Abstract
The normal development and physiological functions of multicellular organisms are regulated by complex gene transcriptional networks that include myriad transcription factors, their associating coregulators, and multiple chromatin-modifying factors. Aberrant gene transcriptional regulation resulting from mutations among these elements often leads to developmental defects and diseases. This review article concentrates on the Atrophin family proteins, including vertebrate Atrophin-1 (ATN1), vertebrate arginine-glutamic acid dipeptide repeats protein (RERE), and Drosophila Atrophin (Atro), which we recently identified as nuclear receptor corepressors. Disruption of Atrophin-mediated pathways causes multiple developmental defects in mouse, zebrafish, and Drosophila, while an aberrant form of ATN1 and altered expression levels of RERE are associated with neurodegenerative disease and cancer in humans, respectively. We here provide an overview of current knowledge about these Atrophin proteins. We hope that this information on Atrophin proteins may help stimulate fresh ideas about how this newly identified class of nuclear receptor corepressors aids specific nuclear receptors and other transcriptional factors in regulating gene transcription, manifesting physiological effects, and causing diseases.
Introduction
Atrophin proteins are conserved transcriptional corepressors that, as we recently demonstrated, are involved in nuclear receptor signaling. This review article provides an overview of our current understanding of this new class of nuclear receptor corepressors, which includes vertebrate Atrophin-1 (ATN1), vertebrate arginine glutamic acid repeats encoded protein (RERE), and Drosophila Atrophin (Atro).
This review discusses the identification of atrophin proteins, their expression and cellular patterns, their connections with nuclear receptor and epidermal growth factor signaling pathways, functional domains, associations with histone modifying factors, roles in animal development, and their implications in human diseases. Deepening our understanding of the Atrophin proteins will expand our insights about how these nuclear receptor corepressors help transcriptional factors, such as nuclear receptors, regulate gene transcription, exert biological effects, and cause diseases.
The identification of atrophin proteins
The Atn1 gene was initially cloned by Christopher Ross’s group at Johns Hopkins University as CTG-B37 in their screens for human genes containing CTG/CAG or CCG/CGG triplet repeats [Li et al., 1993]. In 1994, the Atn1 gene was assigned to the short arm of chromosome 12 and was simultaneously reported by Masao Yamada (National Children’s Medical Research Center, Japan) and Shoji Tsuji (Niigata University, Japan) to be a causative factor for the neurodegenerative disease DRPLA (dentatorubral-pallidoluysian atrophy) when the CAG repeat within the coding region of the Atn1 gene is expanded [Koide et al., 1994; Nagafuchi et al., 1994]. In addition to the glutamine-repeat (coded by the CAG repeat), ATN1 also contains two arginine-glutamic acid dipeptide-like repeats (RE-repeats) that reside at its C-terminus. In search of potential ATN1 homologues, Masao Yamada’s group identified RERE [Yanagisawa et al., 2000], named after the RE-like repeats present at the C-terminus of the protein. Owing to its resemblance with ATN1, RERE has also been referred to as Atrophin-2.
In 2002, Drosophila Atro was reported by two groups independently: Tian Xu’s group at Yale University, using a dominant female sterile-FRT/FLP approach, identified Atro during their search for lethal mutations that affect growth and patterning [Zhang et al., 2002]; and Steve Kerridge and Laurent Fansano’s group at the IBDML (Marseille, France), isolated Atro through an EMS mutagenesis in their screening for mutations that affect the expression of the region-specific pattern gene teashirt [Erkner et al., 2002]. In the latter study, Atro was also referred to as Grunge. Drawing on their characterization of Atro mutants, both groups reported that Atro is an essential gene involved in multiple developmental pathways.
