Abstract
DNA encoding facilitates the construction and screening of large chemical libraries. Here, we describe general strategies for the stepwise coupling of coding DNA fragments to nascent organic molecules throughout individual reaction steps as well as the first implementation of high-throughput sequencing for the identification and relative quantification of the library members. The methodology was exemplified in the construction of a DNA-encoded chemical library containing 4,000 compounds and in the discovery of binders to streptavidin, matrix metalloproteinase 3, and polyclonal human IgG.
Keywords: drug discovery, immunoglobulins, streptavidin, selection, affinity chromatography
The isolation of small organic molecules, capable of specific binding to proteins of interest, is a central problem in chemistry, biology, and medicine. The conjugation of a unique DNA fragment to each compound of a chemical library, serving as an amplifiable “bar code” for the identification and relative quantification of individual library members, represents an attractive avenue for the synthesis and screening of large combinatorial libraries (1–4). When performing selections with DNA-encoded chemical libraries of small size (comprising a few hundreds of compounds), for example, encoded self-assembling chemical libraries in which each DNA strand carries a unique sequence for each pharmacophore (5–7), the identification and relative quantification of library members before and after selection can often be achieved by using DNA-microarrays (5–10). By contrast, selections of binding molecules from larger DNA-encoded chemical libraries (comprising several thousand to millions of compounds) may require the use of high-throughput sequencing technologies to assess the relative abundance of library members before and after selection against a target protein of interest.
Herein, we describe the construction of a DNA-encoded chemical library consisting of 4,000 compounds covalently attached to unique DNA fragments serving as amplifiable identification bar codes. Similar to our previous experiments with DNA-encoded libraries consisting of several hundreds of members (7), we have initially assessed the relative composition of the new library and its functionality by performing selection experiments on Sepharose resin coated with streptavidin. Because a variety of ligands were known with dissociation constants ranking from the millimolar to the femtomolar range (7) the challenge was to investigate whether binders with various affinities could be easily and rapidly isolated from a library containing 4,000 members. We have found that selections can conveniently be decoded by using a recently described high-throughput DNA sequencing technology (termed “454 technology”) developed for genome sequencing (11), revealing chemical structures with submicromolar dissociation constants toward streptavidin. In addition, we have performed selections to against the target polyclonal human IgG and the catalytic domain of matrix metalloproteinase 3. To our knowledge a high-throughput sequencing application for decoding of DNA-encoded chemical libraries has not been reported previously. Furthermore, we have devised strategies for the construction and decoding of DNA-encoded chemical libraries containing up to 106 compounds built on the basis of multiple independent sets of building blocks.
Results
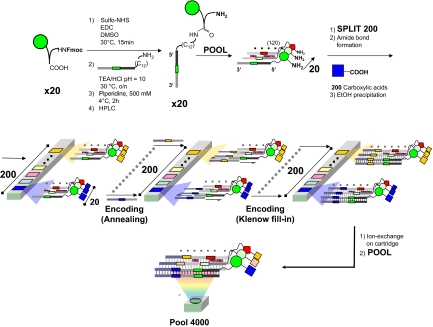
Fig. 1 describes the strategy for the construction of a DNA-encoded chemical library consisting of 20 × 200 modules (i.e., 4,000 compounds), joined together by the formation of an amide bond. Initially, 20 Fmoc-protected amino acids were chemically coupled to 20 individual amino-tagged oligonucleotides. After deprotection and HPLC purification, the 20 resulting DNA-encoded primary amines were coupled to 200 carboxylic acids, generating a library of 4,000 members. To ensure that each library member contained a different DNA code, a split-and-pool strategy was chosen, which also minimizes the number of oligonucleotides needed for library construction. As indicated in Fig. 1, the 20 primary amines covalently linked to individual single-stranded oligonucleotides were mixed and aliquoted in 200 reaction vessels, before coupling with the 200 different carboxylic acids (1 per well). The identities of the carboxylic acids used for the coupling reactions were encoded by performing an annealing step with individual oligonucleotides, partially complementary to the first oligonucleotide carrying the chemical modification. A successive Klenow fill-in DNA-polymerization step yielded double-stranded DNA fragments, each of which contained 2 identification codes (1 corresponding to the initial 20 compounds and 1 corresponding to the 200 carboxylic acids; see Fig. 1). The 200 reaction mixtures were then purified on an anion exchange cartridge and pooled. Model reactions performed before library construction had shown that the yields of the amide bond forming reaction ranged between 51% and 98%. The resulting DNA-encoded chemical library, containing 4,000 compounds, was aliquoted at a total DNA concentration of 300 nM and stored frozen before further use. Detailed structures of the 20 Fmoc-protected amino acids and of the 200 carboxylic acids used can be found in supporting information (SI) Dataset S1. Even though the concentration of an individual library member is <1 nM, binding compounds can efficiently be recovered by selection with biotinylated target protein in solution at concentrations above the dissociation constant Kd, followed by streptavidin capture. Similarly, the selection can be performed with the protein of interest immobilized at high surface density on a solid support (e.g., CNBr-activated Sepharose), in full analogy to the procedures commonly used for the selection of antibodies from phage display libraries (12).
