGTAATG |
| PhetF-BamHI-F | ATATAGGATCCTGTAGCTCTGTGTCTCTTG |
| hetR-SmaI-R | ATATACCCGGGAATCTTCTTTTCTACCAAACACC |
| nir1-SacI | GCGCGCGAGCTCAGCTACTCATTAGTTAAGTGTAATG |
| hetRcf-NdeI | CATATGAGTAACGACATCGATCTGATC |
| hetR6H-r | TTAGTGATGGTGATGGTGATGATCTTCTTTTCTACCAAACACCATTTG |
| hetRH6-SacI-R | TTACTGAGCTCTTAGTGATGGTGATGGTGATGATCTTCTTTTCTACCAAACACCATTTG |
| hetRcr-SacI | GAGCTCTTAATCTTCTTTTCTACCAAACACC |
| PpetEF-SacI | TTTGGATCCTGCAGTCGGTAATTTGGTGTTTTTCC |
| PhetF-Seq-1 | AAATTGCATCAAAGGTATGCTG |
| hetF-con-R | GTATGGACAAGATTACCAGC |
| 696newII | AATCGCACAATCTCTCCTTGAGGTG |
| 184newI | CATTGCACTGGGGCCAAGACGCTT |
| 696newI | TTTGCTCTTATGGCAGTGTAGGTCG |
| New184nesrev | AAAGTAGTTGGTATACACCCATAGGGGG |
| PpetE-F | GCTGAGGTACTGAGTACACAGCTAAT |
| PpetE-gfp-R | TTCTCCTTTACTCATGGCGTTCTCCTAACCTGTAG |
| Txp-LacZ-F | CGCGGATCCATCCCGGGAGGAAACAGCTATGACCATGATTAC |
| Txp-LacZ-R | CGCGGTACCTTATTTTTGACACCAGACCAACTGG |
| gfp-PpetE-F | TAGGAGAACGCCATGAGTAAAGGAGAAGAACTTTTCACTG |
| gfpKPNI-R | GGTACCTTATTTGTACAATTCATCCATACC |
Oligonucleotides are shown in the 5′-to-3′ direction.
pDR267 is a mobilizable shuttle vector containing the hetF promoter and coding region. A 4,763-bp region containing the hetF promoter and coding region was amplified from the chromosome (with primers PhetF-SacI-F and hetF-BamHI-R) and cloned into pAM505 (23) as a SacI-BamHI fragment by using restriction sites introduced on the primers to create pDR267.
pDR282 and pDR284 are mobilizable shuttle vectors containing the hetF promoter and coding region with nucleotide substitutions resulting in H201Y and C246A amino acid substitutions, respectively. A 4,763-bp fragment containing the hetF promoter and coding frame was amplified from the chromosome (with primers hetFH201Y-F and hetFH201Y-R or hetFC246A-F and hetFC246A-R, along with hetF-Up-F and hetF-BamHI-R) via overlap extension PCR as previously described (12). The resulting products were cloned into pDR267 as KpnI-BamHI partial fragments to create pDR282 and pDR284.
pDR281 is a mobilizable shuttle vector containing the hetF promoter and coding frame with six histidine codons added to the 3′ end of the coding frame. A 4,778-bp fragment containing the hetF promoter and coding region plus six histidine codons [hetF(H6)] was amplified from the chromosome (with primers hetF-Up-F and hetF-H6-BamHI-R). The resulting product was cloned into pDR267 as a KpnI-BamHI partial fragment to create pDR281.
pDR255 is a pET21a (Novagen) derivative used to overexpress hetF(H6) in E. coli. A 2,502-bp region containing the hetF coding region plus six histidine codons added to the 3′ end was amplified from the chromosome (with primers hetF-NdeI-F and hetF-H6-BamHI-R) and cloned into pET21a as an NdeI-BamHI partial fragment to create pDR255.
pDR204, pDR216, and pDR226 are mobilizable shuttle vectors containing Pnir transcriptionally fused to hetR(H6), hetR, and hetR(S179N), respectively, and were constructed as previously described (21).
pSMC138 is a mobilizable shuttle vector containing a PpetE-gfp transcriptional fusion. The petE promoter and gfp were amplified from chromosomal DNA and pAM1956, respectively, and fused by overlap extension PCR (with primers PpetE-F, PpetE-gfp-R, gfp-PpetE-F, and gfpKPNI-R). The 1,062-bp fragment containing PpetE-gfp was cloned into pGEM-T (Promega) and subsequently moved into pAM505 as a KpnI-SacI fragment to create pSMC138.
pSMC148 is an expression vector used for production of HetR with an N-terminal polyhistidine epitope tag in E. coli. The coding region of hetR was amplified from chromosomal DNA with primers hetRcf-NdeI and hetRcr-SacI, cloned into pGEM-T, and subsequently moved to pPROEX-1 (Life Technologies) as an NdeI-SacI fragment to generate pSMC148.
pDR293 is a mobilizable shuttle vector containing the gene for the green fluorescent protein (GFP), gfp, translationally fused to the 3′ end of hetR, the transcription of which is under the control of the copper-inducible petE promoter. A 758-bp region containing the gfp coding frame and a linker region 5′ of the gfp start codon was amplified from pAM1956 (with primers TNL-GFP-F and TNL-GFP-R) and cloned into pAM504 (23) as a BamHI-SacI fragment by using restriction sites introduced on the primers to create pSMC232. A 1,239-bp region containing the petE promoter transcriptionally fused to the hetR coding region was amplified from pDR120 (19) (with primers PpetE-BamHI-F and hetR-SmaI-R) and cloned into pSMC232 as a BamHI-SmaI fragment with sites introduced on the primers to create pDR293.
pDR309 and pDR310 are mobilizable shuttle vectors containing a PpetE-gfp transcriptional fusion and either a Pnir-hetR(H6) transcriptional fusion or the hetF promoter and coding region, respectively. A 1,074-bp region containing a PpetE-gfp transcriptional fusion was amplified from pSMC138 (with primers PpetEF-SacI and TNL-GFP-R) and cloned into pDR204 and pDR267 as a SacI fragment by using restriction sites introduced on the primers to create pDR309 and pDR310, respectively.
pDR311, pDR312, and pDR350 are mobilizable shuttle vectors carrying transcriptional fusions between the reporter gene lacZ and the nir, hetF, and petE promoters, respectively. The lacZ gene was amplified from E. coli strain K-12 with primers Txp-LacZ-F and Txp-LacZ-R and cloned directly into pAM504 as a BamHI-KpnI fragment with sites on the primers to yield pNC101. The promoter regions of the nirA (primers Pnir-BamHI-F and Pnir-SmaI-R), hetF (primers PhetF-BamHI-F and PhetF-SmaI-R), and petE (primers PpetE-BamHI-F and PpetE-SmaI-R) genes were generated by PCR and cloned directly into pNC101 to make plasmids pDR311, pDR312, and pDR350, respectively.
