Abstract
The molecular details of glycolipid presentation by CD1 antigen-presenting molecules are well studied in mammalian systems. However, little is known about how these non-classical MHC class I (MHCI) molecules diverged from the MHC locus to create a more complex, hydrophobic binding groove that binds lipids rather than peptides. To address this fundamental question, we have determined the crystal structure of an avian CD1 (chCD1–2) that shares common ancestry with mammalian CD1 from ≈310 million years ago. The chCD1–2 antigen-binding site consists of a compact, narrow, central hydrophobic groove or pore rather than the more open, 2-pocket architecture observed in mammalian CD1s. Potential antigens then would be restricted in size to single-chain lipids or glycolipids. An endogenous ligand, possibly palmitic acid, serves to illuminate the mode and mechanism of ligand interaction with chCD1–2. The palmitate alkyl chain is inserted into the relatively shallow hydrophobic pore; its carboxyl group emerges at the receptor surface and is stabilized by electrostatic and hydrogen bond interactions with an arginine residue that is conserved in all known CD1 proteins. In addition, other novel features, such as an A′ loop that interrupts and segments the normally long, continuous α1 helix, are unique to chCD1–2 and contribute to the unusually narrow binding groove, thereby limiting its size. Because birds and mammals share a common ancestor, but the rate of evolution is slower in birds than in mammals, the chCD1–2-binding groove probably represents a more primordial CD1-binding groove.
Keywords: evolution, glycolipid
All jawed vertebrates possess an adaptive immune system that is based on the highly conserved group of genes that are involved in antigen processing, presentation, and recognition (1). At the core of antigen presentation by the immune system are the genes that comprise the major histocompatibility complex. MHCI and MHC class II (MHCII) genes encode a large family of cell surface receptors involved in the presentation of peptide antigens to T cells. In addition to the “classical” MHCI and MHCII molecules, which present peptides to cytotoxic CD8+ and helper CD4+ T cells, respectively, an extended set of genes that are evolutionarily related to MHCI also are present in most mammals. These “non-classical” MHCI genes include ZAG, FcRn, HFE, HLA-E, MICA, CD1, and others (2). Most of these genes are not linked to the MHC locus and are thought to have arisen by gene duplication and neofunctionalization of primordial MHCI (3). The MHC gene products thus are highly diverse and have adapted to serve a range of functional roles both inside and outside the immune system.
The CD1 family of non-classical MHCI genes in humans is comprised of 5 non-polymorphic members designated CD1A, -B, -C, -D, and -E, with CD1D being the sole isoform in mice (4–6). Virtually all mammals that have been investigated to date possess 1 or more of these prototypic isoforms (3). The proteins encoded by CD1 retain the capacity to present antigens to specific T cells in a manner that is analogous to the presentation of peptides by MHC (7). However, the CD1 proteins have undergone significant structural evolution to bind and present lipid molecules instead of peptides (8). The CD1-restricted T cells have been implicated in a diverse array of immune functions including host defense against pathogens, immune regulation, and autoimmunity (9, 10). In addition, foreign lipids derived from bacteria, as well as self-lipid antigens, have been described for CD1 (11–16). Although the CD1 lipid antigens are chemically diverse, the general motif for these antigens corresponds to 2 hydrophobic acyl chains that are connected to a polar head group, which usually is a carbohydrate moiety (17). The structural adaptation that allows CD1 to bind lipids is reflected in the nature of the antigen-binding pocket that is defined by the α1 and α2 superdomain. The shallow, usually charged or hydrophilic, groove that is nestled between the α helices and accommodates peptide antigens in MHCI and MHCII has been replaced in CD1 by a narrow but deep hydrophobic crevice that is essentially shielded from the solvent. These features endow CD1 with the capability to bind the hydrophobic alkyl chains of lipids and glycolipids for antigen presentation to T cells. Narrow pockets in the CD1 groove restrict the lipid chains and enhance the presentation of the antigenic head group (usually carbohydrate) for interaction with the few polar residues on the surface of the CD1-binding groove, as well as recognition by the T-cell receptor (TCR).