Fig. 1.
Schematic representation of the strategy used for the synthesis and encoding of the DEL4000 library. Initially, 20 different Fmoc-protected amino acids were coupled to unique oligonucleotides derivatives, carrying a primary amino group at the 5′ extremity. Subsequently, these derivatives were pooled and coupled to 200 carboxylic acids in parallel reactions. The identity of each carboxylic acid was encoded by means of a Klenow polymerization step, by using a set of partially complementary oligonucleotides. This procedure resulted in a 4,000-member library (DEL4000), in which each chemical compound was covalently attached to a double-stranded DNA fragment, containing 2 coding domains that unambiguously identify the compound's structure (i.e., the 2 chemical moieties used for compound synthesis).
Selections were performed by incubating the library DEL4000 with streptavidin–Sepharose resin (7). The resin, containing the retained DNA-encoded binding molecules was washed 4 times with 400 μL of PBS and finally resuspended in 100 μL of water for a subsequent PCR amplification step, followed by high-throughput sequencing.
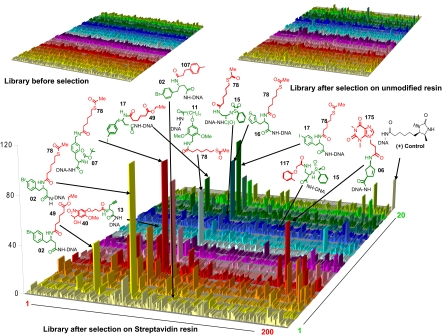
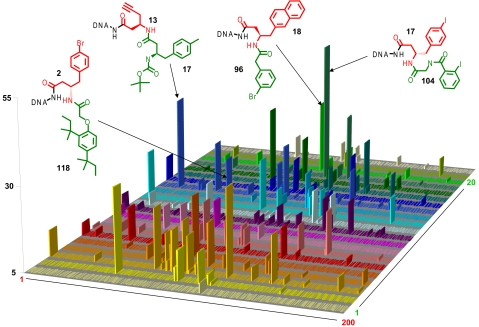
Fig. 2 shows the results of the high-throughput sequencing analysis performed on the library before selection, after selection on unmodified Sepharose resin used as negative control, and after selection on streptavidin-coated Sepharose. High-throughput sequencing of the library containing 4,000 DNA-encoded compounds yielded up to 40,000 sequences per sample. The counts for individual library codes (z axis of the 20 × 200 matrices in Fig. 2) indicate the abundance of the corresponding oligonucleotide–compound conjugate. As expected, in the library before selection, compounds were found to be represented in comparable amounts. When analyzing 7,336 individual codes from the library before selection, the average counts and the standard deviations for the 4,000 compounds were found to be 1.72 ± 1.42. Similarly, no striking enrichment was observed for selections on unmodified resin. By contrast, the decoding of the streptavidin selection revealed a preferential enrichment of certain classes of structurally related compounds (Fig. 2). In addition to desthiobiotin, a biotin analogue with nanomolar affinity to streptavidin, which had been spiked into the library as positive control before selection, we observed an enrichment of derivatives of the thioester moiety 78, of the ester moiety 49 as well as of other pharmacophores (e.g., 175). Fluorescent amide derivatives of compounds 49 and 78 had previously been found to bind to streptavidin with dissociation constants in the millimolar range, as assessed by fluorescence polarization assays (7), whereas others (e.g., 175) had not previously been reported as streptavidin binders.
