Abstract
We have expressed A-FOS, an inhibitor of AP-1 DNA binding, in adult mouse striatal neurons. We observe normal behavior including locomotion and exploratory activities. Following a single injection of cocaine, locomotion increased similarly in both the A-FOS expressing and littermate controls. However, following repeated injections of cocaine, the A-FOS expressing mice showed increased locomotion relative to littermate controls, an increase that persisted following a week of withdrawal and subsequent cocaine administration. These results indicate that AP-1 suppresses this behavioral responses to cocaine. We analyzed mRNA from the striatum before and 4 and 24 hours after a single cocaine injection in both A-FOS and control striata using Affymetrix microarrays (430 2.0 Array) to identify genes mis-regulated by A-FOS that may mediate the increased locomotor sensitization to cocaine. A-FOS expression did not change gene expression in the basal state or 4 hours following cocaine treatment relative to controls. However, 24 hours after an acute cocaine treatment, 84 genes were identified that were differentially expressed between the A-FOS and control mice. 56 gene are down regulated while 28 genes are up regulated including previously identified candidates for addiction including BDNF and Per1. Using a random sample of identified genes, quantitative PCR was used to verify the microarray studies. The chromosomal location of these 84 genes was compared to human genome scans of addiction to identify potential genes in humans that are involved in addiction.
Keywords: A-FOS, addiction, sensitization, gene expression
Psychostimulants can produce a persistent state of compulsive drug use known as addiction. Although, there is much evidence that both the rewarding and addictive properties of these drugs are mediated by increased synaptic dopamine (Di Chiara and Imperato, 1988; Hurd and Ungerstedt, 1989; Kalivas and Duffy, 1990; Robbins and Everitt, 1996), the downstream effects are not well understood at the molecular level. Psychostimulants produce acute and long-term gene expression changes in the striatum and cortex, as well as changes in neuronal morphology (Berke et al., 1998; Graybiel et al., 1990; Robinson et al., 2001; Robinson and Kolb, 1999). This has led to the hypothesis that changes in gene expression underlie the progressive long term behavioral alterations that are critical components of drug addiction (Berke and Hyman, 2000; Nestler et al., 1993).
One of the earliest molecular events following psychostimulant exposure is the activation of the transcription factor CREB, resulting in the expression of a variety of immediate early genes (IEGs) including members the transcription factor activator protein-1 (AP-1) family (Konradi et al., 1994). AP-1 is a heterodimer consisting of Fos family (c-Fos, Fos-B, Δ Fos-B, Fra-1, and Fra-2) and Jun (Jun-B, c-Jun, and Jun-D) family members (Morgan and Curran, 1995; Vinson et al., 2002). Some of these proteins are induced by acute cocaine treatment including c-Jun, Jun-B, c-Fos and Fos-B while others are induced by chronic cocaine treatment including Δ-Fos-B, Fra-1 and Fra-2 (Hope et al., 1992; Hope et al., 1994; Moratalla et al., 1996; Moratalla et al., 1993; Young et al., 1991). These varying patterns of subunit expression suggest that AP-1 complex activity and composition could differentially regulate gene expression and may be important for producing long–term changes associated with drug use.
The function of the AP-1 complex in the development of psychostimulant-related behaviors has been examined using a variety of knockout and transgenic mice and has given various results. The Fos-B knockout mice (lacking Fos-B and its truncated variant Δ-Fos-B) had reduced AP-1 complexes following chronic cocaine exposure and increases cocaine mediated hyperlocomotion and conditioned place preference (CPP) (Hiroi et al., 1997). The long-term over expression of Δ-Fos-B increases AP-1 complexes but also increases cocaine mediated hyperlocomotion and CPP (Kelz et al., 1999). In contrast, short term over expression of Δ-Fos-B produced the opposite response by decreasing CPP (McClung and Nestler, 2003). The lack of the transactivator domain in Δ-Fos-B might be expected to block activity at AP-1 binding sites, however, both increases and decreases in transcription of specific genes are observed. Transgenic mice expressing a transactivation deletion of c-Jun (Δ-c-Jun) would also be expected to block activity at the AP-1 binding site, yet produces different behavioral and gene expression results to Δ-Fos-B transgenic mice (Peakman et al., 2003).
To examine the consequence of inhibiting all AP-1 activity, we used a transgenic mouse line expressing an AP-1 dominant negative termed A-FOS (Gerdes et al., 2006). This dominant negative inhibits AP-1 DNA binding by blocking endogenous AP-1 formation. A-FOS contains the c-Fos dimerization domain and an artificial acidic extension that replaces the DNA binding domain (Olive et al., 1997). The acidic extension interacts with the basic region found in all c-Fos dimerizing partners (typically, Jun family members). Thus A-FOS has a 10,000 fold greater affinity than the endogenous c-Fos protein (Olive et al., 1997) for Jun family members (a required component of AP-1 complexes (Cohen et al., 1989; Nakabeppu et al., 1988). Thus A-FOS blocks Jun proteins from forming AP-1 complexes. Secondly, the acidic extension, by clamping shut the DNA binding α-helix, blocks the DNA binding of A-FOS/Jun containing complexes (Bonovich et al., 2002; Olive et al., 1997; Vinson et al., 2002) thereby leaving the AP-1 binding site free of AP-1 complexes. A-FOS expression was controlled by using the tetracycline-controlled transactivator (tTA) to act on the TetOp promoter of A-FOS. Using the neuron-specific enolase (NSE) promoter to regulate tTA expression, we limited A-FOS expression primarily to adult striatal neurons, including the nucleus accumbens.
Experimental Procedures
Animals
Generation and initial characterization of mice
All animal procedures were performed according to NIH guidelines. Animals were housed 3–5 per cage on a 12:12 hour light:dark cycle with free access to food and water. The TetOp-A-FOS mice have been previously described (Gerdes et al., 2006). These mice were produced and maintained on a FVB/N background. To produce A-FOS expression in the brain the TetOp-A-FOS line was crossed with mice containing the NSE-tTA transgene that uses the neuron-specific enolase promoter to drive expression of the tTA and have been previously described (Chen et al., 1998). The NSE-tTA line was backcrossed 2 times onto the FVB/N background before mating to TetO-A-FOS mice. Control animals were single transgenic littermates of bitransgenic experimental animals and were comprised of similar numbers of NSE-tTA and TetOp-A-FOS only mice too control for the variation in background. For all behavioral experiments, the two single transgenic control lines were first compared to each other to make sure that there were no statistical nor qualitative differences between them, before they were combined into one control group for further analysis. No differences in behavior were observed between the two control lines in either the home cage environment or during the behavioral experiments. All mice used in these experiments were males and between 50 and 100 days of age unless otherwise stated.
Tail clips and ear-taggings were performed at weaning and the DNA isolated using standard techniques. Genotyping was performed by PCR using primers AACAACCCGTAAACTCGCCC and GCAACCTAAAGTAAAATGCCCCAC to detect the NSE-tTA transgene and primers CCACGCTGTTTTGACCTCCATAG and ATTCCACCACTGCTCCCATTC to detect the A-FOS transgene. To verify the sex of newborn pups, used for in situ hybridization experiments that looked at the onset of A-FOS expression, PCR was performed to identify the presence of the Y-linked zinc finger protein-1 (zfy1) using CCTATTGCATGGACTGCAGCTTATG and GACTAGACATGTCTTAACATCTGTCC as primers.
in situ hybridization
Procedures for ribonucleic acid probe labeling and in situ hybridization (Gerfen et al., 1995) and fluorescent in situ hybridizations. (Paletzki, 2002) were as previously described. Briefly, plasmids containing the sequence of the A-FOS construct or the preproenkephalin gene were used for in vitro transcription to make either 35S, digoxigenin or fluorescein labeled antisense probes. Sections hybridized with radioactively labeled probes were opposed to Biomax MR film (Eastman Kodak) for 5 to 30 days and the images digitized and gray density measured using the NIM Image software. The tyramide signal amplification system (TSA™ Fluorescence System, NEN,MA) was used to fluorescently label the non-radioactive probes which where then visualized using a Olympus fluorescent microscope.
RNA Isolation
Striatal tissue samples, rapidly dissected and frozen to −80°C, were used to obtain mRNA initially purified using Trizol reagent (Invitrogen) following their protocol. The supernatant from the Trizol separation was combined with ethanol to 53% and added to an RNAeasy column (Qiagen). The supplied RNAeasy protocol was used to DNase treat and further purify the mRNA, which was then eluted in 10mM Tris HCl pH 8.
Microarray hybridization and analysis
Animals received a single injection of saline or cocaine (30 mg/kg i.p.) and the striatum harvested at 4 hour or 24 hours. Six control and six A-FOS expressing mice were used at each time point, and the mRNA from two animals were combined into one mRNA sample and used to hybridize a single Affymetrix GeneChip® Mouse Genome 430 2.0 Array. Three GeneChips were used for each of the six groups. The striatal mRNA was used to synthesize probe, hybridize onto arrays, washed and scanned by the microarray core facility at Gene Expression Profiling Core, Johns Hopkins University (Baltimore, MD). CEL files produced by the Affymetrix software, where uploaded into Avadis Microarray analysis software (Strand Genomics) along with CDF and annotation files provided by Affymetrix. Robust Multi-Chip Averaging algorithm (RMA) was used for background correction, probe set summarization and normalization. The data was then filtered by producing a present or absent calls for each data set using the microarray analysis software, and removing sets with less then 75% present calls among all arrays leaving 17,979 probe sets for further analysis. Standard T-test was used to identify genes differentially expressed between control and A-FOS striatal samples at the saline, 4 and 24 hour time points. To identify genes regulated by cocaine over 24 hours after an acute injection, a One-way-Analysis of Variance test was used to compare the conditions, saline, 4 and 24 hours. For all statistical analysis, a p<0.001 was used as a cutoff for the selection of genes. Affymetrix probe set IDs or official gene symbols from the National Center for Biotechnology Information Entrex Gene Database (www.ncbi.nlm.nih.gov/) are used to identify genes.
