Abstract
Heart failure is a complex, complicated disease that is not yet fully understood. We used the Module Map algorithm to uncover groups of genes that have a similar pattern of expression under various conditions of heart stress. These groups of genes are called modules and may serve as computational predictions of biological pathways for the various clinical situations. The Module Map algorithm allows a large-scale analysis of genes expressed. We applied this algorithm to 700 different mouse experiments downloaded from the Gene Expression Omnibus database, which identified 884 modules. The analysis reconstructed partially known principles that play a role in governing the response of heart to stress, thus demonstrating the strength of the method. We have shown a role of genes related to the immune system in conditions of heart remodeling and failure. We have also shown changes in the expression of genes involved with energy metabolism and changes in the expression of contractile proteins of the heart following myocardial infarction. When focusing on another module we noted a new correlation between genes related to osteogenesis and heart failure, including Runx2 and Ahsg, whose role in heart failure was unknown so far. Despite a lack of prior biological knowledge, the Module Map algorithm has reconstructed known pathways, which demonstrates the strength of this new method for analyzing gene profiles related to clinical phenomenon. The method and the analysis presented are a new avenue to uncover the correlation of clinical conditions to the molecular level.
Keywords: heart, microarray, gene expression, myocardial infarction
heart failure is a prevalent life-threatening condition, yet the underlying pathophysiological processes are not completely understood. In patients suffering from ischemic heart disease, cardiomyopathy occurs due to necrosis of cardiomyocytes that do not regenerate and are replaced by fibroblasts that form a noncontractile scar tissue, leading to regional contractile dysfunction. If the dysfunctional area is large enough, it may impact the contractile ability of the entire heart, which leads to heart failure (5). Heart failure is a common disease in the US, with five million patients and 500,000 newly diagnosed each year. Approximately 1% of the population older than 65 yr suffers from heart failure, which may reach an incidence of 1 yr mortality as high as 45% in some clinical subtypes (45). Understanding the molecular processes that underlie the clinical conditions is a prerequisite for future treatments improving cardiomyocyte function. To focus treatment on specific genes, proteins, and pathways, further research that will characterize their behavior in various clinical conditions is necessary.
To better understand the heart response to stress, we used a bioinformatic approach to identify groups of genes that are expressed together under certain conditions. Coexpression of genes can indicate that the groups of genes share a common function or that they are related to a common physiological condition. The identified groups of genes can also share transcriptional regulation principles and thus highlight novel pathways and regulators related to heart stress. The identified groups of genes are called modules and can tentatively point out previously unknown genes that play a role in biological processes. However, identifying modules is not trivial and requires the use of specialized bioinformatic algorithms. There are several algorithms for discovering modules, which have been used to process information obtained from yeast (40, 74, 90) and humans (73). In a manner similar to the evolutionary conservation of genes, modules have been discovered to be conserved in multiple distant organisms including Saccharomyces cerevisiae, Caenorhabditis elegans, Escherichia coli, Arabidopsis thaliana, Drosophila melanogaster, and Homo sapiens (9, 55, 86). Some of the computational predictions generated by these experiments have been verified at the experimental level, mainly by PCR analyses (40, 74, 86).
Regarding heart failure, there are small-scale bioinformatic studies aimed at analyzing gene expression in the heart and only a few larger-scale experiments. One such large-scale experiment used 230 series from the GEO (Gene Expression Omnibus) (35) and identified 57 modules (called biclusters). However, the biological implications of the modules identified were not discussed, thus weakening the value of the analysis. Other studies reviewed the common biological trends present in multiple articles, either by a textual review of the different articles (76) or by computationally combining the different datasets and analyzing them together (8). In these studies, the data sets were combined for further analysis, but this was only done on a small scale, with no more than a few tens of samples in total in each paper.
In the current study, we used the Module Map algorithm to perform a large-scale characterization of the mouse cardiac tissue under multiple conditions, and we discuss some of the biological implications. This algorithm was previously used to analyze human cancer samples (73) and is suitable for processing information on mammalian transcriptomes. Simple clustering algorithms attempt to detect groups of genes, where these genes are expressed in a similar manner in all samples. Such algorithms would fail in our case, since the chance of consistent expression across all samples decreases with the number of samples examined. Biclustering algorithms, like Module Map, attempt to find groups of genes which are expressed in a consistent manner in a group of samples. This is a task much more likely to succeed (Supplementary Fig. S11 ). Searching for modules (biclusters) using all possible starting points is computationally impossible (91), so this algorithm uses a starting point of a group of defined gene sets whose definition is based on known biological properties (see materials and methods). These initial gene sets are expanded by the algorithm to include unrelated and unknown genes. In addition, the Module Map algorithm searches for clinical attributes that are enriched, allowing us not only to discover groups of similarly expressed genes, but to link clinical attributes to biological pathways. Thus, the Module Map algorithm appears to be the optimal choice for our analysis.
The current analysis is the largest study ever performed on the heart and included 700 microarray samples and 10,978 gene sets, resulting in hundreds of modules describing the response of the mouse cardiac tissue to varied biological conditions. The computational results presented here identified biological processes without having prior knowledge of the behavior of the heart under the specific conditions analyzed. The results presented in this study point out the involvement of the immune system in heart remodeling and failure after conditions such as pressure-induced overload. We also highlighted changes in the energy metabolism and in the contractile proteins of the heart, changes that were known in the literature. The changes demonstrated the existence of two trends of expression in the contractile proteins, trends that behaved in an opposite manner. Thus, we have shown that the Module Map algorithm is able to validate main pathophysiological findings in heart failure that are reported in the literature, which can increase the readers' confidence in this complex bioinformatic approach. In addition, we identified a new correlation between two biological pathways related to osteogenesis and to heart failure. Exploring the possible connection between these pathways pointed out two genes, Runx2 and Ahsg, whose role in heart failure was previously unknown.
MATERIALS AND METHODS
Gene expression data.
A series is a group of microarray samples that are biologically similar and whose expression was assessed in the same experiment. Thirty-eight series of mouse expression arrays were retrieved from the GEO database (7), for a total of 700 samples that were summarized in 26 previously published articles. These samples represent all experiments on the mouse heart publicly available in the GEO as of November 2006. For the purpose of this study, we unified series that were separate in the GEO, if they were originally performed by the same lab, using the same control samples (see Supplementary Table S1). Each series was processed and prepared individually as follows: 1) Probe sets (PS) were retained if their expression value was >32 and if they were marked present by Affymetrix MAS algorithm (if used) in at least 5% of the samples in the series. PS with only some of the samples lower than this value had the expression value in these samples floored to 32. It is necessary to filter and floor MAS5 expression values to remove noise and achieve an accurate picture of gene expression changes (65). This previously published article used the Absent/Present detection calls of the Affymetrix algorithm to filter gene expression. Since not all samples used in our analysis had the detection calls, a threshold value of 32 was determined empirically on a small subset of samples with detection calls and then used for all samples. The expression value of retained PS was log-transformed using base 2. PS were then assigned to genes (identified by Entrez Gene IDs) using the Affymetrix annotations, released on 15 November 2006. PS that matched no genes or matched more then one gene were omitted from the analysis. 2) We filtered the data to remove genes with “noise,” defined as large differences of expression levels between their PS. Genes that had half or more of their PS removed due to “noisiness” were not included in the analysis of this series. 3) Gene expression values were transformed into log-space, by taking log2 of their PS expression values (averaging multiple probes in genes with more than one probe). The log-space expression value of genes in each sample was normalized relative to the median expression in all samples in the same series, by subtracting its median in that series from each of its expression value measurements. After this normalization, the median value of each gene, in each series, is zero. 4) All the processed series were unified horizontally into one matrix based on Gene IDs. Genes that were present in <36 samples (<5% out of the total 700 samples) were omitted from the final analysis. A total of 18,677 genes passed all filtering steps and were used in our analysis.
