Abstract
By the analysis of thermodynamic RNA secondary structure predictions, we previously obtained evidence for evolutionarily conserved large-scale ordering of RNA virus genomes (P. Simmonds, A. Tuplin, and D. J. Evans, RNA 10:1337-1351, 2004). Genome-scale ordered RNA structure (GORS) was widely distributed in many animal and plant viruses, much greater in extent than RNA structures required for viral translation or replication, but in mammalian viruses was associated with host persistence. To substantiate the existence of large-scale RNA structure differences between viruses, a large set of alignments of mammalian RNA viruses and rRNA sequences as controls were examined by thermodynamic methods (to calculate minimum free energy differences) and by algorithmically independent RNAz and Pfold methods. These methods produced generally concordant results and identified substantial differences in the degrees of evolutionarily conserved, sequence order-dependent RNA secondary structure between virus genera and groups. A probe hybridization accessibility assay was used to investigate the physical nature of GORS. Transcripts of hepatitis C virus (HCV), hepatitis G virus/GB virus-C (HGV/GBV-C), and murine norovirus, which are predicted to be structured, were largely inaccessible to hybridization in solution, in contrast to the almost universal binding of probes to a range of unstructured virus transcripts irrespective of G+C content. Using atomic force microscopy, HCV and HGV/GBV-C RNA was visualized as tightly compacted prolate spheroids, while under the same experimental conditions the predicted unstructured poliovirus and rubella virus RNA were pleomorphic and had extensively single-stranded RNA on deposition. Bioinformatic and physical characterization methods both identified fundamental differences in the configurations of viral genomic RNA that may modify their interactions with host cell defenses and their ability to persist.
The genetic material of the three domains of life is double-stranded DNA. In contrast, many of the viruses that parasitize these organisms have RNA-based genomes. In addition to their capacity to directly encode proteins, RNA genomes have the ability to form secondary and higher-order RNA structures that contribute to their stability and can form sequence motifs in a structural context that may function during the virus life cycle. For example, several virus groups have evolved internal ribosome entry sites that function in ribosome recruitment and the initiation of translation. A range of different types of internal ribosome entry site elements are recognized, all characterized by extensive (200- to 500-nucleotide [nt]) internal RNA-RNA base pairing (5). Similarly, genome replication per se or the packaging of the genome into capsid precursors can involve the interaction of the virus polymerase and/or other viral proteins with defined RNA structural elements (9, 16, 18, 28, 36, 46). Typically, RNA structures involved in these replication functions take the form of discrete stem loops of various sizes, frequently with evolutionarily highly conserved sequences in terminal loops that are involved in RNA-protein or RNA-RNA interactions.
Of the many methods devised to predict RNA secondary structures, the algorithm used in MFOLD and RNAFold (48) is probably the most widely used, and it is based on an energy minimization algorithm using empirically determined values for the various base pairs that form simple stem loops (29). In a recent investigation of folding free energies of RNA sequences using MFOLD (40), we obtained evidence for extensive RNA structure formation in many families and genera of both positive-strand animal and plant RNA viruses. There was remarkable variability in the occurrence of what we termed genome-scale ordered RNA structure (GORS) in different virus genera; for example, hepatitis C virus (HCV) in the family Flaviviridae showed thermodynamic evidence for an extensive RNA structure within the polyprotein-coding region that was absent in both the related Pestivirus and Flavivirus genera. Similar genus-associated variability was observed in the Picornaviridae, the Caliciviridae, and many plant virus families. Since replication strategies usually are conserved within a family, we considered that this genus-specific characteristic was unlikely to have a role in a fundamentally conserved aspect of replication or genome encapsidation. However, the presence of GORS was invariably associated with the ability of the virus to persist in their natural hosts, raising the intriguing possibility of a role for GORS in the subversion or avoidance of innate intracellular defense mechanisms. We speculated that this may take the form of modulating or blocking defense pathways triggered by double-stranded RNA, analogous in function to the expression of structured RNA transcripts by large DNA viruses (11, 30, 37). GORS is not restricted to viruses that infect animals and is indeed widespread in many plant virus groups/genera. Again, we considered that this may be involved in shielding functions, for example, from Dicer-mediated defense pathways (6, 27).
In the work we have carried out to date, the detection of GORS was based on quantifying differences in the minimum free energy (MFE) of native sequences from the same sequences scrambled in sequence order by a variety of algorithms that preserve different organization features of the nucleotide sequences, e.g., dinucleotide biases, codon structure, protein coding, or all three (40). The aim of the current study is to apply a wider range of bioinformatic RNA structure prediction programs to substantiate our previous evidence for large-scale structural differences between different RNA viruses. A number of methods have been developed recently for the large-scale screening of human and other large genome sequences for secondary structure elements, as they often locate to functional regions (e.g., controlling the transcription and processing of coding and noncoding mRNA sequences) and therefore can contribute to their annotation. In the current study, we have used two algorithmically independent methods. RNAz uses thermodynamic predictions that are weighted by their phylogenetic conservation and the occurrence of covariant sites (13). A second method, Pfold, applies pairing rules to a reconstructed evolutionary history and stochastic context-free grammar to give a probability distribution of structures (22).
We coupled these bioinformatic approaches with an experimental analysis using an oligonucleotide probe accessibility assay and atomic force microscopy (AFM) to investigate the link between the predicted RNA structure and physical solution-phase structures of full-length genomic strands of viruses with and without GORS. Our results strongly support our previous bioinformatic analysis and provide biophysical evidence and further insights into the existence of fundamental structural differences between the RNA genomes of persistent and nonpersistent mammalian RNA viruses.
MATERIALS AND METHODS
Datasets.
Sequence databases for different virus groups analyzed in the study were created using the Simmonic Sequence Editor v. 1.6 (http://www.picornavirus.org/software/) (39) from complete genome sequences downloaded from GenBank. Sequences used for structure conservation studies were aligned using ClustalW with default settings and then were manually edited. Codon-based alignments were used for coding regions of virus genomes (Table 1). All analyses were carried out on coding regions with 5′- and 3′-untranslated regions (UTRs) excluded or the terminal 300 bases of the genomes for viruses with UTRs shorter than this. Large and small subunit rRNA (LSU and SSU, respectively) sequences were downloaded from http://www.psb.ugent.be/rRNA/ (44). Subsets used for analysis were gap-stripped and realigned by ClustalW and manually edited as required (Table 1). Sequences were screened for sequence identity, and those showing less than 1% sequence divergence were excluded from the analysis.
TABLE 1.