Quantitative real-time RT (qPCR)
20 genes identified in the microarray experiments as differentially expressed were randomly chosen and primmer pairs were selected from 2 different regions of each gene using the Primer 3 software (Rozen and Skaletsky, 2000). All PCR primmer pairs were first tested using standard PCR procedures for the production of a single product of the expected length before they were used for qPCR. Primer pairs for 4 genes failed these standard PCR tests and were not used for qPCR analysis. Only one primer pair was used per gene for the qPCR reactions. As an additional check all qPCR reaction products were checked using standard agarose gel electrophoreses to verify that only a single PCR product of the expected length was produced. As positive controls, 3 additional genes identified that change under our conditions were chosen and used in qPCR, Bdnf, Egr1 and c-Fos.
All qPCR were performed using individual striatal mRNA samples from 5 to 6 animals per group that were treated identically to the animals used in the microarray studies. cDNA was generated from 1µg mRNA using the Superscript III Reverse Transcriptase Kit (Invitrogen) and a dT20-VN primer. The qPCR reactions were performed using 1µl cDNA and the Platinum SYBR green qPCR Supermix-UDG (Invitrogen) and carried out on an ABI PRISM 7900HT. The PCR program consisted of a single incubation of 50 °C for 2 min then 95°C for 10 min and 45 cycles of 95°C for 15 sec, 60°C for 60 sec. The fluorescence was measured at each cycle at 60°C and the threshold cycle value (Ct) identified. ΔCt was determined by normalizing the Ct values for each sample to those of a reference gene (either β-Actin or Gapdh), which were amplified in parallel. Standard T-tests were used to determine if the ΔCt values were significantly different between groups of animals.
The sequences of the qPCR primers were as follows: Actb AGGTCATCACTATTGGCAACG & ATCTCCTTCTGCATCCTGTCA, BDNF GGTATCCAAAGGCCAACTGA & CTTATGAATCGCCAGCCAAT, BB471698 TTTGCACTACGGACATCGAG & GCCAGCCTGGTCTACAAAGT, Camk4 GCTTCAGCCGATGAGATGAG & GGTCCATTTCTTCCTCCACA, Clk1 AGAAAACCAGGAAACGCAGA & TTCTTTTGGCGGGATCATAC, Ctsb CAGCCTCCAGGTTTTGATGT & GGCATGGAGACTTCATTGGT, Fos GGAATTAACCTGGTGCTGGA & AACACGCTATTGCCAGGAAC, Egr1 AAGGGAGAGGCAGGAAAGAC & GGCAGGGATGGTAAGTGAAA, Eif4b TTTGATTTCTGCCTGCTCCT & AGCGATTTGCTGTGCCTACT, Entpd3 ACCTGTCCCGTGCTTAAATG & AGACAGAGTGAAGCCCCTGA, Fndc1 CTACCAGGCATCCCACAACT & ATCTTCCTCGGGGTAGCACT, Gapdh CATCTTGGGCTACACTGAGGA & CCAGGGTTTCTTACTCCTTGG, Gpr123 GTACAAAGCCCCACCTCAAA & TGGGAAGAGAAAGCTGGAGA, Gnb5: ATGAAACGGACACATGCAGA & AGGAATGGCAACCTGTTTTG, Homer2 ACCTCAGCTCATGTCCGAGT & CATGGAGGTCGTCGATCTTT, Hspa1b GAGACATGGACAAGCAAGCA & TCTCAAGAGAGGCGGGATAA, Odz4 GGCTGCTCATCAAAGCCTAC & CTCAGGCGTTTCCAGAGTTC, Pde10a TCATCACCCCACTTGACTGA & AGCATCAGCACCACACAAAG Per1 CCCTGCTACCTTCCCTTCTC & CGAGTTGAACAGTGGGGAGT, Plcb1 CCCTGGATGCAGAAATGACT & CTTGAGAGCTTGAGGGTTGG, Sbsn GCAGACTTGGCAAGAAGGAG & TGCTGCAACCTGTTCATTTC.
Western Blots
Striatal tissue samples were rapidly collected from fresh whole brains and quickly frozen on dry ice. Protein extracts were purified by sonication in 2%SDS with 50mM Tris-HCl pH 7 (10µl/mg tissue), briefly boiled, centrifuged at 14,000g for 15 minutes and the supernatants collected. Protein concentration was determined in triplicates using a bicinchoninic acid (BCA) assay kit (Pierce, Rockford IL). 10 to 25 µg protein samples were separated by SDS-PAGE and Western blots using the NuPAge Novex (Invitrogen, Carlsbad, CA) electrophoresis system following the supplied protocols. Immunohistochemistry was performed using the Jun-B #J106 or c-Jun #J104, (Sigma, St Louis, MO) primary antibodies at 1:1000. Antibody binding was detected using anti-rabbit-IgG-HRP secondary antibodies (#7074 Cell Signaling Technology, Beverly MA) and visualized using chemiluminescence (Supersignal, Pierce) and autoradiographic film (Kodak, Rochester NY). Films were digitized and analyzed using NIH-Image to determine the size of the band using the area under the plot profile for each band. Two-way analysis of variance was used to determine significant effects of genotype and drug treatment or an interaction between these two factors.
Cocaine Sensitization
A 35×35 cm boxes were used with the video based tacking system Ethovision (Nuldus, Netherlands) to determine locomotion following drug administration. Animals received a single injection per day of saline, cocaine 5 mg/kg, or cocaine 10 mg/kg i.p. and were immediately placed into the test chambers for 15 min. These treatments were performed on days 1,2,3,4 & 5. For 6 days, day 6 through 11, the animals received no treatments and were not removed from the colony room. On day 12, all animals received a single injection of cocaine (10mg/kg) and were placed in the same test boxes for 15 min while their locomotion was measured.
Construction of the genome linkage map
The map was designed as described previously (Mir et al., 2003). The human orthologs of the 84 genes differentially expressed in AP-1 expressing animals 24 hours after cocaine, were identified and their coordinates in the human genome were obtained from the UCSC Genome Database (http://genome.ucsc.edu, May 2004 version of the human genome). The map data were managed using Microsoft Excel (Microsoft, Seattle Washington) and plotted using SigmaPlot 9.0 (Systat, Point Richmond, California). The positions for the genetic linkage and association peaks were collected from the genome scan publications referenced in the Results section. The coordinates for the maximum of the peak and the median (left and right borders at the half-height) were plotted. The coordinates of the peaks were interpolated using reference markers from the publications. LOD scores or P-values for the peaks are provided on the chromosomal map as reported by authors.
Statistics
Prism 4 (Graphpad Software, San Diego, CA) software was used to perform T-tests, 2-way-ANOVA with Bonferroni Post-Test and 1-way-ANOVA with Tukey’s multiple comparison post hoc test for all data except for the microarray expression data which was analyzed with the Avadis Software described above.
Results
Expression of A-FOS in the brain
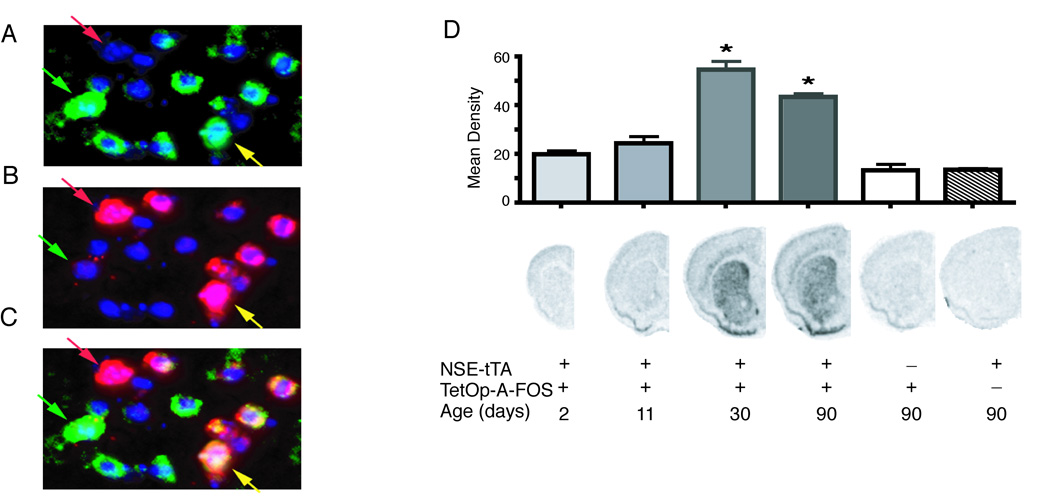
To express A-FOS in the striatum we used the two-transgene tetracycline transactivator system. The TetOp-A-FOS line(Gerdes et al., 2006) that expresses A-FOS using the Tet-O promoter was crossed with a line using the neuron-specific enolase (NSE) promoter to drive expression of tTA (NSE-tTA Line A) (Chen et al., 1998). Coronal brain sections were analyzed by in situ hybridization to confirmed the A-FOS mRNA expression patterns (Fig. 1). A-FOS expression was strong and homogenous throughout the striatum including the dorsal and ventral/nucleus accumbens regions. Weaker expression was also seen in some neocortical layers. To identify which population of striatal projection neurons express A-FOS mRNA, qualitative double fluorescent in situ hybridization was performed using enkephalin (Enk) as a marker of D2-dopamine receptor expressing indirect projecting neurons (Gerfen and Young, 1988). Qualitative assessments from 2 double transgenic mice identified A-FOS mRNA labeling in about 70% of cells, equally distributed to both Enk positive (yellow arrow) and Enk negative cells (green arrow) (Fig. 2) indicating that A-FOS is expressed in indirect projecting neurons and likely the direct projecting neurons as well. No A-FOS labeling was seen in control single transgenic animals.
Fig. 1.
