Abstract
Background
'Omics' tools provide novel opportunities for system-wide analysis of complex cellular functions. Secondary metabolism is an example of a complex network of biochemical pathways, which, although well mapped from a biochemical point of view, is not well understood with regards to its physiological roles and genetic and biochemical regulation. Many of the metabolites produced by this network such as higher alcohols and esters are significant aroma impact compounds in fermentation products, and different yeast strains are known to produce highly divergent aroma profiles. Here, we investigated whether we can predict the impact of specific genes of known or unknown function on this metabolic network by combining whole transcriptome and partial exo-metabolome analysis.
Results
For this purpose, the gene expression levels of five different industrial wine yeast strains that produce divergent aroma profiles were established at three different time points of alcoholic fermentation in synthetic wine must. A matrix of gene expression data was generated and integrated with the concentrations of volatile aroma compounds measured at the same time points. This relatively unbiased approach to the study of volatile aroma compounds enabled us to identify candidate genes for aroma profile modification. Five of these genes, namely YMR210W, BAT1, AAD10, AAD14 and ACS1 were selected for overexpression in commercial wine yeast, VIN13. Analysis of the data show a statistically significant correlation between the changes in the exo-metabome of the overexpressing strains and the changes that were predicted based on the unbiased alignment of transcriptomic and exo-metabolomic data.
Conclusion
The data suggest that a comparative transcriptomics and metabolomics approach can be used to identify the metabolic impacts of the expression of individual genes in complex systems, and the amenability of transcriptomic data to direct applications of biotechnological relevance.
Background
Commercial wine yeast strains have been selected to meet specific requirements of wine producers with regard to phenotypical traits such as fermentation performance, general stress resistance, the profile of aromatic compounds produced, the ability to release enzymes or mannoproteins of oenological relevance and many more [1]. As a result, more than 200 different yeast strains, almost exclusively of the species Saccharomyces cerevisiae are currently produced and sold in the global industry. Many research and development programs have focused on improving specific aspects of wine yeast strains [1]. However, many of the relevant traits are of a polygenic nature, and our understanding of the genetic and molecular regulation of complex, commercially relevant phenotypes is limited [2]. In this paper, we investigate the possibility of using a holistic systems biology approach to identify genes that impact on volatile aroma compound production during fermentation. The approach is based on combining comparative transcriptomics and aroma metabolomics of five commercial wine yeast strains that produce significantly different aroma profiles.
During alcoholic fermentation, Saccharomyces cerevisiae strains convert sugars to ethanol, but also produce a large number of volatile aroma compounds, including fatty acids, higher alcohols and esters (table 1). Many of these compounds are important flavor and aroma compounds in wine and beer, and different strains of S. cerevisiae are well known to impart significantly different aroma profiles to the final product.
Table 1.
Exo-metabolites measured in this study
| Primary metabolites | Organic acids | Higher alcohols | Esters | Acids | Fatty Acids |
| Glucose | Citrate | Methanol | Ethyl Acetate | Valeric Acid | Octanoic Acid |
| Fructose | Malate | Propanol | Hexyl acetate | Propionic Acid | Decanoic Acid |
| Glycerol | Acetate | Isoamyl alcohol | Isoamyl Acetate | Iso-Valeric Acid | Ethyl Caprylate |
| Ethanol | 2-Phenyl Ethanol | 2-Phenylethyl Acetate | Iso-Butyric Acid | Ethyl Lactate | |
| Isobutanol | Butyric Acid | Ethyl Caprate | |||
| Butanol | |||||
The metabolic pathways responsible for the production of these compounds are responsive to many factors including the availability of precursors, different types of stress, the cellular redox potential and the energy status of the cell [3-11]. These pathways are not linear, but rather form a network of interlinked reactions converging and diverging from shared intermediates (figure 1). Moreover, intermediates are not only shared between the different 'branches' of aroma compound production, but also with other pathways related to fatty acid metabolism, glycolysis, stress tolerance and detoxification to name a few.
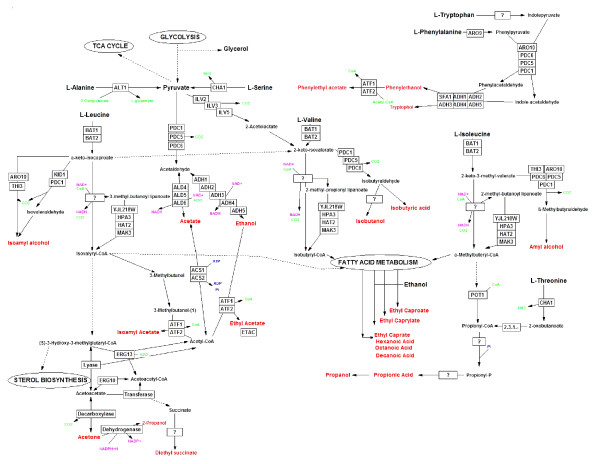
Figure 1.
Diagrammatic representation of pathways associated with aroma production and links to associated metabolic activities. Dashed arrows are used when one or more intermediates or reactions are omitted. Red font is used to identify relevant aroma compounds. Full gene names and functions can be viewed in the appendix. The main pathway for the production of higher alcohols is known as the Erlich Pathway [3]: it involves three basic enzyme activities and starts with the deamination of leucine, valine and isoleucine to the corresponding α-ketoacids. Each α-ketoacid is subsequently decarboxylated and converted to its branched-chain aldehyde [4-6]. The final step is an alcohol dehydrogenase-catalyzed step which could potentially be catalyzed by the seven putative aryl alcohol dehydrogenase genes [7], and the seven alcohol dehydrogenase genes [8]. Finally ester formation involves the enzyme-catalyzed condensation reaction between a higher alcohol and an activated acyl-coenzyme A [9-11]. Fatty acids are derived from fatty acid biosynthesis, but can also be produced as intermediates of the higher alcohol and ester producing pathways [9].
Most of the genes encoding the enzyme activities of the aroma network are also co-regulated by transcription factors that are related to total nitrogen and amino acid availability [12]. Thus the nutritional status of the cell as well as the nutrient composition of the growth media throughout fermentation plays a vital role in determining the aroma profile produced by the fermenting yeast. A further complication is due to the fact that very little is known about the kinetics of individual enzymes involved in these pathways. What is clear is that a number of these enzymes are capable of catalyzing both the forward and reverse reactions, depending on the ratios of substrates to end products, as well as the prevailing redox balance of the cell [13-15]. The various dehydrogenase- catalyzed reactions which are integral to most branches of aroma production are particularly sensitive to the ratios of enzyme co-factors such as NAD and NADH, with obvious ramifications regarding the directionality of various key reactions [16]. This intricate lattice of chemical and biological interactions makes interpretation of individual gene and enzyme contributions problematic in the context of aroma compound production as a whole (figure 1). Indeed, individual parts of the system can combine and interact in unexpected ways, giving rise to emergent properties or functions that would not be anticipated by studying a single part of the system. Such systems are thus irreducible, and cannot be understood by dissection and analysis of a single part at a time. In recognition of the complex and intricate nature of this process we have sought to follow an 'omic' approach in the study of aroma compound production.
In the present study our goal was to compare the aroma-relevant exo-metabolomes of five industrial yeast strains at three different stages of fermentation, and to align these data with gene expression data obtained through microarray-based genome-wide transcription analysis. This enabled the incorporation of gene expression levels and aroma compound production into multivariate statistical models. By using these models as a predictive tool various genes were identified as potential candidates for overexpression in order to increase/decrease the levels of key aroma compounds during fermentation. To verify whether genes whose differential regulation appeared most strongly linked to the differences observed in the aroma profiles of different strains were indeed impacting on aroma compound metabolism, five of these genes were individually overexpressed in one of the industrial strains. The data indicate that these genes indeed impacted significantly on the aroma profiles produced by the modified strains. Moreover, the pattern of changes observed was significantly correlated to the pattern predicted through the comparative analysis of transcriptome and metabolome. The data therefore clearly support our hypothesis that direct comparative analysis of transcriptomes and metabolomes can be used for the identification of genes that affect specific metabolic networks and for predicting the impact of the expression of such genes on these networks.
