Abstract
The c-Abl tyrosine kinase and the p53 tumor suppressor protein interact functionally and biochemically in cellular genotoxic stress response pathways and are implicated as downstream mediators of ATM (ataxia-telangiectasia mutated). This fact led us to study genetic interactions in vivo between c-Abl and p53 by examining the phenotype of mice and cells deficient in both proteins. c-Abl-null mice show high neonatal mortality and decreased B lymphocytes, whereas p53-null mice are prone to tumor development. Surprisingly, mice doubly deficient in both c-Abl and p53 are not viable, suggesting that c-Abl and p53 together contribute to an essential function required for normal development. Fibroblasts lacking both c-Abl and p53 were similar to fibroblasts deficient in p53 alone, showing loss of the G1/S cell-cycle checkpoint and similar clonogenic survival after ionizing radiation. Fibroblasts deficient in both c-Abl and p53 show reduced growth in culture, as manifested by reduction in the rate of proliferation, saturation density, and colony formation, compared with fibroblasts lacking p53 alone. This defect could be restored by reconstitution of c-Abl expression. Taken together, these results indicate that the ATM phenotype cannot be explained solely by loss of c-Abl and p53 and that c-Abl contributes to enhanced proliferation of p53-deficient cells. Inhibition of c-Abl function may be a therapeutic strategy to target p53-deficient cells selectively.
The c-Abl protein is a ubiquitously expressed nonreceptor tyrosine kinase that was discovered originally as the cellular homologue of the oncogene v-Abl carried by Abelson murine leukemia virus. It is also involved in the pathogenesis of some forms of human leukemias by formation of the Bcr–Abl chimeric fusion protein through chromosomal translocation (1, 2). c-Abl and its relative Arg (Abl-related gene; ref. 3) contain SH3, SH2, and tyrosine kinase domains in common with other Src-family members but, in addition, contain a unique large C-terminal segment with multiple domains such as nuclear localizing signals (4), DNA- and actin-binding domains (5, 6), a nuclear export signal (7), and proline-rich sequences involved in binding to adaptor proteins (8). Although the presence of these functional domains suggests that c-Abl is involved in regulation of signal transduction pathways, a comprehensive understanding of c-Abl's cellular functions has been elusive. Mice homozygous for targeted disruptions in c-Abl suffer from multiple defects, including runting, neonatal lethality, morphological abnormalities, splenic and thymic atrophy, reduced B lymphocytes, and susceptibility to infections (9, 10). A true null mutation (10) and a truncation mutation with intact kinase activity (9) result in a similar phenotype, pointing to the importance of the multiple functional domains in the C-terminal fragment of c-Abl.
Recent evidence suggests that c-Abl plays a role in regulation of the cell cycle and the cellular genotoxic stress response pathways. Overexpression of c-Abl produces cell-cycle arrest in G1, which requires kinase activity and nuclear localizing signals and depends on the wild-type p53 tumor suppressor (4, 11, 12). In addition, c-Abl binds p53 and enhances the DNA-binding and transcriptional activity of p53 (12–15). These studies suggest that c-Abl acts as a negative regulator of growth. Other studies suggest that c-Abl has growth-promoting activity, based on activation of its kinase during S phase and its ability to abrogate Rb-mediated growth suppression in SAOS-2 cells, which are deficient in Rb and p53 (16, 17). In addition, c-Abl is activated by growth factor treatment and is involved in the cytoskeletal rearrangement and mitogenic response to platelet-derived growth factor (18).
c-Abl has also emerged as a potential intermediate in the cellular DNA damage response (19). DNA damage from ionizing radiation (IR) or chemotherapeutic agents leads to transient activation of c-Abl kinase and the p53 protein (20, 21). Activation of both c-Abl and p53 in response to IR-induced DNA damage depends on the presence of ataxia-telangiectasia mutated (ATM) kinase, which phosphorylates c-Abl on Ser-465 and p53 on Ser-15 (22–26). Lack of ATM results in defective cell-cycle checkpoint controls, a slower growth rate, and hypersensitivity to irradiation (27–29). Activation of c-Abl by IR also requires DNA-dependent protein kinase (30). Substrates of c-Abl activated by DNA damage include Rad51, involved in recombination repair, and p73, a homologue of p53 (31–35). The fact that ATM, c-Abl, and p73 may biochemically define a DNA damage response pathway has led to the proposal that p53 and p73 may represent distinct effectors of ATM, with c-Abl serving to link ATM and p73 (36).