Plasmids pDR324 and pDR266 are mobilizable shuttle vectors containing transcriptional fusions between the promoter regions of nirA or hetF and gfp, respectively. The promoter region of nirA was amplified by PCR with primers nir1-SacI and Pnir-SmaI-R and cloned directly into pAM1956 (28) as a SacI-SmaI fragment to make pDR324. The promoter region of hetF was amplified by PCR with primers PhetF-SacI-F and PhetF-SmaI-R and cloned directly into pAM1956 as a SacI-SmaI fragment to make pDR266.
Strain construction.
Deletion of the hetF coding region was performed as previously described (3), by using plasmid pDR256 to delete cleanly the hetF coding region in Anabaena sp. strains PCC 7120, UHM100, UHM101, UHM103, and UHM109 to create strains UHM130, UHM131, UHM135, UHM132, and UHM134, respectively. The resultant strains were screened via colony PCR with primers PhetF-Seq-1 and hetF-con-R, which anneal outside of the region of DNA used on plasmid pDR256 to make the deletion, and tested for streptomycin and spectinomycin sensitivity.
Generation of a polyclonal antibody to HetR.
HetR with an N-terminal polyhistidine epitope tag was purified from E. coli strain BL21 (Stratagene) carrying plasmid pSMC148 as previously described (21). Five milligrams of protein at a concentration of 1 mg ml−1 was digested with tobacco etch virus protease according to the manufacturer's (Invitrogen) specifications to remove the epitope tag and subsequently dialyzed against TSEG buffer (21), incubated with 1 ml Ni-nitrilotriacetic acid agarose (Qiagen) for 1 h at 4°C, and added to a column, and the eluate was collected to separate it from the tobacco etch virus protease, which has the epitope tag, and the epitope tag was cleaved from the HetR protein. The resulting protein, which is predicted to be wild-type HetR with the addition of residues GAH preceding the N-terminal methionine, was sent to New England Peptide (Gardner, MA) for the generation of a polyclonal antibody in rabbits.
Western blot analysis.
Two milliliters of cultures collected at the indicated time points was centrifuged at 2,000 × g for 3 min, resuspended in 8% sodium dodecyl sulfate (SDS), incubated at 100°C for 10 min, and centrifuged at 13,000 × g for 10 min to remove insoluble cell debris. Protein was precipitated from the supernatant by the addition of acetone to 80% and centrifuged at 13,000 × g for 10 min. Pellets were air dried and dissolved in a final volume of 25 μl 8% SDS. Protein concentrations were determined by the DC Protein Assay (Bio-Rad Laboratories). A 3.5-μg sample of protein was then subjected to SDS-polyacrylamide gel electrophoresis and electrophoretic transfer onto polyvinylidene difluoride membrane. HetR and HetF were detected with Penta-His antibodies (Qiagen) or a polyclonal antibody directed against HetR, followed by chemiluminescence detection (Western Breeze; Invitrogen). Detection of Penta-His antibodies was performed by exposure to Lumi-Film Chemiluminescent Detection Film (Roche), and detection of polyclonal anti-HetR antibodies was performed with the GeneGnome BioImaging System (Syngene). Coomassie staining of the membrane was performed with Simply Blue Safe Stain (Invitrogen). All Western blot analyses were performed in duplicate.
β-Galactosidase assays.
Cells for β-galactosidase assays were treated as previously described (22), except that in place of treatment with chloroform, cavitation for 4 min in a Branson 1510 cavitator was used to lyse cells. After centrifugation to remove cell debris, β-galactosidase activity of the aqueous phase was measured according to Miller (17) and protein concentration was measured with the DC Protein Assay. Activity was expressed as nanomoles of o-nitrophenol per minute per milligram of protein.
Rapid amplification of cDNA ends (RACE).
One-hundred-milliliter cultures were harvested 18 h after removal of combined nitrogen by centrifugation at 2,000 × g, and the cells were frozen in a liquid nitrogen bath. Frozen cell pellets were ground with a mortar and pestle while mixed with liquid nitrogen for 45 min and then mixed with 5 ml TRIzol (Invitrogen) and incubated at room temperature for 1 h. The mixture was then centrifuged at 10,000 × g for 10 min at 4°C. One milliliter of chloroform was added to the supernatant, and the mixture was shaken, incubated for 15 min at room temperature, and centrifuged at 10,000 × g for 10 min at 4°C. RNA was precipitated from the supernatant by the addition of 2.5 ml isopropanol and incubated overnight at −20°C. The RNA was collected by centrifugation at 10,000 × g for 10 min at 4°C, washed with 5 ml of 75% ethanol, dried, and resuspended in diethyl pyrocarbonate-treated water. The RNA was digested with RNase-free DNase I (Roche) with the addition of Protector RNase Inhibitor (Roche) for 3 h at 37°C. An acid phenol-chloroform extraction was performed, and the RNA was precipitated with 3 M sodium acetate and isopropanol, washed with 70% ethanol, and dried (ToTTALY RNA; Ambion Inc.). The pellet was resuspended in diethyl pyrocarbonate-treated water, and 5 μg of total RNA was used for the RACE procedure according to the 5′/3′ RACE kit, 2nd generation (Roche), with primers 696newII and 184newI for synthesis of cDNA for the −728 and −696 and the −271 and −184 transcriptional start sites, respectively. Primers 696newI and New184nesrev were used for amplification of cDNA from the −728 and −696 and the −271 and −184 start sites, respectively. Products were analyzed with a 3% agarose gel. For all RNA preparations, PCRs with primers hetRcf-Nde1 and hetR6H-r were done as negative controls to detect contaminating DNA.
Creation of “mosaic” filaments.
To create and observe mosaic filaments in which only one cell contained a plasmid, modifications were made to the standard conjugation protocol (8). E. coli cells were grown to an A600 of 0.5, washed twice with BG-11, and used either concentrated 30-fold, concentrated 3-fold, diluted 3-fold, or diluted 30-fold. E. coli aliquots of 150 ml were combined with 150 μl of Anabaena at an A750 of 0.6 and spread onto 40 ml of solid BG-11 medium containing 5% LB by volume in a petri dish (15 by 100 mm). After 24 h, cells were resuspended in 2 ml BG-11, washed twice in BG-11 with centrifugation at 2,000 × g to collect primarily Anabaena cells, and resuspended in 2 ml of BG-11 for filaments with the plasmid containing hetR, pDR309, or 2 ml of BG-110 for filaments with the plasmid containing hetF, pDR310. Filaments were examined at this point for GFP fluorescence in isolated cells and the presence of heterocysts. The remainder of the filaments were incubated under standard growth conditions for an additional 24 h before microscopic examination. To induce expression of gfp from the petE promoter, 3 μM copper was added to all culture media.
