SUMMARY
We utilized a red chromophore formation pathway, in which the anionic red chromophore is formed from the neutral blue intermediate, to suggest a novel rational design strategy to develop blue fluorescent proteins with a tyrosine-based chromophore. The strategy was applied to red fluorescent proteins of the different genetic background such as TagRFP, mCherry, HcRed1, M355NA, and mKeima, which were converted into blue probes. Further improvement of a blue variant of TagRFP using random mutagenesis resulted in an enhanced monomeric protein, mTagBFP, characterized by substantially higher brightness, faster chromophore maturation and higher pH stability than blue fluorescence proteins with a histidine in the chromophore. Detailed biochemical and photochemical analysis indicates mTagBFP is the true monomeric protein tag for multicolor and lifetime imaging as well as the outstanding donor for green fluorescent proteins in FRET applications.
INTRODUCTION
Green fluorescent protein (GFP) from Aequoria victoria and its homologues have been cloned from various marine organisms. A number of optimized blue, cyan and yellow mutants of GFP have been also developed [1]. While the cyan and yellow variants exhibit enhanced photochemical properties, existing blue fluorescent protein (BFP) variants are characterized by moderate intrinsic brightness (defined as the product of extinction coefficient and quantum yield) and low photostability. Recently, two new improved BFP variants such as Azurite and EBFP2 were introduced [2,3]. Both Azurite and EBFP2 exhibited significant improvement in the photostability compared to original BFP (GFP with Y66H/Y145F substitutions) and EBFP (GFP with F64L/Y66H/Y145F/V163A substitutions) [2,3]. The 1.6-fold and 2-fold enhancement in the brightness were achieved in Azurite and EBFP2, respectively. However, the brightness still was 0.43 and 0.56 of that of widely used green fluorescent probes such as TagGFP or EGFP [1].
The improvement in the brightness was mainly due to the increase in the quantum yields but not in the molar extinction coefficients. Because all BFPs have His66 in the chromophore we speculated that their low absorbance would be attributed to this residue, and its change to Tyr66, which is typically observed in green and yellow variants such as EGFP and EYFP, could increase the extinction coefficient. However, earlier attempts to design BFP variant with Tyr66 in a protonated state on the basis of GFPs by blocking an excited-state proton transfer (ESPT) pathway, and in this way preventing the GFP chromophore from deprotonation, had only limited success such as in the case of mKalama1 [3] and mutants of PSCFP [4]. In both cases the mutants had the brightness and/or photostability lower than that of EBFP2. Therefore we turned our attention to red fluorescent proteins with anionic chromophores where ESPT has not been observed [5, 6]. Earlier we have shown that the DsRed-like red chromophore is mainly formed from a blue protonated GFP-like chromophore as an intermediate [7]. The intermediate has been detected during the maturation of several tetrameric red-shifted proteins such as DsRed, hsriCP and asulCP. We speculated that the same red chromophore formation mechanism should be preserved in other RFPs. Here, we use a structure-based directed evolution in combination with random mutagenesis to develop monomeric blue fluorescent proteins on the basis of the red fluorescent proteins of different genetic background such as TagRFP, mCherry, HcRed1, M355NA and mKeima.
RESULTS AND DISCUSSION
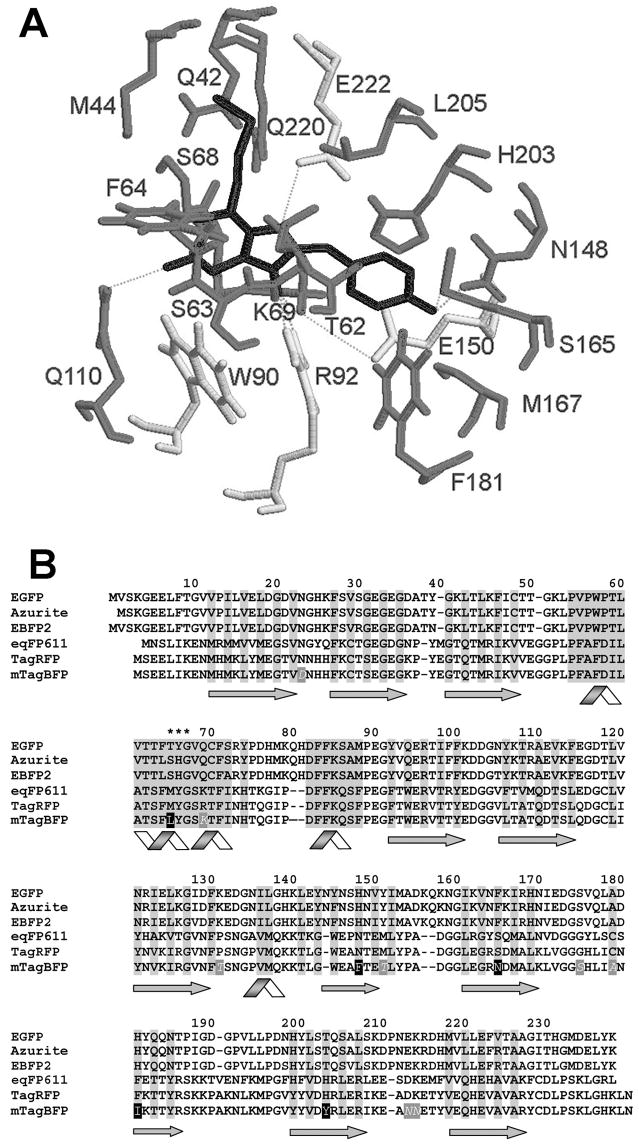
We selected several monomeric RFPs such as TagRFP [8], mCherry [1], mKeima [9] and tetrameric red FPs HcRed1 [10] and M355NA [11] to introduce site-specific mutations preventing maturation of the chromophore beyond the blue protonated intermediate and also stabilizing it. Our primary choice of positions for amino acid substitutions was based on several crystal structures including those for eqFP611 [12], which shares 76% homology with TagRFP [8] and has similar chromophore environment (Figure 1A), mCherry [13], HcRed [14], and KFP [15] variant of asulCP, which is the precursor for M355NA.
