Abstract
The origin recognition complex (ORC) has an important function in determining the initiation sites of DNA replication. In higher eukaryotes, ORC lacks sequence-specific DNA binding, and the mechanisms of ORC recruitment and origin determination are poorly understood. ORC is recruited with high efficiency to the Epstein–Barr virus origin of plasmid replication (OriP) through a complex mechanism involving interactions with the virus-encoded EBNA1 protein. We present evidence that ORC recruitment to OriP and DNA replication function depends on RGG-like motifs, referred to as LR1 and LR2, in the EBNA1 amino-terminal domain. Moreover, we show that LR1 and LR2 recruitment of ORC is RNA dependent. HMGA1a, which can functionally substitute for LR1 and LR2 domain, can also recruit ORC in an RNA-dependent manner. EBNA1 and HMGA1a RGG motifs bound to structured G-rich RNA, as did ORC1 peptides, which interact with EBNA1. RNase A treatment of cellular chromatin released a fraction of the total ORC, suggesting that ORC association with chromatin, and possibly cellular origins, is stabilized by RNA. We propose that structural RNA molecules mediate ORC recruitment at some cellular and viral origins, similar to OriP.
Keywords: EBNA1, ORC, replication, RNA binding
Introduction
Eukaryotic DNA replication is a highly regulated process that guarantees that the genome is faithfully duplicated every cell cycle (Blow and Dutta, 2005; Stillman, 2005; DePamphilis et al, 2006). Initiation sites, referred to as origins, must be spatially and temporally coordinated to ensure that the genome is completely replicated during a single cell cycle (Machida et al, 2005). Origins are established by the formation of a pre-replication complex nucleated by the origin recognition complex (ORC) (Bell, 2002; Sasaki and Gilbert, 2007). ORC is an evolutionarily conserved multiprotein complex, which consists of several subunits that possess ATPase activity. In Saccharomyces cerevisiae, ORC binds DNA in an ATP-dependent manner and recognizes a specific DNA consensus sequence (Bell and Stillman, 1992). However, in most other organisms, ORC lacks sequence-specific DNA-binding activity. Schizosaccharomyces pombe ORC lacks sequence-specific binding, but the spORC4 subunit possesses a species-specific AT-hook domain that confers non-specific binding to AT-rich DNA (Chuang and Kelly, 1999). In higher eukaryotes, replication can initiate at discrete sites (Abdurashidova et al, 2000) or within diffuse zones (Dijkwel et al, 2002), and the mechanisms through which ORC establishes a functional origin have not been simple to determine.
The Epstein–Barr virus (EBV) origin of plasmid replication (OriP) provides an attractive model to study ORC recruitment in human cells (Lindner and Sugden, 2007). EBV is a human herpesvirus that establishes latent infections in multiple cell types and contributes to various malignancies (Young and Rickinson, 2004). The latent form of the virus exists as an episomal minichromosome that replicates once per cell cycle and segregates faithfully, similar to the cellular chromosome (Kanda et al, 2007; Nanbo et al, 2007). OriP is composed of two regions, the family of repeats (FR) and the dyad symmetry (DS). A viral-encoded protein, EBNA1, binds to both FR and DS, but ORC recruitment and replication initiation occur primarily at the DS region (Chaudhuri et al, 2001; Dhar et al, 2001; Schepers et al, 2001; Ritzi et al, 2003). Other cellular factors bind to DS, including the telomere repeat factor 2 (TRF2) protein, which we have shown can interact with ORC and contribute to OriP replication activity (Deng et al, 2002; Atanasiu et al, 2006). However, TRF2 by itself cannot efficiently stimulate DNA replication at OriP and requires EBNA1 for replication and plasmid maintenance function. The precise contribution of EBNA1 to replication function is not completely understood.
In some circumstances, recruitment of ORC to DNA may be sufficient to establish an active origin of replication. Recruitment of ORC1, ORC2, or CDC6 by a Gal4 tethering system is sufficient to establish an active origin of replication on plasmids (Takeda et al, 2005). ORC can also be recruited and an active origin can be established at the rat aldolase B gene by specific interactions with transcription factor AIF-C1 (Saitoh et al, 2002; Minami et al, 2006). Other sequence-specific DNA-binding proteins have been reported to interact with replication initiation factors, including Drosophila myb (Beall et al, 2002), human c-myc (Dominguez-Sola et al, 2007), and human TRF2 at OriP (Deng et al, 2007). Architectural proteins, such as HMGA1a protein, have also been implicated in ORC recruitment and origin selection in human cells (Thomae et al, 2008).
The ability of DNA-binding proteins to recruit ORC to specific genome locations is a first step to explain origin site selection in higher eukaryotes. However, the molecular mechanisms through which these factors recruit ORC and regulate origin function have not been clearly elucidated. We present data indicating that EBNA1 and HMGA1a protein recruit ORC through an RNA-dependent mechanism. A function of RNA molecules in mammalian origin formation and initiation of DNA replication has been suggested through biochemical reconstitution studies (Christov et al, 2006). Our findings suggest that RNA may mediate ORC recruitment and origin formation at various chromosomal locations.
Results
EBNA1-linking domains LR1 and LR2 recruit ORC
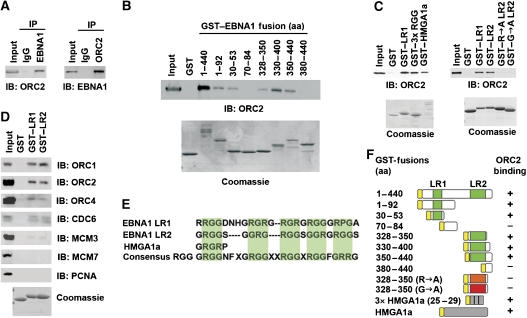
It has been previously established that ORC is recruited and replication is initiated in an EBNA1-dependent manner at the DS of OriP (Chaudhuri et al, 2001; Dhar et al, 2001; Schepers et al, 2001; Ritzi et al, 2003). However, the interaction between EBNA1 and ORC has been difficult to demonstrate by gentle immunoprecipitation (IP) methods (data not shown). We found that when Raji cell extracts were generated rapidly using more disruptive RIPA buffer, EBNA1 could efficiently co-IP with ORC2 (Figure 1). ORC2 was identified in the IP with EBNA1-specific antibody, but not with control IgG (Figure 1A, left panel). Similarly, EBNA1 was detected in the IP with ORC2-specific antibody, but not with control IgG (Figure 1A, right panel). EBNA1 could also be detected in ORC2 IPs from 293T cells transfected with EBNA1 transgenes (Supplementary Figure S1). This indicates that ORC2 and EBNA1 can form a stable complex in latently infected Raji cell, as well as in transfected 293T cell extracts.
Figure 1.