Virus and rRNA sequence alignments
| Virus or rRNA sequence alignment | Genus or rRNA subunit | Groupa | No. of sequences | No. of bases | % MPDb | % G+C |
|---|---|---|---|---|---|---|
| Virus family | ||||||
| Caliciviridae | Lagovirus | Rabbit hemorrhagic fever virus | 18 | 6,636 | 7.7 | 51.3 |
| Norovirus | MNV (GGV) | 31 | 6,593 | 10.4 | 56.8 | |
| Norovirus GG2 | 33 | 6,809 | 21.5 | 49.7 | ||
| Sapovirus | Sapporovirus | 21 | 6,654 | 38.5 | 52.1 | |
| Vesivirus | Feline calicivirus | 14 | 6,880 | 20.7 | 45.5 | |
| Ocean group viruses | 8 | 7,488 | 16.7 | 48.2 | ||
| Flaviviridae | Flavivirus | Dengue virus | 64 | 9,905 | 24.4 | 46.2 |
| Japanese encephalitis virus | 30 | 10,200 | 21.9 | 51.1 | ||
| TBEV | 16 | 10,130 | 20.0 | 54.1 | ||
| Yellow Fever virus | 19 | 10,051 | 12.5 | 49.7 | ||
| Hepacivirus | HCV genotype 1 | 28 | 8,843 | 16.8 | 58.6 | |
| HCV genotype 2 | 35 | 8,906 | 17.1 | 57.1 | ||
| HCV genotype 4 | 20 | 8,735 | 13.2 | 56.6 | ||
| HCV genotype 6 | 42 | 8,886 | 24.0 | 56.2 | ||
| Pestivirus | Bovine viral diarrhea virus | 12 | 11,155 | 17.2 | 45.7 | |
| Classical swine fever virus | 23 | 11,511 | 9.7 | 47.0 | ||
| Unassigned | HGV/GBV-C | 39 | 8,437 | 12.1 | 59.1 | |
| Picornaviridae | Aphthovirus | FMDV serotype A | 43 | 6,638 | 10.8 | 53.7 |
| FMDV serotypes B, C | 17 | 6,619 | 12.3 | 53.8 | ||
| FMDV serotype O | 44 | 6,662 | 10.6 | 54.0 | ||
| FMDV SAT serotypes | 18 | 6,654 | 16.6 | 53.8 | ||
| Enterovirus | Species A | 25 | 6,292 | 24.3 | 47.5 | |
| Species B | 40 | 6,285 | 24.0 | 47.6 | ||
| Species C | 45 | 6,341 | 20.6 | 46.1 | ||
| Rhinovirus A | 13 | 6,217 | 25.8 | 39.5 | ||
| Rhinovirus B | 33 | 6,147 | 27.8 | 38.1 | ||
| Hepatovirus | Hepatitis A virus | 32 | 6,358 | 12.1 | 37.4 | |
| Kobuvirus | Aichi virus | 4 | 7,135 | 7.1 | 59.4 | |
| KobuvirusB | Aichi virus | 14 | 2,616 | 7.2 | 61.1 | |
| Parechovirus | Human parechovirus | 11 | 6,257 | 22.0 | 39.1 | |
| Teschovirus | Porcine enterovirus | 29 | 6,286 | 17.7 | 44.0 | |
| (Unassigned) | Duck hepatitis virus | 21 | 6,682 | 14.4 | 42.8 | |
| Togaviridae | Alphavirus | New World alphaviruses | 25 | 11,080 | 31.4 | 49.7 |
| Old World alphaviruses | 18 | 11,080 | 37.8 | 51.0 | ||
| Rubivirus | RV | 15 | 8,976 | 5.5 | 70.1 | |
| Other | Astrovirus | Human astrovirus | 11 | 5,964 | 14.9 | 44.0 |
| Hepevirus | Hepatitis E virus | 71 | 6,465 | 21.5 | 56.1 | |
| rRNA | Subunit | |||||
| LSU | Archaeobacteria | 20 | 2,834 | 26.1 | 59.1 | |
| Eubacteria | 32 | 2,546 | 17.6 | 51.0 | ||
| Viridoplantae | 30 | 3,039 | 12.9 | 54.9 | ||
| Vertebratae | 15 | 3,706 | 13.1 | 62.6 | ||
| Primate mitochondrial | 8 | 1,560 | 12.3 | 43.0 | ||
| Rodent mitochondrial | 7 | 1,564 | 27.5 | 36.7 | ||
| SSU | Archaeobacteria | 91 | 1,384 | 25.9 | 57.9 | |
| Cyanobacteria | 69 | 1,407 | 13.2 | 55.0 | ||
| Dicotyledon | 182 | 1,691 | 4.3 | 49.5 | ||
| Nematodae | 160 | 1,580 | 23.1 | 47.6 | ||
| Insectae | 353 | 1,927 | 20.7 | 48.2 | ||
| Vertebratae | 72 | 1,741 | 4.5 | 55.0 | ||
| Primate mitochondrial | 26 | 925 | 18.6 | 43.0 | ||
| Rodent mitochondrial | 78 | 762 | 25.8 | 38.6 |
Alignments are available from http://www.picornavirus.org/alignments/. FMDV, foot-and-mouth disease virus.
MPD, mean pairwise distance between sequences.
Bioinformatic methods.
Sequence distances and nucleotide and dinucleotide compositions for each data set were computed using built-in functions within the Simmonic Sequence Editor. MFEs were calculated for consecutive 300- or 2,100-base fragments (in 150- or 1,700-base increments, respectively) of each sequence from each alignment using the hybrid-ss-min program from a locally compiled copy of UNAfold (26) running on Mac OS X. Perl scripts were used to automate sequence handling and the statistical analysis of data stored in a MySQL database. For each sequence, MFE differences from the null expectation (MFEDs) and Z scores (43) were calculated by the parallel submission of 100 control sequences scrambled using the NDR algorithm in the Simmonic Sequence Editor, which retains biases in dinucleotide frequencies (40). Results were expressed as MFEDs {i.e., the percent difference in MFEs of native and scrambled sequence, calculated as [(MFENATIVE/MFESCRAMBLED) − 1] × 100]} and as Z scores (i.e., the position of the native sequence in the distribution of control values) (43).
Each sequence data set was analyzed by Pfold (22) and RNAz (10) using default settings. The automated submission of thousands of sequences to the Pfold server (http://www.daimi.au.dk/∼compbio/rnafold/) was achieved using Perl scripts, and the results were similarly retrieved and analyzed. Results from Pfold were expressed as the mean frequency of bases in alignments for each virus or control that were predicted to be paired with a confidence level of 95% or greater. RNAz was locally compiled, and Perl scripts again were used to automate the submission, retrieval, and analysis of data. Due to the limitations of RNAz, which can handle a maximum of six sequences of 400 nt simultaneously, input sequences were randomly sampled in batches of six before being windowed (using the associated script rnaz-window.pl provided by the authors of RNAz) and then analyzed by RNAz. Results were expressed as the mean number of predicted stem loop structures per 1,000 bases for each set of alignments.
Hybridization accessibility assay.
DNA oligonucleotide probes were designed to be complementary to viral genomic RNA sequences. A series of 21-nt oligonucleotide sequences complementary to each transcript were specified throughout the viral genomes at approximately 300-nt spacing and with G+C contents between 48 and 52% (see Table S1 in the supplemental material). Additionally, two oligonucleotide probes (PVcre-A and PVcre-B) were specifically designed to be complementary to single-stranded (loop) and double-stranded (stem) regions of the poliovirus (PV) cis-acting replication element (CRE) RNA structure, respectively.
Genome-length virus RNA was produced by in vitro transcription from full-length clones of hepatitis G virus/GB virus-C (HGV/GBV-C) (45), HCV (21), PV (4), rubella virus (RV) (42), murine norovirus type 3 (MNV3), and the L segment of bunyavirus (BV) (7). The transcription template for HCV/BV chimeric RNA was cloned by inserting the 3.5-kb NsiI fragment of pT7RiboBUNL(+) into the unique NsiI restriction site in pJFH-1_GND in the sense orientation, creating the clone pJFH-1_GNDΔ3kbBV. Biotinylated RNA was generated from plasmid DNA templates linearized with the appropriate restriction enzyme (Table 2). The tick-borne encephalitis virus (TBEV) transcription template was generated by PCR using the Expand Long Range dNTPack kit (Roche) from pIC Hypr 3157-11167 (containing TBEV nonstructural genes and the 3′UTR) using the primers TAATACGACTCACTATAGGGATCGATAATGCTGACGTGGTGG (the T7 promoter is in boldface) and CGAGTCACACATCACCTCCTTG. RNA transcriptions (except HCV/BV chimeric RNA) were performed using the MegaScript T7/SP6 kit (Ambion) with the inclusion of biotin-11-UTP at a molar ratio of 1:4 with unlabeled UTP and incubated at 37°C for 6 h. HCV/BV chimeric RNA was synthesized using the T7 RiboMAX express kit (Promega) with the inclusion of 50 nmol biotin-11-UTP and was incubated at 37°C for 1 h. DNA template was removed by DNase I digestion, and biotinylated RNA was purified using the RNeasy mini kit (Qiagen) with elution in nuclease-free water. RNA integrity was confirmed by electrophoresis through a 1% agarose gel. The RNA concentration was determined by measuring the A260, followed by aliquoting and storage at −80°C.