Typically, the conjugations with E. coli cultures concentrated threefold or diluted threefold yielded many filaments with a single fluorescent cell. Conjugations with E. coli cultures concentrated 30-fold yielded filaments with multiple fluorescent cells per filament, occasionally as many as 50% of the cells, and the 30-fold dilution yielded very few filaments with fluorescent cells. For strains that formed heterocysts in response to the plasmid, filaments with a single fluorescing heterocyst were common, presumably because heterocyst formation precluded further cell division. In contrast, strains that did not form heterocysts had primarily multiple contiguous fluorescent vegetative cells, with isolated fluorescent cells representing less than 10% of the total of fluorescing cells.
RESULTS
hetF from Anabaena sp. strain PCC 7120 is similar to hetF from N. punctiforme.
To determine if hetF from Anabaena sp. strain PCC 7120, designated open reading frame all1730, is functionally equivalent to that of N. punctiforme, the hetF coding region was deleted cleanly from the chromosome of the wild-type strain, Anabaena sp. strain PCC 7120. Compared to the wild type, hetF deletion strain UHM130 grew poorly in BG-11 and BG-110 liquid media, displayed an aberrant cell morphology characterized by enlarged and elongated cells in both BG-11 and BG-110 (the latter lacks a source of fixed nitrogen), and was unable to form heterocysts even after several days in BG-110 (Fig. 1A and B). When the hetF promoter and coding region were introduced on replicating plasmid pDR267 to complement UHM130, the aberrant cell morphology was resolved and multiple contiguous heterocysts were observed 24 h after nitrogen step-down and thereafter (Fig. 1C). UHM130 complemented with hetF on a plasmid did not form extra heterocysts in BG-11, which contains nitrate, and vegetative filaments were slightly irregular (data not shown). Cells were also significantly smaller than those of the wild type in both BG-11 and BG-110. When a fusion between the promoter of hetF and gfp was introduced on plasmid pDR266 into the wild-type strain, fluorescence appeared moderate and unpatterned and showed no noticeable induction upon nitrogen step-down (data not shown). Several aspects of hetF from N. punctiforme and Anabaena sp. strain PCC 7120 appear to be very similar (26).
FIG. 1.
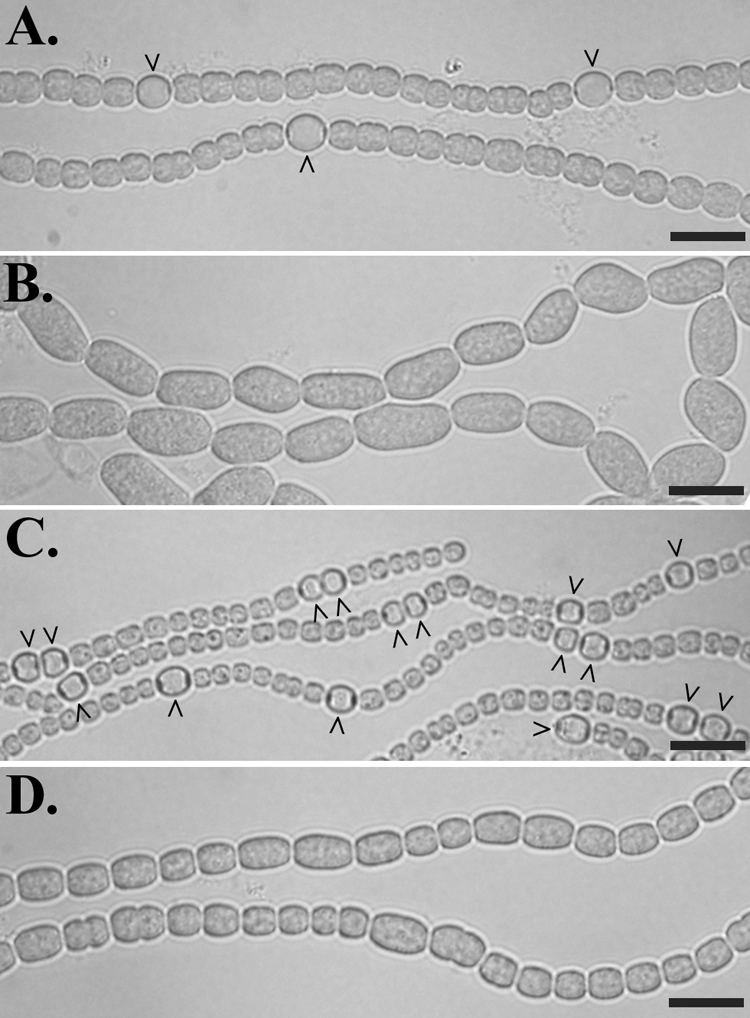
Phenotype of a hetF deletion strain, UHM130, 48 h after the removal of combined nitrogen. Anabaena sp. strain PCC 7120 (A); hetF deletion strain UHM130 (B); UHM130 complemented with pDR267, a plasmid bearing PhetF-hetF (C); and UHM130 carrying pDR282, a plasmid bearing PhetF-hetF(H201Y) (D), are shown. Carets indicate heterocysts. Bars = 10 μm.
Conserved histidine and cysteine residues required for HetF activity.
Since the discovery of HetF, a bioinformatic study indicated that a region of HetF is homologous to the CHF class of cysteine proteases and is most closely related to the eukaryotic separases (1), but to date this relationship has not been demonstrated functionally. As a first step toward determining if HetF is indeed a protease, sequence alignments were used to predict the active-site cysteine and histidine residues of HetF that correspond to the catalytic dyad of known CHF proteases (data not shown), and changes to conservative substitutions with alanine and tyrosine, respectively, were made in hetF. Plasmids pDR282 and pDR284, bearing hetF with H201Y and C246A substitutions, respectively, failed to complement the heterocyst deficiency of the hetF mutant, UHM130, consistent with the idea that HetF is a CHF protease that requires these conserved active-site residues (Fig. 1D). Although pDR282 and pDR284 did not restore the ability of UHM130 to form heterocysts, some alleviation of the aberrant cell morphology was observed.
Altered cell morphology of a hetF mutant requires hetR and patA.
The aberrant cell morphology of hetF deletion strain UHM130 is similar to that of the wild type in which patA is overexpressed ectopically (S. S. Young and S. M. Callahan unpublished data). Therefore, hetF patA and hetF hetR double-deletion mutants were constructed to determine if either patA or hetR contributes to the aberrant cell morphology of hetF deletion strain UHM130. Filaments of both double-mutant strains consisted of all vegetative cells with a wild-type morphology in both BG-11 and BG-110, indicating that both patA and hetR are necessary for the aberrant cell morphology observed in the hetF mutant (data not shown).
HetF and PatA affect HetR protein turnover.
Increased fluorescence from a hetR-gfp translational fusion in a hetF deletion strain suggested that mutation of hetF affects HetR protein levels in N. punctiforme (26). To characterize the effects of hetF and patA on HetR levels in Anabaena sp. strain PCC 7120, a polyclonal antibody directed against HetR was used to assess protein levels in strains with or without each gene. Strains with one or both of the genes deleted from the chromosome had substantially higher levels of HetR than the wild type (Fig. 2A). The band for HetR from the wild type was very faint, whereas in the mutant backgrounds prominent bands were observed. In addition, the hetF patA double-deletion mutant had protein levels similar to those of the corresponding single mutants. Addition of a plasmid containing hetF to the mutant strains restored HetR to a level similar to that of the wild-type strain (Fig. 2A).