EBNA1-linking regions 1 and 2 are necessary and sufficient for ORC interaction. (A) Raji cell extracts were immunoprecipitated with anti-EBNA1antibody or IgG control (left panel) or anti-ORC2 antibody and IgG control (right panel). IPs were analysed by immunoblot with anti-ORC2 (left panel) and anti-EBNA1 (right panel). Input represents ∼5% of the total input for each IP. (B) GST–EBNA1 fusion peptides were assayed for their ability to recruit ORC2 from HeLa nuclear extracts by immunoblot with anti-ORC2. Purified GST fusion proteins were assayed by Coomassie Blue staining of SDS–PAGE gels (lower panels). EBNA1 amino-acid residues fused to GST are indicated above each lane. (C) GST fusion proteins (GST, GST–LR1 (aa 30–53), GST–3 × RGR, GST–HMGA1a, GST–LR2 (aa 328–350), GST–LR2 (R → A), or GST–LR2 (G → A) were assayed for their ability to recruit ORC2 from HeLa nuclear extracts. ORC2 recruitment was assayed by immunoblot with anti-ORC2 antibody. GST fusion protein expression and purity was assayed by Coomassie blue staining (lower panels). (D) GST–LR1, GST–LR2, or GST alone were incubated with HeLa nuclear extract and assayed by immunoblot for ORC1, ORC2, ORC4, CDC6, MCM3, MCM7, or PCNA, as indicated. Coomassie stain of GST fusion peptides is shown in the lower panel. (E) Sequence alignment of EBNA1 LR1, LR2, HGMA1a RGR, and consensus RGG domain. (F) Summary of GST fusion proteins and their ability to bind ORC2 as represented by experiments shown in panels B and C.
To map the ORC interaction domain in EBNA1, we generated a series of GST–EBNA1 fusion peptides and assayed them for their ability to recruit ORC2 from HeLa nuclear extracts (Figure 1B). Previous studies have shown that RGG-like motifs in the EBNA1 amino-terminal domain, referred to as linking regions 1 and 2 (LR1 and LR2), are necessary for replication and plasmid maintenance function (Yates and Camiolo, 1988; Wu et al, 2002; Sears et al, 2004). We found that that EBNA1 amino-terminal domain (aa 1–440) could efficiently recruit ORC2, whereas GST alone demonstrated no ORC-binding activity (Figure 1B). Further deletion mapping revealed that either LR1 (aa 30–53) or LR2 (aa 328–350) alone was sufficient to recruit ORC2, although not as robust as EBNA1 (aa 1–440) (Figure 1B). Deletion of LR1 and LR2 from EBNA1 eliminates ORC binding in co-IP experiments, indicating that these domains are necessary for ORC recruitment in the context of the EBNA1 DNA-binding domain in vivo (Supplementary Figure S1).
The amino terminus of EBNA1 can be functionally substituted with the high-mobility group A1a protein (HMGA1a) in replication and plasmid maintenance assays (Hung et al, 2001; Sears et al, 2004). To test whether ORC recruitment correlated with replication function, we assayed the ability of GST–HMGA1a to recruit ORC in vitro. We found that GST–HMGA1a efficiently recruits ORC from HeLa nuclear extracts (Figure 1C, left panel). We also found that three copies of the six amino-acid RGR-like motif of HMGA1a, GST-3 × RGR, was sufficient for ORC recruitment (Figure 1C, left panel), suggesting that this common minimal motif also found in LR1 and LR2 constituted an ORC interaction surface. To determine if arginines or glycines in this motif were essential for ORC recruitment, we assayed mutations in LR2 that substitute all arginines (R → A) or glycines (G → A) to alanine. We found that both mutations abrogated ORC recruitment, indicating that both arginines and glycines in the RGG motif in LR2 contribute to ORC interactions (Figure 1C, right panel).
As ORC2 may exist as a single polypeptide separate from other ORC subunits, we tested whether LR1 and LR2 were capable of recruiting other ORC subunits and replication initiation factors. We found that LR1 and LR2 were capable of recruiting multiple subunits of ORC, including ORC1, ORC2, and ORC4. LR1 and LR2 could also recruit the ORC1-related protein CDC6, but did not efficiently recruit other components of the replication initiation complex, including MCM3, MCM7, or PCNA (Figure 1D). These experiments indicate that one copy of either LR1 or LR2 is sufficient for recruitment of multiple ORC subunits, and that the interaction of EBNA1 and HMGA1a with ORC is mediated through a common RG-rich motif (summarized in Figure 1E and F).
LR1 or LR2 stimulates DNA replication and plasmid maintenance
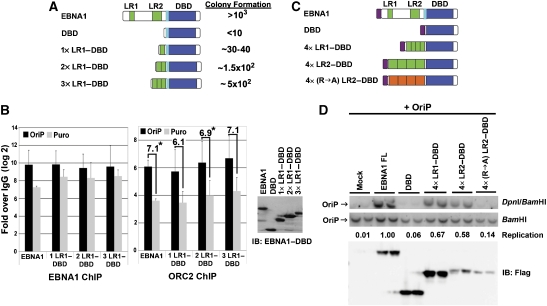
We next asked whether LR1 and LR2 were sufficient for ORC recruitment in vivo and OriP-dependent DNA replication (Figure 2). As a first approach, we determined whether 1, 2, or 3 copies of LR1 when bound to the EBNA1 DNA-binding domain (DBD) could confer stable plasmid maintenance using a colony formation assay (Gerhardt et al, 2006). Stable 293 cell lines expressing either full-length EBNA1, the EBNA1 DBD, or tandem fusions of 1 × , 2 × , or 3 × LR1 to the EBNA1 DBD were transfected with OriP-plasmid containing the puromycin resistance gene (Figure 2A). After 2 weeks selection in puromycin, the number of stable episomes were quantified by plasmid rescue and colony formation in E. coli. We found that EBNA1 DBD yielded between 0 and 1% of the colonies formed by wt EBNA1, whereas 1 × LR1 yielded ∼3.8%, 2 × LR1 yielded ∼15.6%, and 3 × LR1 yielded ∼50% of EBNA1 wt (Figure 2A). EBNA1 protein levels were expressed to similar levels in all cell lines generated (Figure 2B, right panel). These findings indicate that a single copy of LR1 fused to the EBNA1 DBD can promote colony formation of OriP-containing plasmids, and increasing numbers of LR1 greatly enhance colony formation. We next determined if a single copy of LR1 fused to the EBNA1–DBD was capable of recruiting ORC2 in vivo as measured by chromatin IP (ChIP) assay (Figure 2B). ORC2 and EBNA1 were assayed for their binding to the stable plasmids recovered from the colony formation assays described in Figure 2A (and Supplementary Figure S2). In cells containing wt EBNA1, ORC2 was enriched at OriP by ∼64-fold relative to the control IgG. In cells with 1 × LR1–DBD, ORC2 was enriched ∼32-fold at OriP relative to IgG, whereas in cells containing 2 × and 3 × LR1–DBD, ORC2 enrichment was closer to ∼64-fold similar to that found in wt EBNA1. All EBNA1 constructs were highly enriched at OriP relative to IgG control (>600-fold). EBNA1 and ORC2 were both enriched at least four-fold more at OriP relative to the puromycin gene (PURO) (Supplementary Figure S2). The background ChIP binding of EBNA1 and ORC to PURO is likely a result of its close proximity to OriP on the stable episome (∼2 kB). No OriP DNA could be amplified from the cells containing the DBD alone (data not shown). Thus, a single copy of LR1 is sufficient for stable plasmid maintenance and ORC recruitment to OriP, and multiple copies enhance this maintenance and ORC recruitment in vivo.
Figure 2.