To study the link between c-Abl and p53 in vivo in mice, we analyzed the phenotype of mice lacking both genes. We postulated that mice lacking both c-Abl and p53 may resemble the ATM phenotype, because both genes lie on putative signaling pathways downstream of ATM. Unlike ATM−/− mice, we found that c-Abl−/− p53−/− mice are not viable, indicating previously unknown essential roles for c-Abl and p53 during development. DNA damage studies of c-Abl−/− p53−/− embryo fibroblasts exposed to IR indicated that loss of both c-Abl and p53 does not cause more severe defects in G1/S checkpoint control or decreased clonogenic survival than loss of p53 alone. However, cell growth and colony formation assays of c-Abl−/− p53−/− cells revealed an unexpected dependence of p53-deficient cells on c-Abl for enhanced proliferation, suggesting that pharmacologic inhibition of Abl may have therapeutic value in the cancer cells lacking p53.
Materials and Methods
Mice.
A breeding pair of c-Abl+/− mice in the 129Sv/Ev × C57BL/6J mixed background, described in Hardin et al. (37), was kindly provided by Stephen Goff (Columbia University, New York). In this line, originally constructed by Tybulewitz et al. (10), the exon encoding the kinase domain of c-Abl was disrupted, and the c-Abl−/− mice are null for protein expression and kinase activity. A p53+/− breeding pair (129S3/SvImJ strain) was obtained from The Jackson Laboratory. The c-Abl+/− and p53+/− mice were mated to produce compound heterozygotes, which were then mated together. Some matings also involved c-Abl+/− p53−/− or c-Abl−/− p53+/− mice to increase the probability of producing c-Abl−/− p53−/− mice. Genotyping of c-Abl and p53 loci was performed by PCRs of tail DNA as described by Hardin et al. (37) or according to directions provided by The Jackson Laboratory.
Radiation Response and Growth Studies on Embryo Fibroblasts.
Fibroblasts were isolated from day-14 embryos by standard methods and cultured in DMEM medium and 10% (vol/vol) FCS. DNA isolated from embryos was used for genotyping as described above. For determination of the cell-cycle response after IR, low passage embryo fibroblasts were synchronized in G0/G1 by growing to confluence and then given 0 or 10 Gy of IR. Cells were replated subconfluently, and the cell-cycle profile was determined by flow cytometry analysis of propidium-iodide-stained cells. Percentages of cells in G1, S, and G2 were calculated with cellfit software (Becton Dickinson). The cumulative labeling index method for analyzing progression to S phase by labeling cells with [3H]thymidine was performed as described (38).
For clonogenic survival after IR, embryo fibroblast cells were detached by trypsinization, exposed to indicated doses of IR, and then replated at low density (500 or 1,000 cells per plate, each density in duplicate). After 10 days of culture, cells were fixed in 70% (vol/vol) ethanol and stained with crystal violet, and colonies were counted. p53−/− ATM−/− embryo fibroblasts were kindly provided by Philip Leder (Harvard Medical School, Boston; ref. 39).
For growth curves, cells at low passage were plated at 3.0 × 105 per 6-cm dish, and viable cells were counted at 24-h intervals for 7 days. Medium was changed on day 3. For colony formation assays, cells were plated at 3.5 × 103 cells per 10-cm dish. Medium was changed on day 3. On day 8, cells were fixed with 70% (vol/vol) ethanol for 15 min and then stained with crystal violet. Visible colonies greater than 1 mm in diameter were scored.
Expression of c-Abl by Retroviral Transduction.
Retrovirus stocks of control neomycin virus or virus encoding wild-type c-Abl or kinase inactive c-Abl K290R mutant were produced by transient cotransfection in 293T cells of the ecotropic packaging plasmid and the retroviral vector pSRαMSV tk neo encoding the appropriate c-Abl transgene (40). Embryo fibroblasts at low passage were incubated with virus stock in the presence of polybrene (8 μg/ml) overnight and then plated for colony formation assay 2 days after the start of infection. Transduction of c-Abl was verified by immunoblotting of cell extracts with a monoclonal antibody specific for c-Abl.
Results
Mice Deficient in Both c-Abl and p53 Are Not Viable.