TABLE 2.
Full-length template sequences for RNA transcripts
| Virus | Accession no.a | Plasmid | Linearization enzyme | Promoter | Transcript length (kb) | % G+C |
|---|---|---|---|---|---|---|
| RV | NA | pRobo502 | EcoRI | SP6 | 9.8 | 70 |
| GBV-C | AF121950 | pGBV-C | SpeI | T7 | 9.4 | 59 |
| HCV | AB047639 | pJFH-1 | XbaI | T7 | 9.7 | 58 |
| MNV | DQ223041 | pMNV3 | XhoI | T7 | 7.4 | 57 |
| PV | NC_002058 | pJM1 (Mah) | MluI | T7 | 7.4 | 46 |
| BV | NC_001925 | pT7BUNL(+) | RsrII | T7 | 6.9 | 33 |
| TBEV | NA | pIC Hypr 3157-11167 | 8.0 | 55 | ||
| HCV/BV | NA | pJFH-1_GNDΔ3kbBV | SbfI | T7 | 13.2 |
NA, not assigned.
Hybridizations were performed between full-length RNA transcripts and their complementary oligonucleotide arrays under nondenaturing conditions in order to maintain the RNA structure. All solutions were prepared in diethyl pyrocarbonate-treated water, and all incubations were performed at room temperature unless otherwise stated. Oligonucleotides (50 and 5 pmol) were spotted onto Hybond-N membranes (GE Healthcare) using a dot blot apparatus attached to a vacuum pump followed by UV cross-linking using a UV Stratalinker 1800 (Stratagene). Membranes were washed for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sarcosyl and for a further 5 min in 2× SSC, followed by staining in 0.3 M sodium acetate (pH 5.5), 0.02% (wt/vol) methylene blue for 5 min. Stained blots were imaged using a Genegenius imager (Syngene) and destained using four 5-min washes in 0.2× SSC, 1% sodium dodecyl sulfate (SDS). Membranes were blocked in 6× SSC, 10× Denhardt's reagent, 1% SDS, 100 μg/ml salmon sperm DNA at 50°C, followed by being washed in 1× Hyb buffer (20 mM HEPES, pH 7.8, 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol) for 15 min at 37°C. Hybridizations were performed in 1× Hyb buffer supplemented with 50 μg/ml poly(A), 50 μg/ml yeast tRNA, 0.01% SDS, 40 U RNasin (Promega), and 2 μg/ml biotinylated RNA at 37 or 65°C for 2 h. Membranes were washed twice in 1× Hyb buffer, 0.2% SDS for 15 min at 37 or 65°C, followed by two washes in 1× Hyb buffer for 15 min. Oligonucleotide/RNA hybridization was detected by developing the membranes using the BrightStar BioDetect kit (Ambion) followed by exposure to Hyperfilm (GE Healthcare).
AFM.
RNA transcripts were diluted in AFM imaging buffer (20 mM HEPES, 10 mM MgCl2, 3 mM NiCl2, pH 7) to 0.5 to 2 ng/μl. Prior to RNA deposition, freshly cleaved mica (grade V2; Ted Pella Incorporated, Redding, CA) was treated with 4 mM NiCl2 for 1 min at room temperature and washed twice by being soaked in 10 ml nuclease-free water for 10 and 1 min, respectively. The NiCl2-treated mica then was dried under a stream of nitrogen. Ten microliters of diluted RNA was dropped on the surface of the NiCl2-treated mica and allowed to adsorb for 5 min at room temperature. Nonadsorbed RNA was removed by two washes with 10 ml of nuclease-free water as described above, after which the sample was dried under a stream of nitrogen and imaged. Imaging was performed at room temperature (22 ± 1°C) on a PicoSPM atomic force microscope (Molecular Imaging) in AC mode using aluminum-coated silicon tips with spring constants of ∼40 N/m and resonance frequencies between 250 and 325 kHz. Fields of 0.5 to 3.0 μm were scanned at 1 to 1.5 Hz. Images were flattened, and approximate cluster diameters were determined using PicoScan 5.3.3 software (Molecular Imaging). Two or three independently prepared samples were imaged for each RNA transcript, and several areas were scanned for each sample.
RESULTS
Bioinformatic methods for prediction of GORS.
GORS originally was identified in RNA virus genomes from observations of consistent differences between MFEs of native sequences and those of sequence-order-scrambled controls (MFEDs) (40). In the current study, we have investigated the use of additional bioinformatic RNA structure prediction methods developed and used extensively for large-scale scanning and structure prediction in genome sequences (RNAz and Pfold). All three methods show moderate to high specificity when used for structure predictions in viruses selected to show a wide range of G+C contents and previously determined to be predominantly unstructured (RV, TBEV, and hepatitis A virus) (40) (Fig. 1).
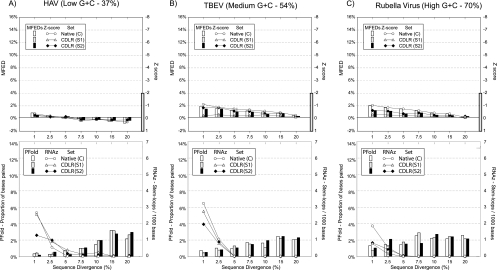
FIG. 1.
Specificity of different RNA structure prediction methods. RNA structure prediction methods using thermodynamic (MFOLD; upper panels), stochastic context-free grammar (Pfold), and phylogenetic methods (RNAz; lower panels) methods were analyzed. Each data set was initiated with coding sequences of previously predicted unstructured RNA virus genomes of different G+C contents and two independently sequence-order-randomized sequences using a codon-based method that also retains dinucleotide frequencies (CDLR). For each of the three viruses, native and scrambled sequences were subjected to artificial sequence drift (x axis) to generate increasingly diverse aligned sequences that were assayed for RNA structure. For MFOLD, both MFEDs and Z scores were recorded. HAV, hepatitis A virus.
Folding energy (MFEs) differences between native and sequence-order-randomized controls (MFEDs) ranged from 0 to 2%, irrespective of the degree of artificially generated sequence divergence within each data set. Similarly low values were obtained for each of the two native sequence scrambled by CDLR and their derivatives, a method that preserved both coding order and dinucleotide frequencies (43). Z scores represent the position of a native-sequence MFE in the distribution of MFEs for sets of sequence-order-randomized controls (n = 100); the MFEs for all control sequences, irrespective of G+C content and sequence divergence, were substantially above the value of −2, indicative of folding free energies consistently within the range of MFEs of scrambled controls.
Other methods for RNA determination were assessed. Multiple random subsets of six sequences in each alignment were selected and analyzed by RNAz. Multiple sampling of the alignments ensured that all sequences were analyzed. For alignments containing sequences with 2.5% or less divergence, a number of false-positive structure predictions were made. However, for datasets with divergence values in the range of the native RNA virus alignments (7.7 to 38.5%) (Table 1), specificity was high, with little if any RNA structure predicted, irrespective of G+C content (apart from a low frequency of structure predicted for hepatitis A virus). In contrast, mean frequencies of base pairing predicted by Pfold showed consistent nonspecificity that increased with sequence divergence irrespective of G+C content (Fig. 1).