Fig. 2.
Plots representing the frequency (i.e., sequence counts) of the 4,000 library members before selection, after selection on empty resin, and after selection on streptavidin resin, as revealed by high-throughput 454 sequencing. The chemical structures of some of the most relevant straptavidin binders are indicated. The building blocks used in the 2 synthetic steps are indicated in green and red, respectively, together with the respective identification number. A known streptavidin binder (desthiobiotin) had been mixed with the library at low concentration before the selections serving as positive control.
To evaluate whether the extensions of the pharmacophore 49 and 78 moieties within the 4,000-member chemical library (02, 07, 11, 15, 16, and 17 depicted in green in Fig. 2) contribute to an increased affinity toward streptavidin, we measured the dissociation constants of the most enriched compounds by fluorescence polarization at 25 °C, after conjugation to fluorescein (Fig. 3; see also SI Text). To assess the specificity of preferentially enriched compounds, we also determined the binding affinities toward 2 unrelated proteins (bovine carbonic anhydrase II and hen egg lysozyme) serving as negative controls, and we included 4 nonenriched compounds (15-117, 02-107, 13-40, and 15-78) in the analysis.
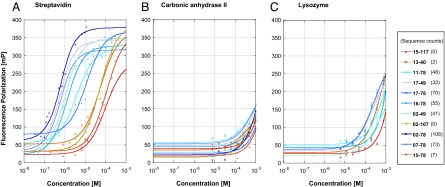
Fig. 3.
Dissociation constants of the selected compounds determined by fluorescence polarization. Individual compounds identified in the streptavidin selection experiments were synthesized as fluorescein conjugates (SI Text) and incubated with different concentrations of target proteins (streptavidin, bovine carbonic anhydrase II, and hen egg lysozyme). (A) The top streptavidin binding molecules (various shades of blue), identified with at least 30 counts (see also Fig. S1C), exhibited a preferential binding toward streptavidin, with Kd values ranging between 350 nM and 11 μM. By contrast, nonenriched compounds (shades of red) did not exhibit an appreciable binding to streptavidin (Kd > 50 μM). (B and C) Neither the streptavidin binders nor the nonenriched compounds exhibited an appreciable binding to carbonic anhydrase II or to lysozyme. The structures of the 11 compounds can be found in Fig. 2 and in SI Text).
The dissociation constants toward streptavidin of the most enriched compounds ranged between 350 nM and 11 μM [Kd (17-49) = 350 nM; Kd (02-78) = 385 nM; Kd (17-78) = 374 nM; Kd (02-49) = 804 nM; Kd (16-78) = 1.1 μM; Kd (11-78) = 3.5 μM; Kd (07-78) = 11 μM; Fig. 3A]. These compounds, each represented at least 30 times in the high-throughput sequencing results, were found at least 10 times more frequently after selection on streptavidin, compared with their occurrence in the unselected library and with what would be predicted by a random statistical distribution (for a simulation, see Fig. S1). By contrast, 4 randomly chosen negative-control compounds, experimentally found <7 times after sequencing, exhibited Kd values to streptavidin >50 μM (Fig. 3A). Importantly, all compounds exhibited no appreciable binding affinity (Kd > 200 μM; Fig. 3 B and C) toward lysozyme and carbonic anhydrase serving as negative control proteins, thus confirming the specificity of the streptavidin selection. A complete list with all the dissociation constants towards the different targets can be found in Table S1.