To date, CD1 crystal structures from 2 mammalian species have been described. The structure of mouse CD1d was described initially by Zeng, et al. (18) and later was analyzed with bound lipid antigens (19). Crystal structures of human CD1a (hCDa), hCD1b, and hCD1d isoforms with various lipid antigens have advanced our understanding the nature and specificity of the CD1 antigen-binding pocket (20–23). The structure and function of the mouse and hCD1d proteins are highly homologous, illustrating the highly conserved nature of the CD1 antigen-presentation pathway among mammals (24). Analysis of the structural data has revealed that each of the CD1 isoforms has a unique binding-groove architecture that probably is an adaptation to facilitate presentation of a diverse array of self and foreign lipids that possess alkyl chains of different sizes (6, 25). These structural data have provided valuable insights into the mode and mechanism of lipid antigen-binding and presentation by CD1 (8, 17, 26).
The evolution of the MHC antigen-binding superdomain into a diverse family of receptors capable of binding 2 distinct classes of antigens—lipids and peptides—underscores the adaptability of this structure. A better understanding of the structural evolution of the MHC and CD1 families of molecules may provide insights into the selective pressures that have diversified these gene products and enabled them to acquire new functions. Recent studies have shown that 2 homologs of CD1 are present in avian species (27, 28). Investigation of these non-mammalian CD1 homologs may shed light on the evolution of CD1. Here, we describe the crystal structure of the chCD1–2 protein of the chicken (Gallus gallus) with a bound fatty acid ligand. These data provide an opportunity to compare CD1 structures from 2 highly divergent vertebrate groups that are separated from a common ancestor by at least 310 million years of evolutionary history.
Results
Structure Determination of Chicken CD1–2.
The fully glycosylated chCD1–2/human β2-microglobulin (β2M) heterodimeric receptor (residues 1–283 heavy chain and 1–99 β2M) was secreted from SF9 insect cells and purified to homogeneity using column chromatography (see Methods for details). Crystals were grown in 20% polyethylene glycol 4000, 100 mM sodium citrate pH 5.5 and 10% isopropanol, and the structure was determined by molecular replacement (MR), using a stripped-down version of HLA-E (1MHE) (29) as the starting model. The structure was refined to a final resolution of 2.0 Å with crystallographic R (Rcryst) and free R (Rfree) values of 21.6% and 26.6%, respectively, and with 96.3% of the residues in the favored region of the Ramachandran plot. One chCD1–2/β2M heterodimer occupies the asymmetric unit of the crystal.
Overview of Chicken CD1–2 Structure.
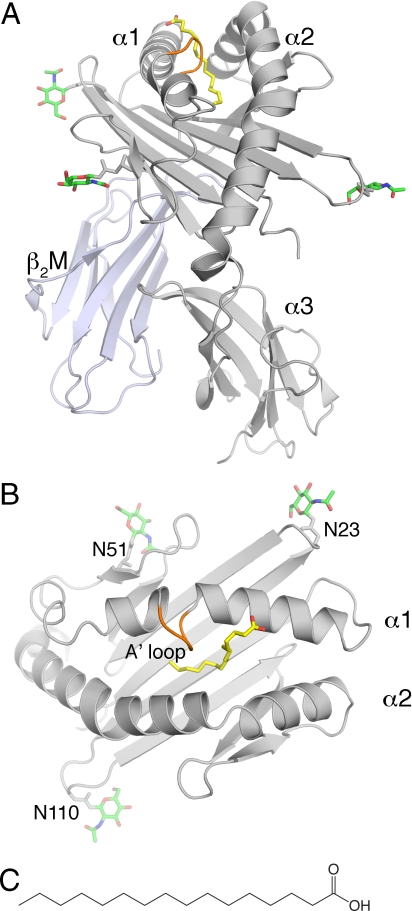
The crystal structure of chCD1–2 with an endogenously bound ligand, presumably palmitic acid, was determined to a resolution of 2.0 Å [supporting information (SI) Table S1]. The overall structure of chCD1–2 resembles that of mammalian CD1 molecules (8, 17, 18, 21, 26, 30–33). Briefly, 2 α helices (α1 and α2) sit atop a central 6-stranded, anti-parallel, β-sheet platform, thus forming the α1-α2 superdomain. The α3 domain is located below the β-sheet platform, where it associates non-covalently with β2M (Fig. 1). The groove width, as measured by the distance between the opposing α1 and α2 helices, is even narrower than in mammalian CD1. A striking difference, in comparison with mammalian CD1 or MHC molecules, is the acquisition of a novel A′ loop (Ser-73, Met-74, Val-75, Gly-76) that interrupts the long α1 helix toward its N-terminal half (Fig. 1). This loop forms a roof above the A′ pocket by making intimate van der Waals contact across the groove with the Val-158 backbone of the α2 helix (Fig. 2). This scenario is reminiscent of hCD1a, in which the extended side chain of Arg-73 (α1 helix) forms a salt bridge with Glu-155 (α2 helix) (23). In addition, Val-75, located at the bottom of the loop, limits the extent and height of the A′ pocket, similar to Phe-70 (A′ pole), which is present in all other CD1 crystal structures thus far investigated (Fig. 2 and Fig. S1).