Gene sets.
A gene set is a group of genes that share a common property, such as expression in a specific tissue, being a similar type of enzyme, or being involved in a specific process. A total of 10,978 gene sets describing the properties of 23,158 genes, of which 15,583 were expressed in the data processed, were compiled from the following sources: 3,451 gene sets from all three branches of the Gene Ontology (4), 2,735 gene sets based on the mouse Gene Expression Database describing expression in the mouse embryo (80), 184 gene sets from the Kyoto Encyclopedia of Genes and Genomes (48), 2,810 gene sets from the MGI Mammalian Phenotype database (79), 1,460 gene sets based on motif-containing genes as presented by Xie et al. (97), 217 gene sets based on calculated targets of miRNA, as present in the miRNAMap database release 1.0 (one gene set per miRNA sequence, where we removed target genes below the 40th percentile in either minimum free energy score or miR score) (38), and 122 tissue-specific gene sets (one set was defined for each sample in this experiment by taking all genes above absolute expression of 600 and removing genes whose absolute expression was above 600 in >60 of the 122 samples) (87). All data were downloaded on February 2007, except for the miRNAMap data, which were released on July 2005. When processing all these gene sets with Genomica (“Creating Modules”), we ignored sets containing >1,000 genes or <3 (out of genes expressed in the tested samples). Gene sets with >1,000 genes are usually so general as to be biologically irrelevant (for example: protein located in the cytoplasm), and their large size leads to extremely high significance of enrichment, obscuring more focused and relevant data. Gene sets with <3 genes are too biologically focused, usually have no statistical significance, and needlessly complicate the statistical space the algorithm works in. Using such small gene sets would have caused a massive increase in the amount of time necessary to run the algorithm, without noticeably changing the results.
Sample attributes.
To describe the properties of the 700 samples, we used 364 sample attributes, based on the design of the original experiments, using the distinctions made in each original article. Sample attributes can be considered analogous to gene sets, defining groups of samples that share a property instead of groups of genes. Example attributes included: myocardial infarction (MI), MI infarcted region, age, gene knockout. Each series was normalized relative to its own median, and the control samples are unique to each series. Control animals in each series were marked with an attribute of their own. When the response to treatment in a series is present in all treated tissues in the same direction, the treated samples will be highly expressed and the control samples will be expressed at a low level (or vice versa). Because of our normalization, a reaction to treatment that was observed with a similar pattern but not with the same intensity in all experimental samples can be more clearly seen when observing the relative change of the gene expression in the control sample. Such change in the control samples indicates the opposite trend in the noncontrol samples. In some cases, when treatment was tested at two time points, at two dosages, or under two different treatments, it is likely that the response to treatment will not be at the same intensity, even if it is in the same direction. In such cases it becomes more important to look at the expression of the control and observe the relative change in those samples.
Creating modules.
To create modules of genes, we applied the Module Map algorithm (73), as implemented in the program Genomica (http://genomica.weizmann.ac.il, version 2.071206). This algorithm searched for samples that contain gene sets whose expression is significantly induced or repressed relative to the median. Induced or repressed are defined as having a normalized expression value >1 or <−1 (i.e., raw data were at least twice as large as the median or half as small). For each gene set and each sample, the fraction of genes induced or repressed in this gene set for this sample was calculated. The significance of this fraction was calculated using the hypergeometric distribution (92) and corrected with a false discovery rate (FDR) of 5% (85). Each gene set was then associated with an expression fraction - a vector (indexed by samples), where each value is the fraction of genes from this gene set induced or repressed in the appropriate sample. Repressed genes were represented with a negative fraction; induced genes were represented by a positive fraction. The fraction was set to zero if not significant.
Gene sets were clustered based on similarity in their expression fractions (significant induction or repression as calculated before). Each cluster of gene sets had a virtual gene set defined as a union of all gene sets in the cluster. Then, each individual gene in this virtual gene set was separately tested for consistency of direction: is the individual gene induced or repressed in a statistically significant manner in the samples in which this virtual gene set was significantly repressed or induced? A module was created out of the significantly consistent genes, when correcting for multiple testing with FDR of 5%. For each module and each attribute, the fraction of samples belonging to this sample attribute that was induced or repressed in this module was tested for significance, by hypergeometric distribution. Significant attributes were noted as candidates for further analysis and discussion.
The algorithm may be repeated more than once, when each new iteration treats the previously identified modules as existing gene sets and starts over (Fig. 1C). To select the optimal number of iterations, we repeated the algorithm 1–4 times and observed the following: The number of modules decreased sharply with one and two iterations (1,430 & 884 modules, respectively) but then decreased in a much more gradual manner with three and four iterations (742 and 724 modules, respectively). The distribution of attributes in the modules with the different number of iterations remained constant (Supplementary Table S2). Based on these observations, we decided to analyze the 884 modules created after two iterations.
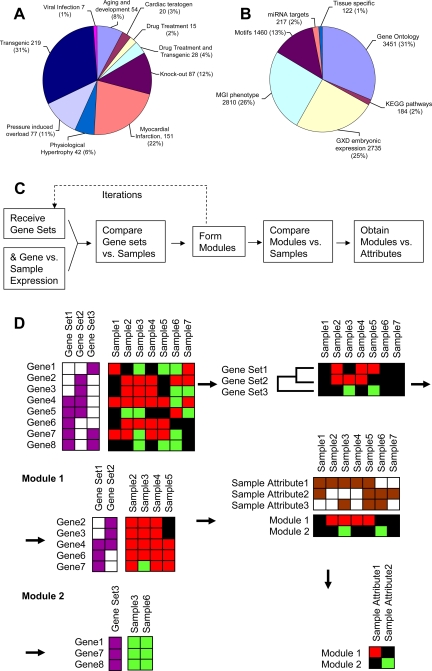
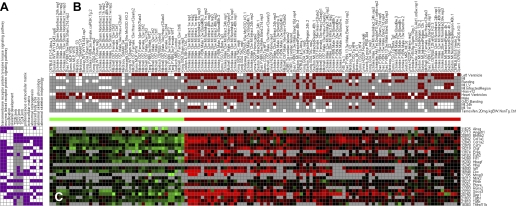
Fig. 1.
Overview of the Module Map analysis procedure. This figure shows the categories of the samples used (A), the gene sets used (B), a flowchart of the algorithm (C), and a graphical representation of the algorithm (D), where red, green, and black represent induced, repressed, and no change, respectively. Purple indicates a gene belongs to a gene set, brown indicates that the sample belongs to a sample set. D is adapted from Segal et al. (73). The algorithm starts with expression values of genes and gene sets (dividing the genes into groups). It uses the expression values of the genes to determine if a gene set is induced or repressed in each sample. Gene sets that have a similar pattern of expression are unified to form modules, for example unifying gene set 1 and gene set 2 to module 1. The modules are formed using only the genes that contributed to the expression of the gene set (gene 5 is not included in module 1, since it was repressed where the other genes in these gene sets were induced). The modules are tested to see in which samples they are repressed or induced. Samples attributes (the sample equivalent of gene sets) that are significantly induced or repressed are noted and presented.