Results
Fermentation kinetics and metabolite formation
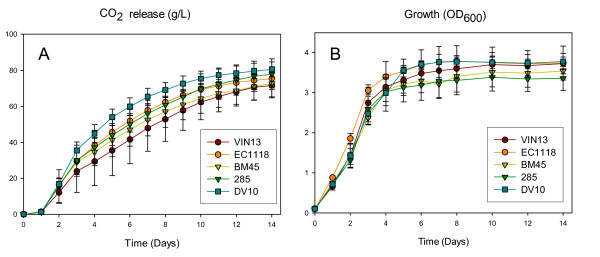
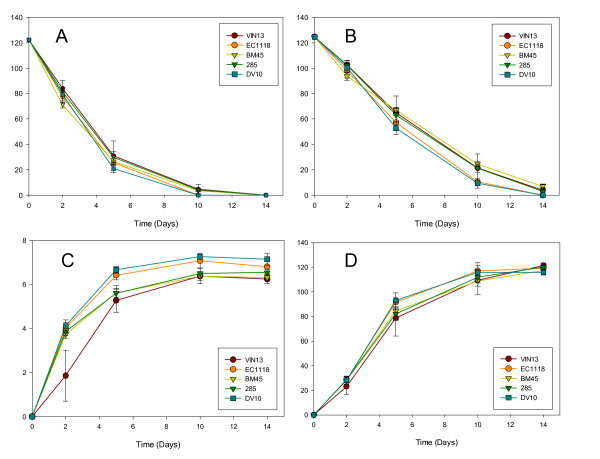
Fermentation behaviour of all five strains in our conditions followed typical wine fermentation patterns. All five strains fermented the synthetic must to dryness within the monitored period, broadly followed similar growth patterns (figure 2) and showed similar rates of fructose and glucose utilization as well as ethanol and glycerol production (figure 3). This is to be expected, as all five strains are widely used in the wine industry and are optimized for fermentation performance.
Figure 2.
Growth rate (frame A) and CO2 release (frame B) of the five commercial wine yeast strains during alcoholic fermentation. Values are the average of 4 biological repeats ± standard deviation.
Figure 3.
Fermentation kinetics of the five yeast strains used in this study: Glucose utilization (A), fructose utilization (B), glycerol production (C) and ethanol production (D). All y-axis values are in g.l-1 and refer to extracellular metabolite concentrations in the synthetic must. Values are the average of 4 biological repeats ± standard deviation.
On the other hand, the strains did show significant variability regarding the volatile organoleptic compounds produced during fermentation (tables 2, 3, 4), suggesting that these 'secondary' pathways of higher alcohol and ester production are less conserved between different strains.
Table 2.
Volatile alcohols and esters present in the fermentation media at day 2 of fermentation
| Day2 | Threshold | Odor | VIN13 | EC1118 | BM45 | 285 | DV10 |
| Ethyl Acetate | 12 | Apple, Pineapple, fruity, solvent, balsamic | 4.87 ± 1.22 | 7.38 ± 0.72 | 7.32 ± 1.07 | 5.43 ± 1.27 | 7.32 ± 1.94 |
| Methanol | 0 ± 0.00 | 0 ± 0.00 | 0 ± 0.00 | 42.59 ± 3.08 | 41.72 ± 4.08 | ||
| Propanol | 306 | Alcohol, ripe fruit, stupefying | 27.16 ± 11.35 | 21.04 ± 4.26 | 20.87 ± 2.08 | 19.24 ± 2.60 | 23.15 ± 6.42 |
| Isobutanol | 74 | Alcohol, nail polish | 5.7 ± 1.25 | 6.77 ± 0.63 | 8.3 ± 0.81 | 7.28 ± 0.40 | 7.04 ± 0.61 |
| Isoamyl Acetate | 60 | Solvent marzipan, malt | bd | bd | bd | bd | 0.05 ± 0.01 |
| Butanol | 50 | Medicinal, wine-like | bd | bd | bd | bd | bd |
| Isoamyl alcohol | 26.54 ± 5.51 | 31.23 ± 2.74 | 30.39 ± 3.12 | 27.31 ± 1.87 | 31.09 ± 4.74 | ||
| Hexyl acetate | 0.67 | Apple, cherry, pear, floral | bd | bd | bd | bd | 0.01 ± 0.01 |
| Ethyl Lactate | 150 | Fruity, buttery | bd | bd | 4.08 ± 0.07 | bd | bd |
| Ethyl Caprylate | 0.58 | sweet pear banana | bd | bd | bd | bd | 0.05 ± 0.03 |
| Acetic Acid | Vinegar | 549 ± 56 | 766 ± 19 | 841 ± 22 | 756 ± 32 | 733 ± 60 | |
| Propionic Acid | 1.59 ± 0.62 | 1.55 ± 0.11 | 1.78 ± 0.32 | 1.7 ± 0.35 | 1.74 ± 0.15 | ||
| Iso-Butyric Acid | 8.1 | Rancid butter cheese | 0.75 ± 0.11 | 0.6 ± 0.03 | 0.62 ± 0.05 | 0.56 ± 0.07 | 0.7 ± 0.04 |
| Butyric Acid | 2.2 | Rancid cheese, sweet | 1.10 ± 0.27 | 1.19 ± 0.06 | 1.03 ± 0.06 | 0.96 ± 0.09 | 1.23 ± 0.08 |
| Ethyl Caprate | 0.5 | Brandy fruity grape floral | 0.02 ± 0.01 | 0.07 ± 0.03 | 0.09 ± 0.00 | 0.09 ± 0.01 | 0.11 ± 0.01 |
| Iso-Valeric Acid | 0.7 | Rancid cheese putrid | 0.36 ± 0.08 | 0.33 ± 0.13 | 0.28 ± 0.04 | 0.27 ± 0.04 | 0.340.05 |
| Diethyl Succinate | 1.2 | Fruity, melon | bd | bd | bd | bd | bd |
| Valeric Acid | SWEATY | 0.22 ± 0.05 | 0.29 ± 0.04 | 0.26 ± 0.08 | 0.29 ± 0.06 | 0.29 ± 0.06 | |
| 2-Phenylethyl Acetate | 1.8 | rosy honey fruity | 0.22 ± 0.05 | 0.51 ± 0.03 | 0.32 ± 0.03 | 0.33 ± 0.03 | 0.57 ± 0.03 |
| 2-Phenyl Ethanol | 200 | Rose, honey | 9.07 ± 2.37 | 12.8 ± 0.41 | 13.38 ± 1.07 | 14.88 ± 1.33 | 12.67 ± 1.36 |
| Octanoic Acid | 10 | Fatty, rancid, oily, soapy faint fruity | 0.81 ± 0.04 | 1.57 ± 0.05 | 1.01 ± 0.05 | 1.11 ± 0.07 | 1.74 ± 0.08 |
| Decanoic Acid | 6 | Fatty, rancid | 0.66 ± 0.07 | 1.46 ± 0.02 | 1.06 ± 0.12 | 1.22 ± 0.13 | 1.7 ± 0.11 |
* Values are expressed as mg.L-1 and are the average of 4 biological repeats ± standard deviation. Metabolites present at concentrations below the detection limit are indicated by "bd".
Table 3.
Volatile alcohols and esters present in the fermentation media at day 5 of fermentation
| Day5 | Threshold | Odor | VIN13 | EC1118 | BM45 | 285 | DV10 |
| Ethyl Acetate | 12 | Apple, Pineapple, fruity, solvent, balsamic | 15.8 ± 4.43 | 16.923 ± 4.22 | 14.37 ± 1.49 | 14.54 ± 1.9 | 23.87 ± 2.21 |
| Methanol | 15.80 ± 4.49 | 36.13 ± 6.26 | 0 ± 0.00 | 29.89 ± 34.7 | 34.42 ± 16.84 | ||
| Propanol | 306 | Alcohol, ripe fruit, stupefying | 77.11 ± 20.23 | 55.58 ± 7.76 | 31.67 ± 6.38 | 35.19 ± 7.65 | 58.1 ± 5.38 |
| Isobutanol | 74 | Alcohol, nail polish | 8.96 ± 2.15 | 11.98 ± 0.63 | 13.57 ± 1.60 | 11.54 ± 0.85 | 11.66 ± 0.60 |
| Isoamyl Acetate | 60 | Solvent marzipan, malt | 0.11 ± 0.04 | 0.09 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.10 ± 0.01 |
| Butanol | 50 | Medicinal, wine-like | 0.95 ± 0.11 | 0.89 ± 0.05 | 0.7 ± 0.06 | 0.77 ± 0.05 | 0.98 ± 0.02 |
| Isoamyl alcohol | 57.02 ± 10.92 | 61.46 ± 6.52 | 51.46 ± 5.98 | 49.32 ± 3.95 | 67.15 ± 8.09 | ||
| Hexyl acetate | 0.67 | Apple, cherry, pear, floral | 0.01 ± 0.02 | 0.16 ± 0.01 | 0.1 ± 0.01 | 0.04 ± 0.01 | 0.16 ± 0.01 |
| Ethyl Lactate | 150 | Fruity, buttery | 4.33 ± 0.14 | 4.24 ± 0.12 | bd | bd | 4.48 ± 0.06 |
| Ethyl Caprylate | 0.58 | sweet pear banana | 0.08 ± 0.02 | 0.11 ± 0.02 | 0.07 ± 0.01 | 0.07 ± 0.00 | 0.09 ± 0.01 |
| Acetic Acid | Vinegar | 1306.24 ± 74.24 | 879.5 ± 22.00 | 1000.58 ± 27.72 | 703.33 ± 43.78 | 1105.93 ± 24.80 | |
| Propionic Acid | 2.18 ± 0.35 | 2.32 ± 0.28 | 2.15 ± 0.29 | 2.01 ± 0.22 | 2.52 ± 0.19 | ||
| Iso-Butyric Acid | 8.1 | rancid butter cheese | 0.58 ± 0.12 | 0.58 ± 0.04 | 0.59 ± 0.04 | 0.54 ± 0.04 | 0.76 ± 0.04 |
| Butyric Acid | 2.2 | rancid cheese, sweet | 1.97 ± 0.41 | 2.50 ± 0.15 | 1.53 ± 0.30 | 1.57 ± 0.31 | 2.77 ± 0.62 |
| Ethyl Caprate | 0.5 | brandy fruity grape floral | 0.25 ± 0.05 | 0.32 ± 0.03 | 0.18 ± 0.03 | 0.2 ± 0.02 | 0.33 ± 0.05 |
| Iso-Valeric Acid | 0.7 | rancid cheese putrid | 0.59 ± 0.11 | 0.62 ± 0.04 | 0.53 ± 0.08 | 0.54 ± 0.16 | 0.66 ± 0.05 |
| Diethyl Succinate | 1.2 | Fruity, melon | bd | bd | bd | bd | bd |
| Valeric Acid | SWEATY | 0.57 ± 0.25 | 0.38 ± 0.07 | 0.23 ± 0.02 | 0.26 ± 0.03 | 0.39 ± 0.03 | |
| 2-Phenylethyl Acetate | 1.8 | rosy honey fruity | 0.33 ± 0.03 | 0.63 ± 0.02 | 0.35 ± 0.02 | 0.32 ± 0.03 | 0.68 ± 0.05 |
| 2-Phenyl Ethanol | 200 | Rose, honey | 12.71 ± 2.47 | 14.98 ± 0.58 | 16.35 ± 1.68 | 17.21 ± 0.83 | 16.33 ± 0.87 |
| Octanoic Acid | 10 | Fatty, rancid, oily, soapy faint fruity | 0.95 ± 0.2 | 1.43 ± 0.05 | 0.9 ± 0.08 | 0.88 ± 0.09 | 1.59 ± 0.09 |
| Decanoic Acid | 6 | Fatty, rancid | 1.5 ± 0.41 | 2.86 ± 0.38 | 1.32 ± 0.15 | 1.42 ± 0.13 | 3.01 ± 0.22 |
* Values are expressed as mg.L-1 and are the average of 4 biological repeats ± standard deviation. Metabolites present at concentrations below the detection limit are indicated by "bd".