Judging from the findings that the DNA damage response pathway downstream of ATM involves both c-Abl and p53, which are both activated and form a complex in response to DNA damage, and other data suggesting functional interactions between c-Abl and p53 (12–15, 22, 23), we wished to investigate genetic interactions between c-Abl and p53 in vivo. Mice deficient in both c-Abl and p53 may be expected to produce a phenotype similar to mice deficient in ATM if c-Abl and p53 mediate much of the events downstream of ATM, at least with respect to the DNA damage response. Therefore, we attempted to generate mice homozygous for targeted disruptions in both c-Abl and p53 loci. In the mixed background used in this study, c-Abl−/− mice survive longer than previously reported (9, 10) and therefore are represented approximately at the expected Mendelian ratio at 3 weeks after birth, although their life span is significantly decreased (data not shown). However, no pup with the genotype of c-Abl−/− p53−/− was found of 143 pups genotyped at 3 weeks of age, significantly less than the 12 predicted by Mendelian inheritance (Table 1). These results suggested that mice lacking both c-Abl and p53 are not viable and die either in utero or shortly after birth, before 3 weeks. To pinpoint the time of their demise, the genotype distribution was determined at embryonic day 14 or immediately after birth. At embryonic day 14, c-Abl−/− p53−/− embryos were represented at the expected frequency and showed no gross abnormality. Of 62 pups genotyped at birth, only one c-Abl−/− p53−/− pup was found, rather than the four predicted, and this c-Abl−/− p53−/− pup died shortly after birth. Taken together, these results suggest that mice deficient in both c-Abl and p53 are not viable, and they probably die late in development, between embryonic day 14 and the time of birth.
Table 1.
Distribution of genotypes
| Genotype
|
Embryos at 14 days
|
Pups at birth
|
Pups at 3 weeks
|
||||
|---|---|---|---|---|---|---|---|
| c-Abl | p53 | Observed | Expected | Observed | Expected | Observed | Expected |
| +/+ | +/+ | 2 | 3.68 | 10 | 3.63 | 6 | 6.01 |
| +/+ | +/− | 4 | 7.38 | 3 | 7.25 | 13 | 15.50 |
| +/+ | −/− | 2 | 3.68 | 4 | 3.63 | 15 | 9.51 |
| +/− | +/+ | 16 | 7.38 | 13 | 7.75 | 19 | 14.38 |
| +/− | +/− | 13 | 14.75 | 14 | 15.50 | 44 | 35.75 |
| +/− | −/− | 6 | 7.38 | 6 | 7.75 | 19 | 21.38 |
| −/− | +/+ | 6 | 3.68 | 5 | 4.13 | 7 | 8.39 |
| −/− | +/− | 6 | 7.38 | 6 | 8.25 | 20 | 20.25 |
| −/− | −/− | 4 | 3.68 | 1 (NV) | 4.13 | 0 | 11.89 |
| Total | 59* | 59 | 62† | 62 | 143‡ | 143 | |
Embryos and pups were genotyped with PCR-based methods at the indicated time points. The expected distribution of genotypes was calculated, assuming independent, Mendelian inheritance of each allele. Because some of the crosses involved homozygous mice (c-Abl−/− p53+/− or c-Abl+/− p53−/−) to increase the probability of producing c-Abl−/− p53−/− mice, the expected number of c-Abl−/− p53−/− mice is greater than 6.25% expected from crosses with c-Abl+/− p53+/− mice alone. P values are derived from χ2 analysis and show that the difference between observed and expected is statistically significant for pups at birth and 3 weeks but not for embryos at 14 days. *Not significant (P > 0.05); †, P < 0.01; ‡, P = 0.0131; NV, nonviable.
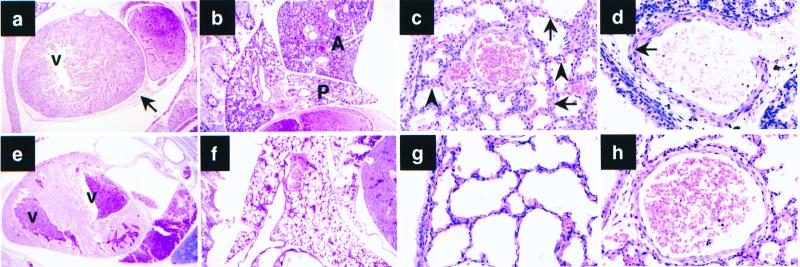
The nonviable c-Abl−/− p53−/− pup showed multiple abnormalities in the lungs and the heart (Fig. 1). The lungs were irregularly and poorly expanded. The walls of the alveolar sacs were thicker than those in wild-type controls. The media of the arterial walls were thick, showing that the thinning of this layer that normally follows breathing after birth had not occurred. The heart of the c-Abl−/− p53−/− mouse was displaced toward the left and although the atria were distended with blood, both ventricular cavities were small and empty. The pericardial sac was filled with blood. This later change suggests that the cause of death could have been cardiac tamponade, which had consequently impeded the filling of the ventricles. Of note, similar pericardial hemorrhage also occurs in c-Abl−/− Arg−/− mice (41).