RESULTS
In this study we analyzed expression profiles compiled from 38 series (each comprising biologically related samples from the same experiment, see Supplementary Table S1), measuring the expression of 18,677 mouse genes in 700 samples spanning 10 clinical categories (Fig. 1A). To analyze the 10,978 gene sets (Fig. 1B) we applied the Module Map algorithm (73). This algorithm organizes genes into higher-level modules and identifies attributes (Fig. 1, C and D), representing clinical conditions, in which different modules are induced or repressed. The application of this algorithm resulted in 884 modules.
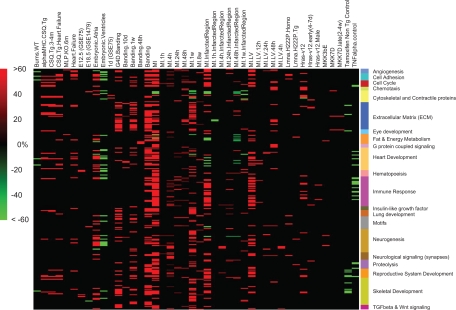
Next, we focused on 266 modules whose genes were differentially expressed in clinical conditions related to MI and pressure-induced overload, conditions that lead eventually to hypertrophy and heart failure (17, 21, 22). The modules relevant to these clinical conditions demonstrated common changes in biological properties such as metabolism of energy and contractile proteins, described below. The relationship between these modules and clinical conditions is shown in Fig. 2 in a matrix form, where each row represents a module, each column represents a sample attribute, and each individual cell represents a module-attribute combination. A positive value indicates that samples with the attribute (the column) were induced in the module (the row), and the value represents the percentage of the induced samples out of all samples in this module-attribute combination. Similarly, a negative value represents the percentage of samples repressed. The color of each cell represents the percentage of its samples that were induced or repressed, where red indicates induced and green indicates repressed. We created a concise label for each module that describes the biological functions of the gene sets associated with it. These labels may be shared between modules, when they have a common biological function, such as fat and energy metabolism (Fig. 3). The rows in the matrix were then grouped by this concise label, which is shown in Fig. 2 in colored bars.
Fig. 2.
Matrix of modules related to myocardial infarction (MI) and pressure-induced overload. This figure presents the 266 modules that had their expression change in the clinical conditions of MI and pressure-induced overload. The figure presents the sample attributes directly related to MI and pressure-induced overload (banding) such as MI infarcted region or banding after 48 h. We also present additional sample attributes that represent a variety of clinical conditions, including embryonic stages, transgenic causes of heart stress and failure (CSQ transgenic, MLP knockout, G4D transgenic), transgenic activation of TNF-α or three MAP kinases (ERK, p38, and JNK). Each row in this matrix represents a module; each column represents a sample attribute. Each cell represents the percentage of samples that were induced (red) or repressed (green) samples out of all samples belonging to one specific module-attribute combination. Color intensity corresponds to the size of the percentage, where black represents zero and indicates that the amount of samples with this attribute that were repressed or induced was not statistically significant. Concise biological labels were added (colored bars, right side), and modules are grouped by these labels.
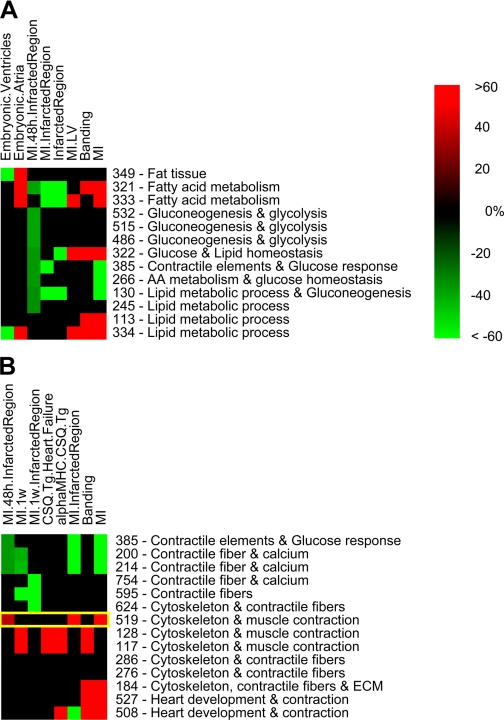
Fig. 3.
Changes in fat metabolism (A) and contractile elements (B) during MI. This figure details the changes in fat metabolism (A) present in MI and pressure-induced overload and the changes in contractile proteins (B) present during MI and heart development. The yellow rectangle indicates module 519. As in Fig. 2, each cell represents the percentage of samples that were induced (red) or repressed (green) samples out of all samples belonging to one specific module-attribute combination. Color intensity corresponds to the size of the percentage, and a color bar is present. The numbers to the right of the figure are the number of the module in our analysis, with a concise biological label for each module.
This matrix allows an overview of the clinical conditions and the pathways activated in them that can provide biological insights even without looking at the specific genes in these modules. For example, the modules whose expression changed in the attribute of Embryonic Atria generally, but not always, change in the opposite direction from the changes portrayed in the attribute (column) of Embryonic Ventricles. Such expression makes sense, since the attributes Embryonic Atria and Embryonic Ventricles are opposites; samples marked with the attribute Embryonic Atria were negative for the attribute of Embryonic Ventricles and vice versa. The modules that were induced and repressed in stressed and failing hearts were related to biological functions including angiogenesis, proteolysis, cell cycle, extracellular matrix (ECM) deposition and remodeling, immune reactions, heart, skeletal, nervous and reproductive system development (Fig. 2). This figure demonstrates a similarity between gene expression patterns of the modules representing MI, banding, and other situations leading to heart stress and failure. The existence of modules with similar pattern of gene expression indicates a common biology between these clinical conditions.
Energy metabolism.
To explore the energy metabolism of the heart under stress we selected a subset of 13 modules, labeled by various aspects of metabolism and energy in a significant manner. These metabolic aspects were represented by multiple gene sets including fat tissue (P < 1E-12), gluconeogenesis and glucose metabolism (P < 1E-9), and lipid metabolism (P < E-10). The results for this subset of modules are shown in Fig. 3A. We can see that modules related to fat metabolism were induced in the embryonic atria and were repressed in the embryonic ventricles. We also see that modules related to metabolism of energy substrates (fat, glucose, and glycogen) were repressed in the infarcted region, and many of these modules were induced in the general attributes of MI, left ventricle of MI, and banding.
Contractile elements.