Table 4.
Volatile alcohols and esters present in the fermentation media at day 14 of fermentation
| Day14 | Threshold | Odor | VIN13 | EC1118 | BM45 | 285 | DV10 |
| Ethyl Acetate | 12 | Apple, Pineapple, fruity, solvent, balsamic | 29.56 ± 7.56 | 28.79 ± 1.83 | 23.58 ± 2.42 | 23.24 ± 1.87 | 27.83 ± 1.11 |
| Methanol | 68.39 ± 4.8 | 62.3 ± 20.12 | 70.87 ± 7.02 | 63.92 ± 4.49 | 62.18 ± 6.21 | ||
| Propanol | 306 | Alcohol, ripe fruit, stupefying | 83.23 ± 13.34 | 53.5 ± 7.49 | 39.72 ± 4.13 | 41.63 ± 5.337 | 64.14 ± 6.96 |
| Isobutanol | 74 | Alcohol, nail polish | 11.27 ± 2.80 | 15.44 ± 1.30 | 19.58 ± 2.03 | 15.18 ± 1.15 | 15.33 ± 0.76 |
| Isoamyl Acetate | 60 | Solvent marzipan, malt | 0.10 ± 0.03 | 0.09 ± 0.01 | 0.07 ± 0.01 | 0.06 ± 0.00 | 0.09 ± 0.01 |
| Butanol | 50 | Medicinal, wine-like | 1.36 ± 0.14 | 1.26 ± 0.08 | 1.02 ± 0.18 | 1.03 ± 0.05 | 1.32 ± 0.04 |
| Isoamyl alcohol | 67.29 ± 4.89 | 71.44 ± 2.54 | 68.17 ± 8.81 | 61.59 ± 4.25 | 76.86 ± 6.50 | ||
| Hexyl acetate | 0.67 | Apple, cherry, pear, floral | 0.05 ± 0.04 | 0.14 ± 0.01 | 0.1 ± 0.00 | 0.1 ± 0.00 | 0.14 ± 0.00 |
| Ethyl Lactate | 150 | Fruity, buttery | 3.86 ± 0.19 | 3.96 ± 0.04 | bd | bd | 3.92 ± 0.09 |
| Ethyl Caprylate | 0.58 | sweet pear banana | 0.07 ± 0.00 | 0.11 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.13 ± 0.01 |
| Acetic Acid | Vinegar | 1616 ± 158 | 1061 ± 120 | 1197 ± 133 | 838 ± 114 | 1251 ± 71 | |
| Propionic Acid | 1.96 ± 0.39 | 2.84 ± 0.22 | 2.18 ± 0.47 | 2.19 ± 0.24 | 3.4 ± 0.45 | ||
| Iso-Butyric Acid | 8.1 | rancid butter cheese | 0.74 ± 0.16 | 0.81 ± 0.079 | 0.7 ± 0.09 | 0.63 ± ± | 1.02 ± 0.12 |
| Butyric Acid | 2.2 | rancid cheese, sweet | 2.27 ± 0.66 | 4 ± 0.23 | 2.37 ± 0.62 | 2.35 ± 0.42 | 3.96 ± 0.29 |
| Ethyl Caprate | 0.5 | brandy fruity grape floral | 0.24 ± 0.04 | 0.45 ± 0.01 | 0.21 ± 0.04 | 0.26 ± 0.04 | 0.45 ± 0.03 |
| Iso-Valeric Acid | 0.7 | rancid cheese putrid | 0.74 ± 0.11 | 0.95 ± 0.11 | 0.71 ± 0.15 | 0.67 ± 0.05 | 1.01 ± 0.11 |
| Diethyl Succinate | 1.2 | Fruity, melon | 0.09 ± 0.02 | 0.17 ± 0.02 | 0.05 ± 0.03 | 0.04 ± 0.01 | 0.16 ± 0.03 |
| Valeric Acid | SWEATY | 0.26 ± 0.03 | 0.49 ± 0.11 | 0.26 ± 0.03 | 0.26 ± 0.04 | 0.41 ± 0.04 | |
| 2-Phenylethyl Acetate | 1.8 | rosy honey fruity | 0.35 ± 0.06 | 0.55 ± 0.04 | 0.26 ± 0.04 | 0.27 ± 0.05 | 0.61 ± 0.05 |
| 2-Phenyl Ethanol | 200 | Rose, honey | 11.88 ± 2.91 | 17.24 ± 0.69 | 16.68 ± 2.54 | 17.33 ± 2.28 | 17.98 ± 1.42 |
| Octanoic Acid | 10 | Fatty, rancid, oily, soapy faint fruity | 0.65 ± 0.08 | 1.07 ± 0.12 | 0.51 ± 0.14 | 0.65 ± 0.18 | 1.28 ± 0.09 |
| Decanoic Acid | 6 | Fatty, rancid | 0.84 ± 0.29 | 1.47 ± 0.07 | 0.64 ± 0.11 | 0.89 ± 0.19 | 1.52 ± 0.19 |
* Values are expressed as mg.L-1 and are the average of 4 biological repeats ± standard deviation. Metabolites present at concentrations below the detection limit are indicated by "bd".
In general, the aroma compounds produced all showed a steady increase in concentration in the synthetic must over time, although the most active period of aroma compound accumulation appears to be in the earlier stages of fermentation. For the most part, compounds such as methanol, isoamyl alcohol, butanol, ethyl caprylate are only detectable in the fermentation media by day 5 of fermentation (table 3), whereas others such as diethyl succinate can only be detected at the end of fermentation (table 4). In general, the higher alcohols and their corresponding esters are present throughout fermentation at the highest concentration in the medium (tables 2, 3, 4). The aroma profiles of the DV10 and EC1118 strain are very similar, while the BM45 and 285 strains also produce similar exometabolomic signatures. The aroma compounds that are proportionally the most variable between strains are propanol, isobutanol, ethyl caprylate, acetic acid, propionic acid, butyric acid, ethyl caprate, diethyl succinate, valeric acid, 2-phenylethyl acetate, octanoic- and decanoic acid, as well as ethyl lactate, which is completely absent in the BM45 and 285 strains (table 4).
Microarray analysis
The divergent aroma profiles of the different strains were mirrored by variable gene expression patterns. Since the Affymetrix DNA chips used for the analysis were designed based on the sequence of the laboratory yeast BY4742, a primary concern related to the quality of the microarray data. Both the internal controls and the expression of housekeeping genes were in keeping with international MIAME compliancy standards. Most notably, variation between independent biological repeats was negligible, giving us confidence in the reliability and reproducibility of our microarray analysis. Furthermore, changes in gene expression during the course of fermentation matched up well to data from related microarray analysis for the EC1118 [17] and VIN13 strains [18].