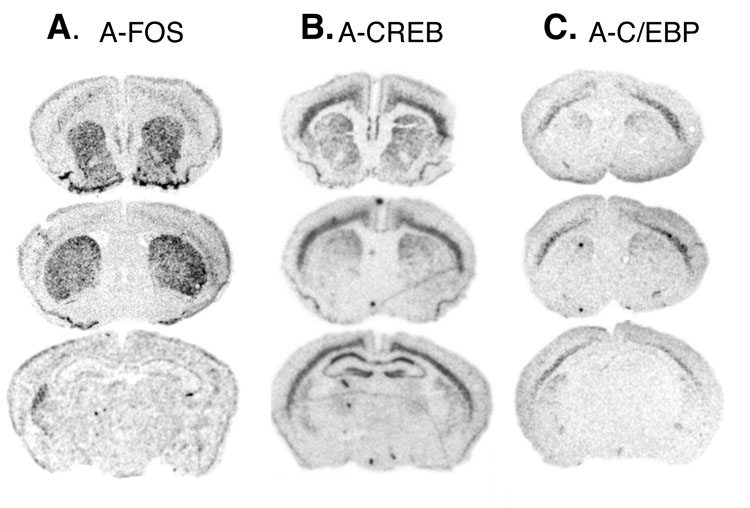
mRNA expression pattern of A-FOS. Adult coronal brain sections from mice containing the NSE-tTA and TetOp-A-FOS transgenes were used for in situ hybridization with radioactively labeled antisense probes complementary to the A-FOS transcripts. Autoradiographic films were digitized and the mRNA labeling is shown as black. A-FOS labeling was observed throughout the striatum including dorsal and ventral regions with little expression in other areas.
Fig. 2.
Striatal localization and developmental expression of A-FOS mRNA. Double fluorescent in situ hybridization was performed on brain sections from adult mice containing both NSE-tTA and TetOp-A-FOS alleles. The A-FOS antisense probe (A) was labeled with Cy3 (green labeling) while the enkephalin antisense probe (B) was labeled with Cy5 (red labeling). (C) The composite image of A-FOS and enkephalin labeling. For all images the nuclei are stained blue using a DAPI counterstained. The green arrows indicate an enkephalin negative neuron (likely a direct projecting striatal neuron) positive for A-FOS whereas the yellow arrow indicates a neuron positive for both enkephalin (indirect projecting striatal neuron) and A-FOS. The red arrows identifies an enkephalin positive neurons that did not express A-FOS. A-FOS expression was observed in a majority of enkephalin positive cells and in a similar number of enkephalin negative cells. (D) A-FOS mRNA expression in the striatum during development was determined by in situ hybridizations with a radiolabeled A-FOS antisense probe of brain sections from transgenic mice at 2, 11, 30 and 90 days after birth. The labeling density over the striatum was determined using digitized images and the mean density of each group (5 to 8 animals each) is indicated in the graph plus standard error. The presence of the NSE-tTA or TetOp-A-FOS allele is indicated with “+” below the example images along with the age. * = p<0.001 relative to NSE-tTA only containing adult controls. (Tukey’s multiple comparison test following 1-way-ANOVA)
NSE is not expressed in the brain until about 2 weeks after birth (Lucas et al., 1988), therefore, the NSE-tTA transgene should regulate TetOp-A-FOS with a similar temporal pattern and thereby produce few developmental adaptations. A-FOS mRNA levels were measured by in situ hybridization using S35 label probes on coronal brain slices from NSE-tTA × TetOp-A-FOS mice at 2, 11, 30, and 90 days after birth. A-FOS expression was not detected at 2 and 11 days after birth (Fig. 2) but became apparent at 30 and 90 days after birth. Furthermore, A-FOS expression was only observed in mice containing both the NSE-tTA and TetOp-A-FOS transgenes.
Behavior of A-FOS expressing mice
Home cage behavior of the A-FOS expressing mice was indistinguishable from littermate controls. They grew at the same rate; they interacted with littermates and breed identically to controls (data not shown). To determine if A-FOS expression in the striatum alters locomotion and habituation in a novel environment, we tested mice by placing them in a novel environment for 15 min on five consecutive days. On the first exposure to the novel environment, there was no difference in locomotion between the A-FOS expressing and control mice (Fig. 3A). For both the control and A-FOS expressing mice, repeated exposure produced a similar habituation to the “novel environment” as measured by a significant decline in locomotion over days (p<0.01) that was independent of genotype.
Fig. 3.
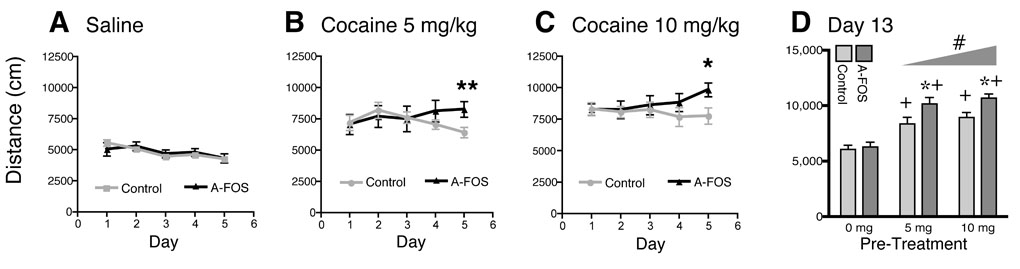
Cocaine induced locomotion and sensitization in A-FOS mice. On five consecutive days, A-FOS expressing and control mice were treated with (A) saline, (B) 5 mg/kg or (C) 10 mg/kg cocaine, immediately placed in a novel environment consisting of a 30×30 cm box and their locomotion measured for 15 minutes using a video tracking system. The graphs display the group means (n=11 to 13 each) and standard errors of total distance traveled for fifteen minutes over the five treatment days. **=p<0.01 and *=p<0.05 relative to day 1 (Bonferroni post-tests). (D) Following six days without treatment, all three groups of animals were tested with a single injection of 10 mg/kg cocaine and immediately placed into the same 30×30 box and their locomotion tracked. The graph displays the group means and standard errors of total distance travel in cm. # significant effect of dose p<0.05 as determined by 2-way-ANOVA. When the control and A-FOS groups were analyzed separately by 1-way ANOVA, the 5mg and 10mg pretreated animals from each group had significantly great (p<0.05) locomotion relative to their respective saline treated controls. *=p<0.05 relative to controls at the same dose. (Bonferroni Post-Test ). +=p<0.05 relative to saline treated animals the same group, control or A-FOS.
To determine whether A-FOS expression in the striatum and nucleus accumbens affected cocaine induced locomotion and sensitization, we injected A-FOS expressing and control mice with saline, 5 mg/kg, or 10 mg/kg of cocaine intraperitoneally on five consecutive days, placing the animals in a novel environment each day, and measured locomotion for 15 minutes. On the first day, cocaine produced a dose dependent increase in locomotion (P<0.001) relative to saline treated animals, in both the A-FOS expressing and control mice (Fig. 3). The increase in locomotion for both groups was not different on the first, second or third day of treatment. However, on the fourth cocaine treatment day, the A-FOS expressing mice exhibited more cocaine induced locomotion relative to control mice at both the 5 mg/kg and 10 mg/kg concentrations. This trend was even greater (P<0.001) on the fifth day of repeated daily cocaine injections. Bonferroni posttest analysis of locomotion on the fifth cocaine treatment day indicated significant differences in cocaine-induced locomotion between A-FOS expressing and control mice at both the 5 mg/kg (p<0.01) and 10 mg/kg (p<0.05) doses, demonstrating that A-FOS expression enhances locomotion after repeated but not single cocaine treatments.
To test for long-term changes in induced locomotion after a drug free period, the mice treated for 5 days, described above, were left untreated for 7 days then retested with a single dose of 10 mg/kg cocaine. A-FOS and control mice pretreated with saline had similar cocaine induced increases in locomotion, again indicating that A-FOS expression does not affect acute responses to cocaine (Fig. 3). As expected, mice pretreated with cocaine had an enhanced locomotion response when treated with cocaine a week later (p<0.001 for drug treatment 2-Way-ANOVA). Additionally, this response was even greater in the A-FOS expressing mice (p<0.001 for genotype). Bonferroni post-tests indicated significant differences in cocaine-induced locomotion between A-FOS expressing and control mice at both the 5 mg/kg and 10 mg/kg pretreatment doses (p<0.05). To be sure that sensitization was seen in both control and A-FOS expressing mice these groups were analyzed separately using 1-way-ANOVA which indicated a significant increase in hyperlocomotion for 5mg and 10mg pretreated animals for both control and A-FOS groups (p<0.05).
A-FOS does not alter AP-1 family member expression
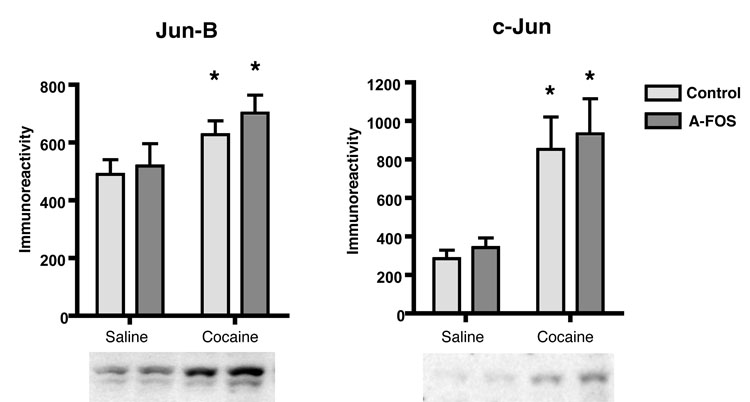
A-FOS can bind to the Jun family of AP-1 proteins including Jun-B and c-Jun that are expressed in the striatum and are up regulated following cocaine injections. We determined if the expression of A-FOS alters the striatal levels of basal or cocaine stimulated Jun-B and c-Jun protein using Western Blots (Fig. 4). Basal levels of Jun-B and c-Jun proteins was not affected by A-FOS expression. Acute cocaine treatment increased the levels of both Jun-B and c-Jun (p<0.001) as has been previously reported (Hope et al., 1992; Moratalla et al., 1993) and this increases was unaffected by A-FOS expression (p>0.05). Thus, these initial molecular responses to cocaine administration were not altered in the A-FOS expressing mice.
Fig. 4.
Jun protein levels in A-FOS expressing mice. Jun-B and c-Jun protein levels in the striatum were determined by western blotting and chemiluminesense. The autoradiographic films were digitized and a density plot for each protein lane was produced. The area under the plot line for each band was used to compare relative protein concentrations in each sample and displayed as the mean immunoreactivity plus standard error for each group. Cocaine produced a significant increase in both Jun-B (*p<0.01) and c-Jun (* p<0.001) protein levels as determined by 2-way-ANOVA. There was no significant affect of A-FOS expression on the levels of Jun-B and c-Jun protein expression.