Fig. 1.
Overview of the chCD1–2 structure with bound palmitic acid. (A) Front view of chCD1–2 (α1, α2, and α3 in gray and β2M in blue-gray) with palmitic acid in yellow. The A′ loop extends from the α1 helix in orange. (B) Top view, looking down into the chCD1–2-binding groove. N-linked carbohydrates are shown as green sticks emanating from 3 Asn positions (N23, N51, and N110), with nitrogen and oxygen colored in blue and red, respectively. (C) Chemical representation of palmitic acid.
Fig. 2.
Conformation of palmitic acid in the chCD1–2-binding groove. (A) Side view of the final 2FoFc map (Fo, observed structure factors; Fc, calculated structure factors) map drawn as a blue mesh around the palmitic acid (yellow) and contoured at 0.8σ. The α2 helix is removed for clarity. Note that the ligand electron density is fairly weak, possibly reflecting less than full occupancy of the palmitate. One hydrogen bond (blue dashed line, 2.9Å) is formed between Arg-82 and the carboxyl group of palmitic acid. Several groove-forming residues are depicted as gray sticks. Met-74 (M74) and Val-75 (V75) are part of the A′ loop (see Fig. 1). (B) Top view (TCR view), looking down onto the slightly transparent molecular surface of the groove. Electrostatic surface potentials were calculated using the APBS program (49). Red is electronegative, and blue is electropositive (−30 to +30 kT/e). Important residues lining the entrance to the groove are labeled. The positive charge of Arg-82 (R82) is ideally situated to neutralize the negative charge of the carboxyl group of the bound palmitate.
The F′ pocket is closed by a lateral wall formed by Phe-83, Met-84, Ile-119, and Met-148 from the α1 and α2 helices (Fig. 2 and Fig. S1). This wall also restricts the size and shape of the hydrophobic groove, allowing occupation only by ligands with a single alkyl chain that contains a maximum of 18 carbons (C18).
Ligand Binding.
The overall shape of the chCD1–2 groove is a continuous, L-shaped tube capable of binding single alkyl chain ligands, such as fatty acids. The entrance to the tube is quite narrow (4 × 6 Å) and is formed by Met-74, Asn-79, and Phe-83 of the α1 helix and by Leu-155 and Val-158 of the α2 helix (Fig. 2 and Fig. S1). Electron density was observed for a bound ligand that must have been acquired during protein expression, because no exogenous ligand was added during protein purification or crystallization (Fig. 2A). Based on its length, shape, and interaction with the surrounding protein residues, we modeled a palmitic acid into the binding groove. This ligand has been identified previously in CD1d as a putative pocket-stabilizing factor that is incorporated in the absence of groove-filling ligands (31, 32, 34). Although most of the palmitic acid is inserted into the narrow binding groove, the carboxylate moiety emerges from the groove entrance where it hydrogen bonds with Arg-82. This situation is reminiscent of mouse and hCD1d, in which the equivalent residue (Arg-79) can interact either with bound glycolipids or directly with the TCR (8, 26, 35). The surface around the entrance of the groove also has a slight positive charge, providing another indication that negatively charged ligands, such as fatty acids, could serve as potential antigens.
Comparison of CD1-Binding Grooves.