Figure 1.
(A) Immediate environment of the chromophore in eqFP611 [12]. The chromophore is showed in black, the conserved amino acid residues are in light gray, and non-conserved residues are in gray. The hydrogen bonds are indicated with dashed lines. (B) Amino acid sequence alignment of mTagBFP with EGFP, Azurite, EBFP2, eqFP611 and TagRFP. Structurally important regions are highlighted in grey, beta-strands are shown with arrows, alpha-helixes are shown with ribbons. The chromophore forming residues are marked with asterisks. Site-specific mutations resulted in conversion of TagRFP into a blue fluorescent predecessor of mTagBFP are shown white on black background. Mutations generated in the course of random mutagenesis are shown white italic on dark grey background. The alignment numbering follows that for EGFP.
To convert RFPs into BFPs we first needed to stabilize the tyrosine-based chromophore in a protonated (neutral) state. In all known RFPs position 65 (numbering is in accordance with EGFP alignment, see Figure 1B and Figure S1 online) is occupied with Met, Gln or Glu. We hypothesized that formation of a double acylamine bond specific for the red anionic chromophore would be less probable in the case of totally aliphatic or aromatic side chains, which could cause less polarization of the 65αC-H bond in the chromophore [6]. Positions 148 and 165 are located in the close proximity to the planar chromophores in RFPs. The side chains of residues at the positions 148 and/or 165 form hydrogen bonds with oxygen of the chromophore phenolate and thus stabilize the anionic form of the red chromophore. Substitutions at these positions could lead to prevention of the hydrogen bond formation with phenolate anion that can be favorable for formation of neutral blue chromophore. The side chain of the amino acid residue in the position 181 forms van der Waals contacts with multiple amino acids which are necessary for the formation and stabilization of the chromophore. In eqFP611 [12] and in the suggested fluorescent state of KFP [15] the hydroxyphenyl group of the red chromophore is coplanar with His203 and forms with it π-π interactions. It also develops the hydrogen bonds with Glu150 and Glu222. The latter one is possibly involved in the catalytic process of the formation of DsRed-like chromophore [16]. In the mCherry and HcRed structures the position 203 is occupied with Ile, which contacts chromophore and its surrounding residues. Therefore substitutions at position 203 would simultaneously stabilize the neutral blue chromophore intermediate and possibly cause change in the hydrogen bonding with Glu222.
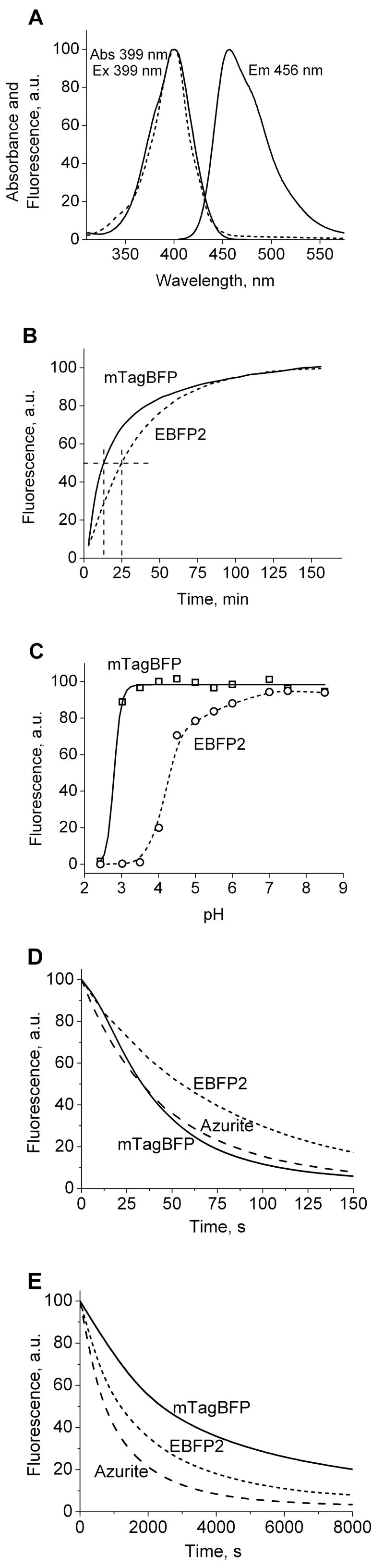
First we tested our structure-based design to convert TagRFP into BFP. Based on the above analysis we performed the site-specific saturated mutagenesis of Met65, Asn148, Ser165, Phe181 and His203 residues of TagRFP using the overlap extension approach [17]. The bacterial library consisting of about 3×107 independent clones (theoretical size was 3×106) has been screened with the fluorescence-activated cell sorter (FACS) followed by selection of the best clones on Petri dishes using fluorescence stereomicroscope. The screening has identified several blue mutants with a blue emission when excited either with 407 nm krypton laser line on FACS or with 390/40 nm filter. Compared to TagRFP the mutants had M65L,H,F/N148F,I,W/S165N,I,A,T/F181I,A,V/H203Y,F substitutions. To increase its brightness and photostability we next used the mixture of the mutants to perform several rounds of random mutagenesis using an error-prone polymerase chain reaction. Bacterial libraries of 5×106–2×107 clones were applied to the high-throughput screening using FACS. After each round 20–50 brightest blue clones with minimal green and red fluorescence have been analyzed by sequencing, and mixture of several best variants were then used as a template for the next round of mutagenesis. If during screening the external mutations at the β-can dimerizing interfaces were observed, such as V106M, T108I, R126I, and K182N [8], they were reversed by the site-specific mutagenesis. After five sequential rounds of random mutagenesis and FACS screening an enhanced bright blue mutant, named mTagBFP, with additional 8 amino acid substitutions was found. Among them R69K/C179A were internal, and N23D/P131T/M151T/G175S/D213N/K214N were external to the β-can (Figure 1B). The mTagBFP exhibited the excitation/emission peaks at 399/456 nm, respectively (Figure 2A and Table 1).