Heart failure is characterized by many changes in gene expression and protein level, including contractile proteins (5, 21, 59, 67). We further examined in detail a group of 14 modules labeled with cytoskeletal components (P < 1E-8 in all but one module) and contractile elements (P < 1E-7). In Fig. 3B, two trends can be observed: The first trend involves a group of modules (lower part of Fig. 3B) that were induced in MI, banding, and CSQ transgenic animals and that were neither induced nor repressed in the infarcted region in the recent MI. The second trend that can be seen in this figure involves a group of modules (appearing in the upper part of Fig. 3B) that were repressed in MI infarcted region at short-term time points (1 wk and 48 h). This group of modules was not noticeably repressed or induced in other conditions of heart stress such as banding or CSQ transgenic animals.
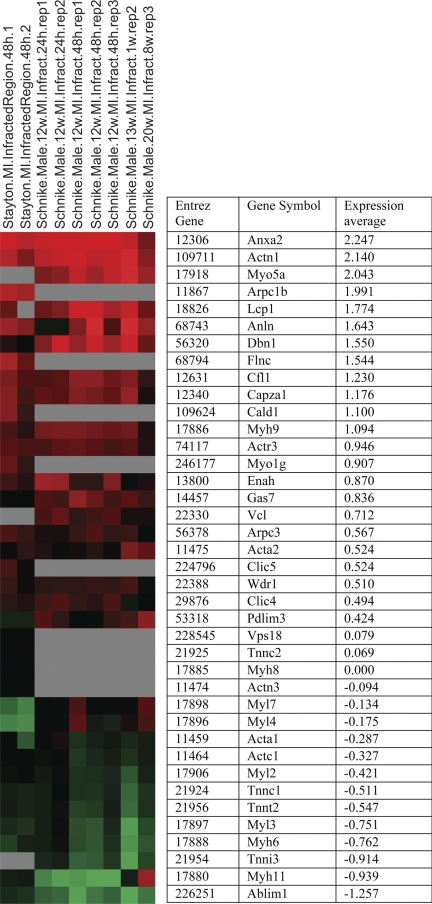
Module 519 (Fig. 3B, yellow rectangle) seems unusual; out of all the modules containing contractile elements in Fig. 3B whose expression changed 48 h after MI, this is the only module that was induced (P = 4E-05). This module is enriched for cytoskeletal and contractile elements with high significance (P value 5E-96 and 1E-30, respectively). The enrichments indicate that this module represents contractile elements, which we would expect to be repressed during the death of cardiac cells. This contradiction can be resolved when reviewing the genes. Module 519 has at least two groups of genes: one group consists of genes that were induced in the infarcted region after MI, while the other group consists of genes that were repressed in this region (Fig. 4). In effect, module 519 represents by itself a mixture of the same two trends of gene expression that are illustrated in Fig. 3B; it includes genes induced in the infarcted region and genes repressed in the infracted region.
Fig. 4.
Expression of genes in module 519 (cytoskeleton and contractile elements). Red indicates positive number (induced vs. median), and green indicates negative expression (repressed). The samples shown include the infarcted region of MI. The average expression of the genes in these samples is written beside the graphical representation. The genes presented are sorted according to their average expression in these samples, which allows us to see the two expression trends: induced in module 519 (leukocyte-related genes) and repressed in this module (heart contractile genes).
Heart stress and matrix formation (module 29).
Figure 2 shows several modules that were labeled with tissues and functions that do not seem related to the heart response, such as skeletal development and reproductive system development. To explain this observation, we focused on a specific module, module 29. We discuss below the genes expressed in this module and explore what role they may have in the cardiac response to MI. Module 29 is induced in samples that have the attributes of (short-term) MI, banding, and Hras (activation of ERK) (P values of 8.82E-9, 1.31E-8, and 3.21E-5, respectively). It is repressed in samples with the attribute of Tamoxifen nontransgenic controls (P = 0.0001). The repression in these control samples is equivalent to induction in activation of all three MAP kinases, apparently more in ERK. The expression patterns in this module and representative gene sets are presented in Fig. 5. The gene sets enriched in module 29 indicate that this module is related to osteogenesis and skeleton formation. The naïve interpretation would be that the heart develops bone-like properties following MI. This conclusion borders on the ridiculous and calls for more investigation, which is present in the discussion.
Fig. 5.
Module 29 (heart stress and matrix formation). This figure shows the gene sets that the genes of this module belong to (A), the attributes that the samples belong to (B), and the expression levels of the genes belonging to this module (C). The red/green bar indicates if the sample was identified as induced or repressed by the algorithm. The expression values are log-normalized, from 3 to −3 (fold change of 8 in the raw data).
DISCUSSION
The results show a similarity in gene expression pattern between MI, banding (representing pressure-induced overload), and other situations leading to heart stress and failure. This suggests a similarity in response, a theory that is supported by the literature, since cardiac hypertrophy and failure follow similar pathways, regardless of the source of the stress (5). Modules enriched in genes related to the immune system were induced in both banding and MI and were repressed in the control samples of TNF-α transgenic animals. Such results may indicate a contamination with immune cells in the transcriptome analysis. However, it is unlikely, since the modules enriched for immune-related genes were induced in a specific manner in MI and banding and were not induced in other clinical conditions (Fig. 2). It is recognized in the literature that immune cells are responsible for the clearance of necrotic cells after MI (17, 25), which directly explains the appearance of immune-related genes in MI. However, the presence of an immune reaction in banding (pressure-induced overload) points to a possible role of the immune cells in ECM remodeling and in the response of the non-infarcted heart tissue to stress. The involvement of immune cells in remodeling is supported by the literature and has been reviewed in previously published studies (18, 47, 98).
Out of the 266 modules presented in Fig. 2, we focused in detail on two groups of modules that are related to energy metabolism and to the expression of contractile proteins. We later summarize module 29, which revealed a possible connection between the biological pathways of osteogenesis and heart failure, connections that were reviewed in detail.
Energy metabolism.
Our results related to energy metabolism (Fig. 3A) indicate that energy sources are utilized in a different manner in embryonic atria and ventricles, with the atria possibly relying more on fat metabolism. Our results also suggest a global decrease in the metabolism of energy in the infarcted tissue after MI, regardless of the source of the energy.
When reviewing the literature, we noted that the normal heart after birth relies mainly on oxidation of fatty acids, while the embryonic heart relies on glycolysis. The contractile performance of the heart is affected by the choice of substrate that serves as an energy source: glucose or fatty acids (83). The energy metabolism in the ventricles is different from the energy metabolism in the atrias both in times of ordinary heart functions and in times of stress. However, the differences between them at rest are not identical to the differences in stressful times (50), indicating the ventricles react to stress differently from the atria. These known differences between the atria and the ventricles might indicate a different maturational process, which can explain the different expression pattern seen between them. It is known that at times of stress, for example chronic MI and pressure overload, the heart shifts to a fetal-like expression profile. This shift changes the expression of metabolism-related genes (67) and is a possible explanation for the correlation seen in our results between the embryonic atria and the MI. This metabolic shift can also serve to explain the changes in metabolism seen after MI and pressure-induced overload. One caveat that should be made is that modules 130, 113, and 334 were labeled in the same manner (lipid metabolic process), but these modules did not behave with the same manner of gene induction or repression. This indicates the complexity of the biological response and the need to review the specific list of genes in each module (see next section for an example of such a review).