LR1 or LR2 confers ORC recruitment and replication at OriP. (A) Schematic of EBNA1 and fusion peptide constructs used in colony formation assay. The light blue box represents the EBNA1 nuclear localization signal (NLS). One, two, or three copies of LR1 (green boxes) were fused to EBNA1 DBD and co-transfected with plasmid-containing OriP and Puromycin-resistance gene (Puro). The number of bacterial colonies transformed with recovered plasmid OriP were quantified as an average of several experiments relative to wt EBNA1 protein. (B) Puromycin-resistant 293T cells pools generated with EBNA1 wt, 1 × LR1–DBD, 2 × LR1–DBD, or 3 × LR1–DBD were harvested for ChIP 14 days post-transfection. Enrichment of EBNA1 and ORC at OriP (black bars) or Puromycin resistance gene (grey bars) was compared with IgG controls by real-time PCR. DBD alone is not shown, as no OriP DNA was detected among the few puromycin-resistant colonies. ChIP data are presented as the log 2 of Ct values relative to IgG controls. The relative enrichment of ORC2 binding at OriP relative to the Puro gene is indicated by the numbers above each set of bars in the panel to the right. P-value of <0.05 using single ANOVA analysis are indicated by *. Expression levels for EBNA1-derived proteins are shown in the western blot to the right. (C) Schematic of fusion peptide constructs used for transient replication assay. The magenta box represents the N-terminal FLAG-epitope. (D) EBNA1 fusion peptides described in C were co-transfected with OriP-containing plasmid. DNA was harvested 72 h post-transfection and visualized with OriP-specific probe by Southern blot hybridization. Replication was measured as resistance to DpnI restriction digestion. Replication efficiency is calculated as DpnI resistance over input (BamHI) and normalized to full-length EBNA1 as quantified by ImageQuant software. Immunoblot below shows protein expression levels for FLAG-tagged EBNA1 proteins.
EBNA1-dependent DNA replication was also measured using a transient transfection assay (Figure 2C and D, and Supplementary Figure S3). HeLa cells were co-transfected with Flag-tagged versions of full-length EBNA1, the EBNA1 DBD, 4 × LR1–DBD, 4 × LR2–DBD, or a mutant form of 4 × LR2–DBD where all arginines were changed to alanines, and an OriP-containing plasmid (Figure 2C). DNA was harvested after 72 h and replication was measured by resistance to DpnI restriction digest (Figure 2D). Full-length EBNA1 supports OriP-dependent replication, whereas the DBD is unable to support replication. Addition of four copies of either LR1 or LR2 fused to the DBD rescues replication, at levels 67% and 58% of full-length EBNA1. Mutagenesis of the LR2 arginines (R → A) abrogate the replication function of 4 × LR2. Protein expression levels of 4 × LR2–DBD was significantly less than the wild type and DBD, but still they stimulated replication to measurable levels, indicating that the LR2 motif is capable of stimulating DNA replication. We failed to detect transient replication of a single copy of LR1 or LR2 (Supplementary Figure S3; and data not shown). This failure may reflect that lack of sensitivity of the transient replication assay, as a single copy of LR1 could generate stable episomes and recruit ORC in ChIP assays (Figure 2A and B). However, the increased replication of 4 × LR1 relative to 1 × LR1 suggests that robust replication requires multiple RG-like interaction surfaces.
RNA-dependent recruitment of ORC by LR1 and LR2
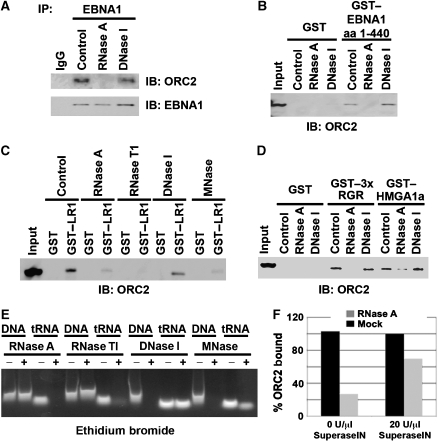
LR1 and LR2 have been referred to as RGG motifs because of their sequence similarity to a family of RNA-binding proteins (Kiledjian and Dreyfus, 1992; Snudden et al, 1994). EBNA1 is known to bind RNA through this domain (Snudden et al, 1994; Lu et al, 2004). Therefore, we asked if RNA was involved in the interaction between EBNA1 and ORC. EBNA1 from Raji cell extracts was immunoprecipitated and then incubated with either RNase A or DNase I during the wash step. We found that RNase A but not DNase I disrupted ORC2 association with EBNA1 in an immunoprecipitated complex (Figure 3A). Recruitment of ORC from HeLa nuclear extracts by GST–EBNA1 amino-terminal domain (aa 1–440), which contains both LR1 and LR2, was also sensitive to RNase A, but not to DNase I treatment I (Figure 3B). To rule out the possibility that RNase A was interfering with EBNA1–ORC2 interactions, we tested several other nucleases for their ability to disrupt the recruitment of ORC by GST–LR1 (Figure 3C). We found that ORC recruitment by LR1 can be inhibited by RNase A, RNase T1, and micrococcal nuclease I (MNase I), all of which have enzymatic activity towards RNA. DNase I, however, did not disrupt ORC recruitment by LR1 nor did the addition of DNA intercalating agent ethidium bromide (data not shown). The activity of each enzyme was monitored in parallel control experiments using single-stranded DNA or tRNA substrates (Figure 3E). RNase A but not DNase I also disrupts ORC recruitment by both the RGR motif of HMGA1a and, to a lesser extent, full-length HMGA1a (Figure 3D). Inhibition of the enzymatic activity of RNase A by SuperaseIN partially reversed the effects of RNase A on ORC recruitment by EBNA1, further indicating that RNA partially mediates this interaction (Figure 3F).
Figure 3.
RNA-dependent recruitment of ORC by EBNA1 LR and HMGA1a RGR domains. (A) EBNA1 IP from Raji cells were incubated with 0.02 mg/ml RNase A, 0.2 U/ml DNase I, or buffer control. ORC2 recruitment was monitored by immunoblot (IB) with anti-ORC2 (top panel) or anti-EBNA1 (lower panel). (B) GST or GST–EBNA1 1–440 (containing LR1 and LR2) was incubated with HeLa nuclear extracts and then treated with 0.02 mg/ml RNase A, 0.2 U/ml DNase I, or buffer control. ORC2 recruitment was monitored by IB. (C) GST or GST–LR1 were incubated with HeLa nuclear extract and then treated with either 0.02 mg/ml RNase A, 2 U/ml RNase T1, 0.2 U/ml DNase I, 0.2 U/ml micrococcal nuclease (MNase I), or buffer control. ORC2 recruitment was monitored by IB with anti-ORC2. (D) GST, GST–3 × RGR, or GST–HMGA1a was incubated with HeLa nuclear extracts and then treated with 0.02 mg/ml RNase A, 0.2 U/ml DNase I, or buffer control and monitored by IB with anti-ORC2. (E) Control digestions were preformed using 50 μg of single-stranded, sonicated, heat-denatured salmon sperm DNA (ssDNA) or 100 μg of tRNA under the same conditions for RNase A, RNase TI, DNase I, and MNase I used in the IPs and GST pull-downs mentioned above. ssDNA and tRNA were separated by agarose gel electrophoresis and visualized after ethidium bromide staining. (F) RNase A-dependent loss of ORC2 binding to GST or GST–LR2 was tested by the addition of SuperaseIN (20 U/μl) in reactions containing either 0 or 90 ng/ml RNase A, as indicated. ORC2 binding was measured by western blot and quantified using ImageQuant and presented as percentage of ORC2 binding in the absence of RNase A.