Figure 2.
Spectral, biochemical and photobleaching properties of the purified mTagBFP. (A) Absorbance, excitation and emission spectra of mTagBFP. (B) Maturation kinetics for mTagBFP and EBFP2. (C) pH dependence for mTagBFP and EBFP2. (D) Photobleaching curves for purified mTagBFP (solid line), EBFP2 (short dashed line) and Azurite (dashed line) under mercury arc lamp illumination. (E) Photobleaching curves for mTagBFP (solid line), EBFP2 (short dashed line) or Azurite (dashed line) expressed in HeLa cells using 405 nm laser scanning. Data represents an average of 8–10 cells per each protein. The time axes in D and E represent the normalized imaging time with an initial emission rate of 1,000 photons/second per molecule.
Table 1.
Properties of mTagBFP in comparison with BFPs containing a histidine in the chromophore.
| Protein | Excitation maximum, nm |
Emission maximum, nm |
Extinction coefficient, M−1cm−1 |
Quantum yield |
Brightness relative to EBFP2 |
Effective pKa |
Fluorescence lifetime, ns |
Maturation half-time, min |
t1/2 bleach (arc lamp), s |
t1/2 bleach (confocal), s |
|---|---|---|---|---|---|---|---|---|---|---|
| EBFP | 383 | 445 | 27,100 [2] | 0.34 [2] | 0.51 | 6.3 [3] | n.d. | n.d. | n.d. | n.d. |
| Azurite | 383 | 447 | 26,200 [2] | 0.55 [2] | 0.80 | 5.0 [3] | 3.4 ± 0.2 | 18 ± 2 | 33 | 710 |
| EBFP2 | 383 | 448 | 32,000 [3] | 0.56 [3] | 1.00 | 4.5 ± 0.2 | 3.0 ± 0.2 | 25 ± 2 | 55 | 1,140 |
| mTagBFP | 399 | 456 | 52,000 ± 500 | 0.63 ± 0.02 | 1.82 | 2.7 ± 0.2 | 2.6 ± 0.1 | 13 ± 2 | 34 | 2,380 |
Next we performed the multiple site-specific mutagenesis of mCherry, HcRed1, M355NA and mKeima. Because during the screening of TagRFP libraries we observed its blue variants with 69R,K and 84W,L,F residues we added these two positions to the positions 65, 148, 165, 181 and 203 suggested on the basis of the protein structures. The semi-random mutagenesis at the seven positions has resulted in several blue variants for each of mCherry, HcRed1, M355NA and mKeima proteins. These variants contained: M65L,F/K69R,K/L84W,L/S148F,I/I165V,I,T/V181A,V/I203Y substitutions in the case of mCherry, E65H,F/R69K,R/F84W/S148F,I/N165A,V,N/H181A,T/I203Y,F in the case of HcRed1, M65H/K69K/F84F/S148F/S165A,V/L181I,V,T/H203F in the case of M355NA, and Q65H/I69K,R/F84W/S148I,F/D165A,V,T/F181A,V,T/R203Y,F in the case of mKeima. The observed mutations were very similar to those found in the TagRFP site-specific blue variants. The purified recombinant mutant proteins had the excitation/emission peaks at 399–410/450–461 nm. The best blue variants, named mCherry-Blue, HcRed1-Blue, M355NA-Blue and mKeima-Blue have mutations and spectroscopic properties summarized in the Figure S1 online and Table 2. Based on the analogous results obtained for five RFPs of the different genetic background we concluded that introduction of 65L,H/69K/84F,W,L/148F,I/165A,I,N/181A,I/203F,Y mutations into other RFPs with the tyrosine-based chromophore most likely will convert them into blue variants. Screening of these mutant libraries should be easy because a number of the distinctive mutants in each library will be limited to 144.
Table 2.
Properties of the blue mutants of mCherry, HcRed1, M355NA and mKeima.
| RFP | Excitation peak, nm | Emission peak, nm | Amino acid substitutions at the indicated positions resulted in the brightest blue mutants | Excitation peak after mutagenesis, nm | Emission peak after mutagenesis, nm | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 65 | 69 | 84 | 148 | 165 | 181 | 203 | |||||
| mCherry | 587 | 610 | L | K | L | F | I | A | Y | 403 | 458 |
| HcRed1* | 588 | 618 | H | K | W | I | A | A | Y | 408 | 455 |
| M355NA** | 576 | 592 | H | K | F | F | A | I | F | 409 | 461 |
| mKeima | 440 | 620 | H | K | W | I | A | A | Y | 399 | 451 |
The amino acid substitutions resulted in the formation of the BFPs are in good agreement with our predictions based on RFP structures [12–16]. First, there are totally aliphatic or aromatic side chains of Leu/His in the position 65 in BFPs. Second, there is Lys69 in combination with the hydrophobic amino acid in the position 84. Third, the hydrophobic Phe/Ile in the position 148, and aliphatic side chain of Ala/Ile in the position 165 are not able to form hydrogen bonds with phenolate anion of the chromophore. The amide group of Asn165 in mTagBFP is a weaker hydrogen bond donor than hydroxyl of Ser in TagRFP and eqFP611 [12]. Forth, there are aromatic side chain of Tyr/Phe in the position 203, and aliphatic Ala/Ile in the position 181. These substitutions promote the formation and stabilization of the tyrosine-based neutral chromophore.
