Abstract
A fundamental concept in development is that secreted molecules such as Wingless (Wg) and Hedgehog (Hh) generate pattern by inducing cell fate. By following markers of cellular identity posterior to the Wg and Hh expressing cells in the Drosophila dorsal embryonic epidermis, we provide evidence that neither Wg or Hh specify the identity of the cell types they pattern. Rather, they maintain preexisting cellular identities that are otherwise unstable and progress stepwise towards a default fate. Wg and Hh therefore generate pattern by inhibiting specific switches in cell identity, showing that the specification and the patterning of a given cell are uncoupled. Sequential binary decisions without induction of cell identity give rise to both the groove cells and their posterior neighbors. The combination of independent progression of cell identity and arrest of progression by signals facilitates accurate patterning of an extremely plastic developing epidermis.
Organizing centers, first discovered by Hans Spemann and Hilde Mangold (Spemann and Mangold, 1924), consist of groups of cells that coordinate patterning by inducing the fate of their neighbors. According to the “organizer model”, the inductive signal carries at once information about position and information that dictates the nature of the cell. Studies in various systems have led to the identification of a number of signaling pathways, mediated by secreted ligands that bind to specific receptors and appear to play instructive roles in determining cellular identity and patterning, in conjunction with cellular history that provides competence to interpret signals in specific ways. Some ligands are thought to execute these functions in a concentration-dependent fashion, determining different cell identities at different levels of ligand concentration. A key feature of this model is that signals induce cell identities. Throughout this discussion, by induction, we mean the eliciting of a cell state different from that in which a cell existed prior to reception of the signal.
Many secreted signals and the components of their signaling pathways have been identified in genetic screens conducted in the Drosophila embryo (Nusslein-Volhard and Wieschaus, 1980; Perrimon et al., 1989; Perrimon et al., 1996). This amenable genetic system provides an effective paradigm to investigate patterning. In particular, various types of denticles and hairs on the larval cuticle provide markers to assess patterning. Distinct models have emerged to explain ventral and dorsal epidermal patterning. Wg and Hh are thought to control the patterning of the ventral epidermis by acting on secondary secreted signals, like the EGF ligand Spitz, constituting a system of induction by relay (Alexandre et al., 1999). In the dorsal epidermis, Hh has been proposed to directly induce and pattern several cell types posterior to the Hh-expressing cells, one of them being distant from the source of secretion (Heemskerk and DiNardo, 1994). Because the data were interpreted to indicate that Hh induces two different cell types at different threshold concentrations, Hh was described as a morphogen. From this first proposal of Hh acting as a morphogen, the Drosophila embryo has provided an elegant system to investigate how inductive mechanisms control patterning.
Positioning of the ligand secreting cells is the primary determinant of cell identity in these models. A single row of cells per segment expresses Wg (Baker, 1987), followed posteriorly by two rows that express Engrailed (En) (Fjose et al., 1985; Kornberg et al., 1985) and secrete Hh (Tabata and Kornberg, 1994; Taylor et al., 1993). The Wg and En cells form what has been referred to as the parasegmental organizer, by analogy to the Spemann organizer and because they form a boundary that does not coincide with the segmental boundary (Martinez-Arias and Lawrence, 1985). The purpose of the segmentation cascade is to specify the location of the parasegmental organizer that in turn was proposed to induce cell fates that give rise to terminally differentiated cells.
The segmentation cascade involves the sequential action of several gene families that define transient domains along the anteroposterior axis (Nusslein-Volhard and Wieschaus, 1980). The pair rule genes control the positioning of the Wg and Hh-secreting cells: sloppy paired (slp) activates wg transcription (Cadigan et al., 1994) and even skipped and fushi tarazu the transcription of en (DiNardo and O’Farell, 1987; Lawrence et al., 1987; Macdonald et al., 1986). Positive feedback between the two signal secreting cells ensures their mutual stabilization at the correct location (van den Heuvel et al., 1993). The position of the parasegmental boundary is therefore defined by the pair rule genes, and maintained by Wg and Hh. According to the current model, the spatial information carried by the pair rule genes is superseded by that provided by En, Wg, and Hh that control intrasegmental patterning. En, Wg and Hh are therefore called segment polarity genes. In this model, signaling by Wg and Hh simultaneously provides spatial information and cell fate specification.
To develop a more precise understanding of this system, we analyzed the development of the groove cells. They differentiate immediately posterior to the En and Hh expressing cells, and their differentiation is Hh dependent (Larsen et al., 2003). We identified Odd skipped (Odd) as a marker of groove cell identity, which allowed us to monitor temporally the influence of Wg and Hh on the development of the groove cells and their neighbors. We report data most consistent with the idea that the groove identity is specified several hours before grooves differentiate. Wg patterns the En stripe of cells by maintaining En, antagonizing a transition toward a more posterior identity. The range of Wg signaling determines the width of the En expressing domain, and therefore the position of the segmental boundary. The cells beyond the reach of Wg, and therefore beyond the boundary, adopt the groove identity. In a distinct subsequent step, Hh, secreted from the En cells, is required to refine the groove cell pattern by blocking another transition toward a more posterior identity. We propose that in this tissue, contrary to the cell fate induction model, signaling acts by blocking transitions of cell identity rather than by instructing cell identity. Cell diversity is generated by a progression of cell identity from one to the next, toward a default fate. The patterning signals act by maintaining spatially selected cells in specific intermediate identities until differentiation occurs.
Material and Methods
We used standard techniques of fluorescence detection and used Leica microscopes SP2 and SP5. 3D image computations were performed with Leica Application Suite Advanced Fluorescence software. Cell tracking was done by painting cells with Adobe Photoshop. The images were exported as a movie with iPhoto.
Fly strains
OreR was taken as wild-type. opa8, run3, odd5, odd01863, hhAC, ptcIN, nkd2, UAS-yanact, are from the Bloomington Drosophila Stock Center. Trh-lacZ (1eve1) was obtained from M. Krasnow, Cadherin-GFP from H. Oda, En-Gal4 and UAS-hh from the Perrimon lab.
Antibodies
Anti-Odd was a generous gift from Jim Skeath, anti-Ptc from Pascal Therond, anti-Slp from John Reinitz. Anti-aPKC was from Santa Cruz Biochemical (sc-216) and mouse anti-β-galactosidase from Promega. Monoclonal antibodies against En, Ena, Dlg, developed by C. Goodman and Wg, Cadherin, Crb, Yan developed respectively by S. Cohen, T. Uemura, E. Knust, G. Rubin were obtained from the Developmental Studies Hybridoma Bank.