Spatial and cellular patterns
Northern blot analysis reveals that Atn1 transcripts are ubiquitously expressed in a variety of neuronal and non-neuronal tissues, including brain, heart, lung, kidney, placenta, skeletal muscle and liver [Kanazawa, 1998; Onodera et al., 1995]. At the cellular level, ATN1 is predominantly localized to the cytoplasm of neuronal cells [Knight et al., 1997], although in cultured HeLa cells, transiently expressed ATN1 was found to be present within the nucleus [Okamura-Oho et al., 1999; Yanagisawa et al., 2000].
The Rere gene codes for two transcripts with estimated sizes of 7.4 and 9.4 kb. Their expression varies considerably in different tissues, being particularly abundant in the cerebellum, testis, uterus, prostate, skeletal muscle and kidney, and relatively lower in lung, colon and leukocytes [Waerner et al., 2001; Yanagisawa et al., 2000]. The cellular localization of RERE protein was reported based on transient overexpression conditions. In transfected HeLa cells, although a low level of RERE can be detected in the cytoplasm, RERE localizes predominantly to the nucleus, where it forms a speckle-like pattern resembling that of promyelocytic leukemia bodies [Waerner et al., 2001; Yanagisawa et al., 2000].
In Drosophila, Atro is at first ubiquitously distributed throughout the embryo during the early stage [Erkner et al., 2002; Zhang et al., 2002]. During later stages of embryogenesis, a higher level of Atro becomes more apparent at the ventral nerve cord region. At the third instar larvae stage, Atro resumes its ubiquitously expressed pattern in various examined imaginal tissues. Throughout different developmental stages, Atro mainly localizes to the nucleus of cells, within which endogenous Atro forms a speckle-like pattern [Zhang et al., 2002] . When overexpressed in human cells, consistently, Atro also forms a nuclear speckle pattern [Wang et al., 2008; Wang et al., 2006], indicating that formation of a nuclear speckle pattern is a shared property of Atro and RERE. Interestingly, this unique cellular feature does not apply to ATN1, probably because it lacks the SANT (SWI3/ADA2/N-CoR/TFIII-B) domain (more information about this domain will be provided below), which is essential for RERE to form nuclear foci [Wang et al., 2006].
Roles of atrophin proteins in animal and Drosophila development
A recent study shows that Atn1 is not an essential gene during mouse development, because Atn1 knockout mouse is indistinguishable from wild-type mouse [Shen et al., 2007]. In contrast, Rere is required for both mouse and zebrafish to develop and to survive [Asai et al., 2006; Plaster et al., 2007; Zoltewicz et al., 2004]. In mouse, mutations of Rere cause a failure in closing the anterior neural tube and fusion of the telencephalic and optic vesicles during early embryogenesis [Zoltewicz et al., 2004]. Other defects include a smaller first branchial arch with a deficit in the mesenchymal component, a fail-to-loop heart tube and somites with irregular shapes and sizes. All homozygous mice die shortly after E9.5, possibly owing to cardiac failure. Accompanying most of these described phenotypes is the disruption of important signaling centers, including the loss of sonic hedgehog in the anterior notochord, and defective expression of fibroblast growth factor 8 (fgf8) in the anterior neural ridge. In zebrafish, modulations of Fgf8 signaling by RERE were also observed in the context of posterior mesoderm formation, midbrain-hindbrain boundary maintenance, and the development of pharyngeal cartilage and inner ear [Asai et al., 2006; Plaster et al., 2007]. The apparently divergent effects of ATN1 and RERE on animal development suggest that their functions are not equivalent. Since ATN1 resembles a truncated form of RERE (Figure 1), the portion of RERE that is missing from ATN1 could account for their functional differences.
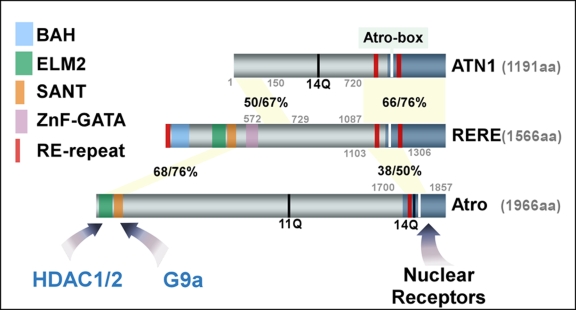
Figure 1. A comparison of ATN1, RERE and Atro, and their functional domains.