We further studied properties of our advanced monomeric mTagBFP and compared it with EBFP2, which is probably the best monomeric BFP reported to date [3]. The mTagBFP, exhibited monomeric behavior at 10 mg/ml in vitro and when expressed as a fusion tag with polymerizing proteins such as β-actin and α-tubulin in mammalian cells (Figure S2 online), that was similar to the parental TagRFP [8]. Compared to EBFP2, mTagBFP is characterized by 1.6-fold higher molar extinction coefficient, as well as a higher quantum yield (Table 1 and Figure S3 online), resulting in more than 1.8-fold brighter fluorescence. These characteristics make an intrinsic brightness of mTagBFP similar to that of commonly used EGFP [1, 18]. Maturation half-time for mTagBFP at 37°C is twice shorter than that for EBFP2, and pH stability of mTagBFP is much better (pKa is below 3.0) (Figures 2B and 2C). Among other mutations, we presume that P131T substitution plays a key role in accelerating mTagBFP folding and/or maturation as it has been shown for a superglo-GFP [19].
To test photostability of mTagBFP in comparison with the most photostable available BFPs such as Azurite and EBFP2 we performed photobleaching experiments both in aqueous drops of the purified proteins in oil using epifluorescence microscope equipped with 100W mercury arc lamp source and DAPI 350/50 filter, and in HeLa cells using 405 nm laser of confocal microscope. Photostability of fluorescence probes can be compared on the basis of a half-time for the decrease of the initial photon emission rates. In the standard photobleaching procedure [1, 20] the photobleaching half-time for both mTagBFP and Azurite was about 0.6 of that for EBFP2 (Figure 2D and Table 1). With this approach, the photostabilities of other FPs such as cyan mCerulean or yellow mVenus are similar to that of mTagBFP [21]. However, the photostability of mTagBFP under laser scanning confocal illumination was 2.1-fold and 3.3-fold higher than that for EBFP2 and Azurite, respectively (Figure 2E). The difference in the photostabilities measured under continuous light source such as arc lamp and periodical source such as scanning laser have been described before [20]. It reflects two distinctive regimes of light illumination when the former one does not but the latter does allow chromophore staying for relatively long time in a ground state before the next cycle of excitation and photon emission.
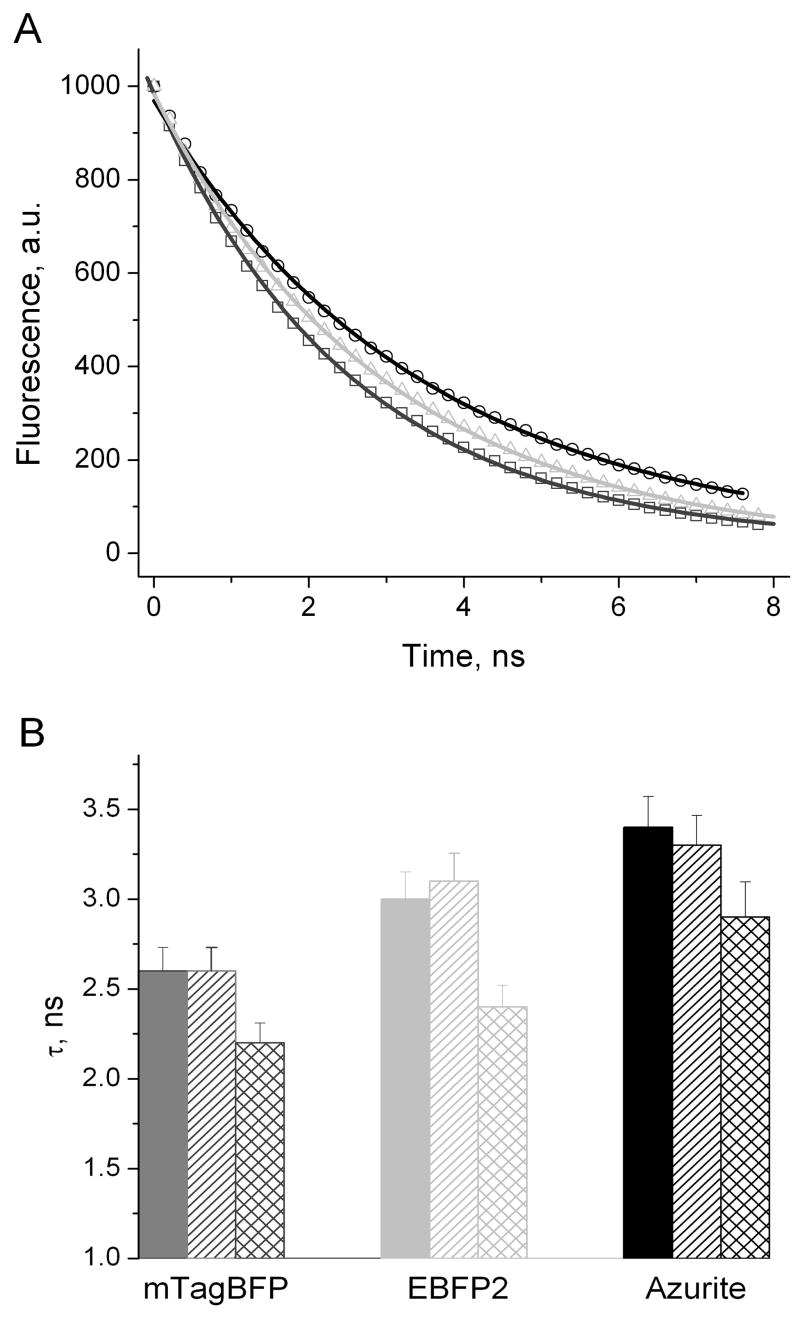
Fluorescence lifetimes for mTagBFP, EBFP2 and Azurite were measured under several conditions. The measured time-resolved fluorescence intensity decays of all these proteins were mono-exponential. At pH 7.4 and 23°C the fluorescence lifetimes of mTagBFP, EBFP2 and Azurite were 2.6, 3.0 and 3.4 ns, respectively (Table 1). The pH change from 7.4 to 6.0 did not change the lifetime for mTagBFP however decreased lifetime for Azurite and increased lifetime for EBFP2 (Figure 3). The temperature increase from 23°C to 37°C resulted in the decrease of all fluorescence lifetimes. At tested conditions mTagBFP exhibited the shortest life-time among available BFPs making it a distinctive lifetime probe for the monochromatic lifetime imaging.