FIG. 2.
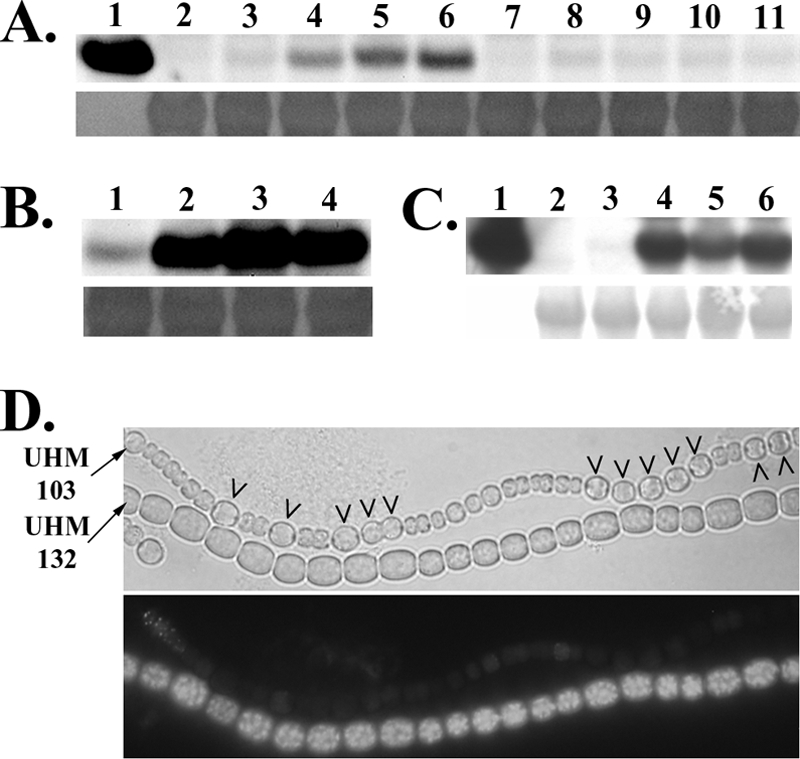
Regulation of HetR levels by patA and hetF. Western blot analysis of HetR from cultures 18 h after the removal of combined nitrogen by a polyclonal antibody is shown in panel A. Shown are 100 ng of recombinant HetR used to generate the antibody (lane 1); UHM103, which is a ΔhetR mutant (lane 2); Anabaena sp. strain PCC 7120, which is the wild type (lane 3); UHM101, which is a ΔpatA mutant (lane 4); UHM130, which is a ΔhetF mutant (lane 5); UHM132, which is a ΔhetF ΔpatA mutant (lane 6); UHM103 carrying hetF on replicating plasmid pDR267 (lane 7); Anabaena sp. strain PCC 7120 with pDR267 (lane 8); UHM101 with pDR267 (lane 9); UHM130 with pDR267 (lane 10); and UHM132 with pDR267 (lane 11). Western blot analysis of HetR expressed from the petE promoter on plasmid pPetHetR (5) 18 h after the removal of combined nitrogen and the addition of 3 μM copper and detected by a polyclonal antibody is shown in panel B. Shown are UHM103, which is a ΔhetR mutant with pPetHetR (lane 1); UHM109, which is a ΔhetR ΔpatA mutant with pPetHetR (lane 2); UHM132, which is a ΔhetR ΔhetF mutant with pPetHetR (lane 3); and UHM134, which is a ΔhetR ΔhetF ΔpatA mutant with pPetHetR (lane 4). Western blot analysis of HetRH6 expressed from the nir promoter on plasmid pDR204 18 h after the removal of 5 mM ammonium and addition of 17.6 mM nitrate with an antibody against the polyhistidine epitope tag is shown in panel C. Shown are 100 ng of recombinant HetRH6 (lane 1), UHM103 carrying untagged HetR on plasmid pDR216 (lane 2), UHM103 carrying epitope-tagged HetRH6 on plasmid pDR204 (lane 3), UHM109 carrying pDR204 (lane 4), UHM132 carrying plasmid pDR204 (land 5), and UHM134 carrying plasmid pDR204 (lane 6). Chemiluminescence detection (upper panels) and a Coomassie-stained section of membrane corresponding to an approximate molecular mass of 50 kDa (lower panels) to show equality of protein loading for Western blot analyses are shown in panels A to C. Light (upper panel) and fluorescence (lower panel) microscopy of a mixture of filaments of UHM103, which is a ΔhetR mutant, and UHM132, which is a ΔhetR ΔhetF mutant, both carrying pDR293, a plasmid bearing a hetR-gfp translational fusion expressed from PpetE, 24 h after the removal of combined nitrogen and the addition of 3 μM copper is shown in panel D. Strains are indicated on the left. Carets indicate heterocysts.
To determine if the increase in HetR in cells lacking hetF or patA was due to either an effect on transcription of hetR or a posttranscriptional regulatory event, hetR was uncoupled from its normal promoter and fused to one of two inducible promoters. The fusions were then introduced on a plasmid into strains that, in addition to having either hetF, patA, or hetF and patA deleted, also had hetR deleted, so that the only source of HetR was from the gene fused to the inducible promoter. Plasmid-borne fusions of the lacZ and gfp reporter genes to the inducible promoters served as controls for potential plasmid or strain background effects on transcription levels, of which none were observed (data not shown). The inducible promoters used were the petE promoter, which is induced by copper in Anabaena sp. strain PCC 7120 (10), and the nirA promoter, which is induced by the presence of nitrate or lack of a fixed nitrogen source (9). When fused to hetR and introduced on a plasmid into either the wild type or a hetR deletion strain, both promoters cause the differentiation of supernumerary heterocysts in BG-11, which contains nitrate, or BG-110 under conditions that promote transcription (5, 21). The nirA promoter has also been used previously to evaluate levels of HetR made from various plasmid-borne mutant alleles of hetR (21). Whether expression of hetR was driven by the petE promoter in BG-110, which contains copper, or the nirA promoter in BG-11, which contains nitrate, levels of HetR were dramatically increased in the genetic backgrounds that lacked hetF, patA, or both genes (Fig. 2B and C), indicating that deletion of hetF or patA affects the regulation of HetR levels posttranscription of hetR.