The boundaries and the percentage of identical/similar residues within the conserved regions between human ATN1 (Atrophin-1), human RERE (arginine glutamic acid dipeptide repeats protein), and Drosophila Atro (Atrophin) are shown. The functional domains, including BAH (bromo adjacent homology), ELM2 (EGL-27 and MTA1 homology 2), SANT (SWI3/ADA2/N-CoR/TFIII-B), RE-repeat (arginine-glutamic acid dipeptide-repeat), ZnF-GATA, and glutamine (Q)-repeat are highlighted in different colors. The mapped nuclear receptor interacting domains at the C-terminus of each protein are marked in dark blue, within which the Atro-box is labeled as a white bar. The ELM2 domain and SANT domain are docking sites where HDAC1/2 (histone deacetylase 1, 2) and G9a (H3-K9 histone methyltransferase) bind, respectively.
As the Drosophila counterpart of RERE, Atro is also crucial for the development of Drosophila. Embryos deficient of maternal or both the maternal and zygotic component of Atro exhibit severe segmentation defects [Erkner et al., 2002; Kankel et al., 2004; Zhang et al., 2002]. Such Atro mutant embryos also lack some ventral patterning elements, and often show holes in the ventral cuticle, suggesting that in addition to its better characterized involvement in anterior-posterior patterning, Atro also participates in dorsal-ventral axis patterning. In embryos devoid of Atro protein (using a null Atro allele), the expression patterns of several segmentation gap genes, such as hunchback, Krüpple and knirps (kni), are altered [Erkner et al., 2002; Wang et al., 2006]. The altered expression of gap genes can partly explain the subsequent abnormal expression of pair-rule genes, such as fushi-tarazu (ftz) and hairy. However, evidence from experiments using hypomorphic Atro alleles reveals that Atro also functions downstream of the gap genes. For example, embryos derived from females carrying hypomorphic Atro alleles do not show loss of continuous segments (gap gene phenotype), but, instead, display loss of even-numbered (ftz-dependent) engrailed stripes [Kankel et al., 2004; Zhang et al., 2002]. Consistently, the cuticles of such Atro mutant embryos also display strong ftz-like segmentation defects. Although these ftz-phenotypes could result from the loss of the repressive activity of a pair-rule protein Even-skipped (Eve), as suggested in [Zhang et al., 2002], other regulatory pathways that control the expression of ftz may be affected by Atro mutations as well.
Atro is also involved in other developmental pathways. For example, mutations of Atro affect planar polarity in the eyes in a non-autonomous manner [Fanto et al., 2003; Zhang et al., 2002], and also cause patterning defects in the legs [Erkner et al., 2002; Wehn and Campbell, 2006]. Atro can antagonize the activity of EGFR (epidermal growth factor receptor) as well [Charroux et al., 2006], since in the Drosophila wing, mutation of Atro or reduced expression of Atro results in ectopic vein formation [Charroux et al., 2006; Kankel et al., 2004; Wang et al., 2008]. (Note that wing vein formation is known to be initiated by activated EGFR [Martin-Blanco et al., 1999]). The possibility that Atro negatively regulates EGFR signaling is further supported by the genetic interactions between Atro and several key genetic components in the EGFR signaling pathways, including argos, rolled, pointed, and yan (anterior open), in both Drosophila wing and eye [Charroux et al., 2006].