EBNA1 binds its own mRNA
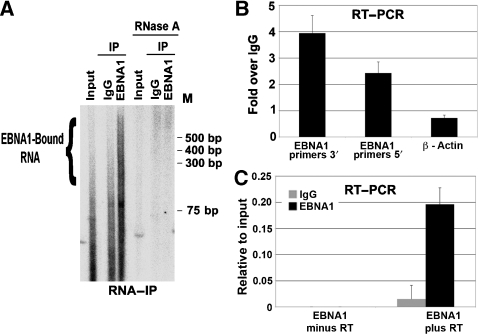
Evidence that EBNA1 associates with RNA in Raji cells was provided by extracting and radiolabelling RNA from EBNA1 immunoprecipitates (Figure 4A). Slower-mobility RNA was isolated from EBNA1 immunoprecipitates, but not from IgG control immunoprecipitates, and all species were sensitive to RNase A treatment (Figure 4A, right panel). These data indicate that EBNA1 interacts with RNA in nuclear extracts, and this RNA contributes to the stable recruitment of ORC. EBNA1 has been reported to bind to its own RNA (Lu et al, 2004). To test this possibility in vivo, we analysed the RNA recovered by EBNA1 immunoprecipitates by reverse transcriptase PCR (RT–PCR) with primers specific for the 5′- or 3′-ends of the EBNA1 mRNA or a control primer set for cellular actin mRNA (Figure 4B and C). We found that both the regions of EBNA1 mRNA were enriched 2.5- to 4-fold in EBNA1 immunoprecipitates relative to control IgG immunoprecipitates (Figure 4B). Actin mRNA was not enriched in the EBNA1 immunoprecipitates. No EBNA1 RNA was detected in reactions that lacked RT, indicating that the PCR amplification was RNA-template dependent. These findings indicate that EBNA1 mRNA is an endogenous substrate for EBNA1 RNA binding in latently infected Raji cells.
Figure 4.
EBNA1 binds its own mRNA. (A) RNA isolated from Raji cell extracts (input) or from immunoprecipitates with anti-EBNA1 or control IgG was radiolabelled with 32P-γATP and polynucleotide kinase. Labelled RNA was visualized by autoradiography of native polyacrylamide gels. RNase A-treated samples are shown on the right, as indicated. (B) RT–PCR with RNA purified from EBNA1 IPs from Raji cells. IP-derived RNA was amplified with primers specific for the EBNA1 ORF 5′ or 3′ or with primers for cellular β-actin mRNA. (C) RT–PCR with RNA purified from EBNA1 or control IgG IP using primers for EBNA1 ORF 5′-region. Reactions were performed with RT (plus) or without RT (minus) as indicated.
EBNA1 binds structured G-rich RNA oligonucleotides
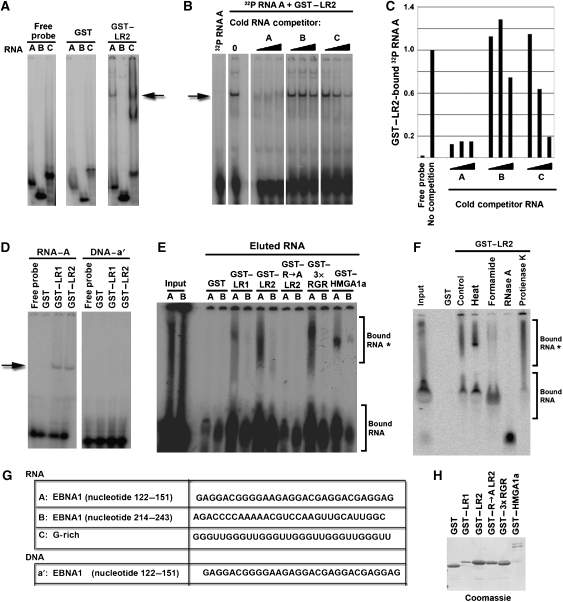
Full-length EBNA1 has been shown previously to exhibit general RNA binding, with a preference for G-rich RNA (Lu et al, 2004). We reasoned that G-rich RNA sequence motifs in the EBNA1 mRNA may serve as substrates for RNA binding by EBNA1 RGG motifs (Figure 5). To test this possibility, we first compared the ability of GST or GST-LR2 to bind to three different 30-nucleotide RNA oligomers (Figure 5A). RNA species A and B are from the EBNA1 mRNA coding region. RNA species A corresponds to a region of the EBNA1 transcript that is G-rich (nucleotides 122–151), whereas RNA species B corresponds to a region of the EBNA1 transcript that is G-poor (nucleotides 214–243) (Figure 5G). RNA species C is G-rich and predicted to form a G-quadruplex structure (Darnell et al, 2001, 2004). We found that purified GST–LR2 peptide bound RNA oligomers A and C, two of the G-rich RNAs that were tested, but not the G-poor RNA species B (Figure 5A). The RNA-binding specificity was further examined by competition with cold competitor RNAs (Figure 5B). GST–LR2 bound to radiolabeled RNA A oligomer was competed effectively by increasing concentrations of RNA A, partially by RNA C, and poorly by RNA B (Figure 5B, and quantified in 5C). We also show that GST–LR1 bound RNA with similar affinity and specificity as GST–LR2 (Figure 5D; and data not shown), and neither peptide bound to a single-stranded DNA oligomer of the same sequence (a′) (Figure 5D, right panel). These experiments suggest that LR1 and LR2 bind G-rich RNA preferentially and may recognize some specific RNA sequence or structure.
Figure 5.
ORC recruiting modules bind G-rich RNA. (A) Purified GST and GST–LR2 were assayed for binding to 32P RNA oligonucleotides A, B, or C using EMSA. RNA-bound complex is indicated by the arrow. (B) Specificity of the interaction between GST–LR2 and 32P-labelled RNA probe A was assayed by addition of 3 × , 9 × , or 27 × fold molar excess of cold competitor RNAs A, B, or C to the binding reaction. The major complex formed between 32P RNA A and GST–LR2 is indicated by the arrow. (C) Graphical quantification of bound RNA A by GST–LR2 from the competition assay in panel B using ImageQuant software analysis. (D) GST, GST–LR1, and GST–LR2 were assayed for binding to 32P-labelled RNA A (left panel), or single-stranded DNA a′ of the same sequence (right panel). (E) 32P-labelled RNA A or B was eluted from GST pull-down experiments with GST, or GST fused to LR1, LR2, R → A LR2, 3 × RGR, and HMGA1a and assayed by gel electrophoresis and visualized by PhosphorImager. Input RNA is indicated in lanes 1 and 2. (F) 32P-labelled RNA A was eluted from GST pull-down with GST or GST–LR2, and then treated with heat (95°C, 3 min), formamide (80%), RNase A (0.2 mg/ml), or Protease K (0.1 mg/ml), as indicated. (G) Sequence of the RNA species A, B, and C and DNA species a′ used in these experiments. (H) Proteins used in the RNA pull-down experiment in E were separated by SDS–PAGE and visualized by Coomassie staining.
The relatively weak binding of GST–LR1 and LR2 in EMSA may have been a result of the poor mobility of structured RNA–protein complexes in polyacrylamide gels. To explore this possibility, we examined the ability of various GST peptides to bind 32P-RNA (G-rich or G-poor) in a GST pull-down assay. Radiolabelled RNA oligomer was incubated with sepharose-bead-bound GST peptides, washed, and then eluted with SDS. Bound and eluted RNA was then analysed by native polyacrylamide gel electrophoresis (Figure 5E). We found that GST–LR1, GST–LR2, GST–3 × HMGA1a RGR, and GST–HMGA1a bound a slower-mobility form (*) of RNA oligomer A but not B, whereas GST alone and GST-R → A LR2 did not bind either RNA oligomer (Figure 5E). To better assess the nature of the slower-mobility RNA-bound species (*), GST-bound and SDS-eluted RNA was subjected to heat (3 min at 95°C), formamide (80%), RNase A, or protease K treatment (Figure 5F). Heat denaturation improved the resolution of the slower-migrating RNA species, whereas fomamide shifted most of the bound species to the faster-migrating species. Protease K had little effect and RNase A eliminated all RNA forms. These finding suggest that the slower-migrating species (*) is a consequence of a secondary RNA structure that can be disrupted by formamide. These results also indicate that EBNA1–LR1, EBNA1–LR2, and HMGA1a RGR motif selectively bind the structured form of the G-rich RNA.