Figure 1.
(a–d) Histological abnormalities in c-Abl−/− p53−/− mice. (e–h) Wild-type newborn sibling control. (a) Section of the heart. Hemorrhage in the pericardial sac (arrow). There is no blood in the left ventricle (V). (e) Both ventricles (V) in the wild-type sibling contain blood. (b) Right lung. Anterior lobe (A) is not expanded. Posterior lobe (P) is minimally expanded. Compare with f. (c) Higher power of the right posterior lobe shows thick walls of the alveolar sacs (arrows) with bulging capillaries (arrowheads). Compare with wild-type sibling shown in g. (d) Arterial wall in the right posterior lung lobe. Note thickness of the media (arrow). Compare with h, which shows the wild-type sibling
c-Abl−/− p53−/− Cells Are Similar to p53−/− Cells in G1/S Cell-Cycle Checkpoint and Clonogenic Survival After DNA Damage.
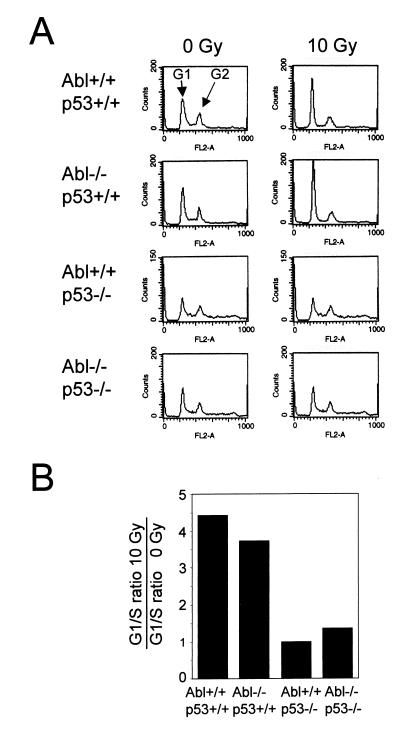
To gain insight into the effect of loss of both c-Abl and p53 at the cellular level, we isolated primary fibroblasts from embryos of defined genotypes and examined the cell-cycle checkpoint in response to DNA damage. Several assays were performed to determine the response to DNA damage induced by IR. In the first assay, the cell-cycle profile of fibroblasts in response to IR was determined. c-Abl−/− embryo fibroblasts underwent G1 cell-cycle arrest after IR similar to wild-type cells, whereas p53−/− fibroblasts and c-Abl−/− p53−/− cells were defective in their ability to undergo G1 cell-cycle arrest after IR (Fig. 2). To measure the delay in progression to S phase by using a different method, a cumulative labeling experiment was performed in which G0/G1-synchronized cells were given 6 Gy of IR, and the cumulative progression of cells into S phase was measured by incorporation of [3H]thymidine into cellular DNA at various time points. p53−/− cells and c-Abl−/− p53−/− cells both showed no delay in progression to S phase after IR (data not shown). These data indicate that both p53−/− and c-Abl−/− p53−/− cells are similarly defective in their ability to undergo DNA damage-induced cell-cycle arrest in G1 phase.
Figure 2.
The cell-cycle arrest after IR is defective in p53−/− and c-Abl−/− p53−/− cells. Confluent embryo fibroblasts were exposed to 0 or 10 Gy of IR and then cultured at a lower density for 19 h. (A) The cell-cycle profile after staining with propidium iodide and flow cytometry is shown for each genotype. Peaks corresponding to G1 and G2 phases of the cell cycle are indicated. (B) The cell-cycle distribution was calculated from data shown in A. The G1/S ratio after 10 Gy of IR relative to 0 Gy controls is shown for each genotype. The ratio substantially greater than 1 indicates G1 cell-cycle arrest after irradiation.