In the basal state, there are no changes in gene expression in A-FOS mice
We determined the effect of A-FOS expression on global mRNA expression profile in the striatum in both the basal state and following cocaine administration. Striatal mRNA from adult control and A-FOS expressing mice was compared using Affymetrix 430 2.0 mouse microarrays that contain a >45,000 probe set. Three control arrays from 6 mice (each array was hybridized with the mRNA from two mice) and three A-FOS arrays (6 mice) were hybridized and the normalized data filtered to remove probe sets that did not produce a signal resulting in 17,979 probe sets for analysis. A standard unpaired T-test was used to determine the probability that the expression of a gene was different between the two groups. A p<0.001 cut off was used which would be expected to produce 18 false positives from the 17,979 probe set analyzed. Only three genes were identified (p<0.001) that were different between the control and A-FOS expressing mice indicating that there is essentially no difference in gene expression in the striatum in the basal state of adult mice.
Acute cocaine administration induces gene expression differences in A-FOS mice at 24 hours
The behavioral experiments indicated that cocaine can produce long-term adaptations rapidly, even after a few days exposure. We wanted to determine what differences in gene expression occur early following cocaine when the animal behaviors are beginning to change. Therefore, we isolated the striatal mRNA from A-FOS and control mice at both 4 and 24 hours following a single injection of 30 mg/kg cocaine and analyzed gene expression using the same Affymetrix 430 2.0 mouse microarrays. At 4 hours, only twelve genes had a P<0.001 indicating that A-FOS expression had little or no effect on cocaine induced gene expression 4 hours after a single injection of cocaine. These 12 genes are different from the 3 identified in the basal case suggesting no systematic differences in these samples.
However, at 24 hours, dramatic differences in gene expression are observed in the striatum between A-FOS and control mice. 84 genes were identified with p<0.001 indicating that A-FOS expression had the greatest effect on gene regulation 24 hours after a single injection of cocaine. These genes are different from the previously identified 15 genes at the basal and 4 hour time points. Of the 84 genes that are different at 24 hours between control and A-FOS mice, 54 changed expression following cocaine administration in control mice but not in the A-FOS mice while 26 genes were different in the A-FOS mice but not in the control mice.
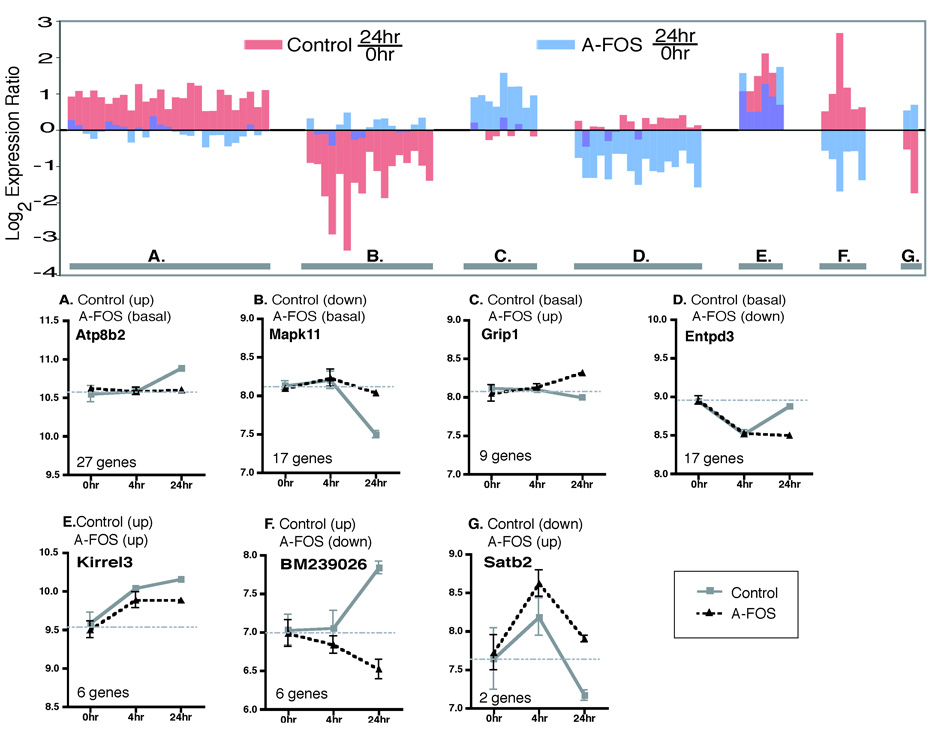
Figure 5 presents the 84 genes organized into seven groups based on both the basal and 24 hour expression profiles. Genes are considered up or down regulated if their expression changed 1.4 fold relative to basal conditions. The two largest groups (Group A (27 genes) and B (17 genes)) contain genes whose expression is either induced or suppressed, respectively, by cocaine in control mice but do not change in the A-FOS mice. These genes are presumably regulated by AP-1 activity and this is blocked by A-FOS. A second set of genes (Groups C (9 genes) and D (17 genes)) contain genes that are not altered following cocaine administration in control mice but are altered in mice expressing A-FOS.
Fig. 5.
A-FOS affected gene expression 24 hours after cocaine. The chart displays the the 84 genes identified as significantly (p<0.001) altered by A-FOS expression along the x axis showing the log2 expression ratios of both the control mice (red) 24 hours/0 hour (saline) and A-FOS mice (blue) 24 hours/0 hour. The 84 genes were divided into 7 groups depending on their 24 hour versus basal expression levels (saline). The 7 groups (A–H) are indicated on the graph by the lower gray bar with individual examples shown below. The lower examples from each group display the mean log expression values plus standard errors from the Affymetrix chip data for both A-FOS expressing and control mice at 0, 4, and 24 hours after a single injection of cocaine. The horizontal gray bar indicates the average expression value of the 0 hour time point (basal expression) for each gene. The example charts also give the official gene symbol for the each gene (from the National Center for Biotechnology Information Entrez Gene Database) shown along with the number of genes in each group. Groups A and B contain genes in which their normal cocaine altered expression was blocked by A-FOS expression. Groups C and D contain genes in which cocaine altered their expression at 24 hours only in A-FOS mice and not in controls. Group E contains genes in which the level of cocaine induced expression was modified by A-FOS expression, while groups F and G contain genes whose expression was reversed by A-FOS expression 24 hours following cocaine.
Table 1 lists the 84 genes organized into the seven groups described above. Using the gene ontology information (http://www.ncbi.nlm.nih.gov/entrez/), 55 of the 84 genes were found to have a known or suspected function. Most of the identified genes are involved in signaling pathways (29), with the others involved with transcription/translation regulation (10), in metabolism (9), cytoskeletal genes (4), and growth and development (3).
Table 1.
List of genes differentially expressed in A-FOS expressing animals 24 hours after cocaine. The table includes the Affymetrix probe set ID for each gene along with its official gene symbol and description/function taken from the NCBI Entrez gene databases (if available). The expression profile group to which a particular gene was assigned is also indicated, using the same letters as in Fig. 6. The number under Ref on the right indicates where a gene has previous been reported in microarray studies of psychostimulants (1= (Yao et al., 2004), 2= (McClung and Nestler, 2003))
| Probe Set ID | Expression Profile Groups | Gene Symbol | Gene Description | Refs |
|---|---|---|---|---|
| 1423126_at | a | Atp1b3 | ATPase, Na+/K+ transporting, beta 3 polypeptide | 1 |
| 1434026_at | a | Atp8b2 | atpase, class I, type 8B, member 2: cation transport ATPase activity | 1 |
| 1428845_at | a | Bclaf1 | BCL2-associated transcription factor 1 | |
| 1455771_at | a | Bzrap1 | benzodiazapine receptor associated protein 1 | |
| 1426124_a_at | a | Clk1 | CDC-like kinase 1:dual specificity protein kinases found in the nucleus | |
| 1417492_at | a | Ctsb | cathepsin B : lysosomal cysteine proteinase | |
| 1424288_at | a | D5Wsu46e | unknown | |
| 1426380_at | a | Eif4b | eukaryotic translation initiation factor 4B | 1 |
| 1426810_at | a | Jmjd1a | jumonji domain containing 1A: encodes a zinc finger protein | |
| 1458428_at | a | Kcnt1 | potassium channel, subfamily T, member 1: cation transport | |
| 1449172_a_at | a | Lin7b | lin 7 homolog b : Veli isoform associates with CASK and Mint1 | |
| 1418763_at | a | Nit2 | nitrilase family, member 2 | |
| 1426548_a_at | a | none | unknown | |
| 1429344_at | a | none | unknown | |
| 1435744_at | a | none | unknown | |
| 1439194_at | a | none | unknown | |
| 1443287_at | a | none | unknown | |
| 1444345_at | a | none | unknown | |
| 1422622_at | a | Nos3 | nitric oxide synthase 3, endothelial cell | |
| 1447640_s_at | a | Pbx3 | pre B-cell leukemia transcription factor 3 | |
| 1417919_at | a | Ppp1r7 | protein phosphatase 1, regulatory (inhibitor) subunit 7 | 1 |
| 1441963_at | a | Prosapip1 | unknown | |
| 1429614_at | a | Prpf18 | PRP18 pre-mRNA processing factor 18 homolog | |
| 1449957_at | a | Ptprv | protein tyrosine phosphatase, receptor type, V | 1,2 |
| 1439630_x_at | a | Sbsn | suprabasin | |
| 1455493_at | a | Syne1 | synaptic nuclear envelope 1 | |
| 1420418_at | a | Syt2 | synaptotagmin 2: membrane bound, calcium ion binding, transporter activity | 1,2 |