ORC1 interacts with EBNA1 and G-rich RNA
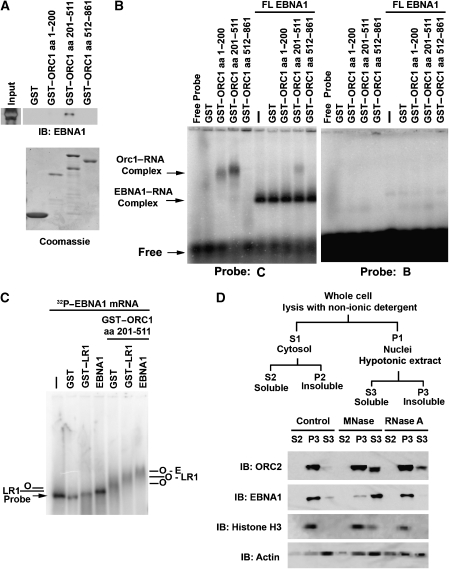
Previous studies have implicated ORC1 in the physical interaction with origin-recruiting proteins, including AIF-C1 and TRF2 (Saitoh et al, 2002; Atanasiu et al, 2006; Minami et al, 2006; Noguchi et al, 2006). We therefore tested the ability of purified ORC1 peptides to interact with purified EBNA1 protein (Figure 6A). We found that baculovirus expressed and purified EBNA1 bound to GST–ORC1 (aa 201–511), but not to the amino-terminal BAH domain (aa 1–200) or to the carboxy-terminal ATPase domain (aa 512–861) or to GST-alone. The ability of ORC peptides to interact with RNA oligomers was assayed using an agarose-gel EMSA. We found that GST–ORC1 (aa 201–511), and to a lesser extent GST–ORC1 (aa 1–200), bound G-rich RNA probe (Figure 6B, left panel, lanes 1–5), but did not bind to the G-poor RNA oligomer, probe B (Figure 6B, right panel). Addition of full-length baculovirus EBNA1 formed a stable complex in the agarose EMSA with the G-rich RNA (Figure 6B, left panel, lanes 6–10), but not with the G-poor probe (Figure 6B, right panel, and Supplementary Figure S4). Addition of GST–ORC1 peptides did not alter the mobility of the EBNA1 complex. However, GST–ORC1 (aa 201–511) retained some binding activity, suggesting that it binds competitively with EBNA1 for similar G-rich oligonucleotides (Figure 6B, left panel, lane 9). The failure to observe an altered EBNA1 mobility in the presence of GST–ORC1 may be due to the RNA oligomer size (30 nt) or to the relatively weak interaction between these purified subdomains. To test whether a larger RNA substrate may reveal a stable ORC1–EBNA1 complex, we generated an ∼1.5 kb RNA probe from the EBNA1 ORF using an in vitro transcription reaction with 32P dUTP (Figure 6C). Using the longer EBNA1 ORF RNA probe, we found that addition of EBNA1–LR1 or baculovirus-expressed full-length EBNA1 caused a small, but reproducible shift in mobility. Surprisingly, addition of GST–ORC1 (aa 201–511) caused a more pronounced, but diffuse mobility shift. Addition of GST–ORC1 (aa 201–511) with EBNA1–LR1 or full-length EBNA1 produced an altered mobility, consistent with the formation of a stable complex between EBNA1–ORC1 (aa 201–511) and the RNA probe. These results indicate that ORC1 (aa 201–511) can bind to G-rich RNA independently of EBNA1 and can form a stable complex with RGG-containing EBNA1 proteins in association with RNA substrates of sufficient size.
Figure 6.
ORC-associated RNA binding and RNA-dependent nuclear retention. (A) GST, GST–ORC1 (aa 1–200), GST–ORC1 (aa 201–511), or GST–ORC1 (aa 512–861) were assayed for binding to purified EBNA1 protein. Input EBNA1 protein is indicated to the left, and bound EBNA1 is detected by IB with anti-EBNA1 antibody (top panel). GST-fusion proteins were detected by Coomassie staining of SDS–PAGE gels (lower panel). (B) RNA binding of GST–ORC1 peptides or FL-EBNA1 or combinations of both proteins was assayed by agarose gel EMSA. G-rich RNA probe C (left panel) or G-poor probe B (right panel) was incubated with GST alone, GST–ORC1 (aa 1–200), GST–ORC1 (aa 201–511), and GST–ORC1 (aa 512–861) in the absence (left lanes) or presence (right lanes) of FL-EBNA1, as indicated above each lane. ORC1–RNA complex and the EBNA1–RNA complex are indicated by the arrows to the left of the EMSA. (C) EMSA in 1.5% agarose gel using 32P-labelled EBNA1 mRNA probe of ∼1.5 kb incubated with GST, GST–LR1, baculovirus-expressed EBNA1 without or with GST–ORC1 (aa 201–551) as indicated. Complexes formed by ORC1 (O), LR1, EBNA1(E), or combinations of these are indicated. (D) Raji cell nuclear pellets (P1) were incubated with 600 U/ml MNase I, 1.0 mg/ml RNase A, or control buffer, and then subject to subcellular fractionation as indicated in the schematic above (Mendez and Stillman, 2000). Fractions were assayed by immunoblot with antibodies to ORC2 (top panel), EBNA1, histone H3, or Actin (lower panel), as indicated to the left of each panel.
RNA stabilizes ORC association with cellular chromatin
The general requirement for RNA in mediating ORC recruitment to cellular chromosomal sites was investigated by examining ORC2 binding to bulk chromatin using the methods of Mendez and Stillman (2000) (Figure 6D). ORC2 is typically strongly associated with the chromatin fraction, but can be released from chromatin after MNase I treatment. As expected, we found that MNase I treatment released ∼40% of the ORC2 protein (Figure 6D). Next, we tested the ability of RNase A treatment to cause the release of ORC2 using the same conditions as that for MNase I treatment. We found that RNase A treatment caused ∼20% of ORC2 release into the soluble S3 fraction (Figure 6D). MNase I treatment also caused the release of EBNA1 and histone H3 from nuclear extracts, but did not cause a significant change in actin, which was present in all chromatin fractions. Interestingly, RNase A did not cause a significant release of EBNA1 from the chromatin fraction, perhaps not disrupting the highly stable DNA-binding activity of EBNA1 to viral DNA. Similarly, RNase A did not cause a loss of histone H3 or nuclear actin. These findings indicate that a fraction of ORC can be released from chromatin by RNase A treatment, supporting our model that RNA mediates ORC interactions with a subset of sites in chromatin that may include viral and cellular origins (Figure 7).
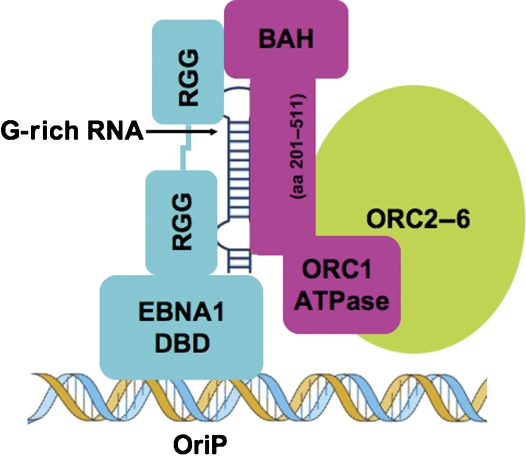
Figure 7.