The function of Atro has also been connected with microRNA. Steve Cohen’s group at Temasek, Singapore, reported recently that Atro is a target of miR-8, after they identified four potential miR-8 binding sites at the 3’ untranslated region (3’UTR) of the Atro gene [Karres et al., 2007]. In support of their finding, the leg phenotype displayed by mir-8 mutants is similar to that caused by the overexpression of Atro [Charroux et al., 2006; Karres et al., 2007]. Additionally, reducing Atro activity can rescue the survival rate of these mir-8 mutant flies [Karres et al., 2007]. Regulation of Atro by miR-8 appears to be a conserved feature, because the same group found that 3’UTR of the Rere gene also contains three potential targets of miR-429 and miR-200b, which are the vertebrate counterparts of miR-8. Therefore, a key function of miR-8 and miR-429/miR-200b may be to fine-tune the expression levels of Atro and RERE in Drosophila and in vertebrates. Consequently, altered expression of miR-8, -200b, or -429 is expected to affect the developmental pathways that are regulated by Atro or RERE.
Roles in nuclear receptor signaling
Our lab recently defined Atrophin proteins as corepressors of nuclear receptors through our work with Drosophila Tailless (Tll) and vertebrate Tlx [Wang et al., 2006], which are two closely related nuclear receptors belonging to the 2E subgroup of the nuclear receptor family (NURSA, http://www.nursa.org/). An interaction between Tlx and ATN1 was also reported by Ron Evans’ lab at the Salk Institute [Zhang et al., 2006].
Tll is a gap protein involved in specifying terminal cell fate during early embryogenesis [Mahoney and Lengyel, 1987; Pignoni et al., 1990], and it is also implicated in neurogenesis [Younossi-Hartenstein et al., 1997]. In parallel with the roles of Tll in Drosophila nervous system development, Tlx participates in regulating vertebrate retinal and forebrain development [Miyawaki et al., 2004; Roy et al., 2002; Zhang et al., 2006] and in maintaining adult neural stem cells [Shi et al., 2004; Sun et al., 2007; Zhang et al., 2008]. Both Tll and Tlx have long been known to exert potent transcriptional repression [Hoch et al., 1992; Pankratz et al., 1992; Pankratz et al., 1989; Yu et al., 1994] and have been defined as dedicated transcriptional repressors [Moran and Jimenez, 2006]. The underlying molecular mechanisms, however, remained unclear until our discovery of their association with Atrophin proteins.
Our results show that Tll/Tlx-Atrophin interactions are mediated through the ligand binding domain (LBD) of Tll/Tlx and the C-terminal regions of the three Atrophin proteins [Wang et al., 2006] (regions marked in blue, Figure 1). In keeping with these observed physical interactions between Tll/Tlx and Atrophin proteins, our in vivo studies in Drosophila demonstrated that Atro indeed assists Tll in repressing the expression of kni, a segmentation gap gene known to be a direct target of Tll [Pankratz et al., 1992]. The following data indicate that Atro is involved in the Tll-regulatory pathways: (1) depletion of Atro from Drosophila embryos leads to derepression of kni similar to that in tll mutant embryos; (2) mutation of Atro in tll mutant embryos enhances the kni derepression; and (3) Atro is naturally present in a region within the kni promoter containing a defined Tll-binding site, as demonstrated by chromatin immunoprecipitation assays.
A recent report reveals that regulation of kni by Atro also involves another nuclear protein, called Brakeless (Bks, also referred to as Scribbler, Sbb). Mattias Mannervik’s group (Stockholm University, Sweden) reported that Atro and Bks/Sbb physically and genetically interact with each other to regulate kni expression [Haecker et al., 2007]. This finding resonates with an earlier report from Gerard Campbell’s group at the University of Pittsburgh, which showed that Atro interacts genetically with sbb to repress the expression of several genes, including thickveins, runt, aristaless and Bar, in fly imaginal discs [Wehn and Campbell, 2006]. Mannervik’s group further demonstrated that a human homolog of Bks/Sbb, zinc finger protein 608 (ZNF608), can also interact with ATN1 directly [Haecker et al., 2007]. Based on this finding, it is possible that ZNF608 and another Bks/Sbb-related protein, ZNF609, may be involved in nuclear receptor signaling in vertebrates as well.