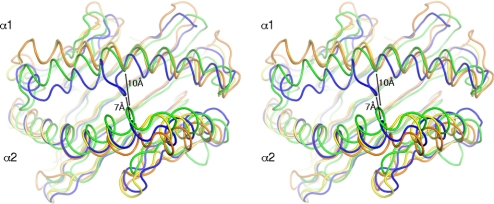
The formation and character of the binding groove is influenced by 3 key factors: (i) by the relative distance and orientation of the 2 α helices with respect to each other; (ii) by the position of the α helices above the β-sheet platform, and (iii), most importantly, by the composition of the groove-forming residues. The relative positioning of the α helices is very similar for CD1 proteins within the same species (Fig. 3); however, single amino acid changes greatly influence the size, shape, and the overall occlusion of the binding groove from the surrounding environment (solvent). Sequence identities between the chCD1–2 ectodomain and other CD1 and MHC molecules are summarized in Table S2. For chCD1–2, we clearly see a more dramatic deviation in the overall distance and orientation of both α helices (Fig. 3). The N-terminal half of the α1 helix, up to the A′ loop, is closer to the α2 helix, thereby substantially restricting the size of the A′ pocket. Also, the A′ loop influences the width of the binding-groove portal. A direct comparison of CD1- and MHC-binding grooves is shown in Fig. 4. The chCD1–2-binding groove is depicted as an orange, semitransparent surface superimposed onto chicken MHC, hCD1d, or hCD1a (Fig. 4 A, B, and C, respectively). Relative to the location of the CD1d groove, the chCD1–2 groove is centrally located and is highly restricted at both N- and C-terminal ends. Its shape and location most resembles hCD1a, but with a narrower groove entrance and a much smaller A′ pocket. In total, 22 residues form the chCD1–2-binding groove with a total volume of 700Å3. Typical volumes of the hCD1-binding groove vary between 1650Å3 for hCD1a (23) and 2200Å3 for hCD1b (20), and the grooves are capable of binding the much larger, 2-chain lipids.
Fig. 3.
Stereo view of the superposition of avian and human CD1 and MHCI α1-α2-binding domains. The α1-α2-binding domains of chicken MHC (yellow) (50), human HLA-E (orange), and hCD1a (green) were superimposed onto the α1-α2 domain of chCD1–2 (blue). Distances between the protein backbone of the α1 and α2 helices of human CD1a and chCD1–2 at the center of the groove are indicated. The chCD1–2 groove is substantially narrower than that of hCD1.
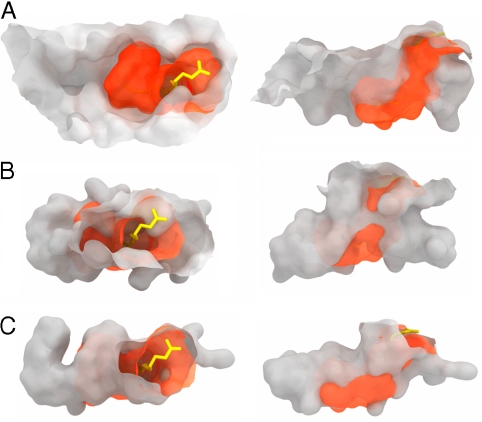
Fig. 4.
Molecular surfaces of MHC- and CD1-binding grooves. The chCD1–2 binding groove (orange) with bound palmitic acid is shown superimposed onto molecular surfaces of chicken MHC (A), hCD1d (B), and hCD1a (C). The left column represents a top view looking down onto the MHC- and CD1-binding grooves as in Fig 1B. The right column shows the binding pockets in a side view. Note that the chCD1–2-binding groove seems to be more similar to CD1a than to CD1d, but it is much smaller.
In silico Evolution of a Lipid-Binding Groove.
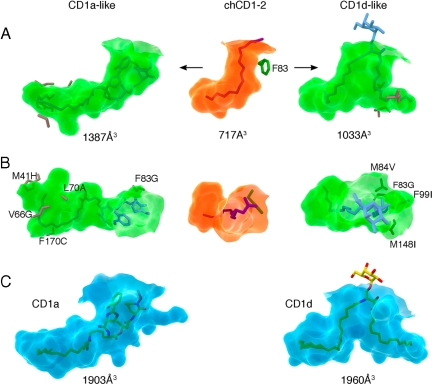
We were interested in exploring whether relatively simple modifications of the groove-lining residues in chCD1–2 to those found in hCD1 proteins, could convert the relatively small chCD1–2 binding pocket into a more “human-like” groove capable of binding larger lipid antigens. Because of its overall resemblance to the more continuous hCD1a-binding groove (Fig. 4), as compared with other CD1 molecules, we carried out in silico mutation of chCD1–2 residues in the binding groove to those of the corresponding hCD1a residues, wherever possible. With only 5 substitutions (M41H, V66G, L70A, F83G, F170C), we were able to create a large and continuous binding groove, very much akin to the hCD1a groove (Fig. 5), that would be capable of accommodating, for example, the didehydroxymycobactin lipopeptide ligand of hCD1a. Two changes within the N-terminal half of the α1 helix (V66G and L70A) were required to create a larger A′ pocket. These substitutions to small (Ala) or no (Gly) side chains are not found naturally in the mammalian CD1 molecules but were necessary to enlarge the groove, because the chCD1–2 α1 helix is much closer to the α2 helix than in mammalian CD1 isotypes.