Model of G-rich RNA-mediating recruitment of ORC by EBNA1 and other RGG-domain-containing proteins. G-rich RNA is bound by the RGG domains and by the ORC1 (aa 201–511) subdomain.
Discussion
Establishment and regulation of replication origins is a fundamental process in eukaryotic chromosome biology (Machida et al, 2005). Origin identity and activity may change during cellular differentiation and may be important for genome organization and stability (Norio et al, 2005). ORC binding is an essential first step in the process of forming an active origin. In higher eukaryotes, ORC lacks sequence-specific DNA binding, so it remains unclear what DNA-binding factors or chromatin environment direct ORC binding and determine the site of origin formation. It is also known that ORC can bind to many locations in the genome without forming an active site of DNA replication (Wyrick et al, 2001). These ORC-binding sites may be dormant origins that have been inactivated by S-phase checkpoint regulation, or they may have alternative functions in the establishment of heterochromatin or sister chromatid cohesion (Shirahige et al, 1998; Early et al, 2004; Sasaki and Gilbert, 2007; Shimada and Gasser, 2007). The precise mechanisms that regulate ORC localization on chromatin and its replication function remain poorly understood. Our findings suggest that structural RNAs presented by RGG-containing motifs contributes to the recruitment of ORC, and consequently, to origin formation.
The EBV OriP has been an instructive model for understanding ORC recruitment and regulation. In this study, we show that the OriP-binding protein, EBNA1, can form a stable complex with ORC. This interaction was mapped to the linking regions 1 and 2 (LR1/LR2) of EBNA1. LR1 and LR2 are well-characterized regions of EBNA1 that have been implicated in most of the functions of EBNA1, including replication, virus episome maintenance, and metaphase chromosome attachment (Sears et al, 2004). Interestingly, LR1 and LR2 can be substituted with several chromatin-associated proteins, including HMGA1a and histone H1 (Hung et al, 2001; Sears et al, 2003, 2004). Sears et al (2004) proposed that the common underlying molecular feature of these proteins is their AT-hook structure and their ability to bind AT-rich DNA. This model suggests that EBNA1 can tether EBV genomes to AT-rich domains of the cellular chromosome to function as a chromosome passenger during metaphase, a process linked to plasmid stability in dividing cells. However, this model did not evoke a function of ORC or of the formation of an origin of DNA replication. Our findings indicate that LR1, LR2, and HMGA1a RGR motif can recruit ORC, suggesting that ORC interaction is a common molecular feature of these different proteins.
One of the more surprising aspects of this study was the requirement of RNA for EBNA1 interaction with ORC. Co-IP of endogenous EBNA1 with ORC was disrupted by RNase A, but not DNase I treatment (Figure 3A). RNA was detected in EBNA1 immunoprecipitates (Figure 4A) and was degraded by RNase treatment, indicating that RNAse A efficiently removed RNA from the EBNA1-bound material. This strongly suggests that RNAse A-dependent disruption of EBNA1–ORC binding was due to the degradation of RNA, and not non-specific interference from the RNAse A protein. The LR1 and LR2 domains of EBNA1 also bound ORC in an RNA-dependent manner, as did the RGR motif found in the HMGA1a protein. Mutations in the EBNA1 LR2 domain disrupted ORC recruitment and RNA binding in EMSA, indicating that the RNA-binding and ORC-recruiting activity are specific for the EBNA1 peptide, and not contaminants from bacterial preparations. The LR1 and LR2 domains bound RNA with similar sequence preference to that of HMGA1a RGR motifs, suggesting that these proteins favour binding to a common RNA structure. In all cases, RNA binding correlated with ORC recruitment and DNA replication activity. We also found that a subdomain of ORC1 (aa 201–511) was capable of binding G-rich RNA in vitro (Figure 6A). This finding provides a simple mechanism for how RNA mediates an interaction between EBNA1 RGG motifs and ORC1.
EBNA1 can bind to a variety of RNA species, with a preference for G-rich oligonucleotides (Snudden et al, 1994; Lu et al, 2004). Our data indicate that endogenous EBNA1 protein from latently infected cells can bind to its own mRNA. The LR1 and LR2 domain of EBNA1 is necessary and sufficient for this RNA binding. Our findings also indicate that LR1 and LR2 recognize G-rich RNA with altered electrophoretic mobility, suggesting that they form higher-order structure. The RGG domain of FMRP binds G-rich RNA that can form G-quadruplex structures (Darnell et al, 2001, 2004). The G-rich RNA oligonucleotides that bound EBNA1 in this study are predicted to have the potential to form G-quadruplex structures on the basis of sequence. We also observed that ORC1 (aa 201–511) could also bind to the G-rich RNA, suggesting that G-rich RNA may mediate the interactions between EBNA1 RGG peptides and ORC1 (Figure 7). The precise length of the RNA species required for stable association has not been determined, but our agarose EMSA studies suggest that a 30-nt RNA may be insufficient to support the binding of both ORC and EBNA1 proteins, whereas larger RNA species (1.5 kb EBNA1 ORF) may be sufficient for stable complex formation. However, further studies will be required to determine the precise RNA identity, length, and conformation involved in complex formation with ORC.
It is also likely that ORC can be recruited to functional origins through non-RNA-dependent mechanisms. RNase A treatment of chromatin fractions released only ∼20% of the chromatin-bound ORC, whereas Mnase I treatment caused the release of ∼40% of ORC (Figure 6D). This is expected, as ORC is almost certainly recruited to many chromatin sites and replication origins through conventional protein–protein interactions. ORC recruitment to OriP is also likely to include additional interactions and mechanisms. Noguchi et al (2006) have shown that the ORC1 amino-terminal domain is important for OriP replication function and interaction with EBNA1. In their study, mutations in the ORC1 BAH domain (E111K) disrupted the recruitment of EBNA1 and replication activity of OriP. They also found that overexpression of the ORC1 amino-terminal domain (aa 1–315), which partially overlaps with the interaction domain that was mapped in our study (aa 201–511), inhibited OriP replication. Our findings are consistent with those of Noguchi et al in that EBNA1 interacts with ORC1, but differs in that we found an RNA-dependent interaction mediated by aa 201–511 in ORC1. The AIF-C1 protein, which also recruits ORC and stimulates replication at the aldolase B origin, was found to interact with aa 210–239 of ORC1 (Saitoh et al, 2002; Minami et al, 2006). Interestingly, the ORC1 interaction domain of AIF-C1 contains RGG motifs similar to EBNA1. We suggest that the ORC1 BAH domain (aa 1–200) facilitates the RNA-dependent interactions of aa 201–511 and may provide additional protein–protein interactions with EBNA1 that are necessary for replication activity in vivo.