Wong and Meeks reported that wild-type N. punctiforme carrying a hetR-gfp translational fusion under the control of the hetR promoter displays fluorescence only in developing heterocysts, whereas the same construct in a hetF mutant has increased fluorescence distributed more uniformly throughout the filament. They suggest that HetF may be responsible for stabilizing HetR in heterocysts while promoting degradation in neighboring vegetative cells (26). However, the hetR promoter is known to be upregulated in developing heterocysts, complicating the interpretation of these results. To better characterize the spatial aspect of HetR turnover within filaments, a hetR-gfp translational fusion was expressed from the copper-inducible petE promoter in order to avoid the spatial pattern of transcription characteristic of the hetR promoter. A plasmid bearing a PpetE-hetR-gfp translational fusion was introduced into a hetR deletion strain and a hetR hetF deletion strain. In BG-110, which contains copper for the expression of hetR-gfp from the petE promoter, fluorescence was much brighter in the strain lacking hetF, consistent with the Western blotting results (Fig. 2D). In the strain with hetF intact, fluorescence was low in both heterocysts and vegetative cells, with only occasional vegetative cells displaying observable fluorescence (Fig. 2D). To control for potential differences in transcription from the petE promoter or GFP turnover in heterocysts and vegetative cells, a plasmid bearing a PpetE-gfp transcriptional fusion, pSMC138, was introduced into both strains. Fluorescence in both strains was similar, and fluorescence resulting from the PpetE-hetR-gfp fusion in the strain with hetF intact was lower in both vegetative cells and heterocysts relative to that from Anabaena sp. strain PCC 7120 carrying the PpetE-gfp transcriptional fusion (data not shown), demonstrating that the low fluorescence in heterocysts generated by the PpetE-hetR-gfp fusion was specifically due to HetR, not lower expression from the petE promoter or more rapid GFP turnover in developing heterocysts. Therefore, hetF limited the levels of HetR in both developing heterocysts and vegetative cells.
Extra copies of hetF can functionally bypass deletion of patA.
A patA mutant strain forms heterocysts primarily at the filament termini (15), a phenotype that can be bypassed by deletion of patS, a negative regulator of differentiation (20), but not by extra copies or overexpression of hetR, the master regulator of differentiation (4, 5). To characterize the relationship between hetF and patA, pDR267, a replicating plasmid that contains hetF behind its native promoter, was introduced into a patA deletion strain, UHM101. UHM101 containing pDR267 formed intercalary heterocysts, and multiple contiguous heterocysts were occasionally observed within filaments when combined nitrogen was removed from the medium (Fig. 3A and B). As for the complemented hetF deletion strain, extra copies of hetF on a plasmid resulted in a decrease in the size of cells in filaments of the patA deletion strain.
FIG. 3.
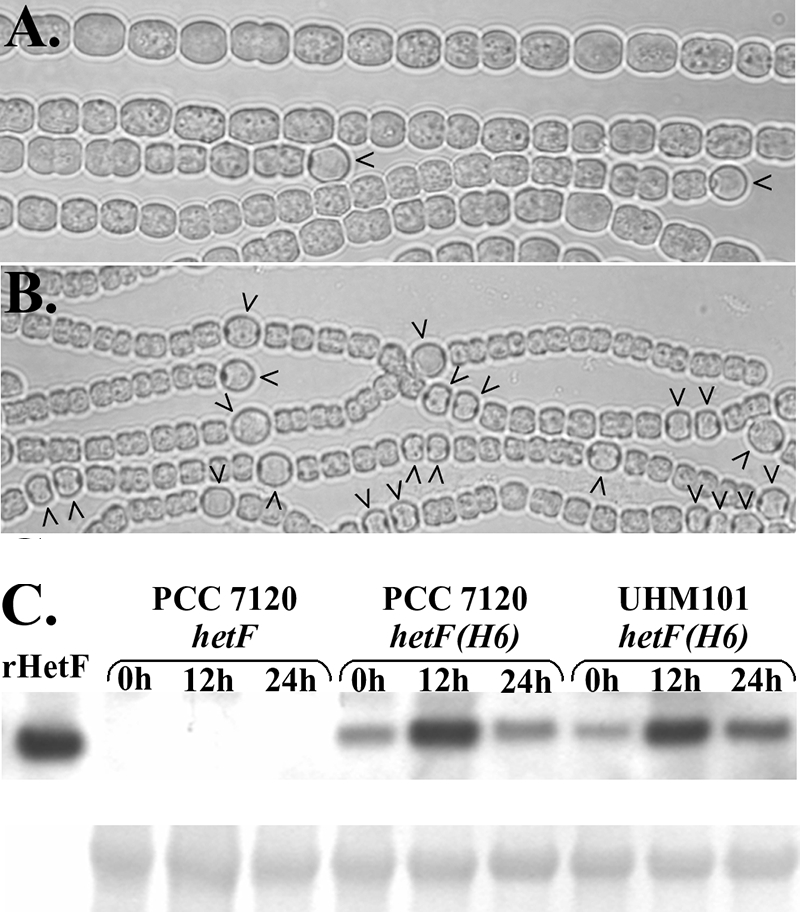
Bypass of deletion of patA by extra copies of hetF and analysis of HetF protein levels. Shown is patA deletion strain UHM101 carrying control plasmid pAM505 (A) or pDR267, a plasmid bearing PhetF-hetF (B), 48 h after the removal of combined nitrogen. Carets indicate heterocysts. Also shown is a Western blot analysis of epitope-tagged HetF in Anabaena sp. strain PCC 7120 and patA deletion strain UHM101 at various times postinduction of differentiation (C). An antibody against the polyhistidine epitope tag was used to detect the protein. Lanes are as indicated. See the text for details. Chemiluminescent detection (upper panel) and a Coomassie-stained section of membrane (lower panel) were used to show equality of protein loading.
Functional bypass of patA by extra copies of hetF could be explained by the regulation of HetF levels by PatA in the wild-type organism. To test if PatA affects HetF protein levels, the level of HetF in a wild-type genetic background was compared to that in one where patA had been deleted. Similar amounts of HetF were detected by Western blot analysis of the wild-type strain, Anabaena sp. strain PCC 7120, and a patA deletion strain, UHM101, containing a plasmid with an epitope-tagged version of hetF under the control of its native promoter (Fig. 3C). Although no effect of deletion of patA on levels of HetF was seen, the level of HetF protein increased in both genetic backgrounds at 12 h after the induction of differentiation and returned to near preinduction levels by 24 h postinduction (Fig. 3C). In contrast, no significant differences in transcription from the hetF promoter region were observed at 0, 12, and 24 h after nitrogen step-down with promoter-gfp or promoter-lacZ fusions on plasmids pDR266 and pDR312, respectively (data not shown).
PatA has been proposed to attenuate patS- and hetN-dependent inhibitors of heterocyst formation based on the ability of strains with patA and patS or patA, patS, and hetN inactivated to form intercalary heterocysts (20). To test if HetF may have the same function, the hetF coding region was deleted from UHM100, a strain with patS deleted and hetN under the control of the copper-inducible petE promoter, to create strain UHM131. If the sole function of hetF is to attenuate one or both of these signals, then the resultant strain should be capable of forming heterocysts in BG-110 lacking copper, which prevents expression of hetN from the petE promoter. Strain UHM131 was unable to form heterocysts under these or any other conditions tested, suggesting that the function of hetF is not attenuation of either a patS- or a hetN-dependent signal (data not shown).