Figure 3.
Fluorescence lifetimes for mTagBFP, EBFP2 and Azurite. (A) Fluorescence decay for mTagBFP (dark grey squares), EBFP2 (light grey triangles) and Azurite (black circles) at pH 7.4 and 23°C. Solid lines indicate fits by the mono-exponential curves; (B) Dependence of fluorescence lifetimes for mTagBFP (dark grey), EBFP2 (light grey) and Azurite (black) on pH and temperature. pH 7.4, 23°C (filled bars); pH 6.0, 23°C (diagonal bars); pH 7.4, 37°C (diamond bars). Error does not exceed 5%.
The emission spectrum of mTagBFP and absorbance spectra of GFPs exhibited substantial overlap suggesting mTagBFP can be a good donor for GFP acceptor in Förster resonance energy transfer (FRET). Among available GFPs, TagGFP was chosen because of the small absorbance below 400 nm where mTagBFP is excited. Construction of the FRET indicators based on fluorescent proteins principally requires that the donor and acceptor are monomeric. Introduction of the well-known monomerizing mutation A206K into TagGFP has resulted in slight decrease of its brightness. To rescue the brightness and to increase the molar extinction coefficient we performed saturated site-specific mutagenesis at positions 99, 153 and 163. It has been shown that a mutant of EGFP, called folding-reporter-GFP [22], with substitutions F99S/M153T/V163A has the molar extinction coefficient 1.5-fold higher than that for EGFP [23]. Indeed, our screening has identified the bright mutant of TagGFP/A203K with L99Q/M153R/V163A mutations, named mTagGFP (Figure S4 online). mTagGFP has the excitation/emission peaks of 483/506 nm (Figure S5 online). Comparison of the purified proteins revealed that mTagGFP has slightly higher molar extinction coefficient and quantum yield compared to parental TagGFP and EGFP (Table S1 online). The introduced mutations did not affect mTagGFP pH stability (pKa=5.0) which is higher than that of EGFP, but resulted in 1.6-fold faster maturation compared to TagGFP (Figure S6 online).
To estimate the potential efficiency of FRET between mTagBFP or EBFP2 as donors, and mTagGFP as an acceptor, we calculated the Förster distances, R0, for the donor-acceptor pairs, as has been described [24]. The calculation yielded values of R0=5.25 nm for mTagBFP-mTagGFP and R0=5.13 nm for EBFP2-mTagGFP, suggesting that the former pair is better FRET pair. Both Förster distances were larger than those reported for the standard ECFP-EYFP and mCyPet-mYPet pairs (Table 3). Then we constructed and purified fusion proteins consisting of a tandem of either mTagBFP and mTagGFP or EBFP2 and mTagGFP, each containing the caspase-3 cleavage sequence, DEVD, within the linker between fluorescent proteins. Emission spectra were measured before and after digestion with trypsin. Cleavage of the fusion proteins led to a decrease in green emission of mTagGFP acceptor and a simultaneous increase in blue emission of mTagBFP (Figure 4A) and EBFP2 (Figure S7 online) donors due to elimination of FRET similar to what has been observed with fusion constructs of other fluorescent proteins. Calculation of FRET efficiencies, E, based on the increase of donor emission upon cleavage of the fusion proteins produced E=0.57 for mTagBFP-mTagGFP and E=0.38 for EBFP2-mTagGFP pairs, respectively. For comparison, FRET efficiencies reported for ECFP-EYFP and mCyPet-mYPet pairs were 0.42 and 0.51, respectively (Table 3). These data suggest the mTagBFP-mTagGFP pair is not only superior to other BFP-GFP pairs but is one of the best among available FRET pairs of the true monomeric fluorescent proteins.
Table 3.
FRET characteristics for several fusion constructs of the fluorescent proteins.
| Fusion construct | Förster distance, R0, nm | FRET efficiency, E | Reference |
|---|---|---|---|
| mTagBFP-mTagGFP | 5.25 | 0.57 ± 0.03 | this paper |
| EBFP2-mTagGFP | 5.13 | 0.38 ± 0.03 | this paper |
| ECFP-EYFP * | 4.86 | 0.42 *** | [24, 31] |
| mCyPet-mYPet ** | 4.93 **** | 0.51 *** | [32, 33] |
Förster distances were calculated under the standard assumption of the random orientation of the chromophores. FRET efficiencies were experimentally measured using the physically linked donor and acceptor proteins.
Proteins of this widely used standard FRET pair have a tendency to form a dimer. Data on monomerized versions of the proteins are not available.
Monomerized versions of CyPet and YPet proteins, which have been specifically optimized for FRET through multiple rounds of site-specific and random mutagenesis [33]. It has been recently shown though that the original CyPet and YPet proteins form hetero- and homo-dimers in solution that substantially enhanced the observed FRET efficiency [33].
Calculated from the data [32].
Figure 4.
Behavior of mTagBFP as the FRET donor. (A) FRET of mTagBFP-mTagGFP fusion construct in vitro. (B,C) FRET imaging of staurosporine-treated HeLa cells: (B) Time course of corrected FRET normalized per intensity of the donor observed in four cells indicated in panels (C). The values are normalized to time of the staurosporine addition. (C) Fluorescent images of the cells after staurosporine treatment are shown as overlaid images of blue and green channels in upper panels. The corrected FRET signals are shown as pseudocolor images in lower panels. Scale bar is 10 μm.