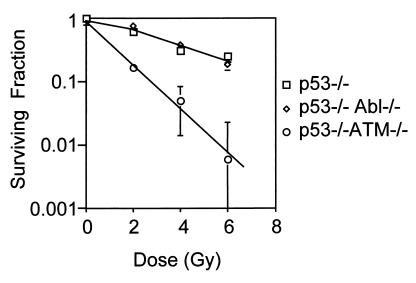
An important feature of ATM deficiency is hypersensitivity to irradiation, and this phenotype seems to be p53-independent, because loss of ATM radiosensitizes even p53-null cells (39, 42). Using the clonogenic survival assay, we next asked whether loss of c-Abl might also sensitize p53-null cells to irradiation, because c-Abl is implicated as a downstream mediator of ATM function (22, 23). There was no difference in clonogenic survival after IR between p53−/− and c-Abl−/− p53−/− cells (Fig. 3). However, as previously reported (39), p53−/− ATM−/− cells were hypersensitive to IR and showed decreased clonogenic survival relative to p53−/− and c-Abl−/− p53−/− cells. Taken together, these data indicate that p53 is the major regulator of the G1/S cell-cycle checkpoint and that loss of c-Abl does not lead to additional effects on the G1/S checkpoint or clonogenic survival after IR. Furthermore, deficiency of both p53 and c-Abl does not re-create the radiosensitive phenotype of ATM-deficient cells.
Figure 3.
Decreased clonogenic survival after IR in ATM-deficient cells but not c-Abl−/− p53−/− cells. Embryo fibroblast cells of defined genotypes as indicated were given graded doses of IR, and clonogenic survival is shown as a fraction of unirradiated control. Data are representative of two (p53−/− ATM−/−) or three (p53−/− and c-Abl−/− p53−/−) independent experiments.
Fibroblasts Deficient in Both c-Abl and p53 Show Reduced Proliferation and Saturation Density in Culture Compared with Cells Deficient in p53 Alone.
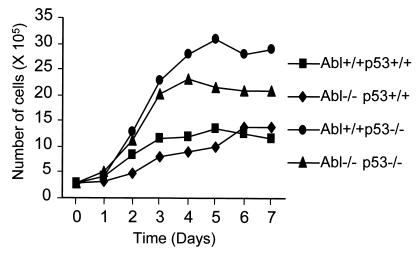
ATM−/− fibroblasts proliferate poorly in culture and undergo premature senescence (27, 28). To assess the role of c-Abl and p53 in the control of cellular proliferation, we examined the in vitro growth characteristics of embryo fibroblasts of defined genotypes. Primary fibroblasts were isolated from embryos at day 14 of gestation, and the growth curve of low passage fibroblasts was determined (Fig. 4). p53−/− cells showed an extremely rapid rate of proliferation and reached a high saturation density as reported (43). c-Abl−/− p53−/− cells proliferated less rapidly than p53−/− cells and also reached a saturation density midway between wild-type and p53−/− cells. Similar results were observed in fibroblasts isolated from two c-Abl−/− p53−/− embryos. c-Abl−/− cells were similar to wild-type cells in saturation density and the rate of proliferation. These data suggust that c-Abl is necessary for enhanced in vitro growth of p53-deficient cells and that c-Abl in the absence of p53 plays a positive role in promoting cell proliferation.
Figure 4.
Growth curves of embryo fibroblasts of defined genotypes. Early-passage embryo fibroblasts were plated (3 × 105 cells per 6-cm dish in duplicate) on day 0. Cells were counted on days 1–7. Each data point is a mean of three or four experiments involving fibroblasts isolated from two separate embryos for each genotype.
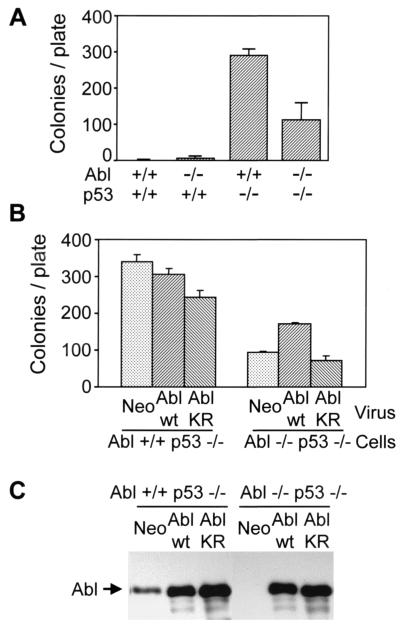
We next determined the ability of embryo fibroblasts to form visible colonies after plating at low density. This colony formation assay measures the proliferative capacity of primary cells and correlates with the ability to overcome senescence. Loss of negative regulators of growth such as p53 and the INK4a locus (p16 and p19ARF) leads to increased colony formation in primary fibroblasts (43, 44). p53−/− cells formed the most colonies, whereas c-Abl−/− p53−/− showed an intermediate phenotype (Fig. 5A). Wild-type and c-Abl−/− cells formed few or no colonies. This result suggests that the enhanced proliferative capacity of p53-deficient cells requires c-Abl and that endogenous c-Abl promotes proliferation in the context of p53 deficiency.