Results
Identification of groove cell markers
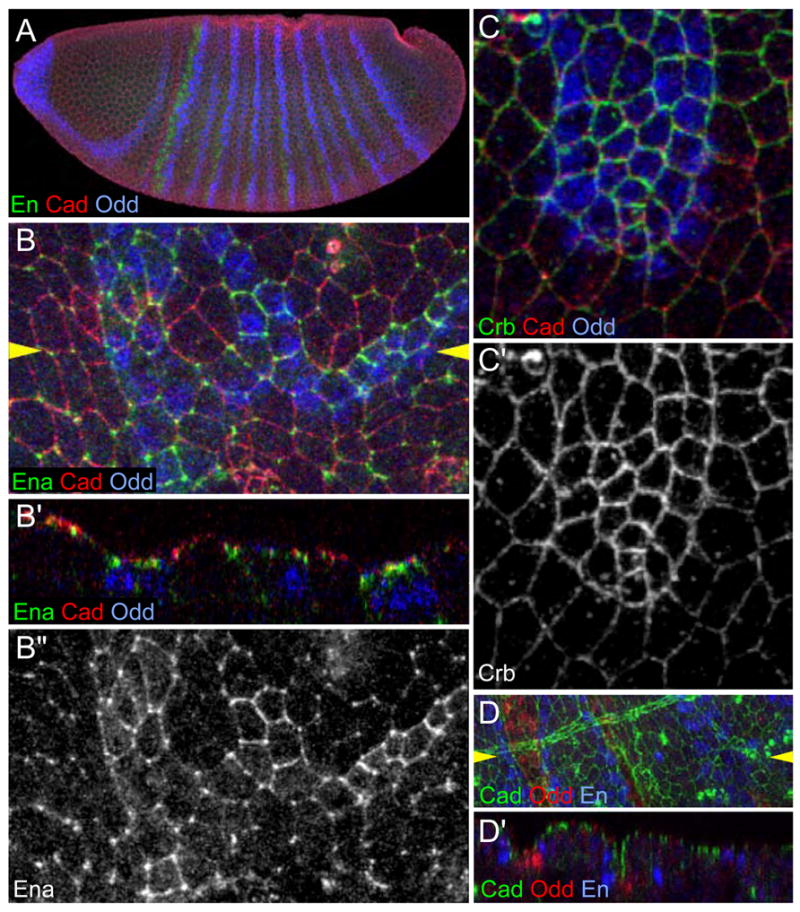
To analyze the determination of cell identity in the embryonic trunk, we examined the development of a highly specialized cell type, displaying specific morphological features and traceable molecular markers. During their morphogenesis, the cells located at the bottom of the segmental grooves (Fig. 1A–C) adopt a characteristic rectangular shape and their cross-sectional area at the level of the adherens junctions decreases to become about 1.5 times smaller than their neighbors (Fig. 1E, E″). Several markers allow their unambiguous identification. The apical proteins Crumbs (Crb) (Tepass et al., 1990) (Fig. 1C, D) and atypical Protein Kinase C (aPKC; Wodarz et al., 2000; Fig. 1E′″) are enriched at the apical circumference of the groove cells at embryonic stage 12, when the epithelium starts folding (Fig. S1). When the groove is fully formed, at stage 13, the actin-binding molecule Enabled (Ena) is enriched at the junctions separating groove cells, whereas it is detected only at the vertices in non-groove cells (Grevengoed et al., 2001) (Fig. 1E′). In the fully deepened grooves at stage 14, the dorsal epidermis contains a single row of about 19 groove cells that terminates with a groove slit, a several cell wide cluster of 8–10 cells in the ventral part of the lateral epidermis. No groove cells are present in the ventral epidermis (Fig. 1D) (Martinez-Arias, 1993). Here we focus on the patterning of the single dorsal row of groove cells.
Figure 1. Groove markers in wild-type embryos.
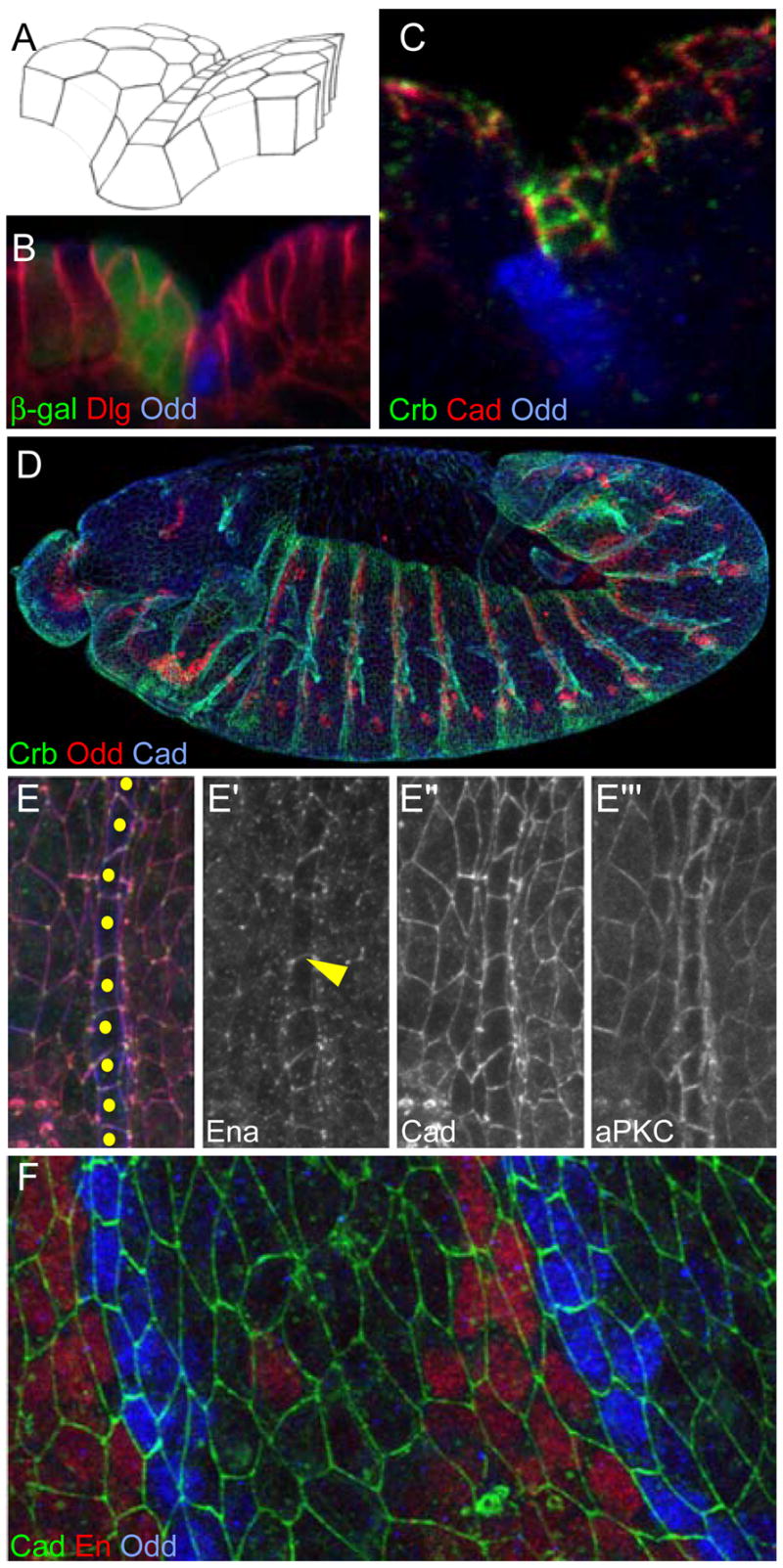
The figures are projections of confocal image stacks. Unless otherwise indicated, anterior is left and dorsal is up. Embryos are ~500μm long and cell diameters after stage 11 are ~4 μm.
(A) Schematic representation of the groove. Groove cells have rectangular junctions and are located at the bottom of the groove.
(B) Cross section of a stage 14 embryo expressing β-gal (green) in the en domain. Odd (blue) marks groove cells and Dlg (red) reveals cell shapes.