Detection of large-scale RNA structure in RNA virus genomes.
The same methods (MFED calculation, RNAz, and Pfold) were used to screen a total of 36 virus and 14 rRNA alignments for large-scale RNA secondary structures (Fig. 2). The virus groups selected represent the principal families, genera, and groups of mammalian positive-stranded icosahedral symmetry RNA viruses for which sufficient comparative sequence data are available.
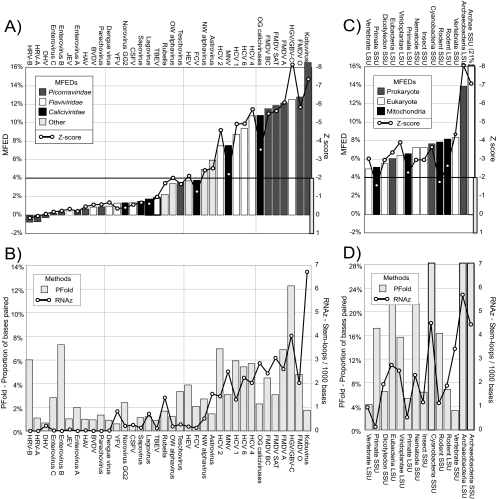
FIG. 2.
Prediction of RNA structure in virus sequence alignments. Shown are the RNA structure predictions in alignments of native sequences for the 36 virus and 14 rRNA alignments listed in Table 1. In both sequence sets, alignments were ordered from left to right by ascending MFEDs. With the exception of Pfold values in panel D, all axes were plotted to the same y axis scale as that used for Fig. 1. DHV, duck hepatitis virus; JEV, Japanese encephalitis virus; HAV, hepatitis A virus; BVDV, bovine viral diarrhea virus; YFV, yellow fever virus; CSFV, classical swine fiver virus; OW, Old World alphaviruses; HEV, hepatitis E virus; FCV, feline calicivirus; NW, New World alphaviruses; OG: ocean group; and FMDV, foot-and-mouth disease virus.
Virus RNA sequences showed a wide range of MFEDs, from around 0 (human rhinoviruses [HRVs]) to greater than 16% (kobuviruses) (Fig. 2A), with 14 of them lying within the range of MFEDs for rRNA sequences. The structure prediction of 2,100-base fragments of each virus alignment produced Z scores ranging from ≈0 to <−8 (HGV/GBV-C); values were tightly correlated with MFEDs (R2 = 0.933). Although absolute values of Z scores are fragment length dependent (longer sequences show tighter distributions of MFEs) (38), the same 14 alignments with high MFEDs all showed mean Z scores below −2. rRNA sequences showed MFEDs ranging from 4.8 to 21% and mean Z scores almost invariably below −2 (Fig. 2C), reflecting the detectable order component in known highly structured RNA molecules.
Each alignment also was analyzed using RNAz and Pfold (Fig. 2B). rRNA sequences contained a variable frequency of strongly predicted stem loop structures using RNAz (expressed as the number of predicted structures/1,000 bases), although it was almost invariably greater than that for control unstructured sequences (Fig. 1), and showed an increasing frequency with greater MFEDs (Fig. 2D). There was a similar good correlation between the RNAz score and MFEDs in virus alignments (R2 = 0.802; P = 2 × 10−12) (Fig. 3A), with few if any structures predicted for viruses showing MFEDs of less than 6%. Pfold found high frequencies of paired bases in each of the rRNA alignments, with values frequently much higher than those found in virus alignments (note the different y axis scales). This may reflect the use of evolutionary and structural parameters by the KH-99 model underlying Pfold originally developed using alignments of tRNA and LSU sequences (22). The correlation between MFEDs and Pfold predictions in virus alignments subsequently was less good, with what are likely to be false-positive results in the HRV-B and enterovirus species B and C alignments (both predicted to be unstructured by RNAz and UNAfold) and failures to detect RNA structures in ocean-group caliciviruses and kobuviruses, both of which are clearly highly structured by other methods. Nevertheless, there was an overall correlation between base-pairing frequencies and MFEDs (Fig. 3B) that achieved statistical significance (R2 = 0.267; P = 0.02).
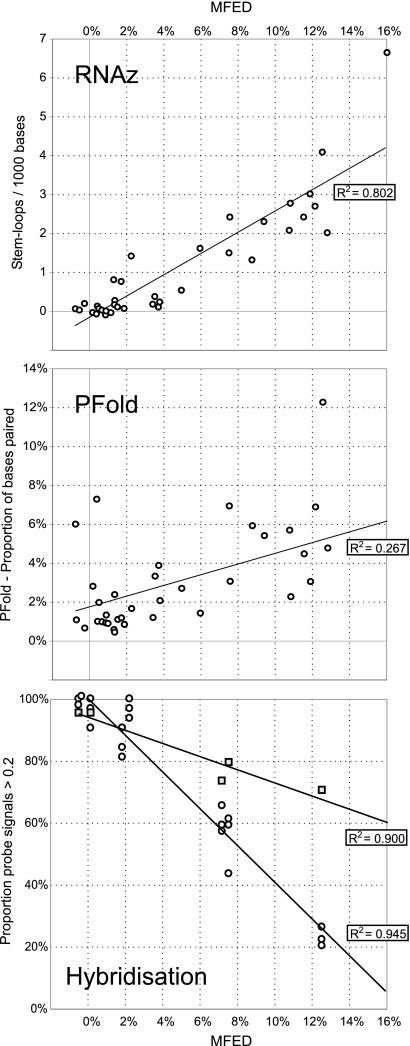
FIG. 3.
Correlation between MFEDs for RNA genomes with structure predictions by (A) RNAz and (B) Pfold. (C) A subset of viral RNA sequences was expressed as labeled transcripts, and filter hybridization to complementary probes was recorded at two temperatures (○, 37°C; □, 65°C) (see the legend to Fig. 5). The proportion of transcripts showing signal intensities of greater than 0.2 was recorded on the y axis. For the hybridization experiment carried out at 37°C, individual points represent results from the three duplicate reactions.
To investigate whether the failure of Pfold to detect structures in kobuviruses was the result of analyzing too small a data set with insufficient covariance data (only four complete genomes of Aichi virus are currently available), a second alignment was creating comprising 14 sequences from the 3Dpol region (Table 1). Despite showing sequence divergence and G+C contents within the range of other virus alignments in which the RNA structure was detected by all three methods, the kobuvirus B data set showed a similarly low frequency of paired bases in Pfold despite high MFEDs (15.9%) and a large number of predicted structures in RNAz (7 per 1,000 bases).
Although large-scale RNA structures were predicted for genera or groups from each of the main virus families analyzed (picornaviruses, flaviviruses, and caliciviruses) (Fig. 2A), detection was highly variable and frequently a genus-associated property (e.g., present in hepaciviruses but absent in pestiviruses and flaviviruses in the Flaviviridae). In the specific case of the Norovirus genus (Caliciviridae), however, genogroup 2 (GG2) (along with GG1, GG3, and GG4; data not shown) showed MFEDs close to zero, while MNV (GG5) showed a mean MFED of 7.6%.
Hybridization accessibility assay.
The bioinformatic predictions of differences in large-scale RNA structures between viral RNAs should be detectable by physical structure determination methods. In the current study, we developed a method to quantify the binding of complementary filter-immobilized probes to transcripts in solutions of viruses (reverse hybridization) with predicted structured and unstructured genomes.