Figure 5.
c-Abl expression stimulates colony formation of c-Abl−/− p53−/− cells. (A) Cells were plated at low density (3.5 × 103 cells per 10-cm plate) and cultured for 8 days. The number of visible colonies per plate after staining with crystal violet is shown for each genotype. (B) p53−/− and c-Abl−/− p53−/− cells were infected with retrovirus encoding neomycin vector only, wild-type (wt) c-Abl, or kinase-inactive c-Abl K290R (KR) mutant. Cells were plated 2 days after infection, and colony formation was determined as described above. (C) Analysis of c-Abl expression. Cell lysates prepared at the time of plating for colony formation after retrovirus infection were immunoblotted with anti-c-Abl monoclonal antibody. c-Abl−/− p53−/− cells lack endogenous c-Abl expression, consistent with their genotype.
Restoration of c-Abl Expression Stimulates Colony Formation in c-Abl−/− p53−/− Cells.
To determine directly the role of c-Abl in increasing colony formation, we investigated the effect of restoring c-Abl expression by retroviral transduction. In c-Abl−/− p53−/− cells, reconstitution of c-Abl expression increased colony formation, whereas kinase inactive c-Abl K290R had no effect (Fig. 5B). Wild-type c-Abl expression had no effect on c-Abl+/+ p53−/− cells, whereas kinase inactive c-Abl expression reduced colony formation of these cells. Because this kinase-inactive c-Abl K209R mutant has been shown to function as a dominant negative mutant (11), this result indicates that inhibition of endogenous c-Abl impairs the growth of p53-deficient cells. Analysis of c-Abl expression confirmed efficient transduction of c-Abl by retrovirus, because infected cells express 5- to 10-fold more protein relative to endogenous c-Abl (Fig. 5C). Taken together, these data establish that c-Abl contributes to enhanced proliferation of p53-deficient cells.
Discussion
Requirement for Both c-Abl and p53 in Development.
Although c-Abl deficiency clearly leads to multiple developmental effects (9, 10), its penetrance is variable, and the postnatal lethality associated with c-Abl deficiency seems to depend somewhat on the background strain. In the strain used in this study, the lethality associated with c-Abl deficiency is moderated, permitting survival for a few weeks after birth. However, mice cannot tolerate the absence of both c-Abl and p53, because no viable c-Abl−/− p53−/− mice could be recovered at 3 weeks of age and just after birth. The c-Abl−/− p53−/− mouse that died shortly after birth has multiple histological abnormalities in the lung and heart, but the underlying developmental defect associated with loss of c-Abl and p53 is not clear. Although the fact that most p53-deficient mice are viable indicates that p53 is not essential for embryonic development (45), there is an increased rate of developmental anomalies such as exencephaly in p53-null embryos (46, 47). To understand why the combined loss of c-Abl and p53 might lead to embryonic lethality, it is important to consider the example of the c-Abl family member Arg. Whereas Arg−/− mice develop normally, c-Abl−/− Arg−/− embryos die from defects in neurulation, suggesting that Arg and c-Abl have redundant or overlapping functions in nervous system development that are apparent only when both genes are missing (41). Our results suggest that c-Abl and p53 may also have unsuspected overlapping or redundant functions in development. Given the fact that c-Abl is a tyrosine kinase and p53 is a sequence-specific transcription factor, the mechanism for potential redundancy is not immediately apparent. One possible explanation is the ability of c-Abl to enhance the transcriptional function of p53 (12–15). More work is needed to examine this issue and to pinpoint the precise nature of developmental deficit leading to death of c-Abl−/− p53−/− embryos.
Role of c-Abl and p53 in DNA Damage Response.
Because ATM is required for activation of c-Abl and p53 in response to DNA damage (21–23), we reasoned that loss of c-Abl and p53 might mimic the phenotype of ATM deficiency. ATM-deficient cells are defective in multiple cell-cycle checkpoints and are hypersensitive to IR (27–29, 42). Like ATM-deficient cells, c-Abl−/− p53−/− cells are defective in the G1/S cell-cycle checkpoint, but this defect seems to be explained fully by loss of p53. In contrast to ATM-deficient cells, c-Abl−/− p53−/− cells are not hypersensitive to IR and are similar to p53−/− cells in clonogenic survival after IR. Therefore, loss of both c-Abl and p53, unlike loss of ATM, does not sensitize cells to IR. Our results suggest that the ATM phenotype is more complex, perhaps because of many additional substrates of ATM kinase (e.g., p95/nibrin, Mre11, Brca1, Rad17, etc.) that may mediate its downstream actions (48).