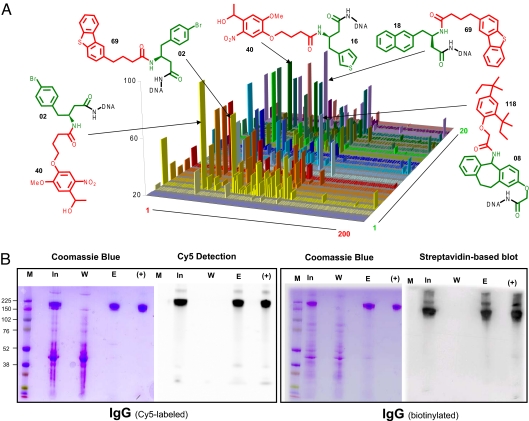
In a second selection, we identified small organic molecules that display binding to polyclonal human IgG, immobilized on CNBr-activated Sepharose. This could be useful for affinity purification of human IgG in industrial manufactures. Fig. 4A reveals that, after selection, compound 02-40 was identified 96 times of a total 39,092 identified sequence tags, whereas >50% of library members were detected between 1 and 10 counts, and ≈10% of the compounds were identified >20 counts. By using the diamino linker O-bis-(aminoethyl)ethylene glycol, compound 02-40 was coupled to CNBr-activated Sepharose, and the resulting resin was evaluated for its performance in the affinity capture of labeled polyclonal human IgG, spiked into CHO cell supernatant (see SI Text. Fig. 4B shows that IgG labeled with both the fluorophore Cy5 and with biotin could be completely and selectively captured from the supernatant and could be eluted by using 100 mM aqueous triethylamine solution.
Fig. 4.
DEL4000 library selection with polyclonal human IgG. (A) Plot representing the frequency (i.e., sequence counts) of DEL4000 library members after selection on polyclonal human IgG resin, as revealed by high-throughput 454 sequencing. The chemical structures of some of the most relevant compounds enriched are indicated. The building blocks used in the 2 synthetic steps are indicated in green and red, respectively, together with an identification number. (B) Affinity chromatography of CHO cells supernatant on resin containing the compound 02-40, spiked with human IgG labeled with either Cy5 or biotin. For antibody purifications, relevant fractions were analyzed by SDS/PAGE both with Coomassie blue staining and with a specific detection method (Cy5 fluorescence and a streptavidin horseradish peroxidase-based blot, respectively). M, molecular weight marker; In, input fraction for the chromatographic process; W, pooled washed fractions; E, pooled eluted fractions. The lane (+) corresponds to Cy-5- or biotin-labeled polyclonal human IgG. In, W, and E fractions were normalized to the same volume before SDS/PAGE analysis. The SDS/PAGE gels containing the fractions from the purification of both Cy5- and biotin-labeled IgG were analyzed by conventional staining with Coomassie blue, or by using a label-specific procedure (fluorescence imaging for Cy5-labeled IgG or streptavidin-based blot for biotinylated IgG; see also SI Text).
Furthermore, we carried out a selection against the catalytic domain of human matrix metalloproteinase (MMP)3. MMPs are zinc-dependent proteases that are involved in tissue remodeling for a variety of physiological and pathological processes (13).
Fig. 5 shows the relative abundance of the individual compounds as obtained from high-throughput sequencing. A different fingerprint compared with the streptavidin and IgG selections was observed. Among the compounds that displayed the highest enrichment in Fig. 5, 4 compounds were selected (02-18, 13-17, 18-96, and 17-104) and tested for MMP3 binding and inhibition. Compound 02-118 exhibited the best dissociation constant (Kd of 11 μM), as assessed by fluorescence polarization with the fluorescein conjugate (see SI Text), yet at a site outside of the catalytic pocket.
Fig. 5.
DEL4000 library selection with human MMP3. The plot represents the frequency (i.e., sequence counts) of DEL4000 library members after selection on human MMP3 resin, as revealed by high-throughput 454 sequencing. The chemical structures of some of the most relevant compounds enriched are indicated. The building blocks used in the 2 synthetic steps are indicated in green and red, respectively, together with an identification number.