Aside from Tll/Tlx, our data also show that Atrophin proteins interact with additional nuclear receptors. For example, Atrophin proteins bind several members of the NR2F group, including human chicken ovalbumin upstream promoter-transcription factor (COUP-TF) and its Drosophila homologue Seven-Up (SVP) [Wang et al., 2006]. This result was somewhat surprising, because NR2F proteins are also known to interact with SMRT family corepressor proteins [Bailey et al., 1997; Shibata et al., 1997], including SMRT (silencing mediator of retinoid and thyroid hormone receptors) [Chen and Evans, 1995], N-CoR (nuclear receptor corepressor) [Horlein et al., 1995], and Drosophila SMRTER (SMRT-related ecdysone receptor interacting factor) [Tsai et al., 1999]. Therefore, certain nuclear receptors, like COUP-TF/SVP, are able to bind two classes of nuclear receptor corepressors, perhaps by means of flexible structural configurations. Whether Atrophin- and SMRT-family proteins bind COUP-TF/SVP in a competitive or a collaborative manner, and whether Atrophin proteins can interact with additional nuclear receptors, are important questions that still wait to be addressed.
Structural and functional domains
The strong transcriptional repressive activity associated with Atrophin proteins has been reported by numerous laboratories. For example, mutations of Atro cause derepression of several segmentation gap genes in Drosophila [Erkner et al., 2002]; Atro binds the transcriptional repressor Even-skipped (Eve) and mediates its transcriptional repressive effect [Zhang et al., 2002]; Atro and ATN1 can provide a heterologous Gal4 DNA-binding domain the ability to repress gene transcription in Drosophila [Zhang et al., 2002]; and the N-terminal regions of both RERE and Atro mediate strong histone deacetylase (HDAC) activities [Wang et al., 2006]. The properties associated with each functional domain of the Atrophin proteins described below thus help explain how Atrophin proteins exert potent repression to regulate gene transcription.
As shown in Figure 1, ATN1 is a smaller protein (1191 aa), compared to RERE (1566 aa) and to Atro (1966 aa) [Erkner et al., 2002; Koide et al., 1994; Nagafuchi et al., 1994; Yanagisawa et al., 2000; Zhang et al., 2002]. Unlike ATN1 and Atro, RERE does not contain any long glutamine-repeat tract. There are two regions located in the C-terminus of RERE that are conserved in ATN1 (the percentages of their identical and similar sequences are shown in Figure 1). Some sequences in the second (the C-terminal) conserved region can also be found in Atro. The functional importance of this conserved region is reflected in its coding of the Atro-box, which is essential for Atrophin proteins to interact with Tll and Tlx [Wang et al., 2006].
ATN1 resembles a truncated version of RERE because it lacks the entire N-terminal region of RERE that harbors several functional domains such as the BAH (bromo adjacent homology) domain, the ELM2 (EGL-27 and MTA1 homology 2) domain, and the SANT (SWI3/ADA2/N-CoR/TFIII-B) domain, as well as a zinc finger (ZnF) GATA-like motif. Among these domains, only the ELM2 and SANT domains can be found in Atro. The conservation of these two domains in both RERE and Atro implies that they are functionally important. Because BAH domain and the GATA-like motif are unique to RERE, it is anticipated that RERE has additional properties that do not apply to Atro.
The RE-repeats
One unique feature in the sequences of Atrophin proteins is their coding of dipeptide repeats. The dipeptide consists of either arginine-glutamic acid (RE), lysine-glutamic acid (KE), arginine-aspartic acid (RD), or lysine-aspartic acid (KD). For convenience, and to be consistent with earlier reports, we refer to these dipeptide repeats as RE-repeats. RERE contains three such repeats: one is located at its very N-terminal end and the other two reside in its C-terminal region. ATN1 has two RE-repeats occupying positions similar to those in the C-terminus of RERE. Interestingly, Atro contains only one RE-repeat. The exact functions of such RE-repeats are currently unclear, although it has been suggested that they are involved in mediating the interaction between RERE and ATN1 [Yanagisawa et al., 2000].