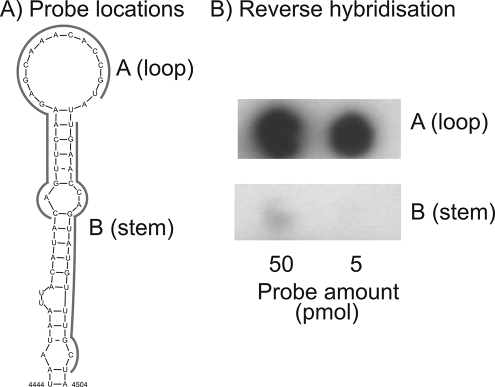
To investigate the ability of the method to differentiate between base-paired and unpaired RNA sequences, we designed probes complementary to the stem and terminal loop of the PV CRE RNA secondary structure element (9) and compared their levels of hybridization to that of a full-length labeled PV transcription in solution (Table 2, Fig. 4A). During the incubation of the probe and target at 37°C (i.e., at an approximately physiological temperature), the probe complementary to predominantly unpaired RNA of the terminal loop and flanking regions (the probe A loop) effectively captured the PV transcript and produced an intense hybridization signal (Fig. 4B), while there was barely detectable binding to the probe (B stem) hybridizing to the predominantly base-paired CRE stem.
FIG. 4.
Analysis of the hybridization of capture probes complementary to predominantly unpaired (PVcreAA) or paired (PVcreB) regions of the PV CRE in the reverse hybridization assay. Filters were spotted with two quantities of capture probe (50 and 5 pmol) to increase the quantitation range.
Using this method, we next analyzed the accessibility of RNA transcripts from a subset of predicted structured and unstructured viruses (n = 7) by quantifying hybridization to complementary probes (Fig. 5). We reasoned that RNA with a high degree of stable internal base pairing should adopt a closed configuration and would be relatively inaccessible to hybridization to external probes; the exposure of single-stranded regions of RNA from predominantly unstructured viral transcripts should, in contrast, lead to a greater degree of hybridization.
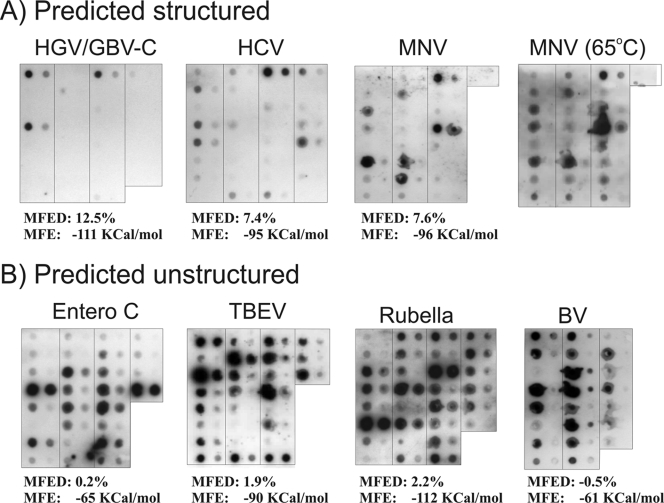
FIG. 5.
Reverse hybridization of filter-immobilized probes to biotin-labeled RNA transcripts of predicted structured (upper) and unstructured (lower) virus genomes. Representative filters are shown with 50 pmol (left) and 5 pmol (right) of each probe spotted onto the membrane. The upper left corner of each filter is the 5′ end of the genome, and the lower right corner is the 3′ end. Entero C, human enterovirus species C.
For the assay, full-length RNA transcripts were generated from viruses with high (MNV, HGV/GBV-C, and HCV) and low (TBEV, PV, and RV) predicted MFEDs. As a further negative control, an RNA transcript (sense orientation) was synthesized from the antisense BV long segment (predicted MFED, −0.5%). The BV RNA was considered a negative control, because a previous thermodynamic analysis of the sequence, along with that of other members of the Bunyaviridae, showed MFEDs close to zero, findings that are consistent with the consistent association of the genomic RNA with the N protein in a ribonucleoprotein complex, a configuration that presumably would prevent any role for a large-scale RNA secondary structure in its genomic RNA (40).
Probes for hybridization were selected from target RNA every 200 to 300 bases down the genome. Duplex regions were specifically selected to have similar G+C contents (48 to 52%) to ensure equal binding strengths irrespective of the overall base composition of the transcript. Filter hybridization intensities of different transcripts were normalized between experiments relative to standard amounts of serially diluted RNA transcripts, where a hybridization intensity of 1.0 corresponds to the spot intensity of 2.5 pg biotinylated RNA transcript following chemiluminescent detection. Where the hybridizations to 50 pmol oligonucleotide probe gave a signal above the linear range of detection, 10 times the signal from hybridization to 5 pmol oligonucleotide probe was used. All filter hybridizations were carried out in triplicate; hybridization intensities, although variable between probes, were highly reproducible between replicates (the mean standard errors for the seven transcripts were within 35% of the mean value; data not shown).
Hybridization data of different transcripts was compared by recording the frequency of probe hybridizations showing signal intensities above 0.2. These ranged from 23% for HGV/GBV-C (i.e., less than a quarter of the probes bound strongly to the target transcript) to 100% (BV L segment) (Fig. 5). For the predicted structured viruses HGV/GBV-C, HCV, and MNV, there was great variability between probes in hybridization intensity, with most failing to hybridize to target sequences in the transcript, while a minority showed very high signal intensities. In the predicted unstructured transcripts, however, hybridization intensities were much more uniform, with the majority of probes hybridizing efficiently to their targets.
Low or absent binding of transcripts to probes indicates that their target sequences are inaccessible to hybridization. To confirm that internal base pairing in the transcript prevented hybridization, transcripts of HGV/GBV-C, HCV, and MNV (all structured) and PV and BV (unstructured) were hybridized to their complementary probes on filters at 65°C. This intermediate temperature was selected to disrupt the transcript secondary structure but to retain the hybridization of target RNA to the oligonucleotide probes (expected melting temperature, ≥70°C). Hybridization at the elevated temperature had no effect on probe binding to PV and BV transcripts, whereas the frequency of probes hybridized to structured transcripts actually was substantially increased (e.g., the MNV transcript at 65°C compared to that at 37°C) (Fig. 5). Similar results were obtained for the other transcripts (Fig. 3C); for example, 70% of probes hybridized to the predicted structured transcript (HGV/GBV-C) at 65°C, whereas <25% hybridized at 37°C.
The mean probe hybridization frequencies for the seven transcripts showed a strong correlation with MFEDs but not with MFEs or other composition variables (R2 = 0.964; P = 8 × 10−5) (Fig. 3C). Probe accessibility was similarly associated with results from other RNA structure prediction methods (RNAz and Pfold) (Table 3), although R2 and P values were lower. Remarkably, there was no correlation between hybridization accessibility and either the G+C content or MFE, although the latter two variables were strongly correlated with each other (R2 = 0.858; P = 0.028). Indeed, the higher values of MFE predicted by MFOLD and other energy-minimizing algorithms that originate from the greater frequencies of base pairing and thermodynamic stability of RNA pairing predicted for G+C-rich genomes were not factors in determining hybridization accessibility. In our analysis, this was almost entirely dependent on the sequence order; the R2 value of 0.964 with the MFED suggests that almost all the observed variability in hybridization is accounted for by sequence order-dependent RNA secondary structure.
TABLE 3.