HetF is required for differentiation exclusively in cells that differentiate.
Wong and Meeks have suggested that HetF is necessary for cell type-specific accumulation of HetR. Differential accumulation of HetR in cells along a filament by HetF could be the result of (i) limiting HetR levels in cells that will not differentiate, (ii) elevating levels of HetR in cells that will differentiate, or (iii) both i and ii. To test if the sole role of HetF in the differentiation of heterocysts is to limit levels of HetR in cells that will not differentiate, mosaic filaments were created that lacked hetF in all cells and had hetR in only one cell of the filament. If HetF is necessary for reduction of HetR only in cells adjacent to a differentiating cell, single cells that have hetR in a filament that otherwise lacks both hetR and hetF should be capable of differentiation, because the neighboring cells lack HetR, thereby obviating the need for HetF. A plasmid containing hetR expressed from the nirA promoter and a transcriptional fusion between the petE promoter and gfp was introduced into a hetF hetR double mutant under conditions that allowed the examination of filaments in which only one cell of the filaments had received the plasmid (see Materials and Methods). Fluorescence generated as a result of the PpetE-gfp fusion was used to identify cells that had received the plasmid. More than 100 filaments with a single fluorescent cell were examined, and none of the fluorescent cells had differentiated into a heterocyst (Fig. 4A). Many of these cells were, however, enlarged, consistent with the phenotype of a hetF mutant strain with an active copy of hetR. The same plasmid introduced into a hetR deletion strain resulted in differentiation of almost all of the cells that were the sole fluorescent cells of a filament (Fig. 4B).
FIG. 4.
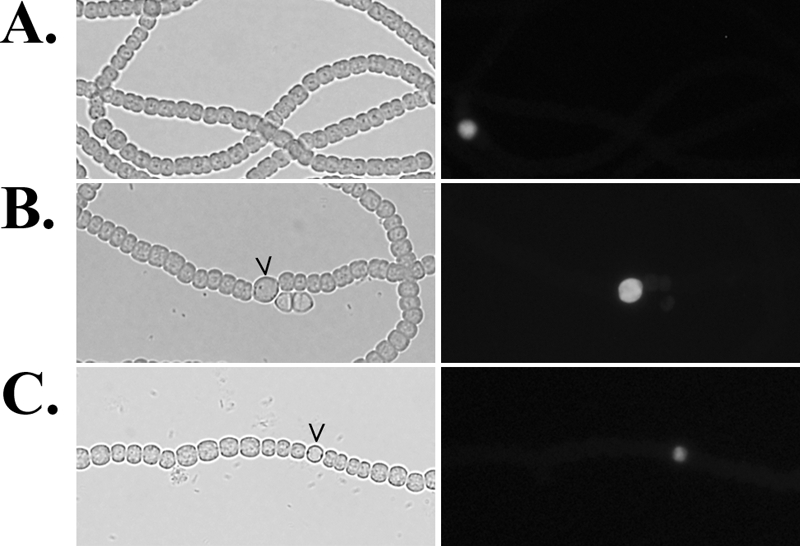
GFP-tagged plasmids bearing hetR or hetF in one cell of a filament. Shown is light (left panel) and fluorescence (right panel) microscopy of UHM132, a hetR hetF deletion strain with hetR and gfp on plasmid pDR309 in a single cell of a filament in BG-11 medium (A); UHM103, a hetR deletion strain with plasmid pDR309 in a single cell of a filament in BG-11 medium (B); and UHM130, a hetF deletion strain with hetF and gfp on plasmid pDR310 in a single cell of a filament in BG110 medium (C). GFP fluorescence identifies cells that have the indicated plasmid. Carets indicate heterocysts.
To test if hetF is required exclusively in cells that will differentiate into heterocysts, mosaic filaments in which only one cell contained an active copy of hetF were constructed. A plasmid containing hetF under the control of its native promoter and a PpetE-gfp fusion was introduced into a hetF deletion strain under conditions that allowed the examination of filaments in which only one cell of the filaments had received the plasmid. In this case, approximately 25% of the 100 cells examined that were the sole fluorescent cells of a filament had differentiated into heterocysts (Fig. 4C). Within these filaments, cells adjacent to the heterocysts often lacked the aberrant cell morphology typical of this strain, although no distinguishable fluorescence could be observed in the adjacent cells, suggesting that a diffusible substance such as that proposed to be produced by patS may be responsible for this phenomenon. This hypothesis is supported by the fact that when added to the medium, the C-terminal pentapeptide PatS-5 can alleviate the aberrant cell morphology of hetF deletion strain UHM130 (data not shown) and that induction of transcription of patS is dependent on hetF (see below). HetF was required for differentiation exclusively in developing heterocysts, and its control of HetR levels in vegetative cells adjacent to a developing cell was not necessary for differentiation.
Transcription from the patS promoter and the −271 TSP of the hetR promoter is dependent on hetF.
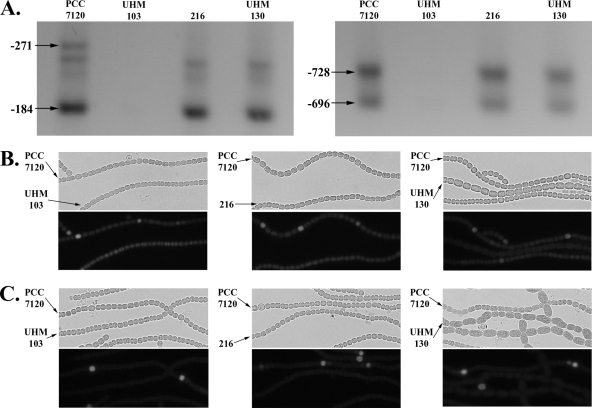
Transcription from the hetR promoter relies on HetR both directly and indirectly. HetR is required for the use of, and binds just upstream of, the −271 TSP of the hetR promoter (13, 18). In addition, HetR is part of an indirect positive feedback loop with NtcA and NrrA, which are necessary for the use of other TSPs in the hetR promoter (7, 18). Therefore, an increase in the levels of HetR in a hetF mutant might be expected to increase the transcription of hetR, yet Wong and Meeks showed with a Northern blot assay that transcription of hetR is reduced in a hetF mutant (26). To test if hetF is required for the regulation of transcription of hetR, a highly sensitive RACE protocol was used to detect transcription from all four TSPs of the hetR promoter in Anabaena sp. strain PCC 7120; a hetF deletion strain; a strain carrying hetR(S179N) in the chromosome, which is incapable of promoting transcription from the −271 TSP; and a strain with both the hetR promoter and coding regions deleted. Transcription from the −728, −696, and −184 start sites was detected from all strains but the hetR deletion strain, indicating that hetF was not required for their use (Fig. 5A). On the other hand, transcripts for the −271 start site were present only in Anabaena sp. strain PCC 7120, indicating that hetF was required for hetR-dependent positive autoregulation of the −271 TSP (Fig. 5A). A third PCR product slightly smaller than that of the −271 TSP was observed for some strains. However, when sequenced, this product corresponded to the −184 TSP, suggesting that nonspecific priming, not another TSP, was responsible for this product.