Fig. 5.
In silico evolution of chCD1–2 to a mammalian-type binding groove. (A) Side view: the semitransparent molecular surface of the chCD1–2-binding groove is shown in the center (green). The predicted shapes and sizes of the binding grooves resulting from the indicated mutations are shown to the left (CD1a-like) and right (CD1d-like). (B) Top view (TCR-view) of the binding groove shown in A. The lipopeptide ligand of CD1a fits well into the mutated chCD1a-like groove, and a truncated form of α-GalCer is modeled into the CD1d-like chCD1–2 groove (blue sticks in A and B). (C) Transparent molecular surfaces (blue) of hCD1a with bound lipopeptide (yellow sticks, left panel) and hCD1d with bound α-GalCer (right panel) for comparison. Groove volumes were calculated using the CASTp server (53).
Furthermore, comparison with human and mouse CD1d sequences (Fig. S1C) revealed that several chCD1–2 residues responsible for the F′ pocket closure have larger side chains than their counterparts in human or mouse CD1d. By the introduction of 4 specific mutations into chCD1–2, we were able to recreate an F′-like pocket with a larger entry portal, similar to that observed in mammalian CD1d. Three of these mutations (M84V, M148I, and F99I) are found either in human or mouse CD1d, whereas F83G, necessary to enlarge the groove portal, is found only in hCD1a. Human and mouse CD1d molecules have an aspartate at position 83, which points upward toward the incoming TCR. However, in chCD1–2, the side chain of residue 83 points toward the groove, and mutation to aspartate would not enlarge the groove portal sufficiently to allow ligand binding of dual alkyl chains. Similar to our approach to convert the chCD1–2 groove into a CD1a-like groove, this second version of a mutated chCD1–2 binding groove, although smaller, now resembles that of mammalian CD1d and would be capable of binding dual alkyl chain ligands with a maximum alkyl chain length of C12 (A′ pocket) and C14 (F′ pocket) (Fig. 5 A and B). Although wholly an in silico study, these models illustrate how, with a minimum number of mutations, a small primordial CD1-binding groove could have evolved to a more complex groove capable of binding larger dual alkyl chain lipids.
Discussion
The CD1 isoforms are relatively non-polymorphic between individuals within a species. This lack of polymorphism is observed in mammals and birds, the 2 extant groups that are known to possess CD1 genes (27). Within mammals, significant conservation of the CD1 isoforms is maintained between disparate orders (e.g., rodents, primates), suggesting that the extended gene family arose early and rapidly during mammal evolution (3). However, no clear evolutionary precursor to the mammal gene family has been found that would indicate whether any of the 5 isoforms could be considered the closest representative to an ancestral archetype. Likewise, because of the large time gap since the existence of a common bird-mammal ancestor, the chCD1–2 protein sequence is sufficiently distant from the mammalian CD1 to preclude its assignment to a specific isoform group. We hypothesized that the structure of avian CD1 would provide information about common features shared among the CD1 structures. These features may have been present in the primordial CD1 of a common ancestor of both birds and mammals around the time of the Synapsid-Diapsid split ≈ 310 million years ago.
The primary feature of the chCD1–2 structure that sets it apart from the known CD1 structures is the smaller overall volume of the lipid-binding pocket. We estimate that no molecule larger than a single-chain C18 lipid molecule could fit within this pocket and still coordinate a charged head group with the conserved Arg-82 residue near the opening of the pocket. Larger head groups that project into the solvent above the pocket opening are possible, as observed for the binding of mycobacterial phosphatidylinositol mannoside with CD1d (30). However, the volume constraints of chCD1–2 protein would limit the length of any bound hydrocarbon chain. All of the mammalian CD1 crystal structures described so far are capable of accommodating antigens with dual acyl chains (with the probable exception of CD1c, whose structure has not yet been determined). The size and shape of the chCD1–2 structure thus would seem to be limited to single-chain lipid antigens.