Correlation between probe hybridization, RNA structure prediction, and composition variablesa
| Variable | Hybridization (R2) | Probabilityc
|
||||
|---|---|---|---|---|---|---|
| MFED | RNAzb | Pfoldb | MFE | G+C | ||
| Hybridization | 8 × 10−5 | 0.015 | 0.024 | 0.169 | 0.439 | |
| MFED | 0.964 | 0.006 | 0.023 | 0.084 | 0.274 | |
| RNAz | 0.805 | 0.874 | 0.060 | 0.119 | 0.423 | |
| Pfold | 0.757 | 0.766 | 0.629 | 0.445 | 0.922 | |
| MFE | 0.341 | 0.480 | 0.494 | 0.152 | 0.028 | |
| G+C | 0.123 | 0.231 | 0.166 | 0.270 | 0.858 | |
Additional composition variables analyzed were mononucleotide frequencies, purine and pyrimidine frequencies, mismatch frequencies between complementary bases [i.e., ABS(C/(G+C); ABS(A/(A+U)], and the underrepresentation of CpG and UpA dinucleotides. None of these composition variables were correlated with probe hybridization frequencies or structure predictions (data not shown).
Correlations between RNAz and Pfold structure predictions with other variables were restricted to six comparisons, because these methods could not be used on the single available BV sequence.
Significant values (P < 0.05) are shown in boldface.
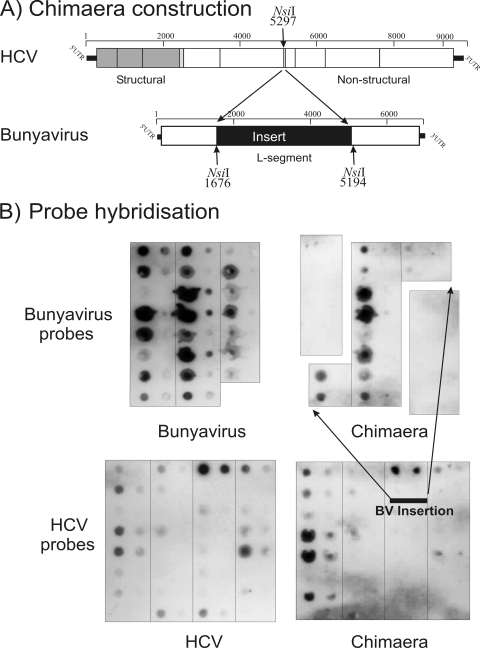
The apparent inaccessibility of the majority of probe targets in structured viruses may be the result of a pattern of the folding of a minority of sequences that sequesters the rest of the genome by forming a shell of base-paired RNA around an unstructured core. Alternatively, it is possible that all of the RNA is structured. To investigate this, we created a hybrid transcript in which a 3,519-base insert of a predicted unstructured BV RNA sequence was inserted in the predicted structured genome of HCV (Fig. 6A). Hybridization to the chimera with both HCV and BV probes was remarkably similar to corresponding targets in native full-length transcripts of HCV and BV. Similar results were obtained by probing a second chimera containing a shorter (2,139-base) insert of BV into the same position in the HCV genome (data not shown). These findings indicate that the insertion of predicted unstructured RNA into the middle of a likely folded, hybridization-inaccessible backbone has little or no effect on its accessibility to external hybridization. Similarly, the BV insert did not increase the accessibility of the surrounding HCV sequence to hybridization. Of the two models, it seems likely that hybridization inaccessibility results from relatively local folding favored by the sequence order-dependent RNA secondary structure. This conclusion is consistent with the distributed nature of predicted structure throughout the genomes of GORS viruses.
FIG. 6.
(A) Strategy for the construction of the HCV (structured RNA)/BV (unstructured RNA) chimera using NsiI restriction sites. (B) Hybridization of BV (upper) and HCV (lower) probes to native BV and HCV transcripts (hybridization controls are at the left) and to transcripts of the chimera (right). The excised parts of the BV probe/chimera filters contain probes that are absent from the chimera sequence.
AFM.
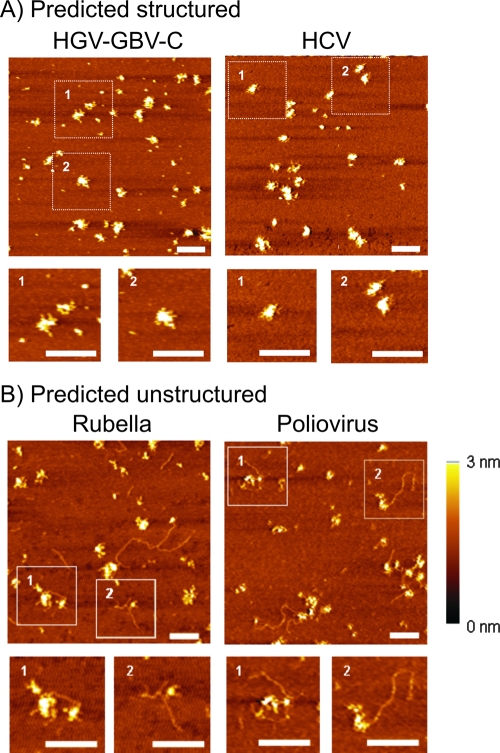
To investigate the physical nature of a bioinformatically predicted large-scale RNA structure, we directly visualized transcripts of predicted structured (HCV and HGV/GBV-C) (Fig. 7A) and unstructured (PV and RV) (Fig. 7B) RNA virus genomes by AFM. Viral RNA transcripts were diluted to 0.5 to 2.0 ng/μl in AFM imaging buffer and deposited onto freshly cleaved mica, washed, and dried under a stream of nitrogen prior to imaging. The imaging of viral RNAs revealed distinct differences in the overall structures between predicted structured and unstructured genomes.
FIG. 7.
AFM analysis of RNA transcripts of (A) predicted structured viral RNA genomes (HGV/GBV-C and HCV) and (B) predicted unstructured viral RNA genomes (RV and PV). All scale bars are 200 nm, and the Z scale ranges from 0 to 3 nm.
HGV/GBV-C and HCV RNA transcripts uniformly adopted a tightly packed condensed state that was largely maintained during the deposition process (Fig. 7A and inset). They showed a regular unit size with a prolate spheroid shape. Although there was some variability in shape, the mean radii for the x and y axes were approximately 30 nm for HCV and 32 nm for HGV/GBV-C, with mean heights of each transcript molecule of 3.9 ± 1.0 nm (n = 10; data not shown) and 3.8 ± 0.9 nm (n = 10), respectively. These measurements predict overall volumes of 7,350 and 8,150 nm3, both only slightly larger than the minimum compacted sizes (for genomic RNAs of 9,600 and 9,400 nt) of 6,900 and 6,550 nm3 predicted by Flory's Law (RG = aN1/3 and volume = 4π/3 × RG3, where RG is the radius of gyration) (17). In contrast, both PV and RV RNA transcripts appeared as pleomorphic globular clusters with frequently observed protrusions of what appeared to be single-stranded RNA often several hundred nanometers in length (Fig. 7B). Transcripts of both viruses were substantially more spread out on the mica solid phase than the relatively compact HCV and HGV/GBV-C transcripts, and they showed a lower mean height (z axis measurements of 2.5 ± 0.9 nm [n = 10] and 2.6 ± 1.1 nm [n = 10], respectively).
DISCUSSION
Structure prediction methods.
This study documents remarkable variability in the extent of RNA secondary structures predicted and observed in the genomes of mammalian positive-stranded RNA viruses. Previous predictions for the existence of GORS have relied on a thermodynamic method in which folding energies of native and sequence-order-scrambled sequences were compared. Although conceptually simple, this method has in the past been criticized for nonspecificity resulting from the use of scrambling methods that failed to preserve certain nonrandom compositional variables, notably the skewed dinucleotide frequencies observed in most mammalian RNA virus genomes (40). Although our use of scrambling methods that preserved biased dinucleotide frequencies should address this problem, we also used two other RNA structure prediction methods based on different bioinformatic principles to further investigate the possibility of major structural differences between viruses with and without GORS.