Between different time points approximately 1000–1500 genes significantly increased or decreased in expression (within the criteria specified in the materials and methods section) for the five yeast strains in our study. At the time points considered, the variation in gene expression between the different strains was in the range of about 50–400 transcripts. Strains that appear to be most similar to one another on a gene expression level were the EC1118 and DV10 strains, as well as the BM45 and 285 strains. The VIN13 strain was least similar to any of the other four strains. This pattern is in line with the differences observed in aroma production for all of these strains.
Numerous and substantial changes in the expression of genes involved in pathways that lead to the production of volatile aroma compounds were evident both between strains at comparable stages of fermentation and for individual strains at different fermentative stages. To identify relevant transcriptional variation in the context of aroma compound production, PCA analysis and PLS1 and PLS2 models were constructed for the compounds in tables 2, 3, 4 using the transcriptomic data as X variables. Transcriptomic data from days 2 and 5 were used for modeling purposes as these time points represent the period when the accumulation rate of most aroma compounds is at a maximum. From these models, transcripts with a strong positive or negative loading were selected for further in depth statistical analysis. The corresponding ORFs, together with a brief annotation, are listed in the additional data files [see Additional data file 1].
The general intrastrain trend revealed a decrease in the transcript levels of enzymes involved in the synthesis of aromatic and branched-chain amino acids, while transcript levels encoding aldehyde and alcohol dehydrogenases, as well as certain acetyltransferases were generally increased. Fold changes for differentially expressed transcripts, both between different strains at either day 2 or day 5 of fermentation and between day 2 and day 5 in individual strains, can be viewed as additional material [see Additional data file 2].
Multivariate analysis of metabolite concentrations and gene expression data
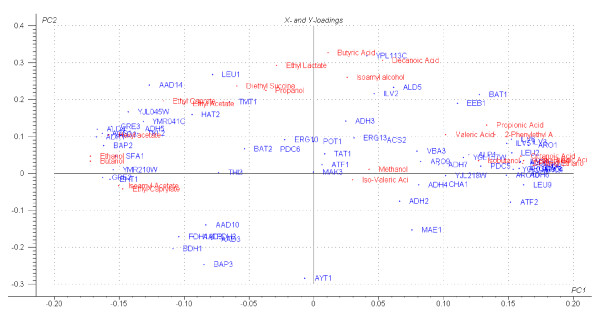
Figure 4 shows a PLS2 plot which depicts the variation/relationships between all the measured aroma compounds as well as the 70 genes selected for multivariate modeling purposes. These genes were selected due to their varying expression levels between different strains as well as different time points during fermentation. Also, we selected genes whose annotation suggested that they may have a role in aroma compound production, such as enzymes whose sequence suggests a role in redox reactions, central carbon metabolism, and amino acid uptake and metabolism (GO and MIPS classification).
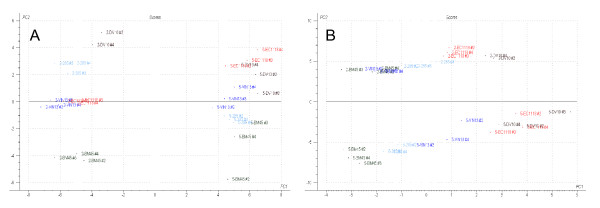
Figure 4.
PLS2 scores and loadings plot of all X and Y variables considered in this study, plotted as coordinates on a PC1 and PC2 plane.
The X-Y scores and loading plots (figure 4) are clearly useful in representing the overall 'structure' of the entire dataset, and are pointing out possible connections between specific compounds/groups of compounds and certain genes. Likewise, scores plots proved a neat way of validating the general design and data generated by our experimental setup/process (figure 5). The samples of independent biological repeats for each of the 5 strains group together closely at both time points. All five strains also clearly segregate into two clusters based on the stage (time point) of fermentation. For example, in the first frame it is clear that the stage of fermentation is the major source of variation (PC1) and strain identity is the source of the second-greatest explained variation (PC2), while this pattern is reversed in frame B.
Figure 5.
Scores plot for the ethyl caprylate (frame A) and octanoic acid (frame B) PLS1 models.
Of the 22 volatile aroma compound measured in this study, 13 were amenable to PLS1 modeling (using transcriptome data) based on our selected criteria for model validation (slope > 0.8; Y-var explained > 75%). The details of these models are summarized in a table that can be viewed as additional material [see Additional data file 3].
Overexpression of selected genes
Of the genes listed in the tables presented in the supplementary material, five were chosen for in-depth analysis due to their significant contributions to the respective prediction models for several of the important higher alcohols and esters, as well as their amenability to easy cloning and vector construction. These genes were BAT1, AAD10, AAD14, ACS1 and YMR210W. AAD10 and AAD14 encode aryl alcohol dehydrogenases which are believed to be responsible for the putative role of degrading the complex aromatic compounds in grape must into their corresponding higher alcohols [7]. BAT1 encodes a mitochondrial branched-chain amino acid aminotransferase that is involved in catalyzing the first transamination step of the catabolic formation of fusel alcohols via the Ehrlich pathway [19]. The YMR210 gene codes for a putative acyltransferase enzyme (similar to EEB1 and EHT1) and is believed to play a role in medium-chain fatty acid ethyl ester biosynthesis. Lastly, the ACS1 gene (encoding an acetyl-coA synthetase isoform) codes for the enzyme responsible for the conversion of acetate to acetyl-coA, which is an intermediate or reactant in several of the aroma compound producing pathways [20].
An in-house BAT1 overexpressing strain was already available for use [21]. For the other 4 genes, a multi-copy overexpression plasmid-based cloning strategy was employed to allow for maximum gene expression and rapid characterization of the transformed VIN13 strains.
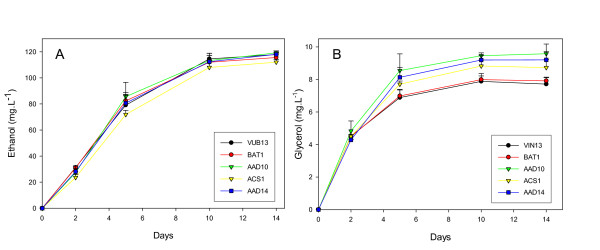
Fermentations were carried out as before with the 5 transformed cell lines and a VIN13 control. Samples for HPLC and GC-FID analysis were taken at the same time points, namely days 2, 5 and 14 of fermentation. No significant differences were observed regarding the glucose and fructose utilization of the overexpression strains during fermentation (Data not shown). Slight differences were found for ethanol production, while some changes in glycerol production were evident for the different strains (Figure 6).
Figure 6.
Concentrations of ethanol (frame A) and glycerol (frame B) in the must during fermentation. Values are the average of 4 independant repeats + standard deviation.
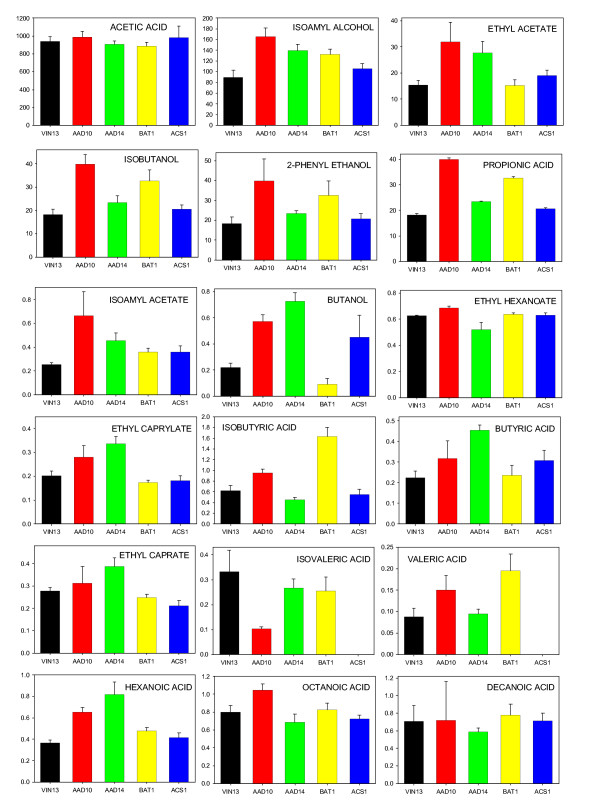
Figure 7 depicts the aroma compound concentrations at the end of fermentation (day 14) only, as this is the most important time point from an enological perspective. Aroma profiles for days 2 and 5 can be viewed as additional data [see Additional data file 4].
Figure 7.
Aroma compound production (μg.L-1) in MS300 fermentations carried out by VIN13 transformed with overexpression constructs. Values are the average of 4 biological repeats + standard deviation.