| 1422168_a_at | b | Bdnf | brain derived neurotrophic factor | |
| 1417606_a_at | b | Calr | calreticulin | 1,2 |
| 1450005_x_at | b | Egfl9 | EGF-like-domain, multiple 9 | |
| 1435473_at | b | Gm347 | unknown | |
| 1424367_a_at | b | Homer2 | homer homolog 2 | |
| 1454784_at | b | Hs3st2 | heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | 1 |
| 1421926_at | b | Mapk11 | mitogen-activated protein kinase 11 | 1 |
| 1417569_at | b | Ncald | neurocalcin delta : calcium ion binding | |
| 1433909_at | b | none | unknown | |
| 1439808_at | b | none | unknown | |
| 1441069_at | b | none | unknown | |
| 1456684_at | b | none | unknown | |
| 1434270_at | b | Nptxr | neuronal pentraxin receptor | |
| 1428393_at | b | Nrn1 | neuritin 1: an immediate-early gene induced by Ca(2+) influx | 1,2 |
| 1450945_at | b | Prkca | protein kinase C, alpha | 1,2 |
| 1436182_at | b | Sat61 | special AT-rich sequence binding protein 1 | |
| 1440128_s_at | b | Sez6l | seizure related 6 homolog (mouse)-like | |
| 1426367_at | c | Cab39l | calcium binding protein 39-like | |
| 1424633_at | c | Camk1g | calcium/calmodulin-dependent protein kinase I gamma | 1,2 |
| 1423286_at | c | Cbln1 | cerebellin 1 precursor protein | |
| 1436529_at | c | Gpr123 | G protein-coupled receptor 123 | |
| 1421350_a_at | c | Grip1 | glutamate receptor interacting protein 1 | |
| 1417435_at | c | Large | like-glycosyltransferase | |
| 1418104_at | c | Nrip3 | nuclear receptor interacting protein 3 | |
| 1434530_at | c | Odz4 | odd Oz/ten-m homolog 4 (Drosophila) | |
| 1425110_at | c | Sorcs3 | sortilin-related VPS10 domain containing receptor 3 | |
| 1434934_at | d | Atpaf1 | ATP synthase mitochondrial F1 complex assembly factor 1 | |
| 1419696_at | d | Cd4 | CD4 antigen | |
| 1428147_at | d | Coro7 | coronin 7: Golgi apparatus, integral to membrane of membrane fraction | |
| 1434049_at | d | Entpd3 | ectonucleoside triphosphate diphosphohydrolase 3 | |
| 1453321_at | d | Fndc1 | fibronectin type III domain containing 1 | |
| 1457189_at | d | Itpr1 | inositol 1,4,5-triphosphate receptor 1 | |
| 1435649_at | d | Nexn | nexilin : nF actin binding protein | |
| 1439724_at | d | none | Ria8: Regulates Golf G-protein signalling | |
| 1418894_s_at | d | Pbx2 | pre B-cell leukemia transcription factor 2 | |
| 1449851_at | d | Per1 | period homolog 1 | 1,2 |
| 1442068_at | d | Pinx1 | PIN2-interacting protein 1 | |
| 1433907_at | d | Pknox2 | Pbx/knotted 1 homeobox 2 | |
| 1422620_s_at | d | Ppap2a | phosphatidic acid phosphatase 2a | |
| 1454902_at | d | Prkcz | protein kinase C, zeta | |
| 1419137_at | d | Shank3 | SH3/ankyrin domain gene 3 | 1 |
| 1423258_at | d | Syt9 | synaptotagmin 9 | 1,2 |
| 1426997_at | d | Thra | thyroid hormone receptor alpha | 1 |
| 1456011_x_at | e | Acaa1 | acetyl-Coenzyme A acyltransferase 1 | |
| 1431402_at | e | Kirrel3 | kin of IRRE like 3 | |
| 1426233_at | e | Map2k4 | mitogen activated protein kinase kinase 4 | |
| 1439502_at | e | none | unknown | |
| 1441753_at | e | none | unknown | |
| 1454487_at | e | none | unknown | |
| 1457536_at | f | Gpc5 | glypican 5 | |
| 1428508_at | f | none | unknown | |
| 1432680_at | f | none | unknown | |
| 1441172_at | f | none | unknown | |
| 1447478_at | f | none | unknown | |
| 1451046_at | f | Zfpm1 | zinc finger protein, multitype 1 | |
| 1418178_at | g | Ina | internexin neuronal intermediate filament protein, alpha | 2 |
| 1427017_at | g | Satb2 | special AT-rich sequence binding protein 2 |
492 Genes regulated by acute cocaine in control mice
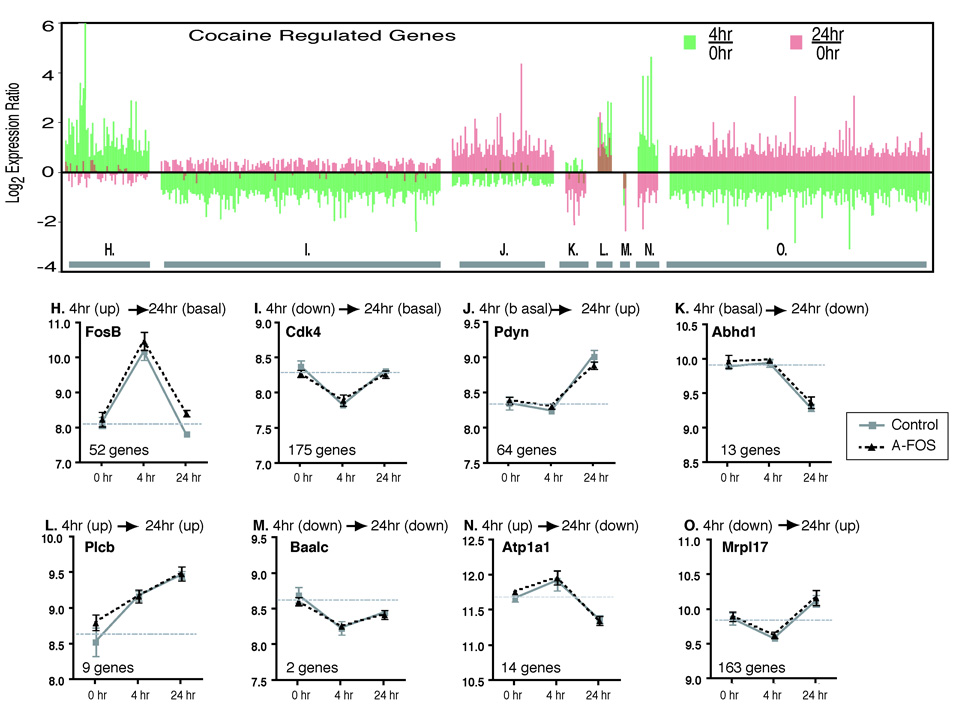
We also identified genes that are regulated by cocaine in wild type mice. Because the mRNA expression experiments were performed on the same set of Affymetrix microarrays, comparisons between the saline, 4 hours and 24 hours groups could be performed using One-Way Analysis of Variance to identify genes that are regulated over time by cocaine. Because fewer genes were identified as differentially expressed then was expected by chance between the saline control and saline A-FOS mice and between the 4 hour control and 4 hour A-FOS mice, these samples were combined into one saline group (6 microarrays) and one 4 hour group (6 microarrays) producing greater statistical power to identify cocaine regulated gene expression changes. 492 genes were identified as having altered expression following cocaine with ≈ 18 genes expected by chance using p<0.001 cutoff. Several general observations are made. At 4 hours, over 4.5 times as many genes go down (340 genes) as go up (75 genes). Conversely, at 24 hours, 8.1 times more genes go up (236 genes) than go down (29 genes).
These 492 genes were divided into 8 groups depending on their expression at 4 hours and 24 hours relative to saline treated controls with the expression considered up or down at each time point if the expression ratio was at least 1.4 times the basal expression (Fig. 6). Group H (52 genes) contain genes that are up regulated at 4 hours and then return to normal at 24 hours. This includes the classic IEGs c-Fos, CREM, Fos-B, and Egr1. In contrast, group J (64 genes) contain genes that are not affected by cocaine at 4 hour but are then up regulated at 24 hours. These include the known cocaine targets dynorphin, and homer1 that are potentially activated by transcription factors in Group H. The remaining groups will be addressed in the discussion. The largest two groups (Group I (175 genes) and Group O (163 genes)) are down regulated at 4 hours which may represent a stress type response were transcription is halted during a challenge and do not contain classically identified cocaine targets.
Fig. 6.
Temporal expression pattern of cocaine regulated genes. The regulation of genes by cocaine was determined by comparing the basal (combined A-FOS and control data), 4 hour (combined A-FOS and control data) and 24 hour (control data only) time points using 1-way-ANOVA to identify 824 genes that were differentially expressed following cocaine (p<0.001). These genes were divided into 8 groups (H–O) based on their temporal expression pattern as described in the methods. The upper chart, organized by group, displays the data for each of the 824 genes showing the log2 expression ratio of 4 hours/saline (green bars) and 24 hours/saline (red bars). The lower charts show an example from each group and displays the mean normalized log expression values and standard errors from the Affymetrix chip data for both A-FOS expressing and control mice at saline, 4, and 24 hours after a single injection of cocaine. The horizontal gray bar indicates the average expression value of the 0 hour time point (basal expression) for each gene. Additionally, the graphs contain the official gene symbol (from the National Center for Biotechnology Information Entrez Gene Database) and the number of genes in each group. The genes are FBJ osteosarcoma oncogene B (Fos-B), cyclin-dependent kinase 4 (Cdk4), prodynorphin (Pdyn), chondroitin sulfate proteoglycan 3 (Cspg3), homer homolog 1 (Homer1), brain and acute leukemia, cytoplasmic (Baalc), fos-like antigen 1 (Fra1) and mitochondrial ribosomal protein L17 (Mrpl17).
To confirm the microarray data, we used quantitative real time-PCR (qPCR) to measure the striatal mRNA expression levels, using independent samples, of some of the genes identified to change in response to A-FOS expression and/or cocaine treatment. A total of 10 genes from the A-FOS regulated gene list and 6 genes from the acute cocaine regulated gene list were randomly selected. 88% or 14 of 16 genes were found to be differentially expressed by qPCR confirming the microarray data (p<0.05) (table 2). As a positive control 3 additional genes identified to be regulated by cocaine (Fos, Egr-1 and Bdnf) were also analyzed and found to be differentially expressed (p<0.05
Table 2.