RNAz determines RNA secondary-structure conservation based on computing a consensus secondary structure and a measure for thermodynamic stability normalized with respect to sequence length and base composition, enabling the identification of RNA secondary structures with high sensitivity and specificity (10). The specificity of the method was demonstrated by an analysis of our control sequences with differing degrees of divergence (Fig. 1), producing results consistent with those of MFED determination.
Pfold is a nonthermodynamic prediction method in which pairing rules and the assessment of the probability of specific pairings were determined using a reconstructed evolutionary history and stochastic context-free grammar to give a prior probability distribution of structures (22). Pfold, however, showed a degree of nonspecificity, with relatively high frequencies of predicted conserved pairings (at >95% probabilities) in all three control datasets, with increasing frequencies with greater sequence divergence. Even taking this nonspecificity into account, Pfold structure prediction using viral datasets (Fig. 2) showed frequent discrepancies (likely false-positive and -negative results in a variety of virus alignments, e.g., those for HRV-B, enterovirus B, and kobuviruses). However, there was still a broad overall agreement between all three methods with which RNA viruses were structured and unstructured.
RNA structure prediction and hybridization.
The hybridization accessibility assay provides a convenient method to probe RNA structure formation in RNA transcripts in solution. The effectiveness of the method in identifying structured and unstructured RNA was demonstrated by the marked difference in hybridization signal between probes complementary to naturally paired and unpaired regions in the PV CRE (Fig. 4).
Intriguingly, it was only the sequence order-dependent component of RNA secondary structure (identified by MFE differences between native and scrambled sequences and by RNAz and Pfold predictions) that correlated with hybridization accessibility results (Fig. 3, Table 3). Particularly informative was the hybridization data for the RV transcript, whose high G+C content (71%) was associated with a very high MFE (−112 kcal/mol for the 300-base fragments) but which appeared virtually unstructured in the hybridization accessibility assay, which correlates with its low MFED. In contrast, the three predicted structured virus transcripts (HGV/GBV-C, HCV, and MNV; MFEDs of 7.5 to 12.5%) with consistently lower MFEs (−95 to −112 kcal/mol) were inaccessible to hybridization with the majority of probes.
The difference between MFEs and MFEDs in their ability to predict the RNA secondary structure in solution may arise through qualitative differences in the nature of the predicted pairings. For example, the type of pairings predicted in unstructured genomes or randomized sequences may be one (metastable) configuration out of many similarly energetically favored structures; dynamic transitions between structures may enable greater probe accessibility than is possible with sequence order-dependent structures identified by high MFEDs and other bioinformatic methods. In contrast, the evolutionary process that created functional structures such as the PV CRE and presumably the large-scale RNA structure in persistent viruses may have created much more stable pairings that shield viral RNA from external hybridization. A numerical analysis of the predicted pairings in structured and unstructured RNA, such as the frequencies of different duplex lengths and pairing distances, would be of value in the future in resolving this issue.
AFM.
The extremely high resolution possible with this method provides the means to directly visualize RNA at the molecular level (1, 12, 23, 31, 35). Although previously used to investigate specific interactions between RNA molecules (such as virus genome circularization [1] or kissing loop interactions [12]), the method also has been used for the visualization of the larger-scale shape of RNA molecules (23) and may thus allow the visualization of the effect of the large-scale secondary structure on the configuration of RNA virus genomes (Fig. 7). In the current study, we indeed found that HCV and HGV/GBV-C RNA transcripts adopted a tightly packed condensed state that was largely maintained during the deposition process (Fig. 7A and insets), in contrast to the irregular, pleomorphic appearance of the PV and RV RNA transcripts and protruding strands of single-stranded RNA (Fig. 7B). The latter appearance of predicted unstructured virus RNAs closely matched that of virion RNA released from PV nucleocapsids after proteolytic digestion in a previous study (23), where the initially compacted genomic RNA rapidly unraveled to leave long strands of unpaired single-stranded RNA visible by AFM. The inclusion of RV was particularly informative; RNA secondary structure formation in its genomic transcript was the most energetically favored of those analyzed (a consequence of its high G+C content), yet both AFM and hybridization accessibility assays predicted an overall open, likely metastable (see the previous section) physical configuration resembling those of other viruses we analyzed with low MFEDs.
Although not the purpose of the investigation, the observed and unexplained differences between PV and several plant viruses in their appearance after capsid dissolution did indeed correlate with their degree of predicted RNA secondary structure. Thus, the predicted unstructured genome of turnip yellow mosaic virus (MFED, 3.4%) (40) resembled PV in rapidly unraveling to single-stranded RNA, while the predicted structured brome mosaic virus, satellite tobacco mosaic virus, and tobacco mosaic virus (all with MFEDs of >9%) (40 and unpublished observations) showed various degrees of compaction and retention of globular shape despite prolonged incubation times in solution, which is comparable to the appearance of HCV and HGV/GBV-C in the current study.
Despite the high resolution of RNA structures possible by AFM, the method does introduce artifacts associated with the deposition process, most notably the flattened appearance of the transcripts (e.g., the measured x and y diameters of HCV and HGV/GBV-C RNA transcripts were approximately 15 times greater than their height) (12, 35). This is caused by the partial dissolution of the condensed state due to interactions with the surface divalent metal ions that become energetically favorable as the buffer is removed. Nonetheless, AFM imaging reveals differences in the overall structures between the RNA transcripts with predicted structured and unstructured genomes that substantiate the bioinformatic and probe hybridization data. Importantly, the estimated volumes of the HCV and HGV/GBV-C transcripts we visualized agree closely with the theoretical size of a compacted RNA calculated by Flory's Law, despite the likely shape distortions arising from the AFM method.
Virus persistence.
The analysis of the expanded data set of mammalian RNA sequences confirmed and extended the previously described association with host persistence (40). All viruses with MFEDs below 6% are naturally nonpersistent in the natural, immunocompetent host, while all of those above this threshold establish persistent infections (where known). This distinction is particularly clear in the families Flaviviridae, Caliciviridae, and Picornaviridae, which comprise clearly separate structured and unstructured categories in the various genera and groups. Major differences in predicted structures were observed between similar viruses; for example, MNV and (human) norovirus GG2 sequences show substantial homology, and coding regions can be readily aligned (20, 41). In this specific example, it is perhaps revealing that human norovirus infections are acute and rapidly cleared (usually in less than 24 h), whereas infections with the predicted structured MNV are persistent and nonpathogenic in immunocompetent mice. (15).
At present, it is difficult to conceptualize what mechanism underlies the association between large-scale RNA structure and host persistence. The RNA secondary structure, calculated as the probability of the internal base pairing of individual bases in an RNA molecule, has been shown to be an important predictive factor for short interfering RNA (siRNA) targeting (24, 25). The formation of GORS and the consequent reduction in the access of genomic RNA to hybridization by guide RNAs or cleavage by Dicer may represent an evasion strategy for viruses infecting organisms that use siRNA for antiviral defense. Intriguingly, the majority of positive-stranded plant viruses show evidence of large-scale RNA structures equivalent in extent to those detected in mammalian RNA viruses (40). It is tempting to imagine that similarly extensive RNA structures and the inaccessibility to hybridization in mammalian RNA viruses plays a similar role in host defense evasion, particularly as the local RNA secondary structure has been shown to similarly prevent access to guide RNAs on human RNA-induced silencing complex and to protect RNA from degradation (2). However, although antiviral resistance mediated by RNA-induced silencing complexes can be induced experimentally in mammalian cells, it is still unclear whether RNA interference (RNAi) actually is used as an antiviral defense pathway at the whole-organism level. Although many DNA viruses were demonstrated to encode and express viral microRNAs (miRNAs) that usurp the host cell miRNA pathway to promote viral replication, none were predicted or experimentally detected in cells infected with HCV or yellow fever virus (32). In further contrast to the established plant and insect siRNA/virus paradigm, the miRNA miR-122 that is expressed abundantly in human liver actually is required for efficient HCV replication, an interaction mediated through the binding of the miRNA to a single-stranded region of the virus 5′UTR (19). In vertebrates, the principal role of RNAi seems to be that of a posttranscriptional regulator of gene expression mediated by endogenously encoded miRNAs.