The results presented in Fig. 3A show modules 321, 532, 322 and 130, which are repressed in the MI 48 h infarcted region, which does not match the generally known response to MI (an increase in metabolism). This apparent contradiction between our results and the literature can be resolved by remembering that previous works focused on the response in advanced (longer term) heart failure, but that most of the cardiac cells that have died as a result of MI have not yet been replaced 48 h after MI (17). The death of most of these cardiac cells is expected to lower metabolic rates of fat and glucose in the short-term time point.
In summary, the computational analysis confirmed both the difference between the metabolism of the atria and the metabolism of the ventricles, and the different responses in their metabolism to MI, as has been previously explored in the literature. This current study also pointed out the decrease of metabolism in the infarcted region 48 h after MI, a decrease that has not been previously noted in the literature, but that matches our physiological knowledge of MI.
Contractile elements.
Our results related to contractile elements have demonstrated two trends of expression: a short-term decrease of contractile elements following MI and a longer-term increase of contractile elements following heart stress such as transgenic animals (Fig. 3B). The short-term decrease in contractile elements seems to indicate an immediate response to stress due to MI; the cells are shocked and dying and have stopped making contractile elements. The longer-term response indicates that the heart eventually increases the amount of contractile elements. The contractile elements repressed in the short term are not identical to those elements induced in the longer term, as will be seen when module 519 is reviewed below.
A review of the literature highlighted that MI, banding (17, 21, 22), and carrying a transgenic version of CSQ (16, 46) cause the heart to respond with (compensatory) hypertrophy, followed by heart failure. Heart hypertrophy leads to heart failure over a period of time, regardless of the source of the heart stress (5). This known long-term reaction matches the longer-term trend seen in the lower part of Fig. 3B, indicating that this compensatory reaction is the end result of MI. It is also known that the early response to MI is characterized by death of heart tissue, followed later by immune cells clearing the damaged tissue (17, 25). This matches the decrease in contractile element containing modules in the infarcted region of the MI, where these contractile modules were repressed specifically at short-term time points.
Reviewing the roles the genes of module 519 are known to play can shed light on the biology of the two gene expression trends. The group of genes that was induced in the infarcted region and that is shown in the upper region of Fig. 4 is expressed in a variety of tissues. Many of these genes are expressed in leukocytes, and some are only expressed in immune cells (1, 15, 33, 43, 57, 63, 66, 68). This group includes genes related to cell adhesion (33, 54, 63, 99), cell motility (6, 10, 28, 33, 37, 63, 70), cell division (cytokinesis) (26, 37), chemotaxis (68), phagocytosis (1, 33), and tissue remodeling (15, 43). This group of genes represents the short-term response of leukocytes and other noncardiac cells to the death of heart tissue; these cells migrate to the area, clear the dead cardiac cells, and create a scar. This explanation matches the time point in which module 519 was induced, since the immune response is known to begin 2 days after MI (17, 25).
The other group of genes in module 519, shown in the lower region of Fig. 4, includes genes that were repressed in the MI infarcted region. Most of the genes represent heart contractile proteins (34, 52, 53, 56, 71, 72). Some of them are known to also be expressed in other types of muscles (71, 93) but typically play a role in the heart (72). The repression of these genes is expected and probably indicates the damage done to the heart tissue and the subsequent death of cardiomyocytes.
Module 519 contained two groups of genes, each demonstrating a different biological process: initial repression of (cardiac) muscle contractile proteins due to the death of cardiac tissue, as well as reactive induction of cytoskeletal and contractile components related to leukocyte clearance of the damaged tissue, remodeling, and the beginning of scar formation. This demonstrates that modules may include genes belonging to different aspects of a known biological function, where each aspect behaves in a unique manner. Module 519 has a low to medium level of overlap in genes with other modules that were repressed in the short-term reaction to MI. Module 519 also shares a similar level of overlap with the modules in this group, which were neither induced nor repressed in this region. Most of the genes that appear in module 519 as well as in the modules that were repressed in MI are genes related to death of cardiac cells, genes that were repressed in the infarcted region after MI (data not shown).
In summary, Figs. 3B and 4 illustrate the biological reactions to MI—the short-term reaction to the death of infarcted myocardial cells, which includes clearance by leukocytes, and the long-term reaction of scar formation and hypertrophy. Figure 3 also demonstrates the strength and utility of the Module Map method; we have reconstructed part of the biological response to MI using only bioinformatic analysis, which was not based on any a priori biological knowledge of these biological processes.
Heart stress and matrix formation (module 29).
Data from the literature on embryonic development teach us that the heart (27) and the skeletal (32) and the reproductive/urinary (36) systems are all derived and develop from the mesoderm germ layer. Their development includes complex interactions of cells with other cells and with the ECM, interactions that are regulated by multiple cytokines (32, 36, 81). Since they share the same embryonic origin, it is logical that modules marked as skeletal development may appear in the heart. The osteogenesis imperfecta cases typically show cardiovascular abnormalities. Additionally, some of the connective tissue disorders may involve the heart and valvular tissue (e.g., Marfan's syndrome, Ehlers-Danlos syndrome, and osteogenesis imperfect). We have also recently performed transcriptome experiments to analyze the potential and plasticity of mesenchymal cells and the connection between bone marrow-derived mesenchymal cells, cardiac muscle, and skeletal muscle. The results have showed similarity and differences in expression pattern that confirmed the shared origin of the cells as well as the differences between myogenic and stromal cells (2, 3). Module 29 is an example of a module that is labeled with functions relevant to bone development but is expressed in the heart. Since the possibility of bone formation in the heart seems unlikely, we went over the genes in this module in depth. When we reviewed the genes in this module we noticed that this module includes genes known to have a role in aspects of heart stress, including tissue remodeling (13) and hypertrophy (58, 88). Example genes are collagen I and II (5, 95), fibronectin (42), Tgfbi (14, 29, 60), and osteopontin, an important protein in bone (20) that is necessary for heart remodeling (62). Many of the other genes in this module are induced by transforming growth factor-β (24, 39, 78), a very important factor in heart remodeling, hypertrophy, and fibrosis (12).
In summary, the results presented in module 29 have demonstrated the futility of blindly accepting the bioinformatic results and the utility of the Module Map method in discovering interesting groups of genes that may have unexpected properties. Accepting the bioinformatic results, at face value, without reviewing the list of genes would have led us to the unlikely conclusion that MI causes the heart to develop bone or bone-like properties. However, reviewing the list of bone-related genes in detail revealed the role most of these genes have in the response to cardiac stress, in addition to their role in skeletal development. The listed genes also included three bone-related genes, Bhlbh2, Runx2, and Ahsg. Bhlb2, also called Dec1, is a gene related to the circadian clock (96), which is expressed in multiple tissues. It is known to be expressed in the heart of the developing (11) as well as the mature animal (69). This gene serves as a regulator in multiple pathways including chondrogenic (77) and osteogenic (41) differentiation of mesenchymal stem cells, lymphocyte activation (75), and cell cycle (51, 75, 89). It is repressed in atrial fibrillation (49), but its exact role in the heart is unknown. There are at least two possible explanations for the presence of this gene in the heart: it may be related to a response of the heart cells to stress or to the attempt of the leukocytes to clear the damaged tissue. The other two bone-related genes are more interesting, since they are not normally expressed in the heart. Runx2, also called Cbfa1, is a transcription factor expressed along the development of bone (23, 64). However, this gene is only known to play a role in the epithelial-to-mesenchymal development (31) and the pathology (30) of heart valves. α-2-HS-glycoprotein (Ahsg, Fetuin) is a serum protein produced by the liver that is not expressed by the normal heart (19). This protein inhibits calcification in small blood vessels (58) and may have a role in regulating hypertrophy (82) and phagocytosis (44). Low levels of fetuin expression have been associated with cardiovascular failure in patients with kidney disease (61, 84, 94). However, these articles assessed the level of circulating serum fetuin protein. Our research is unique in pointing out the expression of fetuin mRNA in the stressed heart; previous work has shown fetuin expression only by the liver (19). The expression of these two genes in the heart samples raised interesting biological hypotheses, that Runx2 is involved in heart stress and that Ahsg is produced by the stressed heart. These hypotheses, in turn, raised the following questions: What is the role of Runx2 in the stressed heart? Which cell in the stressed heart produces Ahsg? The specific involvement of these genes is yet unknown. These hypotheses and unanswered questions merit further inquiry, which will require “wet biology” to fully elucidate.