The ELM2 domain
This domain was first described in a C. elegans protein, EGL-27 [Herman et al., 1999], which is a nuclear protein involved in embryonic patterning, cell polarity, cell migration, and vulval development [Herman et al., 1999; Solari and Ahringer, 2000; Solari et al., 1999]. Based on its sequence, EGL-27 is likely to be an ortholog of Atrophin in the nematode C. elegans. Therefore, EGL-27 may be involved in nuclear receptor signaling in C. elegans. The ELM2 domain can also be found in many other transcriptional regulators, for example, vertebrate and invertebrate MTA1, 2, 3 (metastasis-associated proteins), CoREST (corepressor of REST), and MIER1 (mesoderm induction early response 1) [Andres et al., 1999; Bowen et al., 2004; Ding et al., 2003]. The functions of these different nuclear factors have been under intensive investigation. For instance, MTAs are components of the nucleosome remodeling and histone deacetylase (NuRD) complex [Xue et al., 1998; Zhang et al., 1998], whose other components include ATP-dependent chromatin remodeling protein Mi-2/CHD4 (chromodomain helicase DNA binding protein 4), HDAC 1/2 (histone deacetylase 1 and 2), MBD (methyl CpG binding protein) 2/3, and histone interacting proteins RbAp48/p46. Elevated expression of MTA proteins has been associated with several metastatic cancers [Bowen et al., 2004; Manavathi et al., 2007]; CoREST is required for REST/NRSF (RE1-silencing transcriptional factor, also called neuron-restrictive silencer factor) to silence the expression of several neural-specific genes [Andres et al., 1999]; and MIER1 is expressed at a higher level in several breast carcinoma cell lines and breast tumors [Paterno et al., 2002]. Owing to the studies by Ding et al. and our laboratory, one of ELM2’s key functions has been demonstrated to be the recruitment of HDAC1/2 [Ding et al., 2003; Wang et al., 2006].
The SANT domain
This domain is a 50-amino acid-motif that was first reported in 1996. It is present in many factors involved in transcriptional regulation [Aasland et al., 1996; Boyer et al., 2004]. Sequence and structure analyses indicate that the SANT domain consists of three α-helices [de la Cruz et al., 2005; Grune et al., 2003], and is homologous to the DNA-binding domain (DBD) of c-Myb related proteins. Despite this similarity, no report has shown that the SANT domain can bind DNA. Instead, it codes for diverse functions. For example, (1) a SANT domain of ADA2 and SMRT interacts with histone tails [Boyer et al., 2002; Yu et al., 2003]; (2) a SANT domain of SMRT binds to and activates histone deacetylase 3 (HDAC3) [Guenther et al., 2001]; (3) the SANT domain of MIER1 binds Sp1 and interferes with its transcriptional activating activity [Ding et al., 2004]; (4) a SANT domain enables CoREST to stimulate the histone demethylation activity of LSD1 (lysine specific demethylase 1) [Shi et al., 2005]; (5) the SANT domain is essential for RERE to form nuclear foci [Wang et al., 2006]; and (6) the SANT domain of RERE binds G9a [Wang et al., 2008], which is a major histone H3-lysine 9 methyltransferase [Rice et al., 2003; Tachibana et al., 2002]. These results suggest that SANT is a versatile domain whose properties may be determined not only by its coding sequence, but also by other regions of the proteins within which it resides. Interestingly, many SANT-domain proteins, including vertebrate and invertebrate MTAs, CoREST, MIER-1, also contain the ELM2 domain. The conjoined conservation of both domains in various transcriptional regulators reveals not only their functional importance, but also their potential connection.