FIG. 5.
Effect of mutation of hetF and hetR on transcription from the −271 TSP of hetR and transcription from the hetR and patS promoters. Agarose gel electrophoresis of RACE products from the −271 and −184 (left panel) and −728 and −696 (right panel) TSPs is shown in panel A; lanes are as indicated. Also shown is light (upper panel) and fluorescence (lower panel) microscopy of a mixture of filaments of Anabaena sp. strains PCC 7120 and UHM103, which is a ΔhetR mutant; strain 216, which carries the hetR(S179R) allele, or UHM130, which is a ΔhetF mutant carrying pSMC127, a plasmid bearing PhetR-gfp (B), or pAM1951, a plasmid bearing PpatS-gfp (C). Strain designations are indicated by arrows.
Transcription of patS, as well as hetR, is dependent on hetR. To test the effect of deletion of hetF on patterned induction of patS and hetR transcription in vivo, patterns of fluorescence from transcriptional fusions between gfp and the two promoters were examined in different strains carrying plasmids pAM1951 and pSMC127, respectively. Patterned expression from a PhetR-gfp fusion after nitrogen step-down was observed only in Anabaena sp. strain PCC 7120. Uniform fluorescence at a level similar to that of the vegetative cells of Anabaena sp. strain PCC 7120 was observed in all cells of the hetR deletion, hetR(S179N), and hetF deletion strains (Fig. 5B). Similarly, patterned expression from a PpatS-gfp fusion after nitrogen step-down was observed only in Anabaena sp. strain PCC 7120, with little fluorescence detected in the other three strains (Fig. 5C). A negative feedback loop between hetR and an inhibitor, such as that produced by patS or hetN, could have accounted for the decrease in hetR and patS expression seen in the hetF deletion and hetR(S179N) strains. However, when both fusions were introduced into a hetF patS double-deletion strain with transcription of hetN controlled by a copper-inducible promoter and the strain was examined in BG-110 lacking copper, no increase in expression from the hetR or patS promoter was observed, suggesting that negative feedback between hetR and patS or hetN was not responsible for the lack of induction of the hetR and patS promoters in a hetF deletion strain (data not shown).
DISCUSSION
Since the report of its discovery, HetF has been proposed to be a protease of the CHF class, specifically, the separinoids (1). Our results are consistent with this prediction. Conserved active-site histidine and cysteine residues of CHF proteases were essential for HetF function, and deletion of hetF caused a dramatic increase in the level of HetR. In addition to HetF, deletion of patA caused HetR to accumulate to a similar level. The available evidence suggests that the two proteins work together to regulate levels of HetR, with PatA facilitating the activity of HetF, which, unlike that of PatA, was absolutely required for cellular differentiation. Simultaneous deletion of both patA and hetF did not lead to a further increase in HetR protein, and the addition of hetF to a patA deletion strain functionally bypassed patA to restore intercalary heterocysts and decrease the level of HetR. The role of PatA in this pathway is unclear. PatA was not necessary for wild-type levels of HetF, suggesting that it may upregulate HetF activity, possibly through mediating protein-protein interactions, given that this is the predicted function of the N-terminal PATAN domain found in PatA (16). Alleles of hetR bearing S179N, G36A, and H69Y substitutions have been shown to result in increased HetR protein levels (21, 32), and the level of HetR observed under each of these conditions is similar to that seen after the deletion of hetF or patA in a wild-type hetR background (D. D. Risser and S. M. Callahan, unpublished data), suggesting that they may affect the same HetF-dependent pathway. Unlike HetR accumulation with the mutant forms of HetR, which are nonfunctional, deletion of hetF or patA causes a dramatic increase in the level of wild-type HetR. Therefore, HetF and PatA are required for differentiation even when a large excess of wild-type HetR protein is present.
The role of HetR degradation in heterocyst formation has been somewhat enigmatic. HetR is reported to be a Ca2+-dependent serine protease capable of autodegradation with the active-site serine located at residue 152 and an additional serine at residue 179 also required for protease activity (33). However, the proposed active-site serine is not required for proper HetR turnover or heterocyst formation (21). Also, in vivo, depletion of calcium does not result in an increase in HetR protein similar to that observed for a strain carrying hetR(S179N) in the chromosome, strain 216, even though autodegradation of HetR should be abolished in both backgrounds if calcium is necessary for autodegradation of HetR (31, 32). These inconsistencies cast doubt on the assertion that the serine at residue 179 of HetR is required for protease activity and make it difficult to determine what effect, if any, HetR protease activity may have on hetF-dependent turnover of HetR.
Wong and Meeks speculated that the mechanism by which HetF promotes differentiation may involve cell-specific accumulation of HetR via regulation of HetR autoproteolytic activity (26). Evidence suggesting that HetF is itself a protease suggests that it may have a more direct role in the degradation of HetR. HetF was necessary for limiting HetR levels in both vegetative cells and developing heterocysts, and hetF appears to be expressed at similar levels in the two cell types. The multicellular nature of Anabaena sp. strain PCC 7120 suggests that hetF activity could be required in either or both cell types for differentiation of cells. For instance, HetF may be necessary for decreasing HetR levels in some cells so that others can differentiate. Induction of expression of patS is dependent on hetR (13), and overexpression of both genes prevents the differentiation of any cells in a filament (20). Alternatively, HetF may convert a HetR apoprotein to its active form to promote the differentiation of a cell. Determining which role HetF might play is nontrivial and difficult to do with current methodologies. For example, simply using cell type-specific promoters to drive the expression of hetF and checking for complementation of a mutant will not work because expression from such promoters is dependent on the differentiation process, which is stopped at an early stage in a hetF mutant. Instead, mosaic filaments were examined in which hetF was expressed in only one cell of a filament to show that it can promote the differentiation of that cell even when neighboring cells lack HetF activity. In addition, it was shown that in a hetF deletion background, HetR was not sufficient to promote the differentiation of a cell when the neighboring cells lacked HetR. Taken together, these results indicate that HetF acts to promote the differentiation of heterocysts along a filament in cells that will differentiate. These results do not preclude a possible role for HetF in vegetative cells in patterning.