To explore the benefits of the large Förster distance and enhanced FRET efficiency exhibited by the purified mTagBFP-mTagGFP pair, we applied it to detect apoptosis in living HeLa cells. We used the two-filter method of sensitized FRET measurements [25] on a pixel-by-pixel basis as described in our previous study [24]. The initial mean FRET efficiency in vivo normalized to donor fluorescence was 51.5%. Following 40–80 min exposure to 1 μM staurosporine, the FRET gradually dropped to zero before the shrinking of cells characteristic to apoptosis (Figure 4B and 4C). The large FRET efficiency of mTagBFP-mTagGFP pair enabled the detection of even weak proteolitic activity in each cell at the beginning of the apoptosis when only fraction of the substrate was cleaved. When mTagBFP and mTagGFP free proteins were co-expressed in HeLa cells the cross-bleed corrected FRET normalized to fluorescence of donor was 0.85%. Under our experimental conditions we observed that FRET between ECFP and EYFP free proteins co-expressed in HeLa cells was 6.2% confirming their weak dimerizing tendency. It has been shown the ECFP-EYFP pair linked with 19 amino acid peptide in HeLa cells provided 48% of the FRET efficiency [26]. The same authors documented 5% FRET efficiency between ECFP and EYFP co-expressed free proteins, which is consistent with our data. Thus, mTagBFP and mTagGFP proteins derived from the different marine sources and as the result lucking ability to form heterodimers provide more than 6-fold lower background for FRET analysis than the weakly dimerizing FRET pairs such as ECFP-EYFP.
SIGNIFICANCE
We developed the structure-based directed evolution approach allowing conversion of RFPs into BFPs with a tyrosine chromophore. Among five different BFP variants obtained using this strategy, enhanced mTagBFP protein has the intrinsic brightness similar to that of EGFP. Almost 2-fold increase in brightness of mTagBFP compared to available BFPs with the histidine residue allows using twice less intensities of excitation light and consequently will less harm mammalian cells and tissues, while allowing to acquire images with better signal-to-noise ratio. The higher pH-stability makes mTagBFP suitable for visualization in acidic compartments, and increases reliability of FRET-based and protein translocation assays. The less structured and narrower fluorescence emission spectrum of mTagBFP compared to EBFP2 (semi-widths for the spectra are 59 nm and 70 nm, respectively) provides for more accurate spectral separation with other fluorescent probes that makes it an advantageous tag for multicolor labeling. The spectral characteristics of mTagBFP would allow for simultaneous use of two FRET pairs in a single cell: the first consisting of blue-green and the second consisting of orange-red FPs. The faster mTagBFP maturation will be advantageous to study promoter activities and early stages of protein expression. Altogether these properties make mTagBFP the protein of choice for cell and tissue imaging in the blue part of visible spectrum. mTagBFP will be also useful for in vivo applications such as in transgenic animal models to study neurodegenerative deceases and cancer [27].
EXPERIMENTAL PROCEDURES
Mutagenesis and library screening
TagRFP [8], mCherry [1], mKeima [9], HcRed1 [10] and M355NA [11] were PCR amplified as BglII-EcoRI fragment and inserted into pBAD/His-B vector (Invitrogen). Site specific mutagenesis was performed using QuickChange Mutagensis Kit (Stratagene). For simultaneous mutagenesis at several positions, including a site-specific saturated mutagenesis (i.e., all 20 amino acids were encoded using the mixture of primers), the overlap-extension approach has been applied [17]. Random mutagenesis was performed with GeneMorph II Random Mutagenesis Kit (Stratagene) using conditions resulted in the mutation frequency up to 16 mutations per 1000 base pairs. After mutagenesis a mixture of the mutants was electroporated into LMG194 host cells (Invitrogen)
Typical mutant libraries for each FACS screen consisted of about 106–107 independent clones. Protein expression in the libraries was induced overnight at 37°C with 0.002% arabinose. Next morning the expressing bacteria were washed with Phosphate Buffered Saline (PBS) and then diluted with PBS for FACS sorting to optical density of 0.02 at 600 nm. MoFlo cell sorter (Dako) equipped with standard argon, krypton and argon-krypton mixed-gas lasers was used. Typically about 10 sizes of each library were sorted on the FACS with 407 nm of krypton excitation line and 450/65 nm emission filter. The collected bright blue bacterial cells were rescued in rich SOC medium at 37°C for several hours, and then plated on Petri dishes with 0.02% arabinose. The next day, the dishes were analyzed with Leica MZ16F fluorescence stereomicroscope using a custom blue filter set (390/40 nm exciter, 460/40 nm emitter) from Chroma. For further analysis, 20 to 50 brightest clones were selected and applied for sequencing. Mixture of several best variants was then used as a template for the next round of random mutagenesis.
Protein characterization
Recombinant proteins with polyhistidine tags were expressed in LMG194 bacterial cells grown overnight in RM minimal medium supplemented with 0.002% arabinose and then purified using Ni-NTA agarose (Qiagen). The excitation and emission spectra were measured by using the FluoroMax-3 spectrofluorometer (Jobin Yvon). For absorbance measurements the Hitachi U-2000 spectrophotometer was used.
To study protein maturation, LMG194 bacterial cells were grown at 37°C overnight in RM minimal medium supplemented with ampicillin. Next morning cells were diluted to optical density 1.0 at 600 nm, and 0.2% arabinose was added. Upon induction of protein expression the bacterial cultures were grown at 37°C in 50 ml tubes filled to the brim and tightly sealed to restrict oxygen availability. After 1 hour the cultures were centrifuged in the same tightly closed tubes. After opening the tubes, the proteins were purified using Ni-NTA resin within 30–40 min with all procedures and buffers at or below 4°C. Protein maturation was performed at 37°C in 50 mM NaH2PO4, 300 mM NaCl, 5 mM mercaptoethanol, pH 8.0 using the FluoroMax-3 spectrofluorometer.
To determine extinction coefficients, we relied on measuring of mature chromophore concentrations. For that the purified proteins were either acid- or alkali-denatured. It is known that extinction coefficient of the synthetic compound of the tyrosine-containing GFP-like chromophores is 44,000 M−1cm−1 at 447 nm in 1M NaOH [4] and about 28,500 M−1cm−1 at 382 nm in 1M HCl [28]. Based on the absorbance of the native and denatured proteins, molar extinction coefficients for the native states were calculated.