The demonstration that high-quality DNA-encoded chemical libraries could be synthesized and decoded by using 454 high-throughput sequencing technology encouraged us to investigate methodologies for the construction of even larger DNA-encoded chemical libraries, featuring the stepwise addition of at least 3 independent sets of chemical moieties. Fig. S2 shows that 3 independent identification codes can be added in a stepwise fashion yielding double-stranded DNA fragments, by using experimental procedures based either on the blunt-end ligation of DNA fragments and/or annealing of partially complementary oligonucleotides, followed by Klenow-assisted polymerization.
Discussion
We have constructed a high-quality DNA-encoded chemical library containing 4,000 compounds (DEL4000). This library was selected for the identification of streptavidin, MMP3, and IgG binders. High-throughput sequencing of the library before and after selection revealed the preferential enrichment of binding molecules. In the case of the previously undescribed streptavidin binders, we have observed that both building blocks used for the stepwise synthesis of compounds in the library may contribute to the resulting binding affinity. For example, we observed a >100-fold difference in binding affinity between compounds 02-78 and 15-78, with Kd constants of 385 nM and 78 μM, respectively, in line with their different recovery rates after streptavidin selection (Fig. 3).
We have also shown that the encoding strategy followed for the construction of the DEL-4000 library could be extended, allowing the incorporation of a third set of chemical groups and of the corresponding DNA fragments (Fig. S2). Recent advances in ultrahigh-throughput DNA sequencing with 454 technology indicate that it should be possible to sequence >1 million sequence tags per sequencing run (11). Provided that 2 orthogonal synthetic procedures are used, which feature high coupling yields and which preserve the integrity of the DNA molecule, it should be possible to construct, perform selections, and decode DNA-encoded libraries containing millions of chemical compounds. Such technologies may facilitate the identification of binding molecules (“hits”) for pharmaceutical applications. At present, large pharmaceutical companies typically screen a few hundred-thousand compounds in their high-throughput screening campaigns. Importantly, selections of DNA-encoded chemical libraries such as the one described in this paper do not require target specific assays but only minute amounts of target protein and can be performed in just 1 day.
Among the selections described in this work, the identification of binders to polyclonal human IgG appears to have the most direct application. At present, monoclonal antibodies for therapeutic applications represent the fastest growing sector of pharmaceutical biotechnology (14). Protein A Sepharose, which is used in virtually all industrial purification procedures for monoclonal antibodies, represents the largest cost factor for the manufacture of therapeutic antibodies. In consideration of the substantial costs, these resins are typically regenerated and reused, which complicates certain aspects of good manufacture practice. It could be conceivable to replace protein A-based affinity supports with the affinity purification supports based on IgG-binding molecules, like the ones described in this article.
Materials and Methods
Synthesis of DEL4000 Compounds.
The 4,000 members of DEL4000 library were synthesized in 2 sequential split and pool amide-forming reactions. Initially, 20 Fmoc-protected amino acids were chemically coupled to 5′-amino-modified 42-mer oligonucleotides after activation of the carboxylic acid in 70% (vol/vol) DMSO/water, with N-hydroxysulfosuccinimide, N-ethyl-N′-(3-dimethylaminopropyl)-carbodiimide and, subsequently, addition of aqueous triethylamine hydrochloride solution (pH 9.0). The oligonucleotides had general structures: 5′-GGA GCT TGT GAA TTC TGG XXX XXX GGA CGT GTG TGA ATT GTC-3′, where XXX XXX unambiguously identifies the individual Fmoc-protected amino acid compound. All coupling reactions were stirred overnight at 25 °C; residual activated species were then quenched and simultaneously Fmoc-deprotected by addition of piperidine in DMSO. The reactions were then purified by HPLC, and the desired fractions were dried under reduced pressure, redissolved in water, and analyzed by LC-ESI-MS. Typical coupling yields were >51% overall. Four nanomoles of each DNA–compound conjugate were pooled to generate a 20-member DNA-encoded sublibrary.