(C) Projection oriented as in (A) to display Crb accumulation (green) at the subapical domain of groove cells. Cadherin (red) and Odd (blue).
(D) Stage 12 wild-type embryo showing Crb (green), Odd (red) and cadherin (blue). Grooves are absent from the ventral domain, although Crb accumulation is visible there.
(E) En face view of stage 14 embryo’s rectangular groove cells showing Ena (green; E′), Cadherin (red; E″) and aPKC (blue; E′″). Ena marks junctions between groove cells (arrowhead). Cadherin is uniformly expressed. aPKC is enriched in the subapical domain of groove cells. Yellow dots indicate groove cells.
(F) Stage 12 cadherin-GFP expressing embryo showing Cadherin-GFP (green), En (red) and Odd (blue).
An additional marker, Odd (Nusslein-Volhard and Wieschaus, 1980), is specifically expressed in all groove cells (Fig. 1B, C, D, F). Odd is a zinc finger transcription factor (Coulter et al., 1990) known for its pair rule function (Coulter and Wieschaus, 1988), but has also been reported to have gap and segment polarity properties (Saulier-Le Drean et al., 1998). Odd is expressed in seven stripes at blastoderm stage, and in fourteen stripes when the germ band elongates (Coulter et al., 1990). Significantly, we can follow the Odd protein expression pattern in fixed embryos from early stages (Fig. 2A) until groove morphogenesis. To determine whether Odd expression correlates with groove cell identity, we analyzed groove formation in pair rule mutants, in which antero-posterior patterning defects result in overall spatial disorganization of cell types. In both odd paired (Fig. 2B, B′, B′″) and runt (Fig. 2C, C′) mutant embryos, we find that Odd-expressing cells also express Ena (Fig. 2B″) and Crb (Fig. 2C′) in the characteristic groove cell patterns, and form an indentation in the epidermis (Fig. 2B′), although a full groove is not always present. In odd hypomorphic mutant embryos, the remaining Odd protein is strictly correlated with remaining groove cells (Fig. 2D, D′). Similarly, in other conditions that modify the number or position of Odd cells, there is a strict one-to-one correspondence between Odd expression and groove (see Fig. 4C″, F″). Notably, in odd hypomorphic mutants, segments lacking Odd expression do not make grooves. We conclude that Odd expression faithfully marks groove cell identity after stage 12. As the expression of Odd precedes groove morphogenesis by several hours, we wondered whether the 14 Odd stripes present at germ band extension represent an earlier specification of the groove cell identity.
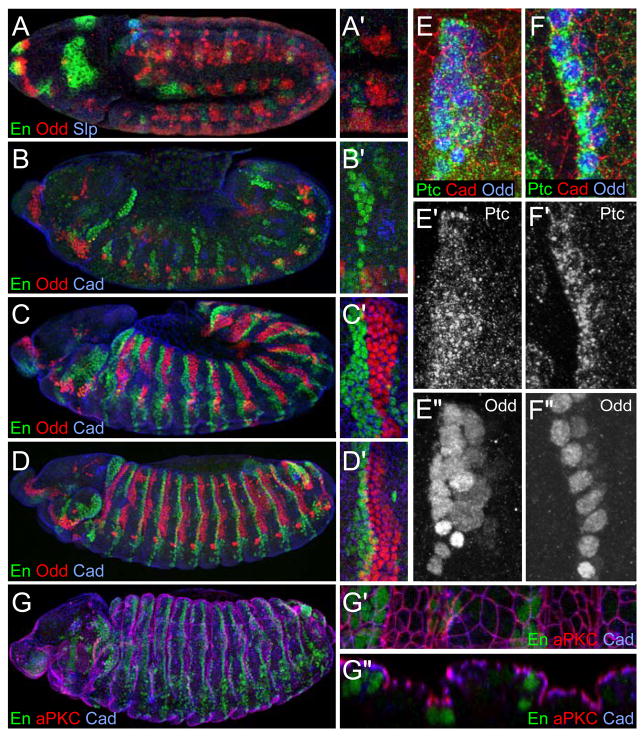
Figure 2. Groove markers in pair rule mutants.
(A) Seven additional stripes of Odd (blue) appear before germ band extension. En stripes (green) also appear at that stage. Cadherin (red).
(B) In stage 13 odd paired8 embryos, Ena (green; B″) accumulates at junctions between Odd cells (blue). Cadherin (red). (B′) cross-section at the position of the yellow arrowheads in (B). Slight depressions are present at the position of the Odd cells.
(C) In runt3 stage 13 embryos, Crb (green; C′) accumulates in Odd expressing cells (blue). Cadherin (red).
(D) Stage 13 Odd-lacZ/odd5 transheterozygous embryo. Residual Odd (red) correlates with remaining groove cells. Cadherin (green) and En (blue). (D′) Cross-section at the position of the yellow arrowheads in (D). No groove forms posterior to En expressing cells where Odd is absent.
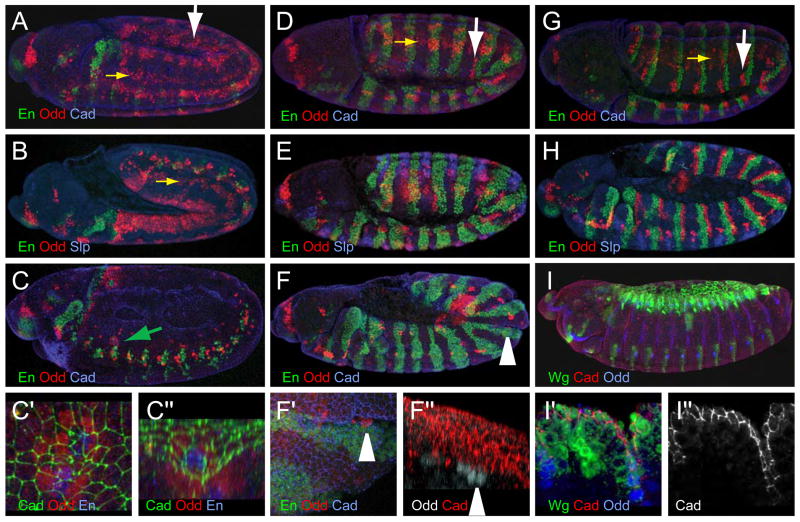
Figure 4. Wg signaling regulates the number of En and Odd cells.
(A–C″) wgIL114 mutant embryos. (A) Stage 10 stained for En (green), Odd (red) and Cadherin (blue). (B) Stage 11 stained for En (green), Odd (red) and Sloppy paired (blue), (C) Stage 15 wg mutant: some En cells (green; green arrow) remain in a thoracic segment, surrounded by Odd cells (red). Cadherin (blue). (C′) Magnification of En cells from (C), with Cadherin (green) showing groove, En, (blue), Odd (red). (C″) 90° rotation of B″. For 3D rotation, see Movie 2 in Supplemental Material.
(D–F″) nkd2 mutant embryos. (D) Stage 10 stained for En (green), Odd (red) and Cadherin (blue). (E) Stage 11 stained for En (green), Odd (red) and Sloppy paired (blue), showing expansion of the En domain and few remaining Odd cells. During retraction, grooves only form where Odd (red) is detected (F; magnified in F′). Cadherin staining (blue) shows that grooves are absent along the En cells (green) not bordered by Odd cells. (F″) 90° rotation of F′ (arrowhead). Odd (gray) and cadherin (red). For 3D rotation, see Movie 3 in Supplemental Material.