For determination of the quantum yields, the fluorescence of the blue and green mutant variants was compared with equally absorbing EBFP2 (quantum yield is 0.56 [3]) and EGFP (quantum yield is 0.60 [18]), respectively. pH titrations were performed using a series of buffers (100 mM NaOAc, 300 mM NaCl for pH 2.5–5.0, and 100 mM NaH2PO4, 300 mM NaCl for pH 4.5–9.0).
Photobleaching measurements with purified proteins were performed using microdroplets of freshly purified proteins at 0.7 mg/ml in PBS under mineral oil in a chamber on the stage of Olympus IX81 inverted microscope equipped with 100W mercury arc light source, 60x oil immersion lens and standard DAPI 350/50 excitation filter (Chroma). Single drops were found at low light levels (0.15% ND filters). Then ND filters were removed, and the protein drops were photobleached. Photobleaching experiments have been performed under identical conditions for mTagBFP, EBFP2 and Azurite. The time to photobleach from 1,000 down to 500 emitted photons per second was calculated according to the standard procedure of measuring photostability of fluorescent proteins [1,20]. In brief, for epifluorescence source such as arc lamp the procedure took into account the spectral output of the source, transmission profile of excitation filter and dichroic mirror, absorbance spectra of the BFPs and their quantum yields. EBFP2, which has been characterized according to this procedure in its original paper [3], was used as the reference.
Fluorescence lifetime measurements were performed on IX71 Olympus microscope using 60x oil immersion lens equipped with a PicoStar (LaVision, Germany) time-gated camera and a Chameleon Ultra Titanium-Sapphire laser (Coherent) with 80 MHz repetition rate tuned to 798 nm output. The TTL signal of the laser was used to synchronize the delay generator of the PicoStar system. Using an ultra fast harmonic generation system (Coherent) the second harmonic of the fundamental beam was generated and separated by a dichroic long pass filter. The beam width of the resulting beam with a wavelength of 399 nm was adjusted using a 12.5x telescope to provide even illumination of the sample and intensity was regulated using ND filter. Laser scattering and background signal was removed from the emission signal with 447/60 band pass filter (Semrock) and the remaining signal passed onto the time gated CCD intensifier of the PicoStar system. Data were acquired using 6–8 mW laser power in front of the telescope. An objective heater and a heated stage inset (Bioptechs, PA) were used for experiments at 37°C. The 30 μl protein samples at concentration 0.1 mg/ml were allowed to equilibrate for at least 10 minutes prior to data acquisition.
FRET analysis
The mTagBFP-mTagGFP and EBFP2-mTagGFP fusion constructs containing N-terminal Strep tag (amino acids -WSHPQFEK-) and C-terminal polyhistidine tag were cloned as NcoI-XbaI fragments into pBAD/Myc-His B vector (Invitrogen). The fusions contained the 20 amino acid linker -EFGGSGSDEVDKLGGSGSGT- with the caspase-3 recognition site (underlined). After overnight expression in LMG194 bacterial cells at 37°C the constructs were isolated with Ni-NTA agarose (Qiagen) followed by additional purification with Strep-Tactin sepharose (IBA). The purified constructs at concentrations 100 μg/ml were digested with 30 μg/ml of trypsin for 2 hours at 37°C.
The Förster distances R0 were calculated as previously described 6. In brief, when donor is excited, the nearby acceptor can receive energy from the donor with efficiency E expressed as [29]: E = R06/(R06 +r6), where R0 is defined by the equation R06 = (8.785 × 1017) × Φ × κ2 × n−4 × J(Φ,ε,λ)6. Φ is a quantum yield of the donor, κ2 is the orientation factor for dipole coupling, n is the refractive index of the media, J(Φ,ε,λ) is the overlap integral of the emission spectrum of the donor and the absorption spectrum of the acceptor. We calculated Förster distances for the pairs of fluorescent proteins with Origin software (Microcal) using the values for the refraction index of water at 25°C n=1.334. The orientation factor for dipole coupling κ2 is a relative orientation factor between the interacting dipoles. Under standard assumption of the random orientation of chromophores in the interacting pairs it has value 2/3. The FRET efficiency E for each pair of fluorescent proteins was experimentally measured from an increase in the donor fluorescence resulted from separation of the donor and acceptor after cleavage of the fusion construct.
Mammalian plasmid construction
To construct pmTagBFP-C1 and pmTagGFP-C1 plasmids, mTagBFP and mTagGFP were PCR amplified as NheI-BglII fragments and swapped with EGFP gene in pEGFP-C1 vector (Clontech). To construct pmTagBFP-N1 and pmTagGFP-N1 plasmids, mTagBFP and mTagGFP were PCR amplified as AgeI-NotI fragments and swapped with EGFP gene in pEGFP-N1 vector (Clontech). The pEBFP2-C1 and pEBFP2-N1 plasmids were generous gifts of R. Campbell and M. Davidson. The pAzurite-C1 and pAzurite-N1 plasmids were kind gifts of P. Daugherty and M. Davidson.
To generate mTagBFP-α-tubulin fusion protein, a PCR-amplified NheI/BglII fragment encoding mTagBFP was swapped with TagGFP gene in pTagGFP-α-tubulin vector (Evrogen). To generate mTagBFP-β-actin fusion protein, a PCR-amplified AgeI/BglII fragment encoding mTagBFP was swapped with TagGFP in pTagGFP-β-actin vector (Evrogen). For FRET in mammalian cells, mTagBFP-mTagGFP fusion containing the 20 amino acid linker -EFGGSGSDEVDKLGGSGSGT- was PCR amplified as AgeI-NotI fragment and swapped with EGFP gene in pEGFP-N1 vector (Clontech).
Mammalian cell imaging
HeLa cells were grown on 23-mm glass-bottom dishes (Bioptechs) in minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone). Transfection was performed with Lipofectamine-2000 (Invitrogen).