Four of the five overexpressing strains showed significant changes in the aroma profiles produced at the end of fermentation. Only the YMR210W overexpressing strain did not show any changes, and is therefore not included in the figures below. We did not further investigate whether this absence of changes in aroma production is due to problems with the expression construct or reflects the absence of aroma–related activity of the gene product.
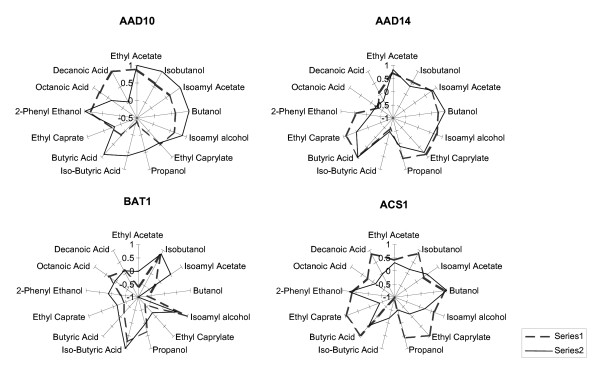
Significant differences were evident in the aroma profiles of the four transformed yeast strains under consideration. We investigated whether the observed changes in aroma compound concentrations at the end of fermentation can be reconciled with the anticipated changes based on multivariate prediction models. Figure 8 represents the qualitative alignment of real vs. predicted changes in aroma compound concentrations. Only aroma compounds with statistically reliable PLS models (test-set validation; slope >0.88; % RMSEP < 20) were taken into consideration. The dashed lines indicate the relative loading weights of each of the four genes (for each of the aroma compound models represented by the plot axes). The solid lines in the figures represent the log ratios of the actual aroma compound concentrations normalized to the VIN13 concentrations of the particular compound.
Figure 8.
Qualitative representation of relative real vs. predicted aroma compound levels in the four transformed VIN13 lines. Dashed gray lines indicate predicted values and solid black lines indicate log-normalised values of real compound concentrations.
To clarify, the predicted influence of a given gene on a particular compound is represented on a scale from -1 to +1, based on statistical projections related to PLS loading weights. On this scale a value of -1 suggests a strong probability of significant concentration decreases of a given compound (for overexpression of the gene), while a value of +1 is indicative of a strong positive correlation between the expression levels of the gene of interest and the compound in question. A value of zero indicates no expected influence of gene expression on the relevant aroma compound.
Likewise, log-normalization was carried out on the actual metabolite concentrations measured in the overexpression strains to represent these values on a scale from -1 to 1, relative to the corresponding concentrations of the control fermentations. Figure 8 clearly shows that predicted and real changes overlapped significantly.
Discussion
The aim of this study was to determine whether the transcription profiles of the various strains during fermentation could be reconciled with the volatile aroma compound production of these strains, and whether this comparative analysis could be used to predict the impact of individual gene expression levels on aroma compounds and profiles.
The data generated by the overexpression of four of the genes whose expression was statistically most significantly linked to the production of aroma profiles suggest that this approach has been successful. Indeed, overexpression of the selected genes had a far reaching impact on the aroma profiles produced by the fermenting yeast, and this impact was generally well aligned with the impact predicted from the comparative omics analysis. Indeed, the data aligned better than we, considering the significant challenges when approaching complex systems, had expected. Our data show that the metabolic changes observed upon overexpression of three of the four genes, AAD10, AAD14 and BAT1, were very significantly aligned with the changes that were predicted from the alignment of transcriptome and metabolome data alone. The predictions, as can be seen from the alignment of predicted vs. observed changes in metabolite levels in a qualitative manner, indeed proved fairly reliable. The model was able to assign positive and negative influences on a particular compound with relative accuracy. Although the extent/magnitude of the increase/decrease is not always well aligned with model values, the absolute direction of the change holds true in most cases. An absolute alignment would not be expected, since the level of expression in a plasmid-based system can not be adjusted to the differences of expression observed between the different strains. In the case of AAD10, only the influence of the overexpression on decanoic acid was not in line with the projection. Predictions for AAD14 and BAT1 were well matched with the observed changes in metabolite profiles. Predicted and real changes did not match satisfactorily in only one case, ACS1. Nevertheless, even in this case, eight out of the thirteen compounds evolved in the predicted direction. It should also be noted that the expression of this gene had generally a less severe impact on changes in the aroma profile than those of the other three genes.
Considering the complexity of the system, the rate of success achieved in this study can be considered as highly significant. To our knowledge, this is the first report to exploit such an intra- and interstrain comparative approach to identify genes that play a significant role in a complex metabolic network.
While we were clearly able to identify genes with significant impact on aroma compound production in a specific industrial environment, and which in some cases had not been previously directly linked to these pathways, the data do not allow a firm conclusion on the exact metabolic role of these genes. Indeed, the vast number of significant changes to metabolite levels makes it difficult to identify the specific 'point of influence' of any overexpressed gene in a given pathway.
The increases/decreases in specific volatile compounds seen for the VIN13(pBAT1-s) strain is in keeping with the results reported in colombar fermentations [21]. The two AAD gene overexpressing strains also showed interesting trends: Both strains produced higher levels (at comparable concentrations) of isoamyl alcohol, ethyl acetate, butanol, ethyl caprylate, ethyl caprate and hexanoic acid. However, noticeable differences can be seen in the levels of isobutanol, 2-phenyl ethanol, propionic acid, isoamyl acetate, ethyl hexanoate, isobutyric acid and isovaleric acid, relative to the control and to one another. This is indicative of the potential for the AAD genes to have overlapping yet distinct functional roles in the pathways leading to higher alcohol and ester production.
Overexpression of the ACS1 gene did not lead to such numerous and substantial increases/decreases in volatile production as was the case for the other three genes. Interestingly, valeric and isovaleric acid were below detection levels in these fermentations. Concentrations of isoamyl acetate, ethyl acetate, butanol and butyric acid were significantly higher, and ethyl caprate lower relative to control fermentations.
On the whole though, our analysis shows that the cross-comparison of gene expression data with metabolite levels has the potential to identify points of interest on a genomic scale. This also opens new possibilities to design improved yeast enhancement strategies for optimized aroma production and fermentation performance.
Other genes of interest
Many other genes showed significant variation in expression between different strains and/or time points, as well as high loadings on PLS models and strong negative or positive correlations with specific aroma compounds. These genes encode enzymes that either are known to participate in aroma compound production, or have activities (either experimentally proven or suggested through sequence alignments) that could suggest such roles. Here we discuss some of the most relevant of these enzymes, which fall into several categories, either according to their place in a specific metabolic pathway such as the metabolisms of branched chain amino acids or of aromatic amino acids, or based on their specific activity such as dehydrogenases (in particular aldehyde and alcohol dehydrogenases) and acetyl transferases.
Of the enzymes involved in branched chain amino acid metabolism, BAT1 has been discussed above. Other genes that encode enzymes in this pathway and that were identified in our study for their strong statistical link between expression levels and the production of specific compounds include LEU2, encoding a beta-isopropylmalate dehydrogenase that catalyzes the third step in the leucine biosynthesis pathway, and, to a lesser degree, LEU1, which encodes an isopropylmalate isomerase [22,23]. Both of these genes showed a significant statistical correlation with compounds such as isobutanol. Of the genes involved in the metabolism of isoleucine and valine (Ilv), only ILV5, which encodes an acetohydroxyacid reductoisomerase involved in branched-chain amino acid biosynthesis [24], showed a very strong positive correlation with almost all of the compounds analysed here, and, interestingly, a negative correlation with ethanol, suggesting that this gene could be an interesting target for metabolic engineering.
While BAT1 expression showed a significant positive correlation with a large number of the volatile compounds measured in our study, the cytosolic isoform (BAT2) of this enzyme showed no significant correlations with any of these aroma compounds. Although this isoform is supposedly highly expressed during stationary phase and repressed during the logarithmic phase, BAT2 expression levels in our study were found to stay constant, if not to decrease slightly upon entry into stationary phase in comparison to the exponential phase at day 2. In addition, BAT2 expression levels were generally considerably lower throughout fermentation when compared to BAT1.
Of the genes involved in aromatic amino acid metabolism, three, ARO1, which encodes a pentafunctional arom protein, ARO7, which encodes a chorismate mutase responsible for the conversion of chorismate to prephenate and ARO8, which codes for an aromatic aminotransferase showed statistically significant correlations between expression levels and metabolite production [25,26]. All three genes showed a modest positive correlation (r2 = 0.7) with 2-phenyl ethanol and mild negative correlations with all the other compounds. Only octanoic acid showed a very strong (r2 = 0.82) negative correlation with ARO8 expression at day 2 of fermentation. Despite its seemingly crucial role, ARO10, which encodes a phenylpyruvate decarboxylase corresponding to the first specific step in the Ehrlich pathway did not show any noteworthy correlations between its expression and any of the volatile compounds in our study [27]. Of course the possibility of translational or post-translational control of activity cannot be excluded.