One hypothesis for multiple CD1 isoforms is that natural selection enhanced the acquisition of mutations that increased the binding affinity and diversity of lipid antigens that could enter the CD1-binding pocket. Lipids are not distributed uniformly throughout the intracellular environment but are sorted actively according to physicochemical parameters, such as acyl chain length (36, 37). Thus, the endosomal localization of a particular CD1 protein determines the subset of the universe of antigens to which CD1 will be exposed (6). Our modeling study is based on known lipid ligands presented by the various mammalian CD1 isoforms. Using these known antigens as size and shape constraints, we selected a minimal set of mutations that would enlarge the chCD1–2 groove to accommodate known mammalian antigens that would, to some extent, emulate some of the major constraints imposed during natural selection. In addition, if the chCD1–2 structure were similar to the primordial precursor, the small size of chCD1–2 pocket could be expanded relatively easily to accommodate the larger antigens that now are found in mammalian CD1. These data, therefore, suggest the possibility that the chCD1–2 lipid-binding pocket could form the basis of a core CD1 receptor upon which further diversification could yield either CD1a-like or CD1d-like antigen-binding pockets. It is important to note that these models are not intended to mimic faithfully actual evolutionary events but rather to illustrate the plasticity of the smaller chCD1–2 groove to attain a significant expansion with a minimal set of amino acid substitutions.
The chCD1–2 protein shares several conserved structural features with known mammal CD1 structures. These features include the overall fold and topology of the α1-α2 superdomain, a hydrophobic antigen-binding pocket, a conserved Arg-82 for coordination of polar antigen head groups and for direct TCR interaction, and, most importantly, the capacity to bind lipids. The conservation of these structural features suggests that they are likely to have been present in a common ancestor, rather than having arisen independently in Synapsids and Diapsids. One question raised by the chCD1–2 structure is whether this extant form represents something closer to the primordial shape of an antigen-binding pocket for the CD1 of a common bird-mammal ancestor. It obviously is impossible to determine the precise sequence or structure of the CD1 that was present at the time of the mammal-bird divergence. However, recent evidence suggests that the rate of evolution is slower in birds than in mammals (38), although some error may be associated with these estimates based on the incomplete fossil records used to calibrate these molecular clock analyses (39). If the tempo of evolution is indeed slower in birds than in mammals, avian CD1 may have preserved more of the ancestral CD1 features. These data therefore would provide a temporal window to examine critical events in the structural evolution of the vertebrate immune system.
Methods
Protein Expression, Purification, and Crystallization.
The G. gallus CD1–2 cDNA (GenBank sequence AY375530) encoding the heavy-chain ectodomain, including a leader sequence and a C-terminal hexa-histidine tag (residues 1–302), and full-length human β2M were cloned into the dual promoter baculovirus transfer vector pBACpHp10, using a strategy similar to that used for mouse CD1d (19). Human β2M was chosen instead of chicken β2M because the chimeric protein could be expressed at amounts suitable for crystallization without affecting the structure of the chCD1–2-binding groove. Expression and purification of chCD1–2 was carried out essentially as described for mouse CD1d (18). Briefly, Spodoptera frugiperda 9 (SF 9) cells (5–8l, shaker flask) were infected with a high titer (1–2 × 108 pfu/ml) of chCD1–2-bearing baculovirus at a multiplicity of infection of ∼ 3 and kept at 27.5 °C on a shaking platform (145 rpm) for 3–4 days. SF9 cells were spun down (1000 × g) for 10 min at 4 °C, and the cell culture supernatant including secreted chCD1–2 protein was exchanged against PBS and concentrated to 300–500 mL of using tangential flow-through concentrators (TFF, Pall Filtron). Ten mM imidazole and 5 mL of Ni-NTA beads (settled volume, Qiagen) were added, and the solution was stirred for at least 4 h or overnight at 4 °C. The Ni-NTA beads were collected using a Buchner funnel (40–60 μM pore size) and were washed briefly with PBS. The Ni-NTA beads were transferred into an Econo colum (Bio-Rad) and were washed with 500 mL of 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM imidazole. chCD1–2 was eluted further with 30–50 mL of 50 mM Tris-HCl pH 8.0, 250 mM imidazole. Eluted chCD1–2 was dialyzed overnight against 10 mM Tris-HCl pH 8.0 and was purified by ion-exchange chromatography on MonoQ, using an AKTA FPLC (GE Healthcare). Eluted chCD1–2 fractions were pooled, concentrated using ultrafiltration devices (Millipore), and purified to homogeneity by size-exclusion chromatography in Superdex S200 16/70 columns (GE Healthcare). chCD1–2/human β2M heterodimeric protein was concentrated to 4 mg/ml in 10 mM Hepes, pH 7.5, 25 mM NaCl. Crystals were grown at 22.3 °C by sitting-drop vapor diffusion by mixing 1 μL of protein with 1 μL of precipitate (20% polyethylene glycol 4000, 100 mM sodium citrate pH 5.5, and 10% vol/vol isopropanol) and were optimized by streak seeding to yield single crystals.