Origin recognition complex recruitment and origin formation are complex processes that are likely to involve multiple components. ORC recruitment to OriP depends on EBNA1, as we have shown here, but it is also dependent on TRF2 and the spacing between EBNA1-binding sites (Bashaw and Yates, 2001; Atanasiu et al, 2006). Alternative arrangements of EBNA1 binding sites, like those present at the FR, is not sufficient for ORC recruitment or origin formation. Epigenetic factors may also influence ORC activity, as some viral genomes recruit ORC to OriP but do not initiate replication every cell cycle (Norio and Schildkraut, 2001, 2004). At most chromosomal origins, the mechanism of ORC recruitment is even more complicated. Some chromosomal proteins, such as HMGA1a, can recruit ORC and promote origin formation (Thomae et al, 2008). HMGA1a is a chromosomal protein that can bind AT-rich DNA through its AT-hook domain and functions in transcriptional regulation and chromatin architecture (Reeves, 2001). We found that the RGR motifs within the HMGA1a AT-hooks can functionally substitute for the RGG motifs of EBNA1 in DNA replication and plasmid maintenance (Sears et al, 2004). Consistent with the fact that the mechanism of ORC recruitment and origin activation is certainly multifactorial, our data support the model that structured G-rich RNA functions as a common cofactor in ORC recruitment to OriP and to some cellular origins.
Structural RNAs have been implicated in replication initiation in several model systems. In tetrahymena, RNA is a structural component of ORC and is involved in origin melting by directly base pairing with DNA (Mohammad et al, 2007). And in human cells, Y-RNA is a structured RNA that is necessary for replication initiation (Christov et al, 2006). Our findings suggest that presentation of structured G-rich RNA by RGG-motif containing proteins may be one mechanism by which ORC is recruited to chromosomal sites. RNA cofactors may provide an additional level of control in the recruitment and activation of ORC. The precise molecular composition of the RNA cofactors and how they regulate replication activity of ORC remain to be determined.
Materials and methods
Plasmids and cell lines
GST–EBNA1 fusion plasmids were generated by PCR amplification of EBNA1 and cloning into the pGex2T (Amersham) vector at the BamHI/ EcoRI sites. For the EBNA1 sequence (LR1—aa 30–53, LR2—aa 328–350, R → A LR2 bp 328–350 with all arginines substituted by alanines, and G → A LR2 bp 328–350 with all glycines substituted by alanines) or HMGA1a (either full-length or an oligomer containing three tandem copies of aa 25–29), oligomers were annealed and cloned directly into the EcoRI/NotI sites of pGex4Ta. GST–ORC1 subdomains GST–ORC1 (aa 1–200), GST–ORC1 (aa 201–511), and GST–ORC1 (aa 512–861) were generated by PCR amplification and cloned as BamHI/EcoRI fragments into pGEX2T, as described previously (Atanasiu et al, 2006).
HeLa and HEK293T cells were grown in DMEM, and Raji cells were grown in RMPI. All cell media were supplemented with 10% fetal bovine serum, 1 × glutamax (GIBCO), and antibiotics. Clonal HEK293T cell lines were established by stable transfection of 293T cells with expression vectors for EBNA1, EBNA1 Δ40–89, EBNA1 Δ1–331, EBNA1 Δ328–377, and EBNA1 Δ1–377 using an EBNA1–IRES–hygromycin cassette. HEK293T cell clones were generated similarly with hygromycin selection (100 μg/ml). Individual cell clones were tested by western blotting for their expression levels. For colony formation assay, oriP-reporter plasmid p2832 carries wild-type oriP and a puromycin expression cassette driven from the CMV reporter.
For the transient replication assay, full-length EBNA1 was generated by cloning EBNA1 (bp 1–1921) into the HindIII/EcoRI sites of the 3 × Flag CMV-24 vector (Sigma). DBD was generated by cloning the DNA-binding domain of EBNA1 (bp 1333–1921) into the BamHI site of the 3 × Flag CMV 24 vector. 4 × LR1–DBD was generated by sequential cloning of an annealed DNA oligomer (IDT) (EBNA1 bp 118–162) into the SalI site of the DBD plasmid. 4 × LR2–DBD (EBNA1 bp 322–350) and 4 × (R → A) LR2–DBD (EBNA1 bp 322–350 with alanine substitutions for all arginines) were generated by the same method. None of these constructs contain the native NLS. The OriP reporter plasmid pHeBo has been described previously (Sugden et al, 1985).
Antibodies
For western blotting, antibodies to EBNA1, rat monoclonal (clone 1H4) (Grasser et al, 1994), rabbit polyclonal to GST (Santa Cruz Biotech), or monoclonal commercial Flag M2 (Sigma), EBNA1 (Advanced Biotechnologies Inc.), ORC2 (MBL International Corp.), ORC1 (AbCam), ORC4 (Transduction Laboratories), CDC6 (Neomarker), MCM3 (MBL International Corp.), MCM7 (AbCam), PCNA (Santa Cruz), histone H3 (AbCam), and βactin (Sigma) were used according to the manufacturer's specifications. For IPs, the following rabbit polyclonal antibodies were used: ORC2 (BD Pharmagen), Orc2 polyclonal (Schepers et al, 2001), and EBNA1 (Deng et al, 2003).
Immunoprecipitations
Exponentially growing Raji cells were harvested, and 10 × 106 cells were used for each IP. Cell pellet was resuspended in 1 ml RIPA buffer (150 mM NaCl, 1% IGPAL, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl, pH 8.0, 50 U/ml SuperaseIN from Ambion) per IP, incubated on ice for 30 min with frequent agitation, and insoluble cell debris was separated by centrifugation at 14 000 r.p.m. for 10 min at 4°C. Soluble cell lysate was precleared for 1 h with protein A sepharose beads (Amersham) by rotation at 4°C. Cell lysate was then added to preconjugated proteinA sepharose beads and incubated overnight at 4°C with rotation. The immunoprecipitated complex was washed three times with 10 volumes of RIPA buffer. Bound proteins were eluted by addition of 6 × Laemmli buffer and boiled for 10 min at 95°C. For RNA isolation, cells were fixed with 1% formaldehyde, lysed with RIPA buffer, and then subject to IP. Immunoprecipitates were washed with RIPA buffer containing 500 mM NaCl. RNA was eluted with 1%SDS, 50 mM Tris (pH 6.5), 5 mM EDTA, 10 mM DTT, 50 U/ml SuperaseIN at 70°C for 1 h, followed by Trizol extraction and isopropanol precipitation. Detection of endogenous RNA associated with EBNA1 immunoprecipitates was performed by RT–PCR using random decamers and amplified with primers specific for EBNA1 (5′-primer is from nt 184–225; 3′-primer is from nt 1487–1505) or Actin mRNA.
GST-interaction assay
GST-protein purification and interaction assays were done as described (Atanasiu et al, 2006). After incubation with the HeLa nuclear extract, enzymatic reactions were performed. To determine RNA sensitivity, RNase A (Roche) in a concentration of 0.02 mg/ml or RNase T1 (Roche) in a concentration of 2 U/ml was added to the wash buffer PBS75. To determine DNA sensitivity, DNase I (Roche) in a concentration of 0.2 U/ml was added to the beads resuspended in DNase I buffer (40 mM Tris–HCl, 10 mM NaCl, 6 mM MgCl2, 1 mM CaCl2, pH 7.9). For micrococcal nuclease I conditions, 0.2 U/ml MNase I (USB Corp.) was added to beads resuspended in MNase I buffer (10 mM Tris–HCl pH 8.0, 1 mM CaCl2). All reactions were incubated at 37°C for 45 min. For the control digestions, either 50 μg single-stranded salmon sperm DNA or 100 μg tRNA was digested under the same conditions and then separated by gel electrophoresis on a 0.8% agarose, 0.5 × TBE gel and visualized by ethidium bromide staining.