In the following split and pool step, the oligonucleotide sublibrary pool was split among 200 reaction vessels, and an amide-forming reaction with a distinct carboxylic acid was performed by following a very similar procedure as reported above. Test coupling reactions were also performed in the same reaction conditions; by using model 42-mer 5′-Fmoc-deprotected amino acid oligonucleotide conjugates and model carboxylic acids. The test reactions were analyzed by HPLC and the masses of the reacted oligonucleotides detected by LC-ESI-MS. Typical HPLC coupling yields on this step were >52% with purity >46%. The identity of the carboxylic acids used for the coupling reactions was ensured by annealing with unique 44-mer oligonucleotides with general structure: 5′-GTA GTC GGA TCC GAC CAC XXXX XXXX GAC AAT TCA CAC ACG TCC-3′ (where XXXX XXXX unambiguously identifies the individual carboxylic acid compound), partially complementary to the first oligonucleotide carrying the chemical modification, and subsequent incubation with Klenow polymerase at 37 °C for a 1-h reaction. After purification on ion-exchange cartridges, the 200 purified reactions were dissolved in 50 μL of water each, and pooled to generate the 4,000-member library DEL4000 to a final total oligonucleotide concentration of 300 nM. Details of the coupling conditions, analytics and structures of the 20 Fmoc-protected amino acids and of the 200 carboxylic acids used can be found in SI Text.
Selection and Decoding Experiments.
For the model selection with streptavidin D-desthiobiotin-oligonucleotide conjugate, synthesized and unambiguously encoded as described above, was added to the library of 4,000 compounds (20 nM total DNA concentration) to a final concentration of 1 pM. Fifty microliters of the library spiked was either added to 50 μL of streptavidin–Sepharose slurry or to Sepharose slurry without streptavidin. Both resins were preincubated with PBS and 0.3 mg/ml herring sperm DNA. After incubation for 1 h at 25 °C, the beads were washed 4 times with 400 μL of PBS (20 mM NaH2PO4, 30 mM Na2HPO4, 100 mM NaCl) and used as template for PCR amplification of the selected codes.
For the selection with polyclonal human IgG and MMP3, 50 μL of DEL4000 library (20 nM total DNA concentration) was added to 50 μL of IgG–Sepharose or MMP3–Sepharose slurry, respectively. After incubation and rinsing of the beads as describe above, they were used as template for PCR amplification.
The PCR primers used in selections with both streptavidin and polyclonal human IgG had the following structure: DEL_P1_A (5′-GCC TCC CTC GCG CCA TCA GGG AGC TTG TGA ATT CTG G-3′) and DEL_P2_B (5′-GCC TTG CCA GCC CGC TCA GGT AGT CGG ATC CGA CCA C-3′). The primers additionally contain, at 1 extremity, a 19-bp domain (italicized) required for high-throughput sequencing with the 454 genome sequencer system. The PCR products were purified on ion-exchange cartridges. Subsequent high-throughput sequencing was performed on a 454 Life Sciences–Roche GS 20 sequencer platform. Analyses of the codes from high-throughput sequencing were performed by an in-house program written in C++. The frequency of each code has been assigned to each individual pharmacophore.
Synthesis of the Binding Molecules as Fluorescein Conjugates.
Details of the synthesis and analytics can be found in SI Text.
Affinity Measurements.
Fluorescein–compound conjugates (500 nM) were incubated with increasing amounts of streptavidin in PBS and 5% DMSO for 1 h at 25 °C. The fluorescence polarization was determined with a TECAN Polarion instrument by excitation at 485 nm and measuring emission at 535 nm (ε = 72,000 M−1cm−1). [All of the curves were fitted by using the Kaleidagraph software package (Synergy Software), by applying the following formula: a + b × 0.5 × (((x + const + c) − ; const = 500 nM.]
Synthesis of IgG Affinity Chromatography Resins (Containing the Compounds 02-40 or 16-40).
Details of the synthesis and analytics can be found in SI Text.
Polyclonal Human IgG Cy5 Labeling and Biotinilated Polyclonal Human IgG.
Details of the preparations can be found in SI Text.
Affinity Chromatography of CHO Cells Supernatant Spiked with Human IgG Cy5-Labeled or Biotinylated Human IgG on IgG Binding.