(G, H), wild-type embryos. (G) Stage 10 stained for En (green), Odd (red) and Cadherin (blue). (H) Stage 11 stained for En (green), Odd (red) and Sloppy paired (blue)
(I) pannier-Gal4 UAS-wg drives ectopic Wg (green; D′) but does not affect Odd expression (blue; I′) or groove formation (Cadherin, red in I′; I″). Yellow arrows, mesodermal Odd staining; white arrows, ectodermal Odd staining.
Odd marks the groove identity
We wished to ask whether the groove cells derive from these early Odd populations. At full germ band extension, Odd is detected in two groups of cells, both posterior to the En stripe. One is in the dorsal domain, and the other in the ventrolateral domain, with the tracheal placode in between (Fig. 3A). No other cell types are present between the En, Odd and tracheal cells. As the tracheal placodes invaginate, significant morphogenetic rearrangement of the epidermis must occur. Analysis of fixed embryos suggests that the dorsal Odd cells may give rise to the dorsal groove cells and the ventral ones to the segmental slit. Using Cadherin-GFP (Oda and Tsukita, 1999) to track cells by live imaging, we found that the epidermal surface lost as the tracheal placode invaginates is replaced by convergence of the cells dorsal and ventral to the placode (Fig. 3B, C; see Movie 1 in supplementary material). Though we do not have a live marker for Odd expression, two cell types could be identified by their morphological features: the tracheal cells that invaginate at stage 10, and the groove cells that adopt their rectangular shapes and align at the bottom of the grooves. Tracing backward, the progenitors of both the tracheal cells and the groove cells could be identified, revealing that the single row of dorsal groove cells originates exclusively from cells dorsal to the placode, in the region where Odd is expressed, but not from cells anterior to the placode, where En is present. We conclude that the groove cells originate from the Odd cells that were previously dorsal to the tracheal placode. These Odd expressing cells are likely to be pulled by the invaginating tracheal cells to occupy the space left by their invagination. Strikingly, the En cells respect the segmental boundary and are not pulled posteriorly by the invaginating trachea, indicating that the segmental boundary is effective before segmental groove morphogenesis and before the border between the En and Odd expressing cells straightens.
Figure 3. Dynamics of Odd expressing cells.
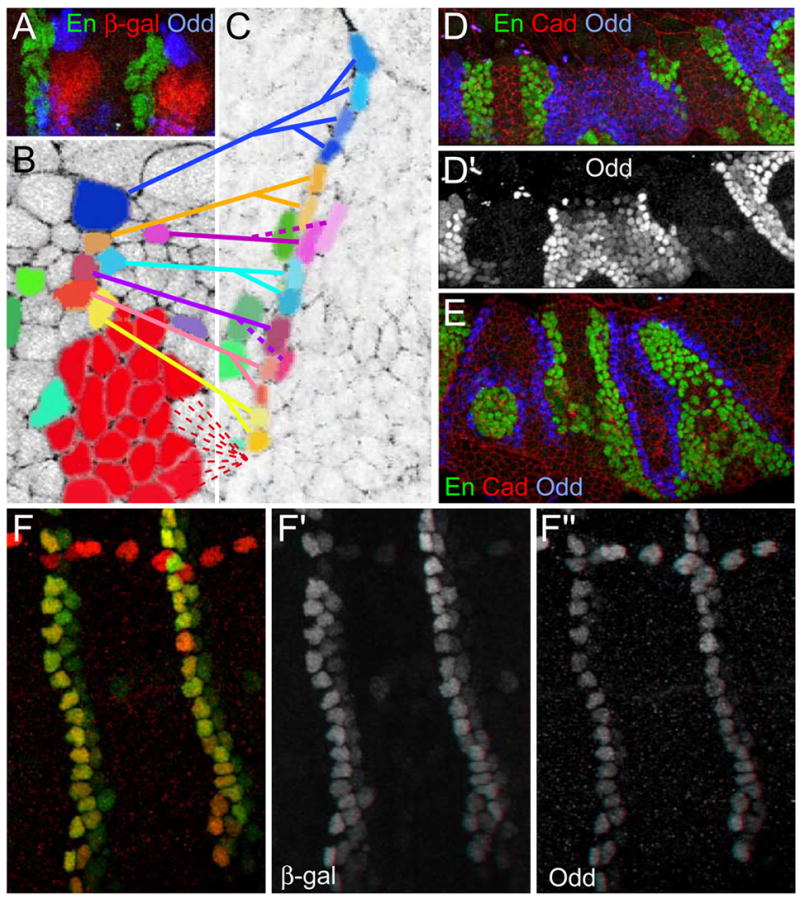
(A) Odd cells (blue) dorsal and ventral to the tracheal placode, marked by β-gal (red) in stage 9 trachealess-lacZ embryos. (B) and (C) Images from a Cadherin-GFP time-lapse movie (see Movie 1 in supplementary material). Cell shape and disposition allows the identification of the groove and tracheal cells. (C) The epithelium after tracheal invagination. Going back in time shows that groove cells originate from the Odd domain. Tracheal precursors (red), groove cell precursors (blue, yellow, pink, burgundy). Some anterior cells (green) reveal that the groove cells abutting the tracheal placode have been displaced ventrally. Two pink sister cells follow different fates as only one becomes a groove cell.
(D, E) runt3 embryos at stage 12 (D) and 14 (E) display refinement of Odd expression. First, Odd expression (blue; D′) decreases away from the En cells (green); later a single cell wide stripe remains (E).
(F) Odd lacZ shows that β-gal remains visible (green; F′) in cells that have lost Odd. (red; F″)
This result contradicts a previous report that the groove cells originate from the posterior-most En cells of each segment (Larsen et al., 2003). In the previous study, progenitors of En cells were marked using En-Gal4 to drive expression of cytoplasmic horseradish peroxidase (HRP), and HRP staining was observed adjacent to groove cell nuclei, suggesting that the groove cells may have derived from cells which had recently expressed En. We repeated this experiment, using En-Gal4 to drive UAS-APC2::GFP to mark the cytoskeleton of En cells. We found that at stage 14, many En cells extend processes labeled with APC2::GFP that can reasonably be mistaken to be within the Odd cells (Fig. S2). Second, a minority of cells posterior to the En stripe express GFP (Fig. S3), but that expression is not restricted to the groove cells (Fig. S4), and time lapse imaging shows that it does not decrease with time, implying a non-physiologic expression due to the Gal4 system rather than residual expression of GFP from a cell that has lost En expression. We conclude that the groove cells originate from the Odd expressing cells and not the En cells.
Odd expression is refined during and after tracheal invagination
Interestingly, a cell division occurs during the morphogenetic movement accompanying tracheal invagination, and we observed that some sisters of Odd-expressing groove cells did not become groove cells (Fig. 3B, C; Movie 1), indicating that the groove cells are selected from among a larger Odd expressing precursor population. Examination of Odd staining in fixed embryos confirms that the number of Odd expressing cells decreases: two rows of Odd cells are present during germ band retraction, and only one remains at stage 14. By following lacZ expression in Odd-lacZ embryos, we verified that only the cells that end up adjacent to the En stripe keep Odd expression, while their posterior neighbors lose Odd expression during stage 13, keeping only the perduring β-galactosidase (Fig. 3F, F′, F″). Pair rule mutants provide additional evidence that maintenance of Odd expression depends on the En cells. In these mutants, the territories of En and Odd are no longer in regular stripes (Fig. 3D, D′, E), yet, as in wild-type, Odd is expressed in regions adjacent to En cells, still in broad territories several cells wide at stage 12, but soon afterward, exclusively in single cell wide stripes that are immediately adjacent to the En cells.