RNA pull-down assay
GST peptides were purified as described previously (Atanasiu et al, 2006). RNA oligomers (IDT) were end-labelled with 32P γ-ATP and polynucleotide kinase. A total of 10 fmol RNA was added to the binding buffer (5 mM beta-mercaptoethanol, 500 μg/ml BSA, 40 μg/ml sonicated salmon sperm DNA) and then added to the bead-bound purified protein, and incubated for 20 min at 30°C. The pellet was washed three times in 1 ml PBS75 (0.05 M phosphate (pH 7.4), 75 mM NaCl supplemented with 0.1% NP-40, 1 mM PMSF, 1 mM EDTA, 1 mM DTT, bacterial protease inhibitors cocktail (Sigma)). RNA was then eluted in elution buffer (1% SDS, 1 mM EDTA, 0.6 U/ml SuperaseIN), and an equal volume of RNA loading buffer was added before the RNA was separated on a 6% acrylamide 0.5 × TBE gel.
Additional methods
Chromatin IP assays were described previously (Ritzi et al, 2003). Colony formation and plasmid recovery assays were performed as reported previously (Gerhardt et al, 2006). Transient DNA replication assays were described previously (Deng et al, 2003). GST-protein purification and interaction assays were described (Atanasiu et al, 2006). Chromatin fractionation of ORC was performed as described previously (Mendez and Stillman, 2000), with the exception that RNase A was added (1.0 mg/ml) to some samples as indicated. Acrylamide gel RNA EMSA was performed essentially as described (Lu et al, 2004). Agarose gel EMSA was performed as described previously, except that magnesium was not included in the gel matrix (Zerby and Lieberman, 1997). EBNA1 ORF mRNA probe was generated using T7 RNA polymerase in vitro transcription reactions (Promega Inc.), with linearized plasmid containing EBNA1 cDNA in pBKSII. Oligonucleotide sequences are available upon request.
Supplementary Material
Supplementary Figure S1
Acknowledgments
We thank F Brad Johnson for many helpful suggestions and Andreas Wiedmer for technical assistance. We acknowledge the services of the Wistar Institute Cancer Center Core Facilities for DNA sequences and protein expression. This work was supported by grants from NIH (CA093606) to PML and an NIH Pre-Doctoral Training Fellowship (5-T32-GM08216-18) to JN.
References
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287: 2023–2026 [DOI] [PubMed] [Google Scholar]
- Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM (2006) ORC binding to TRF2 stimulates OriP replication. EMBO Rep 7: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw JM, Yates JL (2001) Replication from oriP of Epstein–Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J Virol 75: 10603–10611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 [DOI] [PubMed] [Google Scholar]
- Bell SP (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16: 659–672 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL (2001) Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein–Barr virus. Proc Natl Acad Sci, USA 98: 10085–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szuts D, Krude T (2006) Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol 26: 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ (1999) The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA 96: 2656–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107: 489–499 [DOI] [PubMed] [Google Scholar]
- Darnell JC, Warren ST, Darnell RB (2004) The fragile X mental retardation protein, FMRP, recognizes G-quartets. Ment Retard Dev Disabil Res Rev 10: 49–52 [DOI] [PubMed] [Google Scholar]
- Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM (2003) Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein–Barr virus OriP. J Virol 77: 11992–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM (2007) The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr Biol 17: 1989–1995 [DOI] [PubMed] [Google Scholar]
- Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM (2002) Telomeric proteins regulate episomal maintenance of Epstein–Barr virus origin of plasmid replication. Mol Cell 9: 493–503 [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A (2006) Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol 18: 231–239 [DOI] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A (2001) Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell 106: 287–296 [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Wang S, Hamlin JL (2002) Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol Cell Biol 22: 3053–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Early A, Drury LS, Diffley JF (2004) Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage. Philos Trans R Soc Lond B Biol Sci 359: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Jafar S, Spindler MP, Ott E, Schepers A (2006) Identification of new human origins of DNA replication by an origin-trapping assay. Mol Cell Biol 26: 7731–7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser FA, Murray PG, Kremmer E, Klein K, Remberger K, Feiden W, Reynolds G, Niedobitek G, Young LS, Mueller-Lantzsch N (1994) Monoclonal antibodies directed against the Epstein–Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood 84: 3792–3798 [PubMed] [Google Scholar]
- Hung SC, Kang MS, Kieff E (2001) Maintenance of Epstein–Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci, USA 98: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Kamiya M, Maruo S, Iwakiri D, Takada K (2007) Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J Cell Sci 120: 1529–1539 [DOI] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfus G (1992) Primary structure and binding activity of hnRNP U protein: binding RNA through RGG box. EMBO J 11: 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Sugden B (2007) The plasmid replicon of Epstein–Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid 58: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Wu CW, Chang SC, Chen TY, Hu CR, Yeh MY, Chen JY, Chen MR (2004) Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J Gen Virol 85: 2755–2765 [DOI] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A (2005) Right place, right time, and only once: replication initiation in metazoans. Cell 123: 13–24 [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami H, Takahashi J, Suto A, Saitoh Y, Tsutsumi K (2006) Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin. Mol Cell Biol 26: 8770–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MM, Donti TR, Sebastian Yakisich J, Smith AG, Kapler GM (2007) Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J 26: 5048–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Sugden A, Sugden B (2007) The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J 26: 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML (2006) The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J 25: 5372–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL (2005) Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20: 575–587 [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL (2001) Visualization of DNA replication on individual Epstein–Barr virus episomes. Science 294: 2361–2364 [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL (2004) Plasticity of DNA replication initiation in Epstein–Barr virus episomes. PLoS biology 2: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R (2001) Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277: 63–81 [DOI] [PubMed] [Google Scholar]
- Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A (2003) Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein–Barr virus. J Cell Sci 116: 3971–3984 [DOI] [PubMed] [Google Scholar]
- Saitoh Y, Miyagi S, Ariga H, Tsutsumi K (2002) Functional domains involved in the interaction between Orc1 and transcriptional repressor AlF-C that bind to an origin/promoter of the rat aldolase B gene. Nucleic Acids Res 30: 5205–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM (2007) The many faces of the origin recognition complex. Curr Opin Cell Biol 19: 337–343 [DOI] [PubMed] [Google Scholar]
- Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20: 4588–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Kolman J, Wahl GM, Aiyar A (2003) Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by EBNA1. J Virol 77: 11767–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A (2004) The amino terminus of Epstein–Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol 78: 11487–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Gasser SM (2007) The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell 128: 85–99 [DOI] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Snudden DK, Hearing J, Smith PR, Grasser FA, Griffin BE (1994) EBNA-1, the major nuclear antigen of Epstein–Barr virus, resembles ‘RGG' RNA binding proteins. EMBO J 13: 4840–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B (2005) Origin recognition and the chromosome cycle. FEBS Lett 579: 877–884 [DOI] [PubMed] [Google Scholar]
- Sugden B, Marsh K, Yates J (1985) A vector that replicates as a plasmid and can be efficiently selected in B-lymphocytes transformed by Epstein–Barr virus. Mol Cell Biol 5: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A (2005) Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev 19: 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A (2008) Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci USA 105: 1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kapoor P, Frappier L (2002) Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein–Barr nuclear antigen 1. J Virol 76: 2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Yates J, Camiolo SM (1988) Dissection of DNA replication and enhancer activation functions of Epstein–Barr virus nuclear antigen 1. Cancer Cells 6: 197–205 [Google Scholar]
- Young LS, Rickinson AB (2004) Epstein–Barr virus: 40 years on. Nat Rev Cancer 4: 757–768 [DOI] [PubMed] [Google Scholar]
- Zerby D, Lieberman PM (1997) Functional analysis of TFIID-activator interaction by magnesium-agarose gel electrophoresis. Methods: A Companion to Methods in Enzymology 12: 217–223 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1