Role of c-Abl in Control of Proliferation.
The role of c-Abl in growth and cell-cycle progression has been controversial. A growth stimulatory function has been postulated on the basis of the increase in c-Abl kinase activity in S phase and the ability of c-Abl to counteract Rb-mediated growth suppression in SAOS-2 cells (16, 17). In addition, a recent study showed that c-Abl−/− cells have delayed entry to S phase after stimulation of fibroblasts with platelet-derived growth factor, implying that c-Abl plays a role in the mitogenic response to specific growth factors (18). On the other hand, overexpression of c-Abl is associated with growth arrest, and this activity depends on the presence of p53 (4, 11, 12). c-Abl is also activated by DNA damage stimuli that lead to growth arrest (13, 20). The current study shows that the growth of c-Abl−/− p53−/− cells is impaired relative to p53−/− cells, manifested by reduction in the rate of proliferation, saturation density, and colony formation. These data indicate that endogenous c-Abl is required for rapid proliferation associated with p53 loss. This growth stimulatory role for c-Abl in the setting of p53 deficiency was confirmed by enhanced colony formation in c-Abl−/− p53−/− cells after retroviral transduction of c-Abl and inhibition of colony formation in c-Abl+/+ p53−/− cells by expression of dominant negative c-Abl. These data point to the critical dependence on p53 for growth regulation by c-Abl and indicate that c-Abl may play a positive or negative role on growth depending on the p53 status of cells. Whether c-Abl's role in growth is specific to p53 deficiency or applicable to cells driven to excess proliferation because of other genetic factors remains to be seen. Results showing the positive role of c-Abl on the growth of p53-deficient cells raise the possibility that inhibition of c-Abl may retard the growth of malignant cells missing p53 function selectively. Abl-specific kinase inhibitors may prove effective as a treatment for Bcr-Abl-expressing chronic myeloid leukemia (49). Data presented herein provide a rationale for testing the effect of Abl kinase inhibitors on the growth of p53-deficient cancer cells.
Acknowledgments
We are grateful to Stephen Goff for c-Abl+/− mice and helpful discussions; Sharon Boast for advice on genotyping; Phil Leder for p53−/− ATM−/− cells; Naomi Rosenberg and Owen Witte for review of the manuscript; and James Redula, Lisa Humphries, Chloe Chhor, Shobha Castelino-Prabhu, and Jennifer Daigle for technical assistance. This work was supported by National Institutes of Health, Leukemia Society of America, and American Cancer Society grants to C.L.S. and a Scholar Award from the American Society of Hematology to Y.E.W.
Abbreviation
- IR
ionizing radiation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Raitano A B, Whang Y E, Sawyers C L. Biochim Biophys Acta. 1997;1333:F201–F216. doi: 10.1016/s0304-419x(97)00023-1. [DOI] [PubMed] [Google Scholar]
- 2.Van Etten R A. Trends Cell Biol. 1999;9:179–186. doi: 10.1016/s0962-8924(99)01549-4. [DOI] [PubMed] [Google Scholar]
- 3.Kruh G D, Perego R, Miki T, Aaronson S A. Proc Natl Acad Sci USA. 1990;87:5802–5806. doi: 10.1073/pnas.87.15.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen S T, Jackson P K, Van Etten R A. EMBO J. 1996;15:1583–1595. [PMC free article] [PubMed] [Google Scholar]
- 5.Kipreos E T, Wang J Y. Science. 1992;256:382–385. doi: 10.1126/science.256.5055.382. [DOI] [PubMed] [Google Scholar]
- 6.McWhirter J R, Wang J Y. EMBO J. 1993;12:1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y, Hope T J. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren R, Ye Z S, Baltimore D. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 9.Schwartzberg P L, Stall A M, Hardin J D, Bowdish K S, Humaran T, Boast S, Harbison M L, Robertson E J, Goff S P. Cell. 1991;65:1165–1175. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 10.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 11.Sawyers C L, McLaughlin J, Goga A, Havlik M, Witte O. Cell. 1994;77:121–131. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 12.Goga A, Liu X, Hambuch T M, Senechal K, Major E, Berk A J, Witte O N, Sawyers C L. Oncogene. 1995;11:791–799. [PubMed] [Google Scholar]
- 13.Yuan Z M, Huang Y, Whang Y, Sawyers C, Weichselbaum R, Kharbanda S, Kufe D. Nature (London) 1996;382:272–274. doi: 10.1038/382272a0. [DOI] [PubMed] [Google Scholar]