Several specific enzyme activities were also overrepresented in our list. Such enzymes include many dehydrogensases. Aldehyde and alcohol dehydrogenases such as those encoded by ALD5, ALD6, ADH6 and ADH7 showed a substantial decline in expression levels between days 2 and 5 of fermentation, while others (such as ALD3, ALD4, ADH2 and ADH5) increased during this time. The distinct expression patterns during fermentation reflects the different regulatory mechanisms governing the expression of these genes (i.e. expression of ALD3 is glucose-repressed and stress-induced) and suggests that the different ALD gene products have specific roles during different stages of fermentation [28].
ALD4 and ALD5 (mitochondrial), and ALD3 and ALD6 (cytoplasmic) encode aldehyde dehydrogenases involved in the conversion of acetaldehyde to acetate [29].
ALD4 encodes a mitochondrial aldehyde dehydrogenase (utilizing NADP+ or NAD+) that is required for growth on ethanol and conversion of acetaldehyde to acetate [29]. Expression of ALD4 is also glucose repressed, and increases 2–4 -fold from day 2 to 5 of fermentation. ALD4 expression shows a very strong correlation to the amount of hexyl acetate (R2 = 0.82) produced by the fermenting yeast, as well as to ethyl acetate (0.77), isoamyl alcohol (0.91) and isoamyl acetate (0.85).
ALD6 encodes a constitutively expressed cytosolic aldehyde dehydrogenase (utilizes NADP+ as the preferred coenzyme) and is required for conversion of acetaldehyde to acetate [30]. Not surprisingly, ALD6 expression showed a very strong positive correlation to the levels of acetic acid produced by the fermenting cells (0.92). Also, expression was very strongly inversely correlated to ethanol production (R2 = 0.81). Interestingly, fairly strong positive correlations were also evident for 2-phenyl ethanol (R2 = 0.79) and 2-phenyl ethyl acetate (R2 = 0.67).
ADH6 encodes an NADPH-dependent cinnamyl alcohol dehydrogenase family member with broad substrate specificity [31]. Expression was correlated very strongly with isobutanol levels (0.81), isobutyric acid (0.86), propionic acid (0.81), acetic acid (0.87) and 2-phenyl ethanol (0.92). ADH4, ADH5 and ADH7 on the other hand showed only modest correlations with the above-mentioned, or any other aroma compounds for that matter.
With respect to the aryl alcohol dehydrogenase family of genes, the transcripts for AAD3, AAD10 and AAD14 showed the greatest variation in expression, both on an intra- and interstrain level. Expression of AAD10 and AAD14, for example, was increased more than twofold in most of the strains at day 5 relative to day 2 of fermentation. No distinct physiological role has been established for the products of these genes [7], but it is reasonable to suspect that the consistent increase in their respective transcript levels during the course of fermentation could be associated with the increase in one or several of the long chain alcohols or their acid counterparts as fermentation progresses (tables 2, 3).
This hypothesis is supported by the data generated through the overexpression of these genes. Indeed, overexpression yielded changes to the aroma profile that were very similar to those predicted from the alignment of transcriptome and metabolome data sets. The expression of AAD10 showed weak yet significant positive correlations with a number of the aroma compounds. Expression of AAD14 between different strains and time points was also highly variable. Highest expression levels were noted for the DV10 strain, and significant positive correlations with ethyl acetate (0.67) and ethyl caprate (0.74) were observed for this gene.
Acetyl transferases are another family of enzymes of relevance to aroma compound metabolism [32]. However, neither ATF1 nor ATF2, the two most prominent alcohol acetyl transferases, showed statistically strong correlations between expression levels and metabolite production. EEB1, on the other hand, which encodes an acyl-coenzymeA:ethanol O-acyltransferase and is responsible for the major part of medium-chain fatty acid ethyl ester biosynthesis during fermentation [33], showed weak negative correlations with ethanol and other higher alcohols, and a strong positive correlation for 2-phenylethyl acetate (0.9) as well as octanoic acid (0.78). It is tempting to speculate that Eeb1p may thus be largely responsible for the acetylation of 2-phenyl ethanol to produce 2-phenylethyl acetate.
EHT1 encodes an acyl-coenzymeA:ethanol O-acyltransferase that plays a role in medium-chain fatty acid ethyl ester biosynthesis, but also contains a known esterase activity [33]. EHT1 expression increased somewhat as fermentation progressed and inter-strain expression at both day 2 and 5 of fermentation varied significantly. Interestingly, EHT1 expression showed a fairly strong inverse correlated with 2-phenylethyl acetate (R2 = 0.74) and octanoic acid (R2 = 0.75), as well as a weaker yet significant inverse correlation with decanoic acid (R2 = 0.59). This could indicate that the esterase activity of Eht1p could predominate under certain conditions.
YMR210W encodes a putative acyltransferase with similarity to both Eeb1p and Eht1p, and may have a minor role in medium-chain fatty acid ethyl ester biosynthesis [33]. Expression was positively correlated with ethyl acetate (0.74), ethyl caprylate (0.85) and isoamyl acetate (0.78).
In addition to these relatively well studied acetyltransferases, the mRNA levels of the AYT1 gene, encoding a transferase of unknown substrate specificity, also showed considerable variation at different fermentative stages [34].
Conclusion
The impact of these individual genes on aroma compound metabolism has to be assessed individually. However, from the data presented here, it is clear that an analysis based on the comparison of transcriptome and metabolome data derived from different commercial yeast strains can help to identify genes that most significantly impact a metabolic network in specific environmental and industrial conditions. Our over-expression analysis of five genes that were randomly selected from the list of ORFs identified for their statistically significant impact on aroma production also clearly suggests that the method has significant predictive power regarding the reorientation of metabolic flux through the network in response to changes in gene expression levels. Indeed, for four out of five selected genes, BAT1, AAD10, AAD14 and ACS1, the match between predicted and real changes is highly significant. This is the first study linking metabolic networks to transcriptome analysis through the comparative analysis of different wine yeast strains.
Methods
Strains. media and culture conditions
The yeast strains used in this study are listed in table 5. All are diploid Saccharomyces cerevisiae strains used in industrial wine fermentations. Yeast cells were cultivated at 30°C in YPD synthetic media 1% yeast extract (Biolab, South Africa), 2% peptone (Fluka, Germany), 2% glucose (Sigma, Germany). Solid medium was supplemented with 2% agar (Biolab, South Africa).
Table 5.
Strains used in this study
| Strain | Source/Reference |
| VIN13 | Anchor Yeast, South Africa |
| EC1118 | Lallemand Inc., Montréal, Canada |
| BM45 | Lallemand Inc., Montréal, Canada |
| 285 | Lallemand Inc., Montréal, Canada |
| DV10 | Lallemand Inc., Montréal, Canada |
| VIN13(pBAT1-s) | Lilly et al., 2006 |
Fermentation media
Fermentation experiments were carried out with synthetic must MS300 which approximates to a natural grape must as previously described [35]. The medium contained 125 g/L glucose and 125 g/L fructose, and the pH was buffered at 3.3 with NaOH.
Fermentation conditions
All fermentations were carried out under microaerophilic conditions in 100 ml glass bottles (containing 80 ml of the medium) sealed with rubber stoppers with a CO2 outlet. The fermentation temperature was approximately 22°C and no continuous stirring was performed during the course of the fermentation. Fermentation bottles were inoculated with YPD cultures in the logarithmic growth phase (around OD600 = 1) to an OD600 of 0.1 (i.e. a final cell density of approximately 106 cfu.ml-1). The cells from the YPD pre-cultures were briefly centrifuged and resuspended in MS300 to avoid carryover of YPD to the fermentation media. The fermentations followed a time course of 14 days and the bottles were weighed daily to assess CO2 release and the progress of fermentation. Samples of the fermentation media and cells were taken at days 2, 5 and 14 as representative of the exponential, early stationary and late stationary growth phases respectively. It should be stressed that early stationary phase in these conditions is metabolically active, since growth arrest is due to ethanol toxicity. Sugar levels and fermentative activity are still high at this stage.
Growth measurement
Cell proliferation (i.e. growth) was determined spectrophotometrically (PowerwaveX, Bio-Tek Instruments) by measuring the optical density (at 600 nm) of 200 μl samples of the suspensions over the 14 day experimental period.
Analytical methods – HPLC
Culture supernatants were obtained from the cell-free upper layers of the fermentation media. For the purposes of glucose determination and carbon recovery, culture supernatants and starting media were analyzed by high performance liquid chromatography (HPLC) on an AMINEX HPX-87H ion exchange column using 5 mM H2SO4 as the mobile phase. Agilent RID and UV detectors were used in tandem for peak detection and quantification. Analysis was carried out using the HPChemstation software package.