Structure Determination.
Crystals were flash-cooled at 100 K in mother liquor containing 20% glycerol. Diffraction data from a single chCD1–2 crystal were collected at beamline 8.2.1 of the Advanced Light Source (ALS, Berkeley) and processed to 2.0-Å resolution with the Denzo-Scalepack suite (40) initially in tetragonal spacegroup P422 (unit cell dimensions: a = 92.15 Å; b = 92.15 Å; c = 96.69 Å). One chCD1-human β2M molecule occupies the asymmetric unit with an estimated solvent content of 46.0% based on a Matthews' coefficient (Vm) of 2.28 Å3/Da. MR in P41212 was carried out in CCP4 (41) using the program MOLREP (42) from the CCP4 program suite (41) with the HLA-E structure (PDB code 1MHE) (29) as the search model in which all heavy-chain residues were mutated to alanine. The best MR solution had an Rcryst of 56.8% and correlation coefficient of 0.21; in the second-best solution, the Rcryst was 59.3%, and the correlation coefficient was 0.14. Interestingly, when mouse CD1d or hCD1a, -b or -d structures were used as the MR search model, no valid MR solutions were obtained. Subsequent rigid-body refinement followed by 1 round of restrained refinement in REFMAC 5.2 (47) produced an Rcryst of 43.5%. Because the α1 helix did not fit the initial electron density, its coordinates were removed from the model to reduce model bias before proceeding to subsequent refinement. After restrained refinement, the α1 helix was slowly built back into the continually improving σA-weighted, 2Fo–Fc electron-density maps. Initial refinement included several rounds of restrained refinement against the maximum likelihood target in REFMAC 5.2 (47). At a later stage of refinement, carbohydrates were built at 3 N-linked glycosylation sites (N-X-S/T and an atypical N-I-C) in chCD1–2. N-linked glycosylation at the atypical N-X-C motif was shown for bovine protein C in 1982 (43) and since then has been identified in many other proteins, among them hCD69 (44). The refinement progress was judged by monitoring the Rfree for cross-validation (45). The model was rebuilt into σA-weighted 2Fo–Fc and Fo–Fc difference electron-density maps using the program COOT (46). Water molecules were assigned during refinement in REFMAC using the water ARP module for >3σ peaks in a Fo–Fc map and were retained if they satisfied hydrogen-bonding criteria and returned 2Fo–Fc density >1σ after refinement. Final refinement steps were performed using the TLS procedure in REFMAC (47) with 3 anisotropic domains (the α1-α2 domain including carbohydrates, the α3 domain, and the β2M) and resulted in improved electron density maps and a further drop in Rfree. The chCD1–2 structure has a final Rcryst of 21.6% and Rfree of 26.6%, and the quality of the model, as assessed with the program Molprobity (48), was excellent (Table S1).
Structure Presentation.
The program Pymol (49) was used to prepare Figs. 1–5 and Fig. S1. Maxon Cinema4D was used to create the molecular surfaces of Figs. 3–5 and Fig. S1. The PDB2PQR server (50) and the program APBS (51) were used to calculate the electrostatic surface potentials of Fig. 2.
Supplementary Material
Acknowledgments.
I.A.W. is supported by National Institutes of Health grants CA58896 and GM62116 and by the Skaggs Institute. D.M.Z. is the recipient of a Clinical Research Initiative investigator award. This is manuscript MB-19491 of the Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3DBX).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809814105/DCSupplemental.