Conclusions
We performed an analysis of expression trends in 700 samples that cover many physiological conditions of the heart. Utilizing the Module Map algorithm we identified 884 modules, which were based on biological functions and correlated to clinical conditions. Observing the trends of gene expression in various modules has demonstrated interesting biological phenomena, some of which were only partially known previously from the literature. One must note that the module creating algorithm had no biological knowledge of the “rules” governing the samples it was given to analyze, and yet biologically valid processes were highlighted when we discussed modules at a variety of levels, from a large group (266 modules) to medium-size groups to single modules, whose expression changes in MI and banding recapitulated the known connection between the immune reaction and remodeling. This was noted only by comparing the general gene expression of the modules, without deeper reviewing of the specific biological processes expressed and their expression levels. Focusing on the general expression of modules related to energy metabolism and contractile proteins demonstrated biologically significant changes in these two biological functions, highlighting the differences between the energy metabolism of the atria and the metabolism of the ventricles, as well as two different trends in the expression of contractile proteins. Reviewing module 519, as an example of an interesting module from these groups, explained the two different expression trends and recapitulated some of the heart's known responses to MI. Thus, the Module Map algorithm correctly reconstructed biological trends and processes, without a priori knowledge of the biological principles of heart failure. Module 29 was interesting biologically, expressing genes that are related to osteogenesis (bone formation), a biological process not directly related to heart failure. The analysis of module 29 highlighted the genes Runx2 and Ahsg, known to be related to osteogenesis but with a so far unknown role in cardiac stress. Viewing this possible new involvement of these genes in osteogenesis raises interesting biological questions, which can be summarized as “What is the additional role of these two genes? Where they are secreted? Which cell secretes them and what effect do they have?
We have demonstrated the power of reconstructing biological phenomena, using bioinformatics to profile of various cardiac physiological conditions. The Module Map algorithm generates hypotheses that were previously unknown and that need further confirmation by additional experiments. The computational algorithm needs to be supported with experimental results, but the computational methods are of undeniable importance in bringing new view and insights to already existing data.
GRANTS
The study was supported by the CellPRoM grant from the 6th Framework Programme of the European Commission and Schlezak grant from Tel-Aviv Univ. to D. Benayahu.
Supplementary Material
Acknowledgments
Authors' contributions: U. D. Akavia processed the original data, carried out the computational analysis, and performed the literature review. D. Benayahu conceived of the study and coordinated it. Both authors helped to draft the manuscript and read and approved the final manuscript.
This study is a partial fulfillment of U. D. Akavia's requirements toward his PhD thesis at Tel Aviv University, Israel.
Address for reprint requests and other correspondence: D. Benayahu, Dept. of Cell and Developmental Biology, Sackler School of Medicine, Tel-Aviv Univ., Tel-Aviv 66978, Israel (e-mail: dafnab@post.tau.ac.il).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Adachi R, Takeuchi K, Suzuki K. Antisense oligonucleotide to cofilin enhances respiratory burst and phagocytosis in opsonized zymosan-stimulated mouse macrophage J774.1 cells. J Biol Chem 277: 45566–45571, 2002. [DOI] [PubMed] [Google Scholar]
- 2.Akavia UD, Shur I, Rechavi G, Benayahu D. Transcriptional profiling of mesenchymal stromal cells from young and old rats in response to Dexamethasone. BMC Genomics 7: 95, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akavia UD, Veinblat O, Benayahu D. Comparing the transcriptional profile of mesenchymal cells to cardiac and skeletal muscle cells. J Cell Physiol 216: 663–672, 2008. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babick AP, Dhalla NS. Role of subcellular remodeling in cardiac dysfunction due to congestive heart failure. Med Princ Pract 16: 81–89, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Bailly M Connecting cell adhesion to the actin polymerization machinery: vinculin as the missing link? Trends Cell Biol 13: 163–165, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res 35: D760–765, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, Kaab S, Kreuzer E, Steinbeck G, Mansmann U, Poustka A, Nabauer M, Sultmann H. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. J Am Coll Cardiol 48: 1610–1617, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biology 2: e9, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bois PRJ, O'Hara BP, Nietlispach D, Kirkpatrick J, Izard T. The vinculin binding sites of talin and alpha-actinin are sufficient to activate vinculin. J Biol Chem 281: 7228–7236, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev 11: 2052–2065, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bujak M, Frangogiannis NG. The role of TGF-(beta) signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res 74: 184–195, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butt RP, Laurent GJ, Bishop JE. Collagen production and replication by cardiac fibroblasts is enhanced in response to diverse classes of growth factors. Eur J Cell Biol 68: 330–335, 1995. [PubMed] [Google Scholar]
- 14.Cao W, Tan P, Lee CH, Zhang H, Lu J. A transforming growth factor-beta-induced protein stimulates endocytosis and is up-regulated in immature dendritic cells. Blood 107: 2777–2785, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Mocsai A, Zhang H, Ding RX, Morisaki JH, White M, Rothfork JM, Heiser P, Colucci-Guyon E, Lowell CA, Gresham HD, Allen PM, Brown EJ. Role for plastin in host defense distinguishes integrin signaling from cell adhesion and spreading. Immunity 19: 95–104, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Cho MC, Rapacciuolo A, Koch WJ, Kobayashi Y, Jones LR, Rockman HA. Defective beta-adrenergic receptor signaling precedes the development of dilated cardiomyopathy in transgenic mice with calsequestrin overexpression. J Biol Chem 274: 22251–22256, 1999. [DOI] [PubMed] [Google Scholar]
- 17.Cotran RS, Kumar V, Collins T, Robbins SL. Robbins Pathologic Basis of Disease. Philadelphia, PA: Saunders, 1999, p. xv.