Analytical methods – GC-FID
Each 5 ml sample of synthetic must taken during fermentation was spiked with an internal standard of 4-methyl-2-pentanol to a final concentration of 10 mg.l-1. To each of these samples 1 ml of solvent (diethyl ether) was added and the tubes sonicated for 5 minutes. The top layer in each tube was separated by centrifugation at 3000 rpm for 5 minutes and the extract analyzed. After mixing, 3 μl of each sample was injected into the gas chromatograph (GC). All extractions were done in triplicate.
The analysis of volatile compounds was carried out on a Hewlett Packard 5890 Series II GC coupled to an HP 7673 auto-sampler and injector and an HP 3396A integrator. The column used was a Lab Alliance organic-coated, fused silica capillary with dimensions of 60 m × 0.32 mm internal diameter with a 0.5 μm coating thickness. The injector temperature was set to 200°C, the split ratio to 20:1 and the flow rate to 15 ml.min-1, with hydrogen used as the carrier gas for a flame ionisation detector held at 250°C. The oven temperature was increased from 35°C to 230°C at a ramp of 3°C min-1.
Internal standards (Merck, Cape Town) were used to calibrate the machine for each of the compounds measured.
Statistical analysis of metabolite data
T-tests and anova analyses were conducted using Statistica (version 7). HCL and KMC clustering were carried out using TIGR MeV v2.2 [36].
Microarray analysis
Sampling of cells from fermentations and total RNA extraction was performed as described [37]. Probe preparation and hybridization to Affymetrix Genechip® microarrays were performed according to Affymetrix instructions, starting with 6 μg of total RNA. Results for each strain and time point were derived from three independent culture replicates. The quality of total RNA, cDNA, cRNA and fragmented cRNA were confirmed using the Agilent Bioanalyzer 2100.
Transcriptomics data acquisition and statistical analysis
Acquisition and quantification of array images and data filtering were performed using Affymetrix GeneChip® Operating Software (GCOS) version 1.4. All arrays were scaled to a target value of 500 using the average signal from all gene features using GCOS. Genes with expression values below 12 were set to 12 + the expression value as previously described in order to eliminate insignificant variations [38].
Variable (gene) selection is important for the successful analysis of gene expression data since most of the genes are unchanged and irrelevant to the prediction and analysis of phenotypic measurements. These non-informative genes should be removed before further analysis. One approach is by significance analysis of microarrays [39]. Determination of differential gene expression between experimental parameters was conducted using SAM (Significance Analysis of Microarrays) version 2. The two-class, unpaired setting was used and genes with a Q value less than 0.5 were considered differentially expressed. Only genes with a fold change greater than 2 (positive or negative) for inter- or intra- strain comparisons were taken into consideration.
Multivariate data analysis
In terms of design, the samples represent the different fermentations (three independent replicates for each of the five strains) at different time points. The variables considered are the expression levels of the pre-selected genes (genes with a potential and established role in aroma compound metabolism according to GO and MIPS functional classification) as well as aroma compound concentrations in the synthetic must. The patterns within the different sets of data were investigated by principal-component analysis (PCA), while the correlations between different sets of data were determined by using partial least-squares (PLS) regression (The Unscrambler; Camo Inc., Corvallis, Oreg.). PCA is a bilinear modeling method which gives a visually interpretable overview of the main information in large, multidimensional datasets. By plotting the principal components it is possible to view statistical relationships between different variables in complex datasets and detect and interpret sample groupings, similarities or differences, as well as the relationships between the different variables [40].
PLS regression is a bilinear modeling method for identifying the variations in a data matrix for explanatory or predictive purposes [41]. By plotting the first PLS components one can view main associations between X variables and Y variables and also relationships within X data and within Y data. PLS2 analysis was conducted using all X and Y variables considered in our study. For predictive purposes, PLS1 models were constructed for individual Y variables to increase model-specificity and reliability.
The data were analyzed by using test-set validation with centered data and the variables were weighted according to their standard deviations. One strain was used as the test segment at each of the time points. Day 2 and 5 data were considered together as representative of the full scope of fermentation variability as the period from the start of fermentation until day 5 represents the period of maximum aroma compound production.
The Y variables were the respective aroma compounds measured and the X variables were the gene expression levels of the gene set that was pre-selected for analysis [42]. Genes were selected based on known or putative functions related to amino acid transport, metabolism, regulation etc, as well as other enzymatic or regulatory activity in pathways leading to the production of higher alcohols and esters. The same set of genes (X variables) was used for each of the different PLS1 models.
Overexpression constructs and transformation of VIN13
All plasmids used in this study are listed in table 6. Standard procedures for the isolation, cloning and modification of DNA were used throughout this study [43,44]. All enzymes for cloning, restriction digest and ligation reactions were obtained from Roche Diagnostics (Randburg, South Africa) and used according to supplier specifications.
Table 6.
Description of plasmids used in this study
| Plasmid Name | Relevant genotype | Reference |
| pDM-PhR-YMR210W | 2 μ LEU2 TEF1P PhR322 TEF1T PGKP YMR210W PGKT | This study |
| pDM-PhR-AAD10 | 2 μ LEU2 TEF1P PhR322 TEF1T PGKP AAD10 PGKT | This study |
| pDM-PhR-AAD14 | 2 μ LEU2 TEF1P PhR322 TEF1T PGKP AAD14 PGKT | This study |
| pDM-PhR-ACS1 | 2 μ LEU2 TEF1P PhR322 TEF1T PGKP ACS1 PGKT | This study |
The primers listed in table 7 were used to amplify the coding regions of the various genes by the PCR technique. Genomic DNA from the DV10 strain was used as the template. Eshericia coli DH5α (GIBCO-BRL/Life Technologies) was used as the host for the construction and propagation of the plasmids listed in table 6. Sequencing of all plasmids was carried out on an ABI PRISM automated sequencer. All plasmids contain the dominant marker PhR conferring phleomicin resistance (PhR), and were transformed into host VIN13 cells via electroporation [21,45].
Table 7.
Sequences of the primers synthesized in order to amplify genes relevant to the present study
| Primer Name | Sequence (5'-3') |
| PhR322F | GATCCACGTCGGTACCCGGGGGATC |
| PhR322R | GATCGCGATCGCAAGCTTGCAAATTAAAGCC |
| YMR210f | AACGCTGGTAAACTTCCAGAGA |
| YMR210r | GGCGAAGCTTTTCACGTTTT |
| AAD10f | ATGCTTTTTACCAAGCAGGC |
| AAD10r | CATCAAACTGTGTGTGTAAGCG |
| AAD14f | ACCAATTAGCTGAACGGCTTTG |
| AAD14r | ATTTGCACACACTCGGTGGATA |
| ACS1f | AAAGACATTGCCCACTGTGCT |
| ACS1r | CACGAAAAAAAAAAAGTCGTCA |
Authors' contributions
DR carried out the experimental work and contributed to the experimental design and data analysis. She also drafted the manuscript. TN contributed to the statistical analysis of the data. FFB conceived of the study and participated in its design and coordination. All authors read and approved of the final manuscript.
Supplementary Material
Identified ORFs and functional description. The table lists all ORFs whose expression pattern correlated significantly with aroma compound production. A short functional annotation is also provided.
Inter- and intra-strain differences in expression levels of all identified ORFs. The three tables show the differences in expression levels of identified ORFs between different strains at day 2 (Table 1) and at day 5 (Table 2) of fermentation, and differences between day 2 and day 5 in individual strains (Table 3).
Summaries of PLS1 models. The table summarises PLS1 models used for interpretation and selection of genes for overexpression.
Comparison between metabolites produced in the strains overexpressing individual ORFs. The table shows a comparison between the volatile aroma metabolites that are produced in the strains overexpressing individual ORFs at day 2 and day 5 of fermentation.
Acknowledgments
Acknowledgements
Funding for the research presented in this paper was provided by the NRF and Winetech, and personal sponsorship by the Wilhelm Frank Trust. We would also like to thank Jo McBride and the Cape Town Centre for Proteomic and Genomic Research for the microarray analysis, and the staff and students at the IWBT, the analytical laboratory of the Department of Viticulture and Oenology and the Central Analytical Facility, all of Stellenbosch University, for their support and assistance in numerous areas.
Contributor Information
Debra Rossouw, Email: debra@sun.ac.za.
Tormod Næs, Email: tormod.naes@matforsk.no.
Florian F Bauer, Email: fb2@sun.ac.za.