For mammalian cells, the interferon pathway is the principal antiviral defense mechanism driving several antiviral effector pathways such as protein kinase R and 2′-5′oligoadenylate synthetase, as well as activating the acquired immune system (8, 34). As demonstrated for RNAi interactions, it is possible that the physical inaccessibility of structured RNA we found in the current study hinders these effector functions or, alternatively, prevents the recognition of incoming or replicating RNA by retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and toll-like receptor 3 (TLR3) (3, 47). We have very recently obtained preliminary evidence that large-scale RNA structure sequesters the 5′ triphosphate moiety of genomic RNA (R. Blundell, D. J. Evans, and P. Simmonds, unpublished data). Since this is one of the major targets for recognition by RIG-1 (14, 33), GORS may prevent or delay interferon induction during initial infection or replication. While much remains to be done in documenting this phenomenon and other possible consequences of GORS, such as RNA stability within cells, the finding in the current study that a large-scale structure makes RNA genomic transcripts almost entirely inaccessible to external hybridization is likely to be an important, evolutionarily developed attribute of RNA viruses that profoundly influences their interactions within the mammalian cell.
Supplementary Material
Acknowledgments
We thank the following for providing virus cDNA plasmids: J. McLauchlan (pJFH-1), J. Stapleton (pGBV-C), R. M. Elliott [pT7 Ribo BUNL (+)], T. Frey (pRobo502), I. Goodfellow (pMNV3), and A. Tuplin (pIC Hypr 3157-11167). We are grateful to Bjarne Knudsen for the use of the Pfold Web server, to Stefan Washietl for the RNAz source code, and Michael Zuker and Mark Needham for the UNAfold software. We also acknowledge Jenny Cheng for her assistance in AFM imaging.
The work was supported by a project grant from the Wellcome Trust.
Footnotes
Published ahead of print on 17 September 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 796631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ameres, S. L., J. Martinez, and R. Schroeder. 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130101-112. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barclay, W., Q. Li, G. Hutchinson, D. Moon, A. Richardson, N. Percy, J. W. Almond, and D. J. Evans. 1998. Encapsidation studies of poliovirus subgenomic replicons. J. Gen. Virol. 791725-1734. [DOI] [PubMed] [Google Scholar]
- 5.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, E., A. M. Denli, and G. J. Hannon. 2001. The rest is silence. RNA 71509-1521. [PMC free article] [PubMed] [Google Scholar]
- 7.Bridgen, A., and R. M. Elliott. 1996. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc. Natl. Acad. Sci. USA 9315400-15404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 7563-567. [DOI] [PubMed] [Google Scholar]
- 9.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 744590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber, A. R., R. Neubock, I. L. Hofacker, and S. Washietl. 2007. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res. 35W335-W338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunnery, S., A. P. Rice, H. D. Robertson, and M. B. Mathews. 1990. Tat-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded-RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 878687-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansma, H. G., E. Oroudjev, S. Baudrey, and L. Jaeger. 2003. TectoRNA and “kissing-loop” RNA: atomic force microscopy of self-assembling RNA structures. J. Microsc. 212273-279. [DOI] [PubMed] [Google Scholar]
- 13.Hofacker, I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 313429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, C. C., L. K. Riley, H. M. Wills, and R. S. Livingston. 2006. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 56247-251. [PubMed] [Google Scholar]
- 16.Huthoff, H., and B. Berkhout. 2002. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 4110439-10445. [DOI] [PubMed] [Google Scholar]
- 17.Hyeon, C., R. I. Dima, and D. Thirumalai. 2006. Size, shape, and flexibility of RNA structures. J. Chem. Phys. 125194905. [DOI] [PubMed] [Google Scholar]
- 18.Joost Haasnoot, P. C., R. C. Olsthoorn, and J. F. Bol. 2002. The brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 3091577-1581. [DOI] [PubMed] [Google Scholar]
- 20.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2991575-1578. [DOI] [PubMed] [Google Scholar]
- 21.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 1251808-1817. [DOI] [PubMed] [Google Scholar]
- 22.Knudsen, B., and J. Hein. 1999. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics 15446-454. [DOI] [PubMed] [Google Scholar]
- 23.Kuznetsov, Y. G., S. Daijogo, J. Zhou, B. L. Semler, and A. McPherson. 2005. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 34741-52. [DOI] [PubMed] [Google Scholar]
- 24.Li, W., and L. Cha. 2007. Predicting siRNA efficiency. Cell Mol. Life Sci. 641785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, K. Q., and D. C. Chang. 2004. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 318303-310. [DOI] [PubMed] [Google Scholar]
- 26.Markham, N. R., and M. Zuker. 2005. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 33W577-W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110563-574. [DOI] [PubMed] [Google Scholar]
- 28.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 769686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 30.Mathews, M. B. 1995. Structure, function, and evolution of adenovirus virus-associated RNAs. Curr. Top. Microbiol. Immunol. 199173-187. [DOI] [PubMed] [Google Scholar]
- 31.Noestheden, M., Q. Hu, A. M. Tonary, L. L. Tay, and J. P. Pezacki. 2007. Evaluation of chemical labeling strategies for monitoring HCV RNA using vibrational microscopy. Org. Biomol. Chem. 52380-2389. [DOI] [PubMed] [Google Scholar]
- 32.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2269-276. [DOI] [PubMed] [Google Scholar]
- 33.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and R. E. Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 34.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 35.Rivetti, C., M. Guthold, and C. Bustamante. 1996. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 264919-932. [DOI] [PubMed] [Google Scholar]
- 36.Schlesinger, S., S. Makino, and M. L. Linial. 1994. Cis-acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin. Virol. 539-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp, T. V., M. Schwemmle, I. Jeffrey, K. Laing, H. Mellor, C. G. Proud, K. Hilse, and M. J. Clemens. 1993. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 214483-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmonds, P., I. Kakasiliotis, Y. Chaudhry, D. J. Evans, and I. G. Goodfellow. 2008. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 362530-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmonds, P., and D. B. Smith. 1999. Structural constraints on RNA virus evolution. J. Virol. 735787-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds, P., A. Tuplin, and D. J. Evans. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 101337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thackray, L. B., C. E. Wobus, K. A. Chachu, B. Liu, E. R. Alegre, K. S. Henderson, S. T. Kelley, and H. W. Virgin. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 8110460-10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzeng, W. P., and T. K. Frey. 2003. Complementation of a deletion in the rubella virus p150 nonstructural protein by the viral capsid protein. J. Virol. 779502-9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Workman, C., and A. Krogh. 1999. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic. Acids Res. 274816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wuyts, J., G. Perriere, and P. Y. Van De. 2004. The European ribosomal RNA database. Nucleic Acids Res. 32D101-D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang, J., S. Wunschmann, W. Schmidt, J. Shao, and J. T. Stapleton. 2000. Full-length GB virus C (hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J. Virol. 749125-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang, W., A. V. Paul, and E. Wimmer. 1997. RNA signals in entero- and rhinovirus genome replication. Semin. Virol. 8256-273. [Google Scholar]
- 47.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 48.Zuker, M., and P. Stiegler. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9133-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.