- 18.Damas J, Gullestad L, Aukrust P. Cytokines as new treatment targets in chronic heart failure. Curr Controlled Trials Cardiovasc Med 2: 271–277, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denecke B, Graber S, Schafer C, Heiss A, Woltje M, Jahnen-Dechent W. Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem J 376: 135–145, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denhardt D, Guo X. Osteopontin: a protein with diverse functions. FASEB J 7: 1475–1482, 1993. [PubMed] [Google Scholar]
- 21.Ding B, Price RL, Borg TK, Weinberg EO, Halloran PF, Lorell BH. Pressure overload induces severe hypertrophy in mice treated with cyclosporine, an inhibitor of calcineurin. Circ Res 84: 729–734, 1999. [DOI] [PubMed] [Google Scholar]
- 22.Dorn GW, Robbins J, Ball N, Walsh RA. Myosin heavy chain regulation and myocyte contractile depression after LV hypertrophy in aortic-banded mice. Am J Physiol Heart Circ Physiol 267: H400–H405, 1994. [DOI] [PubMed] [Google Scholar]
- 23.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89: 747–754, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Duncan MR, Frazier KS, Abramson S, Williams S, Klapper H, Huang X, Grotendorst GR. Connective tissue growth factor mediates transforming growth factor beta-induced collagen synthesis: down-regulation by cAMP. FASEB J 13: 1774–1786, 1999. [PubMed] [Google Scholar]
- 25.Eaton L, Bulkley B. Expansion of acute myocardial infarction: its relationship to infarct morphology in a canine model. Circ Res 49: 80–88, 1981. [DOI] [PubMed] [Google Scholar]
- 26.Echard A, Hickson GRX, Foley E, O'Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol 14: 1685–1693, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenberg LM Belief vs scientific observation: the curious story of the precardiac mesoderm. Anatomical Record 266: 194–197, 2002. [DOI] [PubMed] [Google Scholar]
- 28.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol 9: 299–309, 2007. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson JW, Mikesh MF, Wheeler EF, LeBaron RG. Developmental expression patterns of Beta-ig (betaIG-H3) and its function as a cell adhesion protein. Mech Dev 120: 851–864, 2003. [DOI] [PubMed] [Google Scholar]
- 30.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature 437: 270–274, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Gitler AD, Lu MM, Jiang YQ, Epstein JA, Gruber PJ. Molecular markers of cardiac endocardial cushion development. Dev Dyn 228: 643–650, 2003. [DOI] [PubMed] [Google Scholar]
- 32.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem 97: 33–44, 2006. [DOI] [PubMed] [Google Scholar]
- 33.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 7: 713–726, 2006. [DOI] [PubMed] [Google Scholar]
- 34.Gulick J, Subramaniam A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem 266: 9180–9185, 1991. [PubMed] [Google Scholar]
- 35.Hong SE, Rho SH, Yeom YI, Kim DH. HCNet: a database of heart and calcium functional network. Bioinformatics 22: 2053–2054, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Horster MF, Braun GS, Huber SM. Embryonic renal epithelia: induction, nephrogenesis, and cell differentiation. Physiol Rev 79: 1157–1191, 1999. [DOI] [PubMed] [Google Scholar]
- 37.Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell 16: 649–664, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res 34: D135–139, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ignotz R, Massague J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem 261: 4337–4345, 1986. [PubMed] [Google Scholar]
- 40.Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377, 2002. [DOI] [PubMed] [Google Scholar]
- 41.Iwata T, Kawamoto T, Sasabe E, Miyazaki K, Fujimoto K, Noshiro M, Kurihara H, Kato Y. Effects of overexpression of basic helix-loop-helix transcription factor Dec1 on osteogenic and adipogenic differentiation of mesenchymal stem cells. Eur J Cell Biol 85: 423–431, 2006. [DOI] [PubMed] [Google Scholar]
- 42.Jane-Lise S, Corda S, Chassagne C, Rappaport L. The extracellular matrix and the cytoskeleton in heart hypertrophy and failure. Heart Fail Rev 5: 239–250, 2000. [DOI] [PubMed] [Google Scholar]
- 43.Janji B, Giganti A, De Corte V, Catillon M, Bruyneel E, Lentz D, Plastino J, Gettemans J, Friederich E. Phosphorylation on Ser5 increases the F-actin-binding activity of L-plastin and promotes its targeting to sites of actin assembly in cells. J Cell Sci 119: 1947–1960, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Jersmann HP, Dransfield I, Hart SP. Fetuin/alpha2-HS glycoprotein enhances phagocytosis of apoptotic cells and macropinocytosis by human macrophages. Clin Sci (Lond) 105: 273–278, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Jessup M, Brozena S. Heart failure. N Engl J Med 348: 2007–2018, 2003. [DOI] [PubMed] [Google Scholar]
- 46.Jones LR, Suzuki YJ, Wang W, Kobayashi YM, Ramesh V, Franzini-Armstrong C, Cleemann L, Morad M. Regulation of Ca2+ signaling in transgenic mouse cardiac myocytes overexpressing calsequestrin. J Clin Invest 101: 1385–1393, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kai H, Kuwahara F, Tokuda K, Imaizumi T. Diastolic dysfunction in hypertensive hearts: roles of perivascular inflammation and reactive myocardial fibrosis. Hypertens Res 28: 483–490, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34: D354–D357, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kharlap MS, Timofeeva AV, Goryunova LE, Khaspekov GL, Dzemeshkevich SL, Ruskin VV, Akchurin RS, Golitsyn SP, Beabealashvilli R. Atrial appendage transcriptional profile in patients with atrial fibrillation with structural heart diseases. Ann NY Acad Sci 1091: 205–217, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Laser A, Ingwall JS, Tian R, Reis I, Hu K, Gaudron P, Ertl G, Neubauer S. Regional biochemical remodeling in non-infarcted tissue of rat heart post-myocardial infarction. J Mol Cell Cardiol 28: 1531–1538, 1996. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Zhang H, Xie M, Hu M, Ge S, Yang D, Wan Y, Yan B. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem J 367: 413–422, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lompre A, Nadal-Ginard B, Mahdavi V. Expression of the cardiac ventricular alpha- and beta-myosin heavy chain genes is developmentally and hormonally regulated. J Biol Chem 259: 6437–6446, 1984. [PubMed] [Google Scholar]
- 53.Lyons G, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol 111: 2427–2436, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, Burlingame AL, Hsuan JJ, Segal AW. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J 328: 105–112, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet 36: 197–204, 2004. [DOI] [PubMed] [Google Scholar]
- 56.McHugh KM, Crawford K, Lessard JL. A comprehensive analysis of the developmental and tissue-specific expression of the isoactin multigene family in the rat. Dev Biol 148: 442–458, 1991. [DOI] [PubMed] [Google Scholar]
- 57.Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279: 527–533, 1998. [DOI] [PubMed] [Google Scholar]
- 58.Merx MW, Schafer C, Westenfeld R, Brandenburg V, Hidajat S, Weber C, Ketteler M, and Jahnen-Dechent W. Myocardial stiffness, cardiac remodeling, and diastolic dysfunction in calcification-prone fetuin-A-deficient mice. J Am Soc Nephrol 16: 3357–3364, 2005. [DOI] [PubMed] [Google Scholar]