Confirmation of a random sample of genes identified by microarrays as differentially expressed. 10 genes from the A-FOS regulated gene list and 6 genes from the cocaine regulated gene list were randomly picked and qPCR used to confirm their differential expression in the striatum from an independent group of animals treated identically to the animals used in the microarray studies. Additionally 3 genes identified to be regulated by these conditions were also tested; Bdnf, Egr1, Fos. The ΔCt value for each gene was determined and a standard T-test was used to identify differences in expression (*= p<0.05). 88% of the randomly chosen genes were confirmed differentially expressed. Table 2 lists the genes using the official gene symbol along with the expression difference based on changes in ΔCt (ΔΔCt) relative to the control or untreated group. A negative number indicates an increase in expression in the A-FOS or cocaine treated animal.
| Gene Symbol | ΔΔCt | |
|---|---|---|
| A-FOS regulated | ||
| Bdnf | −2.24 | * |
| Clk1 | 0.66 | |
| Ctsb | −2.29 | * |
| Eif4b | 1.28 | * |
| Entpd3 | 2.47 | * |
| Fundc1 | 3.84 | * |
| Gpr123 | −2.05 | * |
| Homer2 | −3.21 | * |
| Odz4 | −0.36 | |
| Per1 | 1.74 | * |
| Sbsn | 1.78 | * |
| Cocaine regulated | ||
| BB471698 | −2.38 | * |
| Camk4 | 1.81 | * |
| Egr1 | −3.08 | * |
| Fos | −2.18 | * |
| Gnb5 | 2.00 | * |
| Hspa1a | −1.83 | * |
| Pde10a | −1.29 | * |
| Plcb1 | 3.02 | * |
Gene scan of human addiction: correlation with 84 genes that are mis-regulated by A-FOS
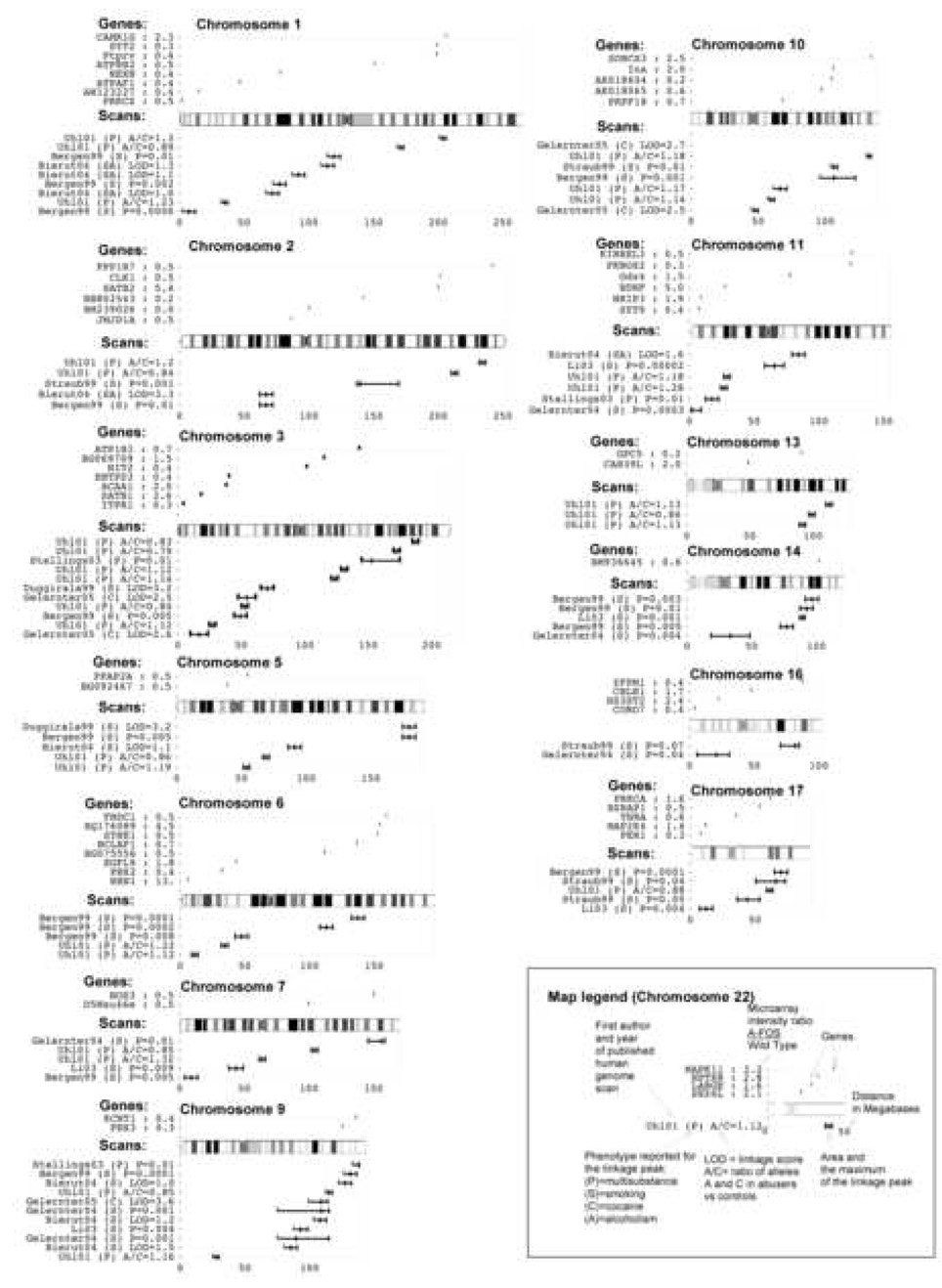
The 84 genes mis-regulated in the striatum of A-FOS expressing animals following cocaine administration potentially mediate the cocaine induced behavioral adaptations in mice and possibly in humans. We have determined if human homologues of the 84 mis-regulated mouse genes are in similar chromosomal positions as loci identified in humans to correlate with addiction. The identification of human genes responsible for addiction comes primarily from family and population linkage studies. To date, nine reports of linkage or association via genome-wide screens for drug addiction genes have been published: (Bergen et al., 1999; Bierut et al., 2004; Duggirala et al., 1999; Gelernter et al., 2004; Gelernter et al., 2005; Li et al., 2003; Stallings et al., 2003; Straub et al., 1999; Uhl et al., 2001). Using polymorphic markers, these researchers genotyped large collections of DNA from human drug addicts and identified genomic locations that correlated with the drug-predisposing genes. This method however can only identify linkage associations with chromosomal regions that contain hundreds of genes. 58 human orthologes were identified from the 84 mis-regulated mouse genes and their chromosomal position was overlaid onto a map of all genetic linage markers (Fig. 7). Mis-regulated genes that occur in the peaks from multiple linkages may have significant affects on behavior in both this animal model and in human drug addiction. Genes that occur within multiple linkages include internexin neuronal intermediate filament protein alpha (Ina) on chromosome 10, brain derived neurotrophic factor (BDNF) on chromosome 11 and protein kinase C alpha (Prkca) on chromosome 17.
Fig. 7.
Linkage Map. The graph displays the physical location on 13 chromosomes of identified loci from human genome scan studies of addiction. Additionally, the location of the human orthologs of the 84 genes differentially expressed in A-FOS animals following cocaine are displayed. The map legend is given using chromosome 22 as an example.
Discussion
We expressed in the adult striatum A-FOS a novel AP-1 dominant negative. Expression does not affect baseline behavior or the initial behavioral response to cocaine. However, following repeated cocaine treatments, A-FOS animals displayed enhanced cocaine induced hyperlocomotion compared with littermates. A-FOS had a similar affect on striatal gene expression, therefore no changes at baseline or 4 hours following a single injection of cocaine, but producing significant effects on gene expression at 24 hours after cocaine. These data indicate that striatal AP-1 activity has little affect on basal gene regulation in the adult striatum and behavior, but that under stimulated conditions such as cocaine exposure; AP-1 activity alters gene expression and blunts long-term behavioral changes mediated by repeated cocaine administration.
Psychostimulants, such as cocaine, produce hyperlocomotion that often increases with repeated treatments, a change in behavior termed sensitization (Kalivas and Stewart, 1991; Robinson and Becker, 1986; Shuster et al., 1977). Over 5 days of cocaine treatment, A-FOS expression produced a greater increase in hyperlocomotion indicating that AP-1 activity in the striatum reduces the behavioral response. Furthermore, these changes are long lasting and do not require daily cocaine treatments in that even 7 days after the last treatment A-FOS animals continued to have a greater response to cocaine. Therefore, our results indicate that inhibiting AP-1 DNA binding in the striatum potentiates this long-term behavioral response to cocaine.
As discussed in the introduction, previous research using various knockout and transgenic mice that alter AP-1 components has given contradictory results concerning the role of the AP-1 in cocaine responses. For example, Δ-Fos-B has been reported to either activate and repress reporter expression constructs containing only consensus AP-1 binding sites (Mumberg et al., 1991; Nakabeppu and Nathans, 1991; Peakman et al., 2003; Wisdom and Verma, 1993; Wisdom et al., 1992; Yen et al., 1991). In a more complex biological context, Δ-Fos-B blocks the expression of a reporter using the collagenase promoter that contains an AP-1 site (Nakabeppu and Nathans, 1991) and blocks cellular transformation of cell lines stimulated by the over-expression of c-Fos or Fos-B (Mumberg et al., 1991; Yen et al., 1991) suggesting that Δ-Fos-B is an inhibitor at the AP-1 site in this context. The long-term over-expression of Δ-Fos-B has been shown to enhance cocaine mediated behavioral changes (Colby et al., 2003; Kelz et al., 1999) similar to what we observe with A-FOS expressing mice suggesting that both might affect behavior by a similar mechanism, the inhibition of AP-1 mediated modification of gene expression.