We conclude that Odd marks the progenitors of the groove cells before reorganization of the epidermis but that the Odd pattern is refined, maintaining expression only adjacent to the En cells, narrowing to a single row of differentiated groove cells. These data indicate that Odd is not a marker of cell competence, as such a marker would remain expressed in cells just posterior to the single row of groove cells at stage 14. Instead, Odd marks a population of cells that, if maintained in their present state, are destined to become groove cells. However, as we will argue below, this cell identity is unstable, and requires maintenance. In the absence of a maintenance signal, which occurs in a single cell wide stripe, we suggest that cells progress toward a different identity.
Early Wg patterns En and Odd expression, and defines the segmental boundary
The presence of Odd in the ancestors of the groove cells suggests that the groove identity may be specified long before the differentiation of the groove. We therefore examined the earlier specification of Odd expressing cells and its effect on groove differentiation. Regulation of the seven stripes of Odd expression before germ band elongation has been studied, but nothing is known about the mechanisms controlling the 14 stripes present during germ band elongation. Two Odd homologues, Bowl and Drumstick, are also expressed in 14 stripes located posterior to the En stripes, and have been shown to be repressed by Wg signaling (Hatini et al., 2005). We therefore asked whether Wg controls the formation of the 14 Odd stripes. We find that Odd expression expands at the expense of En expression when Wg function is impaired, indicating that Wg inhibits Odd expression as the 14 stripes of Odd are established (Fig. 4A, B). At the end of germ band elongation, the 14 Odd stripes appear broader in wgIL114 mutants, and, as expected, only a few En cells remain. naked mutant embryos, in which Wg signaling is derepressed (Zeng et al., 2000), display the opposite phenotype (Fig. 4D, E): the En stripes are wider, and only a few Odd cells are present. Thus, Wg signaling controls the allocation of cells into the En and Odd identities. Wg therefore patterns the position of the segmental boundary that lies between these two cell types, and the boundary is respected during trachea invagination, as Odd cells are displaced along the boundary, but cells on either side do not pass from one side to the other (Fig 3B, C).
The regulation of Odd by Wg influences groove development. The later expression of Odd in wg mutants is highly dynamic. As En expression disappears, the broad domains of Odd, in 14 stripes at stage 9, give way to a pattern in which all the dorsal and ventrolateral cells of the trunk express Odd, forming two longitudinal stripes spanning the length of the trunk at stage 11 (Fig. 4B). However, by stage 12, Odd has disappeared, except in close proximity to the few small groups of En cells that remain. These exceptional small En/Odd territories express groove markers in the Odd cells and form localized indentations centered on the Odd cells (Fig. 4C–4C″). nkd mutants also display a few remaining Odd cells, and these too differentiate as groove cells. In these mutants, no grooves are present at the border of the En cells except when they abut Odd cells (Fig. 4F–F″), supporting our hypothesis that both early specification of Odd cells, as well as their subsequent maintenance, is relevant to groove development. Wg therefore patterns the segment boundary by maintaining the En identity among cells within the reach of the Wg signal, while cells beyond the reach of Wg adopt the groove identity and become part of the next segment.
Wg does not repress groove identity once Odd expression is established
We next tested if Wg controls only the choice between maintaining En and adopting the groove identity, or if it can also repress groove differentiation at later stages as proposed previously (Larsen et al., 2003; Piepenburg et al., 2000). To do this, we analyzed Pnr-Gal4; UAS Wg embryos, in which Wg is ectopically expressed in the dorsal epidermis. Pnr-Gal4 drives expression beginning at stage 10–11, (after tracheal invagination and before groove morphogenesis), until late embryonic stages. Ectopic Wg is therefore delivered after the En/Odd transition has occurred and includes the full period of groove morphogenesis. The ectopic expression is similar in strength to the endogenous Wg expression in the ventral epidermis (Fig. 4I). These embryos show no sign of Odd weakening, and the grooves are not affected (Fig. 4I′, I″). Therefore, Wg does not repress Odd expression or groove identity at later stages, indicating that Wg blocks the specification of the Odd-expressing groove precursor cells during a specific time window. As we demonstrate below, this mechanism is distinct from the mechanism that later refines the population of Odd-expressing groove precursors.
Hh signaling refines the Odd expression domain
Despite its broad expression in wg mutants at stage 10, Odd is maintained after stage 12 strictly next to the residual En cells, consistent with the idea that refinement of Odd expression to a single cell stripe depends on the En cells. As Hh is secreted by En cells (Tabata and Kornberg, 1994; Taylor et al., 1993), and is required for groove formation (Larsen et al., 2003), we investigated whether it controls this refinement of Odd expression. In hh null mutants, Odd expression is normal or slightly wider than wild-type until stage 10 (Fig. 5A, A′), and the dorsal and ventral Odd populations are present on either side of the tracheal placode. At stage 11, Odd expression decreases compared to controls, and disappears by stage 12 (Fig. 5B, B′). When the germ band retracts, no Odd is detected in the epidermis, and no grooves form. The effects of too much Hh activity were first assayed in patched (ptc) mutants; Ptc is the Hh receptor and functions as an antagonist of signaling (Chen and Struhl, 1996; Ingham et al., 1991). In ptc mutants, in which Hh signaling is activated in all non-En cells, Odd is expressed at stage 12 in stripes broader than wild-type stripes (Fig. 5C, C′). The refinement step does not occur, and from stage 13 through stage 15, broad stripes of Odd persist. A similar result was obtained when Hh was overexpressed in the En domain (Fig. 5D, D′). Thus, while Hh absence causes an excessive reduction of the Odd expressing population, Hh excess prevents the refinement of the Odd population, instead resulting in the maintenance of what appears to be the full population of Odd cells previously established when Wg signaling patterns the width of the En stripes.
Figure 5. Hh maintains Odd expression near the En cells.
(A, A′) Stage 11 hhAC embryo stained for En (green), Odd (red) and Slp (blue). Odd is still present whereas En disappears.
(B, B′) Stage 12 hhAC embryo stained for En (green), Odd (red) and cadherin (blue). Some En expressing cells remain, but no Odd cells are in the epidermis. Some Odd staining is visible in the nervous system.
(C, C′) Stage 12 ptcIN embryo stained for En (green), Odd (red) and cadherin (blue). The Odd domain is wider than in the wild-type (compare with Figure 4H)
(D, D′) Stage 13 En-gal4 UASHhWT embryo stained for En (green), Odd (red) and cadherin (blue). Odd is present in a four to five cell wide stripe.
(E) Detail of a stage 11 wild-type embryo stained for Ptc (green), Cadherin (red) and Odd (blue). Both Ptc (E′) and Odd (E″) domains are several cells wide.
(F) Detail of a stage 13 wild-type embryo stained for Ptc (green), Cadherin (red) and Odd (blue). Both the Ptc (E′) and the Odd (E″) domains are a single cell wide.