- 59.Morkin E Control of cardiac myosin heavy chain gene expression. Microsc Res Tech 50: 522–531, 2000. [DOI] [PubMed] [Google Scholar]
- 60.Norris RA, Kern CB, Wessels A, Wirrig EE, Markwald RR, Mjaatvedt CH. Detection of betaig-H3, a TGFbeta induced gene, during cardiac development and its complementary pattern with periostin. Anat Embryol (Berl) 210: 13–23, 2005. [DOI] [PubMed] [Google Scholar]
- 61.Oikawa O, Higuchi T, Yamazaki T, Yamamoto C, Fukuda N, Matsumoto K. Evaluation of serum fetuin-A relationships with biochemical parameters in patients on hemodialysis. Clinical Experimental Nephrology 11: 304–308, 2007. [DOI] [PubMed] [Google Scholar]
- 62.Okamoto H Osteopontin and cardiovascular system. Mol Cell Biochem 300: 1–7, 2007. [DOI] [PubMed] [Google Scholar]
- 63.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton 58: 104–111, 2004. [DOI] [PubMed] [Google Scholar]
- 64.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771, 1997. [DOI] [PubMed] [Google Scholar]
- 65.Pepper S, Saunders E, Edwards L, Wilson C, Miller C. The utility of MAS5 expression summary and detection call algorithms. BMC Bioinformatics 8: 273, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pierce RA, Field ED, Mutis T, Golovina TN, Kap-Herr CV, Wilke M, Pool J, Shabanowitz J, Pettenati MJ, Eisenlohr LC, Hunt DF, Goulmy E, Engelhard VH. The HA-2 minor histocompatibility antigen is derived from a diallelic gene encoding a novel human class I myosin protein. J Immunol 167: 3223–3230, 2001. [DOI] [PubMed] [Google Scholar]
- 67.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation 104: 2923–2931, 2001. [DOI] [PubMed] [Google Scholar]
- 68.Rey M, Vicente-Manzanares M, Viedma F, Yanez-Mo M, Urzainqui A, Barreiro O, Vazquez J, Sanchez-Madrid F. Cutting edge: association of the motor protein nonmuscle myosin heavy chain-IIA with the C terminus of the chemokine receptor CXCR4 in T lymphocytes. J Immunol 169: 5410–5414, 2002. [DOI] [PubMed] [Google Scholar]
- 69.Rossner MJ, Dorr J, Gass P, Schwab MH, Nave KA. SHARPs: mammalian enhancer-of-split- and hairy-related proteins coupled to neuronal stimulation. Mol Cell Neurosci 9: 460–475, 1997. [DOI] [PubMed] [Google Scholar]
- 70.Sandquist JC, Swenson KI, DeMali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem 281: 35873–35883, 2006. [DOI] [PubMed] [Google Scholar]
- 71.Schiaffino S, Gorza L, Ausoni S. Troponin isoform switching in the developing heart and its functional consequences. Trends Cardiovasc Med 3: 12–17, 1993. [DOI] [PubMed] [Google Scholar]
- 72.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev 76: 371–423, 1996. [DOI] [PubMed] [Google Scholar]
- 73.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet 36: 1090–1098, 2004. [DOI] [PubMed] [Google Scholar]
- 74.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176, 2003. [DOI] [PubMed] [Google Scholar]
- 75.Seimiya M, Bahar R, Wang Y, Kawamura K, Tada Y, Okada S, Hatano M, Tokuhisa T, Saisho H, Watanabe T, Tagawa M, O-Wang J. Clast5/Stra13 is a negative regulator of B lymphocyte activation. Biochem Biophys Res Commun 292: 121–127, 2002. [DOI] [PubMed] [Google Scholar]
- 76.Sharma UC, Pokharel S, Evelo CTA, Maessen JG. A systematic review of large scale and heterogeneous gene array data in heart failure. J Mol Cell Cardiol 38: 425–432, 2005. [DOI] [PubMed] [Google Scholar]
- 77.Shen M, Yoshida E, Yan W, Kawamoto T, Suardita K, Koyano Y, Fujimoto K, Noshiro M, Kato Y. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J Biol Chem 277: 50112–50120, 2002. [DOI] [PubMed] [Google Scholar]
- 78.Smith-Mungo LI, Kagan HM. Lysyl oxidase: properties, regulation and multiple functions in biology. Matrix Biol 16: 387–398, 1998. [DOI] [PubMed] [Google Scholar]
- 79.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 6: R7, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res 35: D618–D623, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith TK, Bader DM. Signals from both sides: control of cardiac development by the endocardium and epicardium. Sem Cell Dev Biol 18: 84–89, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivas P, Wagner A, Reddy L, Deutsch D, Leon M, Goustin A, Grunberger G. Serum alpha 2-HS-glycoprotein is an inhibitor of the human insulin receptor at the tyrosine kinase level. Mol Endocrinol 7: 1445–1455, 1993. [DOI] [PubMed] [Google Scholar]
- 83.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev 85: 1093–1129, 2005. [DOI] [PubMed] [Google Scholar]
- 84.Stenvinkel P, Wang K, Qureshi AR, Axelsson J, Pecoits-Filho R, Gao P, Barany P, Lindholm B, Jogestrand T, Heimburger O, Holmes C, Schalling M, Nordfors L. Low fetuin-A levels are associated with cardiovascular death: impact of variations in the gene encoding fetuin. Kidney Int 67: 2383–2392, 2005. [DOI] [PubMed] [Google Scholar]
- 85.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255, 2003. [DOI] [PubMed] [Google Scholar]
- 87.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA 99: 4465–4470, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suleiman MS, Singh RJ, Stewart CE. Apoptosis and the cardiac action of insulin-like growth factor I. Pharmacol Ther 114: 278–294, 2007. [DOI] [PubMed] [Google Scholar]
- 89.Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci USA 97: 4058–4063, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanay A, Sharan R, Kupiec M, Shamir R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc Natl Acad Sci USA 101: 2981–2986, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanay A, Sharan R, Shamir R. Discovering statistically significant biclusters in gene expression data. Bioinformatics 18: S136–144, 2002. [DOI] [PubMed] [Google Scholar]
- 92.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet 22: 281–285, 1999. [DOI] [PubMed] [Google Scholar]
- 93.Vandekerckhove J, Bugaisky G, Buckingham M. Simultaneous expression of skeletal muscle and heart actin proteins in various striated muscle tissues and cells. A quantitative determination of the two actin isoforms. J Biol Chem 261: 1838–1843, 1986. [PubMed] [Google Scholar]
- 94.Wang AYM, Woo J, Lam CWK, Wang M, Chan IHS, Gao P, Lui SF, Li PKT, Sanderson JE. Associations of serum fetuin-A with malnutrition, inflammation, atherosclerosis and valvular calcification syndrome and outcome in peritoneal dialysis patients. Nephrol Dial Transplant 20: 1676–1685, 2005. [DOI] [PubMed] [Google Scholar]
- 95.Weber KT, Janicki JS, Shroff SG, Pick R, Chen RM, Bashey RI. Collagen remodeling of the pressure-overloaded, hypertrophied nonhuman primate myocardium. Circ Res 62: 757–765, 1988. [DOI] [PubMed] [Google Scholar]
- 96.Wu T, Jin Y, Ni Y, Zhang D, Kato H, Fu Z. Effects of light cues on re-entrainment of the food-dominated peripheral clocks in mammals. Gene 419: 27–34, 2008. [DOI] [PubMed] [Google Scholar]
- 97.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu Q, Watson RR, Marchalonis JJ, Larson DF. A role for T lymphocytes in mediating cardiac diastolic function. Am J Physiol Heart Circ Physiol 289: H643–H651, 2005. [DOI] [PubMed] [Google Scholar]
- 99.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci 116: 4605–4613, 2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.