A key difference between the A-FOS mouse and other AP-1 transgenic and knockout mouse models is that A-FOS expression has no detected affects under basal conditions. This is not surprising given that key components of the AP-1 complex are expressed at low levels in untreated adult striatal neurons (Hope et al., 1992; Rosen et al., 1994) and that low levels of AP-1 binding is seen (Couceyro et al., 1994; Hope et al., 1992; Hope et al., 1994). Because basal levels of AP-1 binding are low, the expression of A-FOS has no detectable effects. Only after AP-1 components are induced, such as by cocaine (Hope et al., 1992; Hope et al., 1994; Moratalla et al., 1996; Moratalla et al., 1993; Young et al., 1991), does A-FOS have an affect as demonstrated by altered gene induction 24 hours later and altered behavior that develops over 4 to 5 days of repeated cocaine treatments.
In contrast, the over expression of Δ-Fos-B and Δ-c-Jun, both truncated AP-1 components lacking the transactivation domains, might be expected to produce baseline changes in gene regulation because they still bind the AP-1 site (Colby et al., 2003; Kelz et al., 1999; Peakman et al., 2003) and may interact with other transcription factors at other binding sites and the transcription machinery. Although, A-FOS blocks the DNA binding of endogenous AP-1 complexs (Gerdes et al., 2006), due to the similarity in the recognition sequences of multiple transcription factor binding sites, it is unclear if other transcription factors might bind the empty AP-1 in A-FOS expressing mice and what affect this might have on gene expression.
Our behavioral data and that from others (Cabib, 1993; Robinson and Becker, 1986; Shuster et al., 1977) shows that psychostimulant induced behavioral changes occur quite rapidly, within a few days. Therefore, understanding initial gene expression changes following cocaine administration may give insight into the molecular adaptations that manifest themselves in altered long-term behavior. Affymetrix mouse genome 430 2.0 microarrays were therefore used to measure changes in striatal mRNA expression in A-FOS and control mice following cocaine administration. Similar to the behavioral data, no gene expression differences are observed in the basal state or 4 hours following the initial cocaine administration suggesting that AP-1 does not modulate gene expression at these times. However, 24 hours after a single injection of cocaine, changes in gene expression were observed and 84 genes were differentially regulated in A-FOS expressing animals versus controls. Many of the genes mis-regulated are involved in cell signaling, including cell surface ion channels, receptors, neurotransmitters, intracellular kinases, and phosphatases while, a few genes are scaffold proteins known to interact with other proteins in the postsynaptic density (Table 1). Although it is difficult to make direct comparisons between gene expression changes in previous work on cocaine because of the diverse experimental conditions, 8 of the genes have been previously identified in microarray studies of psychostimulants effects on the striatum while another 9 are family members of proteins previously identified (McClung and Nestler, 2003; Yao et al., 2004). Furthermore, three of the identified genes (BDNF, Per1(period homolog 1), and homer) have previously been shown in mouse knockout experiments to alter the behavioral sensitization to cocaine (Abarca et al., 2002; Ghasemzadeh et al., 2003; Hall et al., 2003; Kalivas et al., 2004). The 84 differentially expressed genes whose activity is altered by A-FOS expression 24 hours after the initial cocaine administration represent candidates for a role in the development of enhanced locomotor sensitization.
The microarray data identified 492 genes that are altered either at 4 or 24 hours after cocaine administration, a much greater number then has previously reported as altered after psychostimulant exposure (Berke et al., 1998; Gonzalez-Nicolini and McGinty, 2002; McClung and Nestler, 2003; Yao et al., 2004). We suggest that the large number of altered genes arises from our use of multiple independent replicates, with 15 arrays used in total, which increase the statistical power of the analysis. The use of a high threshold of p<0.001 to identify differentially expressed genes should lead to very few false positives, approximately 18 expected by chance from a dataset of 17,979 expressed genes. The quality of the microarray data was also supported by the high rate (88%) of confirmed positives using qPCR. Furthermore, the validity of this large microarray dataset is supported by the observations that for some comparisons such as untreated A-FOS expressing animals versus controls, very few differences in gene expression were seen. Additionally, a number of cocaine-affected genes have been previously described as altered by cocaine including but not limited to, Fos, Fos-B, Pdyn, Egr-1. Therefore, we believe that this large dataset of cocaine-affected genes presented here, accurately represents the effects of acute cocaine on transcription in the striatum. The observation that a single injection of cocaine altered the expression of hundreds of genes suggests that cocaine exposure is a potent stimulus to which a neuron produces a complex set of responses.
The mis-regulated genes in the A-FOS expressing mice may be candidates for involvement in human addiction. To identify candidates from the 84 genes mis-regulated by A-FOS, we examined the chromosomal location of their human homologes and compared these data to nine previously published gene linkage studies on addiction in humans. We identified 6 genes whose chromosomal location was implicated in more than one of these linkage studies. Although speculative, these genes represent potential candidates for further study because they may be involved in altered long-term behavioral changes in both our A-FOS expressing mouse model and in human addiction. Of these genes, BDNF has been identified in multiple microarray studies using cocaine with various transgenic mouse models as well as affecting cocaine induced behaviors in BDNF heterozygotic knockout mice (Hall et al., 2003; McClung and Nestler, 2003; Yao et al., 2004). Furthermore, changes in Prkca activity have been shown to alter psychostimulant mediated behavior or to be altered by psychostimulant exposure (Aujla and Beninger, 2003; Narita et al., 2004; Steketee et al., 1998). Therefore, the affects of BDNF and Prkca on psychostimulant-mediated behavior might be of particular interest for further study.
Acknowledgements
We would like to thank Eric Nestler for providing the NSE-tTA mice used in these experiments, and Shari Thomas for her technical support. We would also like to thank Mike Iadarola for his helpful comments on the manuscript. R.P would also like to thank Alexei Morozov (Unit of Behavioral Genetics, National Institute of Mental Health, NIH) and Charles Gerfen (Laboratory of Systems Neuroscience, National Institute of Mental Health,NIH) for providing support for these studies.
Abbreviations
- IEGs
immediate early genes
- AP-1
activator protein-1
- tTA
tetracycline-controlled transactivator
- NSE
neuron-specific enolase
- Enk
enkephalin
- Gnb5
guanine nucleotide binding protein beta 5
- Hspa1a
heat shock protein 1A
- Pde10a
phosphodiesterase 10A
- Ina
internexin neuronal intermediate filament protein alpha
- BDNF
brain derived neurotrophic factor
- Prkca
protein kinase C alpha
- Per1
period homolog 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abarca C, Albrecht U, Spanagel R. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc Natl Acad Sci U S A. 2002;99:9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla H, Beninger RJ. Intra-accumbens protein kinase C inhibitor NPC 15437 blocks amphetamine-produced conditioned place preference in rats. Behav Brain Res. 2003;147:41–48. doi: 10.1016/s0166-4328(03)00136-0. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Korczak JF, Weissbecker KA, Goldstein AM. A genome-wide search for loci contributing to smoking and alcoholism. Genet Epidemiol. 1999;17 Suppl 1:S55–S60. doi: 10.1002/gepi.1370170710. [DOI] [PubMed] [Google Scholar]
- Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–532. doi: 10.1016/s0896-6273(00)81056-9. [DOI] [PubMed] [Google Scholar]
- Berke JD, Paletzki RF, Aronson GJ, Hyman SE, Gerfen CR. A complex program of striatal gene expression induced by dopaminergic stimulation. J Neurosci. 1998;18:5301–5310. doi: 10.1523/JNEUROSCI.18-14-05301.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, Edenberg HJ, Cloninger CR, Begleiter H, Conneally PM, et al. A genomic scan for habitual smoking in families of alcoholics: common and specific genetic factors in substance dependence. Am J Med Genet A. 2004;124:19–27. doi: 10.1002/ajmg.a.20329. [DOI] [PubMed] [Google Scholar]
- Bonovich M, Olive M, Reed E, O'Connell B, Vinson C. Adenoviral delivery of A-FOS, an AP-1 dominant negative, selectively inhibits drug resistance in two human cancer cell lines. Cancer Gene Ther. 2002;9:62–70. doi: 10.1038/sj.cgt.7700409. [DOI] [PubMed] [Google Scholar]
- Cabib S. Strain-dependent behavioural sensitization to amphetamine: role of environmental influences. Behav Pharmacol. 1993;4:367–374. [PubMed] [Google Scholar]
- Chen J, Kelz MB, Zeng G, Sakai N, Steffen C, Shockett PE, Picciotto MR, Duman RS, Nestler EJ. Transgenic animals with inducible, targeted gene expression in brain. Mol Pharmacol. 1998;54:495–503. doi: 10.1124/mol.54.3.495. [DOI] [PubMed] [Google Scholar]
- Cohen DR, Ferreira PC, Gentz R, Franza BR, Jr., Curran T. The product of a fos-related gene, fra-1, binds cooperatively to the AP-1 site with Jun: transcription factor AP-1 is comprised of multiple protein complexes. Genes Dev. 1989;3:173–184. doi: 10.1101/gad.3.2.173. [DOI] [PubMed] [Google Scholar]
- Colby CR, Whisler K, Steffen C, Nestler EJ, Self DW. Striatal cell type-specific overexpression of DeltaFosB enhances incentive for cocaine. J Neurosci. 2003;23:2488–2493. doi: 10.1523/JNEUROSCI.23-06-02488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couceyro P, Pollock KM, Drews K, Douglass J. Cocaine differentially regulates activator protein-1 mRNA levels and DNA-binding complexes in the rat striatum and cerebellum. Mol Pharmacol. 1994;46:667–676. [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci U S A. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Almasy L, Blangero J. Smoking behavior is under the influence of a major quantitative trait locus on human chromosome 5q. Genet Epidemiol. 1999;17 Suppl 1:S139–S144. doi: 10.1002/gepi.1370170724. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Page GP, Stein MB, Woods SW. Genome-wide linkage scan for loci predisposing to social phobia: evidence for a chromosome 16 risk locus. Am J Psychiatry. 2004;161:59–66. doi: 10.1176/appi.ajp.161.1.59. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gerdes MJ, Myakishev M, Frost NA, Rishi V, Moitra J, Acharya A, Levy MR, Park SW, Glick A, Yuspa SH, Vinson C. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 2006;66:7578–7588. doi: 10.1158/0008-5472.CAN-06-1247. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Keefe KA, Gauda EB. D1 and D2 dopamine receptor function in the striatum: coactivation of D1- and D2-dopamine receptors on separate populations of neurons results in potentiated immediate early gene response in D1- containing neurons. J Neurosci. 1995;15:8167–8176. doi: 10.1523/JNEUROSCI.15-12-08167.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Young WSd. Distribution of striatonigral and striatopallidal peptidergic neurons in both patch and matrix compartments: an in situ hybridization histochemistry and fluorescent retrograde tracing study. Brain Res. 1988;460:161–167. doi: 10.1016/0006-8993(88)91217-6. [DOI] [PubMed] [Google Scholar]