References
- Pretorius IS, Bauer FF. Meeting the consumer challenge through genetically customized wine-yeast strains. Trends Biotech. 2002;20:426–432. doi: 10.1016/S0167-7799(02)02049-8. [DOI] [PubMed] [Google Scholar]
- Marullo P, Bely M, Masneuf-Pomarede I, Aigle M, Dubourdieu D. Inheritable nature of enological quantitative traits is demonstrated by meiotic segregation of industrial wine yeast strains. FEMS Yeast Res. 2004;4:711–719. doi: 10.1016/j.femsyr.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Ehrlich F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweissaufbau der Hefe. Berichte Deutsch Chem Gesellschaft. 1907;40:1027–1047. doi: 10.1002/cber.190704001156. [DOI] [Google Scholar]
- Dickinson JR, Lanterman MM, Daner DJ, Pearson BM, Sanz P, Harrison SJ, Hewlins MJE. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 1997;272:26871–26878. doi: 10.1074/jbc.272.43.26871. [DOI] [PubMed] [Google Scholar]
- Dickinson JR, Harrison SJ, Hewlins MJE. An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 1998;273:25751–25756. doi: 10.1074/jbc.273.40.25751. [DOI] [PubMed] [Google Scholar]
- Dickinson JR, Harrison SJ, Dickinson JA, Hewlins MJE. An investigation of the metabolism of isoleucine to active amyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 2000;275:10937–10942. doi: 10.1074/jbc.275.15.10937. [DOI] [PubMed] [Google Scholar]
- Delneri D, Gardner DCJ, Bruschi CV, Oliver SG. Disruption of seven hypothetical aryl alcohol dehydrogenase genes from Saccharomyces cerevisiae and construction of a multiple knock-out strain. Yeast. 1999;15:1681–1689. doi: 10.1002/(SICI)1097-0061(199911)15:15<1681::AID-YEA486>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Dickinson JR, Eshantha L, Salgado J, Hewlins MJE. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem. 2002;278:8028–8034. doi: 10.1074/jbc.M211914200. [DOI] [PubMed] [Google Scholar]
- Nykänen L, Nykänen I. Production of esters by different yeast strains in sugar fermentations. J Inst Brew. 1977;83:30–31. [Google Scholar]
- Peddie HAB. Ester formation in brewery fermentations. J Inst Brew. 1990;96:327–331. [Google Scholar]
- Malcorps P, Cheval JM, Jamil S, Dufour J-P. A new model for the regulation of ester synthesis by alcohol acetyltransferases in Saccharomyces cerevisiae. J Am Soc Brew Chem. 1991;49:47–53. [Google Scholar]
- Vollbrecht D, Radler F. Formation of higher alcohols by amino acid deficient mutants of Saccharomyces cerevisiae. I. The decomposition of amino acids to higher alcohols. Arch Mikrobiol. 1973;94:351–358. doi: 10.1007/BF00769030. [DOI] [PubMed] [Google Scholar]
- Van Dijken JP, Scheffers WA. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- Zoecklein BW, Fugelsang KC, Gump BH, Nury FS. Wine Analysis and Production. New York: The Chapman & Hall Enology Library; 1995. Alcohol and extract; pp. 97–114. [Google Scholar]
- Ribéreau-Gayon P, Dubourdieu D, Donèche B, Lonvaud A. Handbook of Enology. Vol. 1. New York: John Wiley and Sons; 2000. Biochemistry of alcoholic fermentation and metabolic pathways of wine yeasts; pp. 51–74. [Google Scholar]
- Heux S, Cachon R, Dequin S. Cofactor engineering in Saccharomyces cerevisiae: Expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab Eng. 2006;8:303–314. doi: 10.1016/j.ymben.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Rossignol T, Dulau L, Julien A, Blondin B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast. 2003;20:1369–1385. doi: 10.1002/yea.1046. [DOI] [PubMed] [Google Scholar]
- Marks VD, Ho Sui SJ, Erasmus D, Merwe GK van den, Brumm J, Wasserman WW, Bryan J, van Vuuren HJJ. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 2008;8:35–52. doi: 10.1111/j.1567-1364.2007.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Steiner H, Court DA, Rolinski B, Lill R. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem. 1996;271:24458–24464. doi: 10.1074/jbc.271.51.32605. [DOI] [PubMed] [Google Scholar]
- Berg MA Van den, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- Lilly M, Bauer FF, Styger G, Lambrechts MG, Pretorius IS. The effect of increased breanched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res. 2006;6:726–743. doi: 10.1111/j.1567-1364.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- Andreadis A, Hsu YP, Hermodson M, Kohlhaw G, Schimmel P. Yeast LEU2. Repression of mRNA by leucine and primary structure of the gene product. J Biol Chem. 1984;259:8059–8062. [PubMed] [Google Scholar]
- Hsu YP, Schimmel P. Yeast LEU1: Repression of mRNA levels by leucine and relationship of 5'-noncoding region to that of LEU2. J Biol Chem. 1984;259:3714–3719. [PubMed] [Google Scholar]
- Holmberg S, Petersen JG. Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Cur Genet. 1988;13:207–217. doi: 10.1007/BF00387766. [DOI] [PubMed] [Google Scholar]
- Ball SG, Wickner RB, Corrarei G, Schaus M, Tirtiaux C. Molecular cloning and characterization of ARO7-OSM2, a single yeast gene necessary for chorismate mutase activity and growth in hypertonic medium. Mol Gen Genet. 1986;205:326–30. doi: 10.1007/BF00430446. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterization of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new amintransferase subfamily. Mol Gen Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- Vuralhan Z, Morais MA, Tai S-L, Piper MDW, Pronk JT. Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl Environ Microbiol. 2003;69:4534–4541. doi: 10.1128/AEM.69.8.4534-4541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Aviño JP, Prasad R, Miralles VJ, Benito RM, Serrano R. A proposal for nomeclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast. 1999;15:829–842. doi: 10.1002/(SICI)1097-0061(199907)15:10A<829::AID-YEA423>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wang X, Mann CJ, Bai Y, Ni L, Weiner H. Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J Bacteriol. 1998;180:822–830. doi: 10.1128/jb.180.4.822-830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Prix F, Bönquist L, Dequin S. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Aldp6p and Aldp5p isoforms play a major role in acetate formation. Microbiology. 2004;150:2209–2220. doi: 10.1099/mic.0.26999-0. [DOI] [PubMed] [Google Scholar]
- Larroy C, Parés X, Biosca JA. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADH7), a member of the cinnamyl alcohol dehydrogenase family. Eur J Biochem. 2002;269:5738–5745. doi: 10.1046/j.1432-1033.2002.03296.x. [DOI] [PubMed] [Google Scholar]
- Vestrepen KJ, Van Laere SD, Vanderhaegen BM, Derdelinckx G, Dufour JP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol. 2003;69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens SMG, Verstrepen KJ, Van Laere SDM, Voet ARD, Van Dijck P, Delvaux FR, Thevelein JM. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J Biol Chem. 2006;281:4446–4456. doi: 10.1074/jbc.M512028200. [DOI] [PubMed] [Google Scholar]
- Alexander NJ, McCormick SP, Hohn TM. The identification of the Saccharomyces cerevisiae gene AYT1(ORF-YLL063C) encoding an acetyltransferase. Yeast. 2003;19:1425–1430. doi: 10.1002/yea.924. [DOI] [PubMed] [Google Scholar]
- Bely L, Sablayrolles J, Barre P. Description of alcoholic fermentation kinetics: its variability and significance. Am J Enol Viticult. 1990;40:319–324. [Google Scholar]
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. J Comp Biol. 1999;6:281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- Abbott DA, Knijnenburg TA, de Poorter LM, Reinders MJ, Pronk JT, van Maris AJ. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. doi: 10.1111/j.1567-1364.2007.00242.x. Doi: 10. 1111/j. 1567-1364. 2007. 00242. x. [DOI] [PubMed] [Google Scholar]
- Boer VM, de Winde JH, Pronk JT, Piper MD. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus or sulfur. J Biol Chem. 2003;278:3265–3274. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- Tusher CG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardia KV, Kent JT, Bibby JH. Multivariate analysis. UK: Academic Press; 1979. [Google Scholar]
- Martens H, Næs T. Multivariate calibration. UK: J. Wiley and sons; 1989. [Google Scholar]
- Tan Y, Shi L, Hussain SM, Xu J, Tong W, Frazier JM, Wang C. Integrating time-course microarray gene expression profiles with cytotoxicity for identification of biomarkers in primary rat hepatocytes exposed to cadmium. Bioinformatics. 2006;22:77–87. doi: 10.1093/bioinformatics/bti737. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG. Current protocols in molecular biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Wenzel TJ, Migliazza A, Steensma HY, Berg JA van den. Efficient selection of phleomycin-resistant Saccharomyces cerevisiae transformants. Yeast. 1992;8:667–668. doi: 10.1002/yea.320080810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified ORFs and functional description. The table lists all ORFs whose expression pattern correlated significantly with aroma compound production. A short functional annotation is also provided.
Inter- and intra-strain differences in expression levels of all identified ORFs. The three tables show the differences in expression levels of identified ORFs between different strains at day 2 (Table 1) and at day 5 (Table 2) of fermentation, and differences between day 2 and day 5 in individual strains (Table 3).
Summaries of PLS1 models. The table summarises PLS1 models used for interpretation and selection of genes for overexpression.
Comparison between metabolites produced in the strains overexpressing individual ORFs. The table shows a comparison between the volatile aroma metabolites that are produced in the strains overexpressing individual ORFs at day 2 and day 5 of fermentation.