- 14.Sionov R V, Moallem E, Berger M, Kazaz A, Gerlitz O, Ben-Neriah Y, Oren M, Haupt Y. J Biol Chem. 1999;274:8371–8374. doi: 10.1074/jbc.274.13.8371. [DOI] [PubMed] [Google Scholar]
- 15.Nie Y, Li H H, Bula C M, Liu X. Mol Cell Biol. 2000;20:741–748. doi: 10.1128/mcb.20.3.741-748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch P J, Wang J Y. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 17.Welch P J, Wang J Y. Mol Cell Biol. 1995;15:5542–5551. doi: 10.1128/mcb.15.10.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plattner R, Kadlec L, DeMali K A, Kazlauskas A, Pendergast A M. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharbanda S, Yuan Z M, Weichselbaum R, Kufe D. Oncogene. 1998;17:3309–3318. doi: 10.1038/sj.onc.1202571. [DOI] [PubMed] [Google Scholar]
- 20.Kharbanda S, Ren R, Pandey P, Shafman T D, Feller S M, Weichselbaum R R, Kufe D W. Nature (London) 1995;376:785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 21.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 22.Baskaran R, Wood L D, Whitaker L L, Canman C E, Morgan S E, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M B, et al. Nature (London) 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 23.Shafman T, Khanna K K, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Nature (London) 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 24.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, et al. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 25.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 26.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, et al. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 27.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley J N, Ried T, Tagle D, et al. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Baltimore D. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 29.Elson A, Wang Y, Daugherty C J, Morton C C, Zhou F, Campos-Torres J, Leder P. Proc Natl Acad Sci USA. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kharbanda S, Pandey P, Jin S, Inoue S, Bharti A, Yuan Z M, Weichselbaum R, Weaver D, Kufe D. Nature (London) 1997;386:732–735. doi: 10.1038/386732a0. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Yuan S S, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff S P, Wu Y, Arlinghaus R, et al. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 32.Yuan Z M, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, Weichselbaum R, et al. J Biol Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 33.Agami R, Blandino G, Oren M, Shaul Y. Nature (London) 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 34.Gong J G, Costanzo A, Yang H Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y. Nature (London) 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Z M, Shioya H, Ishiko T, Sun X, Gu J, Huang Y Y, Lu H, Kharbanda S, Weichselbaum R, Kufe D. Nature (London) 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 36.White E, Prives C. Nature (London) 1999;399:734–735. doi: 10.1038/21539. [DOI] [PubMed] [Google Scholar]
- 37.Hardin J D, Boast S, Mendelsohn M, de los Santos K, Goff S P. Oncogene. 1996;12:2669–2677. [PubMed] [Google Scholar]
- 38.Syljuasen R G, Krolewski B, Little J B. Cancer Res. 1999;59:1008–1014. [PubMed] [Google Scholar]
- 39.Westphal C H, Hoyes K P, Canman C E, Huang X, Kastan M B, Hendry J H, Leder P. Cancer Res. 1998;58:5637–5639. [PubMed] [Google Scholar]
- 40.Muller A J, Young J C, Pendergast A M, Pondel M, Landau N R, Littman D R, Witte O N. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koleske A J, Gifford A M, Scott M L, Nee M, Bronson R T, Miczek K A, Baltimore D. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- 42.Lavin M F, Khanna K K. Int J Radiat Biol. 1999;75:1201–1214. doi: 10.1080/095530099139359. [DOI] [PubMed] [Google Scholar]
- 43.Harvey M, Sands A T, Weiss R S, Hegi M E, Wiseman R W, Pantazis P, Giovanella B C, Tainsky M A, Bradley A, Donehower L A. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
- 44.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 45.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 46.Armstrong J F, Kaufman M H, Harrison D J, Clarke A R. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 47.Sah V P, Attardi L D, Mulligan G J, Williams B O, Bronson R T, Jacks T. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 48.Kim S T, Lim D S, Canman C E, Kastan M B. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 49.Druker B J, Tamura S, Buchdunger E, Ohno S, Segal G M, Fanning S, Zimmermann J, Lydon N B. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]