- Ghasemzadeh MB, Permenter LK, Lake R, Worley PF, Kalivas PW. Homer1 proteins and AMPA receptors modulate cocaine-induced behavioural plasticity. Eur J Neurosci. 2003;18:1645–1651. doi: 10.1046/j.1460-9568.2003.02880.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Nicolini V, McGinty JF. Gene expression profile from the striatum of amphetamine-treated rats: a cDNA array and in situ hybridization histochemical study. Brain Res Gene Expr Patterns. 2002;1:193–198. doi: 10.1016/s1567-133x(02)00017-0. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Moratalla R, Robertson HA. Amphetamine and cocaine induce drug-specific activation of the c-fos gene in striosome-matrix compartments and limbic subdivisions of the striatum. Proc Natl Acad Sci U S A. 1990;87:6912–6916. doi: 10.1073/pnas.87.17.6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall FS, Drgonova J, Goeb M, Uhl GR. Reduced behavioral effects of cocaine in heterozygous brain-derived neurotrophic factor (BDNF) knockout mice. Neuropsychopharmacology. 2003;28:1485–1490. doi: 10.1038/sj.npp.1300192. [DOI] [PubMed] [Google Scholar]
- Hiroi N, Brown JR, Haile CN, Ye H, Greenberg ME, Nestler EJ. FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine's psychomotor and rewarding effects. Proc Natl Acad Sci U S A. 1997;94:10397–10402. doi: 10.1073/pnas.94.19.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DC. The use of place conditioning in studying the neuropharmacology of drug reinforcement. Brain Res Bull. 1989;23:373–387. doi: 10.1016/0361-9230(89)90224-4. [DOI] [PubMed] [Google Scholar]
- Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci U S A. 1992;89:5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, Nestler EJ. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron. 1994;13:1235–1244. doi: 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Hurd YL, Ungerstedt U. Cocaine: an in vivo microdialysis evaluation of its acute action on dopamine transmission in rat striatum. Synapse. 1989;3:48–54. doi: 10.1002/syn.890030107. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Duffy P. Effect of acute and daily cocaine treatment on extracellular dopamine in the nucleus accumbens. Synapse. 1990;5:48–58. doi: 10.1002/syn.890050104. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Stewart J. Dopamine transmission in the initiation and expression of drug- and stress-induced sensitization of motor activity. Brain Res Brain Res Rev. 1991;16:223–244. doi: 10.1016/0165-0173(91)90007-u. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Szumlinski KK, Worley P. Homer2 gene deletion in mice produces a phenotype similar to chronic cocaine treated rats. Neurotox Res. 2004;6:385–387. doi: 10.1007/BF03033313. [DOI] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Jr., Whisler K, Gilden L, Beckmann AM, Steffen C, Zhang YJ, Marotti L, Self DW, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- Konradi C, Cole RL, Heckers S, Hyman SE. Amphetamine regulates gene expression in rat striatum via transcription factor CREB. J Neurosci. 1994;14:5623–5634. doi: 10.1523/JNEUROSCI.14-09-05623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Cheng R, Dupont RT, Williams NJ, Crews KM, Payne TJ, Elston RC. A genome-wide scan to identify loci for smoking rate in the Framingham Heart Study population. BMC Genet. 2003;4 Suppl 1:S103. doi: 10.1186/1471-2156-4-S1-S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Lamande N, Lazar M, Gros F, Legault-Demare L. Developmental expression of alpha- and gamma-enolase subunits and mRNA sequences in the mouse brain. Dev Neurosci. 1988;10:91–98. doi: 10.1159/000111960. [DOI] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- Mir AA, Myakishev MV, Polesskaya OO, Moitra J, Petersen D, Miller L, Orosz A, Vinson C. A search for candidate genes for lipodystrophy, obesity and diabetes via gene expression analysis of A-ZIP/F-1 mice. Genomics. 2003;81:378–390. doi: 10.1016/s0888-7543(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Moratalla R, Elibol B, Vallejo M, Graybiel AM. Network-level changes in expression of inducible Fos-Jun proteins in the striatum during chronic cocaine treatment and withdrawal. Neuron. 1996;17:147–156. doi: 10.1016/s0896-6273(00)80288-3. [DOI] [PubMed] [Google Scholar]
- Moratalla R, Vickers EA, Robertson HA, Cochran BH, Graybiel AM. Coordinate expression of c-fos and jun B is induced in the rat striatum by cocaine. J Neurosci. 1993;13:423–433. doi: 10.1523/JNEUROSCI.13-02-00423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Immediate-early genes: ten years on. Trends Neurosci. 1995;18:66–67. [PubMed] [Google Scholar]
- Mumberg D, Lucibello FC, Schuermann M, Muller R. Alternative splicing of fosB transcripts results in differentially expressed mRNAs encoding functionally antagonistic proteins. Genes Dev. 1991;5:1212–1223. doi: 10.1101/gad.5.7.1212. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins stimulation by Fos. Cell. 1988;907:907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Narita M, Akai H, Nagumo Y, Sunagawa N, Hasebe C, Nagase H, Kita T, Hara C, Suzuki T. Implications of protein kinase C in the nucleus accumbens in the development of sensitization to methamphetamine in rats. Neuroscience. 2004;127:941–948. doi: 10.1016/j.neuroscience.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Bergson CM, Gultart X, Hope BT. Regulation of neural gene expression in opiate and cocaine addiction. NIDA Res Monogr. 1993;125:92–116. [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- Paletzki RF. Cloning and characterization of guanine deaminase from mouse and rat brain. Neuroscience. 2002;109:15–26. doi: 10.1016/s0306-4522(01)00352-9. [DOI] [PubMed] [Google Scholar]
- Peakman MC, Colby C, Perrotti LI, Tekumalla P, Carle T, Ulery P, Chao J, Duman C, Steffen C, Monteggia L, et al. Inducible, brain region-specific expression of a dominant negative mutant of c-Jun in transgenic mice decreases sensitivity to cocaine. Brain Res. 2003;970:73–86. doi: 10.1016/s0006-8993(03)02230-3. [DOI] [PubMed] [Google Scholar]
- Robbins TW, Everitt BJ. Neurobehavioural mechanisms of reward and motivation. Curr Opin Neurobiol. 1996;6:228–236. doi: 10.1016/s0959-4388(96)80077-8. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Becker JB. Enduring changes in brain and behavior produced by chronic amphetamine administration: a review and evaluation of animal models of amphetamine psychosis. Brain Res. 1986;396:157–198. doi: 10.1016/s0006-8993(86)80193-7. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Gorny G, Mitton E, Kolb B. Cocaine self-administration alters the morphology of dendrites and dendritic spines in the nucleus accumbens and neocortex. Synapse. 2001;39:257–266. doi: 10.1002/1098-2396(20010301)39:3<257::AID-SYN1007>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Kolb B. Alterations in the morphology of dendrites and dendritic spines in the nucleus accumbens and prefrontal cortex following repeated treatment with amphetamine or cocaine. Eur J Neurosci. 1999;11:1598–1604. doi: 10.1046/j.1460-9568.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- Rosen JB, Chuang E, Iadarola MJ. Differential induction of Fos protein and a Fos-related antigen following acute and repeated cocaine administration. Brain Res Mol Brain Res. 1994;25:168–172. doi: 10.1016/0169-328x(94)90295-x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Shuster L, Yu G, Bates A. Sensitization to cocaine stimulation in mice. Psychopharmacology (Berl) 1977;52:185–190. doi: 10.1007/BF00439108. [DOI] [PubMed] [Google Scholar]
- Stallings MC, Corley RP, Hewitt JK, Krauter KS, Lessem JM, Mikulich SK, Rhee SH, Smolen A, Young SE, Crowley TJ. A genome-wide search for quantitative trait loci influencing substance dependence vulnerability in adolescence. Drug Alcohol Depend. 2003;70:295–307. doi: 10.1016/s0376-8716(03)00031-0. [DOI] [PubMed] [Google Scholar]
- Steketee JD, Rowe LA, Chandler LJ. The effects of acute and repeated cocaine injections on protein kinase C activity and isoform levels in dopaminergic brain regions. Neuropharmacology. 1998;37:339–347. doi: 10.1016/s0028-3908(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, et al. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999;4:129–144. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]
- Tzschentke TM. Measuring reward with the conditioned place preference paradigm: a comprehensive review of drug effects, recent progress and new issues. Prog Neurobiol. 1998;56:613–672. doi: 10.1016/s0301-0082(98)00060-4. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Walther D, Hess J, Naiman D. Polysubstance abuse-vulnerability genes: genome scans for association, using 1,004 subjects and 1,494 single-nucleotide polymorphisms. Am J Hum Genet. 2001;69:1290–1300. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R, Verma IM. Proto-oncogene FosB: the amino terminus encodes a regulatory function required for transformation. Mol Cell Biol. 1993;13:2635–2643. doi: 10.1128/mcb.13.5.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R, Yen J, Rashid D, Verma IM. Transformation by FosB requires a trans-activation domain missing in FosB2 that can be substituted by heterologous activation domains. Genes Dev. 1992;6:667–675. doi: 10.1101/gad.6.4.667. [DOI] [PubMed] [Google Scholar]
- Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–638. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- Yen J, Wisdom RM, Tratner I, Verma IM. An alternative spliced form of FosB is a negative regulator of transcriptional activation and transformation by Fos proteins. Proc Natl Acad Sci U S A. 1991;88:5077–5081. doi: 10.1073/pnas.88.12.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ST, Porrino LJ, Iadarola MJ. Cocaine induces striatal c-fos-immunoreactive proteins via dopaminergic D1 receptors. Proc Natl Acad Sci U S A. 1991;88:1291–1295. doi: 10.1073/pnas.88.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]