The resin containing compound 02-40 was loaded on a chromatography cartridge and washed 3 times with PBS before loading a CHO cell supernatant spiked with human IgG Cy5-labeled or biotinilated human IgG. The flow-through, the wash fractions (washing 1 time with PBS; 1 time with 500 mM NaCl, 0.5 mM EDTA; 1 time with 100 mM NaCl, 0.1% Tween 20, 0.5 mM EDTA) and the elutate (elution 3 times with 100 mM aqueous triethylamine solution) were collected and eventually concentrated back to the initial volume by centrifugation in a Vivaspin 500 tube (cut-off 10,000 MW). The samples were then analyzed by gel electrophoresis on a NuPAGE 4–12% Bis-Tris gel by using Mops SDS as running buffer and stained with Coomassie blue. Cy5 activity was detected by a Diana III Chemiluminescence Detection System (Raytest) by excitation at 675 nm and measuring emission at 694 nm (ε = 250,000 M−1cm−1).
Streptavidin-based blot analyses were performed by transferring the proteins to NC membrane with the Xcell II blot module by using standard procedures. The membrane was quickly rinsed with water before soaking it twice in methanol and then dried at room temperature for 15 min and incubated for 1 h with 1:500 dilutions in 4% defatted milk-containing PBS of streptavidin–horseradish peroxidase conjugate. Finally, the membrane was washed 3 times for 5 min with PBS and soaked in a chemiluminescent substrate solution (ECL1plus Western Blotting Detection System) for 5 sec and exposed to BioMax films in an autoradiographic cassette.
Supplementary Material
Acknowledgments.
This work was supported by Eidgenössische Technische Hochschule, Philochem AG, Kommission für Technologie und Innovation Grant 8868.1 PFDS-LS, Gebert-Rüf Foundation Grant GRS-076/06, the Swiss National Science Foundation, and European Union Projects STROMA and IMMUNO-PDT.
Footnotes
Conflict of interest statement: The IgG binders described in this article have been patented by Philochem, a spin-off company of ETHZurich of which D.N. owns shares.
This article is a PNAS Direct Submission. J.A.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805130105/DCSupplemental.
References
- 1.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gartner ZJ, et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halpin DR, Harbury PB. DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2004;2:E174. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melkko S, Dumelin CE, Scheuermann J, Neri D. Lead discovery by DNA-encoded chemical libraries. Drug Discov Today. 2007;12:465–471. doi: 10.1016/j.drudis.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Melkko S, Zhang Y, Dumelin CE, Scheuermann J, Neri D. Isolation of high-affinity trypsin inhibitors from a DNA-encoded chemical library. Angew Chem Int Ed Engl. 2007;46:4671–4674. doi: 10.1002/anie.200700654. [DOI] [PubMed] [Google Scholar]
- 6.Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nat Biotechnol. 2004;22:568–574. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- 7.Dumelin CE, Scheuermann J, Melkko S, Neri D. Selection of streptavidin binders from a DNA-encoded chemical library. Bioconjug Chem. 2006;17:366–370. doi: 10.1021/bc050282y. [DOI] [PubMed] [Google Scholar]
- 8.Dumelin CE, et al. A portable albumin binder from a DNA-encoded chemical library. Angew Chem Int Ed. 2008;47:3196–3201. doi: 10.1002/anie.200704936. [DOI] [PubMed] [Google Scholar]
- 9.Melkko S, Dumelin CE, Scheuermann J, Neri D. On the magnitude of the chelate effect for the recognition of proteins by pharmacophores scaffolded by self-assembling oligonucleotides. Chem Biol. 2006;13:225–231. doi: 10.1016/j.chembiol.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature. 2004;431:545–549. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silacci M, et al. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5:2340–2350. doi: 10.1002/pmic.200401273. [DOI] [PubMed] [Google Scholar]
- 13.Skiles JW, Gonnella NC, Jeng AY. The design, structure, and therapeutic application of matrix metalloproteinase inhibitors. Curr Med Chem. 2001;8:425–474. doi: 10.2174/0929867013373417. [DOI] [PubMed] [Google Scholar]
- 14.Walsh G. Biopharmaceutical benchmarks 2006. Nat Biotechnol. 2006;24:769–776. doi: 10.1038/nbt0706-769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.