(G, G′, G″) Stage 13 ptcIN embryo stained for En (green), aPKC (red) and cadherin (blue). Crb is enriched in the widened grooves. En face (G′) and Z-section (G″) detail from (G).
Note the groove on the left has ectopic En posteriorly, while the groove on the right does not. For 3D rotation movie, see Movie 4 in Supplemental Material.
We next asked whether the additional Odd expressing cells, present when Hh signaling is inappropriately maintained, differentiate into groove cells. In ptc mutants, broad Odd domains are maintained, in which all cells express Crb, aPKC, and Ena in the characteristic groove manner and display smaller diameters (Fig. 5G, G′). All the groove cells of a given segment form a single, wide groove (Fig. 5G″). Edges of the grooves form regardless of the presence or absence of ectopic En cells characteristic of Ptc mutants. Similar wide grooves form when Hh is overexpressed, during which ectopic En cells are never present, ruling out a requirement for direct contact with En cells.
The above results imply that in wild-type, Hh may control the refinement of Odd cells to a single stripe by signaling over a range that becomes progressively limited to a single cell by stage 14. Interestingly, cells that maintain Odd expression also express high levels of the Hh target gene and receptor, Ptc (Forbes et al., 1993), indicating that they are responding to Hh signal (Fig. 5E–E″), while Ptc expression decreases to its basal level in the cells that lose Odd (Fig. 5F–F′″). The synchrony of the reduction of Ptc and Odd expression, corresponding respectively to the pathway activation and the biological output, shows that this system adapts very quickly to the withdrawal of Hh.
Our data show that Hh directs the refinement of a population of groove cells to a single row of cells. If Hh signaling is disrupted, or if it is expanded, either too few or too many cells maintain the groove identity, resulting respectively in the subsequent absence or the widening of the grooves. This mechanism is independent of, and occurs after the specification of the groove identity controlled by early Wg.
Hh does not induce groove identity
Groove cells are not induced by Hh in non-Odd expressing territories, such as the dorsal and ventral Wg expressing cells or the lateral cells on the anterior side of the En stripes. In the ventral domain Hh does not induce groove cells anterior or posterior to the En cells. To rule out that Hh could initiate groove identity if non-Odd cells were present immediately posterior to En cells in the lateral domain, we genetically displaced cells. Activated Yan (Rebay and Rubin, 1995) over-expression compromises the integrity of the En stripe, allowing some cells to cross the segmental boundary and intercalate between the converging Odd populations during tracheal invagination (Fig. 6A, A′). These cells lose En expression, become receptive to Hh and express Ptc at levels similar to their dorsal and ventral neighbors (Fig. 6B–B″). Still, they do not express Odd nor adopt a groove identity (Fig. 6C–C″) but instead interrupt the groove. As a control, ectopic expression of activated Yan in Odd cells before or after groove differentiation does not interfere with groove formation (data not shown). Since the En cells are the nearest lineage to the Odd cells, this is the most stringent possible test of the ability of Hh to induce groove in non-Odd cells. This strongly suggests that Hh signaling cannot induce groove identity in non-Odd cells.
Figure 6. Hh cannot induce grooves in dedifferentiated En cells.
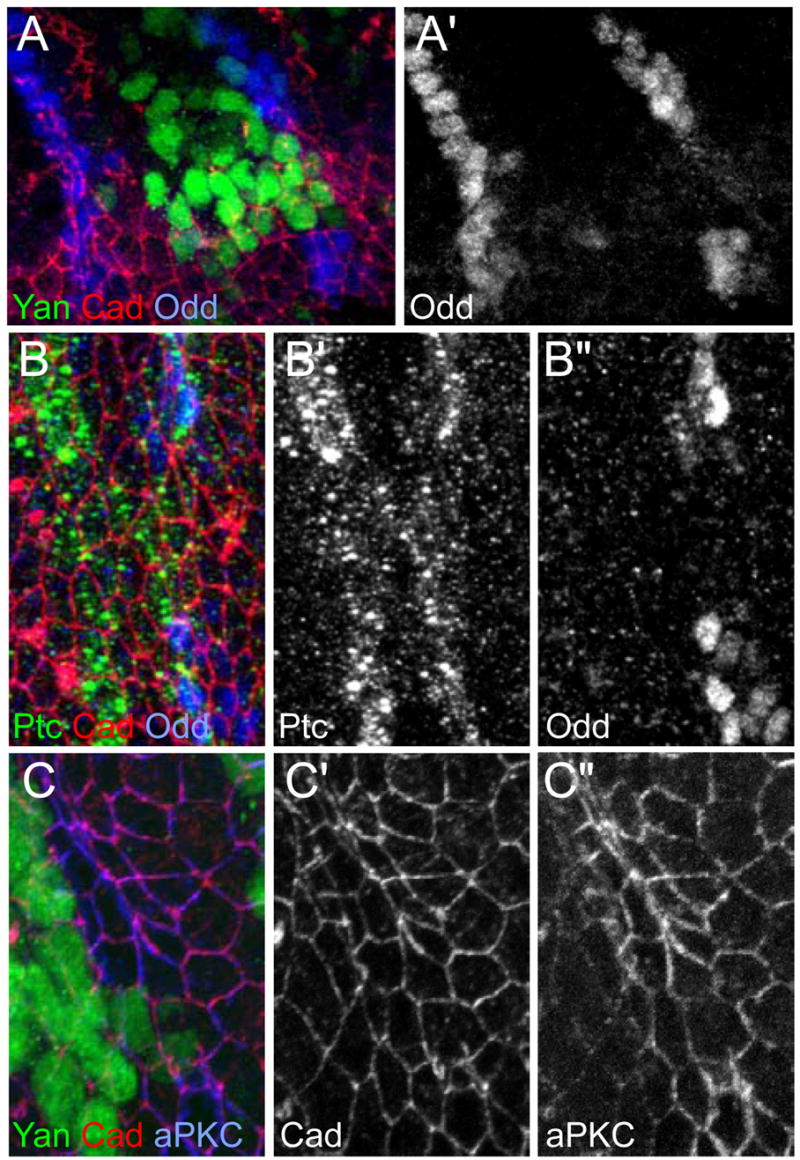
(A) Stage 13 prd-Gal4 UAS-Yanact embryo stained for Yan (green), Cadherin (red) and Odd (blue; A′). Yan expressing cells near the tracheal invagination move posteriorly between the two Odd populations. (B) En-Gal4 UAS-Yanact embryo stained for Ptc (green; B′), cadherin (red) and Odd (blue; B″). Ptc is expressed between the separated Odd populations.
(C) En-Gal4 UAS-Yanact embryo stained for Yan (green), Cadherin (red, C′) and aPKC (blue, C″) showing that the Yan expressing cells do not accumulate aPKC and do not become rectangular. 3D visualization reveals an interruption in the groove (data not shown).
Discussion
We have investigated the specification and patterning of the segmental grooves that develop immediately posterior to the Hh secreting En cells in the lateral and dorsal epidermis. Our findings are most consistent with a model in which Wg and Hh function sequentially, each acting to select from a larger population of cells a group that will maintain an identity, while those that do not receive signal progress toward a different identity. This contrasts with the previous model, in which these signals function to induce cell identities. These signals therefore appear to control binary responses, and the responses entail maintenance of pre-established identities rather than induction of new cell identities.