HetF from Anabaena sp. strain PCC 7120 appears to serve a function similar to that of HetF from N. punctiforme and is required for heterocyst formation. Unlike N. punctiforme, however, deletion of hetF from Anabaena sp. strain PCC 7120 results in aberrant cell morphology as well. It should be noted that when isolating the hetF deletion strain, only three of four isolates tested displayed this aberrant cell morphology, even though all four could be complemented by reintroduction of hetF. Furthermore, a strain isolated from an independent study displayed aberrant cell morphology identical to that of hetF deletion strain UHM130 and was found to have a transposon inserted within the hetF coding region (Risser and Callahan, unpublished). When working with UHM130, presumptive mutants with this phenotype suppressed were also isolated frequently, suggesting a selective pressure to acquire suppressor mutations that abrogate the aberrant cell morphology. Isolation of a suppressor mutant may explain why this phenotype was not observed when hetF was inactivated in N. punctiforme. Transcriptional expression of hetF is constitutive and unpatterned in both N. punctiforme and Anabaena sp. strain PCC 7120, but in Anabaena sp. strain PCC 7120, at least, the level of HetF protein increases transiently after the removal of combined nitrogen, suggesting some temporal, and perhaps spatial, regulation of HetF in response to nitrogen starvation. Unfortunately, an initial attempt to observe HetF in vivo with a hetF-gfp translational fusion was unsuccessful due to a lack of observable fluorescence, although this fusion was capable of complementing hetF deletion strain UHM130 (Risser and Callahan, unpublished). It is unclear if the lack of detectable fluorescence is due to improper folding of GFP or if the level of HetF is simply too low to be observed by such methods.
HetF is necessary for the proper regulation of HetR levels at both the transcriptional and posttranscriptional levels. Transcription from the −271 start point of the hetR promoter and transcription from the patS promoter are dependent on hetR, and HetR has been shown to bind to regions within each of these promoters (13, 18). In hetF deletion strain UHM130, although there is an abundance of HetR protein, no transcription from these sites is observed. This is somewhat surprising given that hetR acts stoichiometrically; overexpression of hetR in a wild-type strain results in an increase in heterocyst formation. This inverse correlation between the effects of HetF on the level of HetR protein and transcription from hetR-dependent promoters suggests two possibilities: (i) that a hetF-dependent product acts with HetR to promote transcription from hetR-dependent promoters, after which HetR is concomitantly degraded, and (ii) that hetF acts directly or indirectly to convert a HetR apoprotein to an active form that can then promote transcription from hetR-dependent promoters. The latter possibility is particularly appealing considering evidence suggesting that HetF is a protease.
Acknowledgments
We thank Ramya Rajagopalan for help with RNA extraction and Nicholas Chang for help with construction of the reporter lacZ backbone vector.
This work was supported by grant MCB-0343998 from the National Science Foundation.
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2002. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins 46355-367. [DOI] [PubMed] [Google Scholar]
- 2.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 977-84. [DOI] [PubMed] [Google Scholar]
- 3.Borthakur, P. B., C. C. Orozco, S. S. Young-Robbins, R. Haselkorn, and S. M. Callahan. 2005. Inactivation of patS and hetN causes lethal levels of heterocyst differentiation in the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 57111-123. [DOI] [PubMed] [Google Scholar]
- 4.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst development in the cyanobacterium Anabaena 7120. Genes Dev. 5321-330. [DOI] [PubMed] [Google Scholar]
- 5.Buikema, W. J., and R. Haselkorn. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. USA 982729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan, S. M., and W. J. Buikema. 2001. The role of HetN in maintenance of the heterocyst pattern in Anabaena sp. PCC 7120. Mol. Microbiol. 40941-950. [DOI] [PubMed] [Google Scholar]
- 7.Ehira, S., and M. Ohmori. 2006. NrrA directly regulates expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1888520-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167747-754. [DOI] [PubMed] [Google Scholar]
- 9.Frías, J. E., E. Flores, and A. Herrero. 1997. Nitrate assimilation gene cluster from the Heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghassemian, M., B. Wong, F. Ferreira, J. L. Markley, and N. A. Straus. 1994. Cloning, sequencing and transcriptional studies of the genes for cytochrome c-553 and plastocyanin from Anabaena sp. PCC 7120. Microbiology 1401151-1159. [DOI] [PubMed] [Google Scholar]
- 11.Golden, J. W., and H.-S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6557-563. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 167351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. USA 1014848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khudyakov, I. Y., and J. W. Golden. 2004. Different functions of HetR, a master regulator of heterocyst differentiation in Anabaena sp. PCC 7120, can be separated by mutation. Proc. Natl. Acad. Sci. USA 10116040-16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, J., L. Scappino, and R. Haselkorn. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc. Natl. Acad. Sci. USA 895655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova, K. S., E. V. Koonin, R. Haselkorn, and M. Y. Galperin. 2006. Cyanobacterial response regulator PatA contains a conserved N-terminal domain (PATAN) with an alpha-helical insertion. Bioinformatics 221297-1301. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 18.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 441377-1385. [DOI] [PubMed] [Google Scholar]
- 19.Nayar, A. S., H. Yamaura, R. Rajagopalan, D. D. Risser, and S. M. Callahan. 2007. FraG is necessary for filament integrity and heterocyst maturation in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 153601-607. [DOI] [PubMed] [Google Scholar]
- 20.Orozco, C. C., D. D. Risser, and S. M. Callahan. 2006. Epistasis analysis of four genes from Anabaena sp. strain PCC 7120 suggests a connection between PatA and PatS in heterocyst pattern formation. J. Bacteriol. 1881808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risser, D. D., and S. M. Callahan. 2007. Mutagenesis of hetR reveals amino acids necessary for HetR function in the heterocystous cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1892460-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer, M. R., and S. S. Golden. 1989. Differential expression of members of a cyanobacterial psbA gene family in response to light. J. Bacteriol. 1713973-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei, T.-F., R. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 1764473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 25.Wolk, C. P., Q. Fan, R. Zhou, G. Huang, S. Lechno-Yossef, T. Kuritz, and E. Wojciuch. 2007. Paired cloning vectors for complementation of mutations in the cyanobacterium Anabaena sp. strain PCC 7120. Arch. Microbiol. 188551-563. [DOI] [PubMed] [Google Scholar]
- 26.Wong, F. C. Y., and J. C. Meeks. 2001. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J. Bacteriol. 1832654-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, X., D. Liu, M. H. Lee, and J. W. Golden. 2004. patS minigenes inhibit heterocyst development of Anabaena sp. strain PCC 7120. J. Bacteriol. 1866422-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon, H.-S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282935-938. [DOI] [PubMed] [Google Scholar]
- 29.Yoon, H.-S., and J. W. Golden. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 1832605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, C.-C., S. Laurent, S. Sakr, L. Peng, and S. Bedu. 2006. Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol. Microbiol. 59367-375. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, Y., Y. Shi, W. Zhao, X. Huang, D. Wang, N. Brown, J. Brand, and J. Zhao. 2005. CcbP, a calcium-binding protein from Anabaena sp. PCC 7120, provides evidence that calcium ions regulate heterocyst differentiation. Proc. Natl. Acad. Sci. USA 1025744-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, R., Z. Cao, and J. Zhao. 1998. Characterization of HetR protein turnover in Anabaena sp. PCC 7120. Arch. Microbiol. 169417-423. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, R., X. Wei, N. Jiang, H. Li, Y. Dong, K.-L. Hsi, and J. Zhao. 1998. Evidence that HetR is an unusual serine-type protease. Proc. Natl. Acad. Sci. USA 954959-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]