References
- 1.Flajnik MF, Kasahara M. Comparative genomics of the MHC: Glimpses into the evolution of the adaptive immune system. Immunity. 2001;15:351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 2.Natarajan K, Li H, Mariuzza RA, Margulies DH. MHC class I molecules, structure and function. Rev Immunogenet. 1999;1:32–46. [PubMed] [Google Scholar]
- 3.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 4.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 5.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 6.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: Insights from comparative genomic analysis. Trends Immunol. 2003;24:412–418. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 7.Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 9.Behar SM, Porcelli SA. CD1-restricted T cells in host defense to infectious diseases. Curr Top Microbiol Immunol. 2007;314:215–250. doi: 10.1007/978-3-540-69511-0_9. [DOI] [PubMed] [Google Scholar]
- 10.Brigl M, Brenner MB. CD1: Antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 11.De Libero G, Mori L. Structure and biology of self lipid antigens. Curr Top Microbiol Immunol. 2007;314:51–72. doi: 10.1007/978-3-540-69511-0_3. [DOI] [PubMed] [Google Scholar]
- 12.Willcox BE, Willcox CR, Dover LG, Besra G. Structures and functions of microbial lipid antigens presented by CD1. Curr Top Microbiol Immunol. 2007;314:73–110. doi: 10.1007/978-3-540-69511-0_4. [DOI] [PubMed] [Google Scholar]
- 13.Moody DB. Polyisoprenyl glycolipids as targets of CD1-mediated T cell responses. Cell Mol Life Sci. 2001;58:1461–1474. doi: 10.1007/PL00000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moody DB, et al. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med. 2000;192:965–976. doi: 10.1084/jem.192.7.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moody DB, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 16.Moody DB, et al. Structural requirements for glycolipid antigen recognition by CD1b- restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 17.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 19.Zajonc DM, et al. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 21.Koch M, et al. The crystal structure of human CD1d with and without α-galactosylceramide. Nat Immunol. 2005;8:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 22.Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 24.Brossay L, Kronenberg M. Highly conserved antigen-presenting function of CD1d molecules. Immunogenetics. 1999;50:146–151. doi: 10.1007/s002510050590. [DOI] [PubMed] [Google Scholar]
- 25.Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol. 2003;3:11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- 26.Zajonc DM, Kronenberg M. CD1 mediated T cell recognition of glycolipids. Curr Opin Struct Biol. 2007;17:521–529. doi: 10.1016/j.sbi.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller MM, et al. Characterization of 2 avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci USA. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salomonsen J, et al. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci USA. 2005;102:8668–8673. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Callaghan CA, et al. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol Cell. 1998;1:531–541. doi: 10.1016/s1097-2765(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 30.Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. [DOI] [PubMed] [Google Scholar]
- 31.Wu D, et al. Design of NKT-cell activators: Structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc Natl Acad Sci USA. 2006;103:3972–3977. doi: 10.1073/pnas.0600285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;8:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giabbai B, et al. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: A molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- 34.Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate Vα14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 36.Mukherjee S, Soe TT, Maxfield FR. Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J Cell Biol. 1999;144:1271–1284. doi: 10.1083/jcb.144.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 38.Pereira SL, Baker AJ. A mitogenomic timescale for birds detects variable phylogenetic rates of molecular evolution and refutes the standard molecular clock. Mol Biol Evol. 2006;23:1731–1740. doi: 10.1093/molbev/msl038. [DOI] [PubMed] [Google Scholar]
- 39.Brown JW, Rest JS, Garcia-Moreno J, Sorenson MD, Mindell DP. Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biology. 2008;6:6. doi: 10.1186/1741-7007-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. HKL: Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.CCP4. Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Vagin AA, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 43.Stenflo J, Fernlund P. Amino acid sequence of the heavy chain of bovine protein C. J Biol Chem. 1982;257:12180–12190. [PubMed] [Google Scholar]
- 44.Vance BA, Wu W, Ribaudo RK, Segal DM, Kearse KP. Multiple dimeric forms of human CD69 result from differential addition of N-glycans to typical (Asn-X-Ser/Thr) and atypical (Asn-X-Cys) glycosylation motifs. J Biol Chem. 1997;272:23117–23122. doi: 10.1074/jbc.272.37.23117. [DOI] [PubMed] [Google Scholar]
- 45.Brünger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 48.Lovell SC, et al. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 49.DeLano W. The PyMOL molecular graphics system. 2002 Available at www.pymol.org.
- 50.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch M, et al. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity. 2007;27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Dundas J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.