The identification of Odd as a marker of groove cells allowed us to trace the groove cell lineage. We observe that in maintaining the Hh/En cells, Wg signaling also inhibits the development of the Odd expressing groove cell precursors. Wg therefore not only stabilizes Hh/En cells, but determines the position of the border between En and Odd expressing cells that becomes the segmental boundary. This boundary is functional even before becoming straight, forming a barrier to prevent En and Odd expressing cells from mixing during tracheal invagination. Because the En cells are specified before Wg action, Wg maintains but does not induce their identity (DiNardo et al., 1988; Martinez-Arias et al., 1988). Our demonstration that Wg patterns the En cells although it does not specify them is consistent with the notion that specification and patterning can be regulated by independent mechanisms (Martinez-Arias and Hayward, 2006).
How might Wg signal the choice to maintain En identity rather than switch to the Odd expressing identity? A previous characterization of the En enhancer (Florence et al., 1997) may help understand the molecular mechanism governing the En/Odd identity switch. The En enhancer contains Ftz and Ftz-F1 binding sites sufficient to establish reporter expression (Fig. S5). A Ftz Ftz-F1 reporter is expressed in a wide domain and is not refined to a two cell wide stripe as is wildtype En, showing that its expression is maintained independently of Wg. A specific region of the En enhancer is needed to refine expression, and was hypothesized to contain Odd binding sites, as Odd limits En expression (Coulter and Wieschaus, 1988). Wg signaling acts through the TCF transcription factor, and TCF binding sites are located between the Ftz, Ftz-F1 binding sites and the repressing region. The TCF sites do not modify expression of the Ftz Ftz-F1 reporter, but do limit refinement to the normal width stripe when the proposed Odd binding sequences are present. In the wildtype enhancer, we propose that, through the TCF binding sites, Wg signaling maintains expression by preventing Odd mediated repression. Consistent with this interpretation, ectopic Wg expression after the switch from En to Odd expression but before grooves differentiate shows that Wg cannot repress either Odd expression or groove morphogenesis (Fig. 4I′, I″). This might therefore provide a molecular model for the ability of Wg to block a switch in cell identity.
Following En patterning, the remaining Odd cells posterior to the En domain are again partitioned into two domains, the anterior domain retaining Odd and the posterior domain losing Odd expression. Our data show that Hh, produced by the En cells, maintains Odd expression within its reach, which in wild-type is progressively narrowed to a single row adjacent to the En domain. The Odd cells differentiate into grooves, while the cells that lose Odd produce a characteristic cuticular hair at later stages. The close temporal relationship between reduction of Ptc expression and loss of Odd expression suggests a relatively close coupling between Hh signal and Odd expression.
Because Odd expression is associated with the groove cell identity, the maintenance of Odd expression in a portion of the Odd expressing cells suggests that Hh maintains the groove identity in cells within reach of the signal. However, because we do not know how to maintain the Odd-associated identity in the absence of Hh signaling, we cannot discern whether Hh is also required to provide a signal in addition to that needed to maintain the groove identity. Whether or not such a hypothetical additional signal might be required, the direct correspondence between Odd maintenance and groove differentiation demonstrates that this hypothetical signal would not provide any information beyond that associated with the binary decision to maintain the Odd/groove cell identity, and would therefore not be instructive in the usual sense of specifying between alternative developmental pathways.
The cell types posterior to the En domain have previously been studied based on the type of cuticle they secrete. The groove cells secrete naked cuticle, whereas their posterior neighbors secrete thick pigmented hairs. Using these markers of cell identity, it was proposed that Hh acts as a morphogen, inducing at high concentration the fate of naked cuticle producing cells, and at lower concentration the fate of thick pigmented hair secreting cells (Heemskerk and DiNardo, 1994). By monitoring Ptc and Odd expression in the wild-type embryo, we observe elevated levels in the Odd (groove) domain, and basal levels as at the groove identity is abandoned as soon as Hh signaling is withdrawn and the groove identity is abandoned in favor of the thick pigmented hair identity. Our data are therefore more consistent with the interpretation that Hh exceeds the threshold for signaling in the naked cuticle (Odd) domain, but drops below threshold in the thick pigmented hair secreting domain. Heemskerk and DiNardo also observed an expansion of the thick pigmented hair domain and a decrease in the posterior fine hair domain with elevated Hh levels (Heemskerk and DiNardo, 1994). Because in wild type we observe equal, basal, Ptc expression in this region, we propose that this boundary is not normally patterned by Hh. We hypothesize that with elevated Hh, decreased cell death in the thick pigmented hair domain and competition with the posterior fine hair domain produce this altered pattern. A single Hh threshold would be incompatible with the morphogen model.
Our data are consistent with the idea that cell identities may be transient, and reveal the existence of a pathway by which cell identity progresses. Although it has not been invoked for the embryonic epidermis, the notion of a default pathway of cell identity is not new. For example, in Drosophila, neuroblasts have been proposed to follow a default neural identity pathway unless the glial cell deficient/missing gene diverts them towards the glial identity (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). In the absence of Wg signaling, dorsal epidermal cells switch from En to groove identity, and subsequently, if Hh is lost, from groove to a third, fine hair identity. It is, as yet, unknown whether these identity switches depend on exogenous signals, or whether they are cell autonomously controlled. Furthermore, how their timing is regulated remains unknown. Wg acts during a specific time window, implying that the patterning signal must persist as long as the transcription factors responsible for the switch are active. The patterning signal has no effect on the pace of the progression. Patterning of the dorsal epidermis is controlled by the persistence and reach of the signal within the time window when the switch is functional and not by the concentration of the signal (Fig. 7).
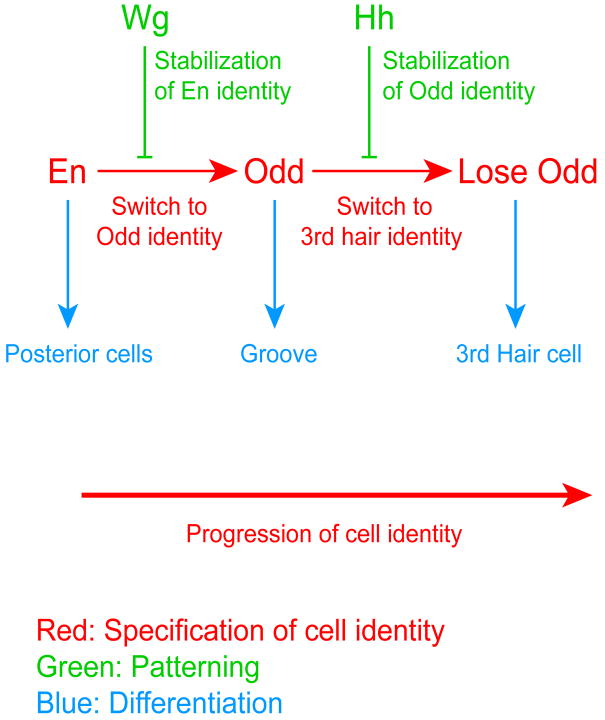
Figure 7. Model of progression of cell identity.