For imaging of mTagBFP-β-actin and mTagBFP-α-tubulin fusion constructs in mammalian cells Leica AFLX 6000 inverted fluorescent microscope equipped with 63x glycerol objective lens was used. For photobleaching experiments with mTagBFP, Azurite and EBFP2 cytoplasmically expressed in mammalian cells, Zeiss LSM 510 META DuoScan microscope equipped with 50 mW 405 nm laser, 460/40 nm emitter and 63x oil objective lens was used. Imaging at the beginning and after every bleaching step was performed with a maximal pinhole, 1% power of 405 nm laser, 512×512 resolution, zoom 1, gain 500 and scan speed 1.61 μs/pixel, which corresponds to 0.986 s per frame. Each bleaching step was performed at 40% laser power and the same scan speed. Calculation of times to photobleach from an initial scan-averaged rate of 1,000 emitted photons per second to 500 photons per second was performed essentially as has been described [20]. In brief, the procedure took into account the output power of the laser, energy of photons, scanned area, extinction coefficients of the BFPs at the laser wavelength and their quantum yields.
For apoptosis analysis, cells 24 hours after transfection were treated with 1 μM staurosporine (Sigma-Aldrich) in a medium buffered with HEPES (20 mM, pH 7.2). Then cells were covered with mineral oil, placed onto DeltaT microscopic stage heating unit (Bioptechs) at 37°C and subjected to fluorescence imaging. The imaging workstation consisted of the Olympus IX81 inverted microscope equipped with 60x oil immersion lens, cooled ORCA-AG camera (Hamamatsu), dual filter wheels, and Xenon arc light source, all controlled with SlideBook 4.1 software (Intelligent Imaging Innovation). Filter sets for image capture consisted of the standard filters from Chroma 74673 (403/12 nm exciter), 66974 (457/50 nm emitter) and 86100 dichroic mirror. After calibration for cross-bleed coefficients, the corrected FRET was calculated on a pixel-by-pixel basis using the FRET option of SlideBook 4.1 software. The details of the calculation procedures have been previously described [24, 25].
Supplementary Material
Supplementary Materials are available on the Chemistry & Biology website.
Acknowledgments
We thank R. Campbell for EBFP2 and M. Davidson for Azurite genes. We grateful to W. King for his assistance with flow cytometry. This work was supported by grants from the National Institutes of Health, GM070358 and GM073913 (to V.V.V.), and AA 013148 (to M.Y.), EC FP-6 Integrated Project LSHG-CT-2003-503259 and NATO Collaborative Linkage Grant CBP.NR.NRCLG 981752. D.M.C. is also supported by Grant of the President of Russian Federation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 2.Mena MA, Treynor TP, Mayo SL, Daugherty PS. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat Biotechnol. 2006;24:1569–1571. doi: 10.1038/nbt1264. [DOI] [PubMed] [Google Scholar]
- 3.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 4.Chudakov DM, Verkhusha VV, Staroverov DB, Lukyanov S, Lukyanov KA. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 5.Hosoi H, Mizuno H, Miyawaki A, Tahara T. Competition between energy and proton transfer in ultrafast excited-state dynamics of an oligomeric fluorescent protein red Kaede. J Phys Chem B. 2006;110:22853–22860. doi: 10.1021/jp064489f. [DOI] [PubMed] [Google Scholar]
- 6.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem Biol. 2004;11:845–854. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 9.Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 10.Fradkov AF, Verkhusha VV, Staroverov DB, Bulina ME, Yanushevich YG, Martynov VI, Lukyanov S, Lukyanov KA. Far-red fluorescent tag for protein labeling. Biochem J. 2002;368:17–21. doi: 10.1042/BJ20021191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulina ME, Verkhusha VV, Staroverov DB, Chudakov DM, Lukyanov KA. Hetero-oligomeric tagging diminishes non-specific aggregation of target proteins fused with Anthozoa fluorescent proteins. Biochem J. 2003;371:109–114. doi: 10.1042/BJ20021796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2.0-A crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 13.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 14.Wilmann PG, Petersen J, Pettikiriarachchi A, Buckle AM, Smith SC, Olsen S, Perugini MA, Devenish RJ, Prescott M, Rossjohn J. The 2.1A crystal structure of the far-red fluorescent protein HcRed: inherent conformational flexibility of the chromophore. J Mol Biol. 2005;349:223–237. doi: 10.1016/j.jmb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Quillin ML, Anstrom DM, Shu X, O’Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 A resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 16.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci USA. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Patterson GH, Day RN, Piston DW. Fluorescent protein spectra. J Cell Sci. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- 19.Flores-Ramírez G, Rivera M, Morales-Pablos A, Osuna J, Soberón X, Gaytán P. The effect of amino acid deletions and substitutions in the longest loop of GFP. BMC Chem Biol. 2007;7:1. doi: 10.1186/1472-6769-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 22.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 23.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 24.Galperin E, Verkhusha VV, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 25.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingo B, Sabariegos R, Picazo F, Llopis J. Imaging FRET standards by steady-state fluorescence and lifetime methods. Microsc Res Tech. 2007;70:1010–1021. doi: 10.1002/jemt.20509. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman RM. Recent advances on in vivo imaging with fluorescent proteins. Methods Cell Biol. 2008;85:485–495. doi: 10.1016/S0091-679X(08)85021-2. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji FI. Chemical nature of the light emitter of the Aequorea green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakowicz JR. Principles of fluorescence spectroscopy. New York: Kluwer Academic, Plenum Publishers; 1999. [Google Scholar]
- 30.Gurskaya NG, Fradkov AF, Terskikh A, Matz MV, Labas YA, Martynov VI, Yanushevich YG, Lukyanov KA, Lukyanov SA. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 2001;507:16–20. doi: 10.1016/s0014-5793(01)02930-1. [DOI] [PubMed] [Google Scholar]
- 31.Patterson GH, Piston DW, Barisas BG. Förster distances between green fluorescent protein pairs. Anal Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 33.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials are available on the Chemistry & Biology website.