Progression of cell identities (red) proceeds independently of the Wg and Hh patterning signals. Patterning signals (green) stabilize already specified cell identities and prevent their switch toward different identities. Differentiation acts on the cells according to their current cell identity (blue). The already differentiated second row of groove cells switch from groove cells to cells producing hairs, showing that differentiation does not block the other two mechanisms. Interplay between cell identity progression and patterning signals ensures that a cell identity is specified when the previous and adjacent one is patterned. Newly specified cells take the patterned cells as a reference, so the anterior and the posterior side of a group of cells is patterned at different stages by distinct signals. The progression of cell identity is therefore visible in the spatial sequence of cell types present in the wild-type embryo. Note that wild-type Odd cells normally receive Hh early. This early influence maintains the population that will later transition to hair producing cells, explaining why these cells are missing in Hh mutants.
Our observations suggest that cell identity and differentiation are independently controlled. Groove differentiation begins before refinement of the Odd domain is complete, as the two remaining rows of Odd cells begin to express some groove differentiation markers (Fig. S1), showing that differentiation depends on the cell’s identity at that time. As the Odd domain is further narrowed to a single cell stripe, differentiation markers are lost along with Odd expression from the row further from the Hh/En cells. Because grooves develop specifically from cells that express the Odd marker well before groove morphogenesis, and because signaling to maintain Odd precedes morphogenesis, we conclude that morphogenesis requires cell prior competence or specification, and does not result simply from the direct influence of signaling on cellular morphogenesis.
How general is this inhibition of cell identity progression by patterning signals? This mechanism may be relevant to the development of stem cells, whose environment provides stabilizing signals that maintain stemness. For example, vertebrate homologues of Wg (Wnt), have been proposed to maintain stemness in hematopoietic stem cells (HSCs) (Reya et al., 2003; Willert et al., 2003). It is likely that Wnt signaling blocks the transition from HSC to the multipotent progenitor identity. Importantly, progenitors may need to escape the Wnt signal in order to proceed through hematopoiesis, since transient Wnt stimulation enhances hematopoietic reconstitution, but constitutive signaling induced by transgenic pathway activation prevents the appearance of progenitors, and hematopoiesis fails (Kirstetter et al., 2006; Scheller et al., 2006). It is unclear why similar experiments using retroviral signal activation produced different result (Reya et al., 2003).
Later stages of hematopoiesis appear to be similarly regulated by Wnt signaling. in vivo constitutive activation of the Wnt pathway blocks development of multiple hematopoietic lineages without affecting already differentiated cells (Kirstetter et al., 2006; Scheller et al., 2006). Transitions between blood cell identities may therefore constitute steps controlled by Wg signaling, analogous to the switch we describe between En and Odd identities. Additional work will be needed to test whether hematopoietic stem cell development is controlled by signal-dependent antagonism of switches governing a progression of cell identity changes.
Supplementary Material
Acknowledgments
We thank J. Skeath, P. Therond, J. Reinitz and the DSHB for antibodies and the Bloomington stock center for flies. We thank T. Clandinin for use of his SP2 microscope and A. Gallet, P. Therond, C. Alexandre, J.P. Vincent, M. Simon, D. Ma and members of the Axelrod lab for critical comments and discussions. This work was supported by NIH R01GM59823 (to JDA). SV was supported by fellowships from EMBO and the Human Frontier Science Program. NP is an investigator of the Howard Hughes Medical Institute.
References
- Alexandre C, Lecourtois M, Vincent J. Wingless and Hedgehog pattern Drosophila denticle belts by regulating the production of short-range signals. Development. 1999;126:5689–98. doi: 10.1242/dev.126.24.5689. [DOI] [PubMed] [Google Scholar]
- Baker NE. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: The spatial distribution of a transcript in embryos. Embo Journal. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994;8:899–913. doi: 10.1101/gad.8.8.899. [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–63. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Coulter DE, Swaykus EA, Beran-Koehn MA, Goldberg D, Wieschaus E, Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. Embo J. 1990;9:3795–804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter DE, Wieschaus E. Gene activities and segmental patterning in Drosophila: analysis of odd-skipped and pair-rule double mutants. Genes Dev. 1988;2:1812–23. doi: 10.1101/gad.2.12b.1812. [DOI] [PubMed] [Google Scholar]
- DiNardo S, O’Farell PH. Establishment and refinement of segmental pattern in the Drosophila embryo: spatial control of engrailed expression by pair rule genes. Genes and Development. 1987;1:1221–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk-Jongens J, Kassis JA, O’Farrell PH. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A, McGinnis WJ, Gehring WJ. Isolation of a homeobox-containing gene from the engrailed region of Drosophila and the spatial distribution of its transcript. Nature. 1985;313:284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- Florence B, Guichet A, Ephrussi A, Laughon A. Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene. Development. 1997;124:839–47. doi: 10.1242/dev.124.4.839. [DOI] [PubMed] [Google Scholar]
- Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev Suppl. 1993:115–24. [PubMed] [Google Scholar]
- Grevengoed EE, Loureiro JJ, Jesse TL, Peifer M. Abelson kinase regulates epithelial morphogenesis in Drosophila. J Cell Biol. 2001;155:1185–98. doi: 10.1083/jcb.200105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, Dinardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 2005;19:709–18. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76:449–60. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–36. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Ingham P, Taylor A, Nakano Y. Role of the Drosophila patched gene in positional signaling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–23. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–56. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Siden I, O’Farrell P, Simon M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Larsen CW, Hirst E, Alexandre C, Vincent JP. Segment boundary formation in Drosophila embryos. Development. 2003;130:5625–35. doi: 10.1242/dev.00867. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Macdonald P, Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skipped genes. Nature. 1987;328:440–2. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–34. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A. Development and patterning of the Drosophila epidermis. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 517–608. [Google Scholar]
- Martinez-Arias A, Baker N, Ingham PW. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:153–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A, Hayward P. Filtering transcriptional noise during development. Nature Reviews Genetics. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A, Lawrence PA. Parasegments and compartments in the Drosophila embryo. Nature. 1985;313:639–42. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Oda H, Tsukita S. Dynamic features of adherens junctions during Drosophila embryonic epithelial morphogenesis revealed by a Dalpha-catenin-GFP fusion protein. Dev Genes Evol. 1999;209:218–25. doi: 10.1007/s004270050246. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics. 1989;121:333–52. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–92. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Vorbruggen G, Jackle H. Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol Cell. 2000;6:203–9. [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–66. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–14. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Saulier-Le Drean B, Nasiadka A, Dong J, Krause HM. Dynamic changes in the functions of Odd-skipped during early Drosophila embryogenesis. Development. 1998;125:4851–61. doi: 10.1242/dev.125.23.4851. [DOI] [PubMed] [Google Scholar]
- Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, Leutz A. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 2006;7:1037–47. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Über induktion von Embryonalagen durch Implantation Artfremder Organisatoren. Roux’ Arch f Entw mech. 1924;100:599–638. [Google Scholar]
- Tabata T, Kornberg TB. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginla discs. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Nakano Y, Mohler J, Ingham PW. Contrasting distributions of patched and hedgehog proteins in the Drosophila embryo. Mech Dev. 1993;42:89–96. doi: 10.1016/0925-4773(93)90101-3. [DOI] [PubMed] [Google Scholar]
- Tepass U, Theres C, Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–99. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Klingensmith J, Perrimon N, Nusse R. Cell patterning in the Drosophila segment: engrailed and wingless antigen distributions in segment polarity mutant embryos. Dev Suppl. 1993:105–14. [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development. 1996;122:131–9. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–52. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150:1361–74. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Wharton KA, Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–95. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.