The BAH domain
This domain was first identified in the chick Polybromo-1 protein, which also contains multiple bromo domains [Nicolas and Goodwin, 1996]. The human homolog of Polybromo-1, called BAF180, is a component of the SWI/SNF chromatin remodeling complex PBAF (polybromo, BRG-1 associated factors) [Xue et al., 2000]. The BAH domain is also present in a wide range of proteins, including all three MTA proteins, a trithorax group protein ASH1 (absent, small, or homeotic discs 1), eukaryotic DNA (cytosine-5) methyltransferases, the Orc1 (origin recognition complex 1) proteins, Sir3 (silent information regulator 3), and yeast RSC1/2 (remodeling the structure of chromatin 1/2) [Callebaut et al., 1999; Goodwin and Nicolas, 2001]. One characterized function of the BAH domain is to mediate the interaction between Orc1 and Sir1, which plays a key role in establishing the silent chromatin structure at the cryptic mating-type loci HMR and HML in yeast [Triolo and Sternglanz, 1996]. The same group (Rolf Sternglanz, Stony Brook University) also reported that the BAH domain is crucial for the function of Sir3 in transcriptional silencing [Connelly et al., 2006]. Recently, the BAH domain of yeast Sir3 was reported to bind to nucleosome and histone tails [Onishi et al., 2007].
The information discussed above suggests that Atrophin proteins, through their multiple functional domains, can act as scaffolds for recruiting various transcriptional cofactors, which, in turn, enable Atrophin proteins to effectively repress transcription. This view is supported by our recent finding that the ELM2 and SANT domains of RERE can recruit multiple proteins in human cells [Wang et al., 2008]. This finding also resonates with two earlier reports that MTA proteins and CoREST (which are ELM2-SANT domain proteins) are components of large protein complexes whose properties range from chromatin remodeling and histone deacetylation to transcriptional repression [Bowen et al., 2004; Manavathi et al., 2007; Xue et al., 1998; Zhang et al., 1998]. Since the transcriptional properties of the Atrophin proteins depend on their associated factors, these factors create pathways through which different developmental or environmental cues can modify the properties of Atrophin proteins in a context-dependent manner (Figure 2).
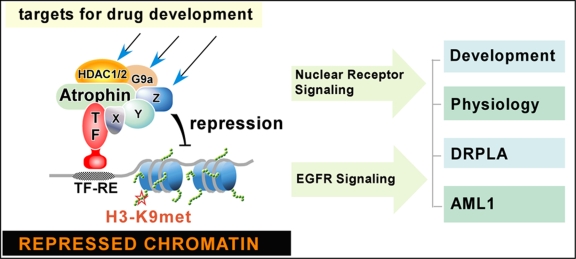
Figure 2. A model depicting the actions of Atrophin-protein complex on chromosomes, signaling pathways, and biological and disease processes.
Atrophin proteins, such as RERE and Atro, use their ELM2 and SANT domains to recruit HDAC1/2, G9a, and additional chromatin-modifying factors (marked as X, Y, and Z). The coordinated actions of these chromatin-modifying factors enable Atrophin proteins to promote methylation of histone H3-K9 (a hallmark of repression) and change local chromatin structures where Atrophin-associated transcriptional factors (TF) bind. Each Atrophin-associated chromatin-modifying factor represents a target that can be modified by EGFR (epidermal growth factor receptor) signaling and that can be used for screening chemical compounds/drugs for treating DRPLA (dentatorubral-pallidolusian atrophy) or AML1(acute myeloid leukemia 1).
Connections with histone modification
Histone tails are subject to different covalent modifications, including acetylation, methylation, phosphorylation and ubiquitination [Strahl and Allis, 2000]. A large body of evidence suggests that controlling the interplay among such modifications is a strategy used by transcription factors or cofactors to regulate gene transcription [Zhang and Reinberg, 2001]. Considerable evidence has already shown that histone deacetylation is responsible for creating specific chromatin structures that favor transcriptional repression. Another major histone modification implicated in transcriptional repression is histone lysine methylation, with methylation of histone H3 lysine 9 (H3-K9) being an important marker of heterochromatin and silenced euchromatin [Jenuwein and Allis, 2001; Rea et al., 2000].
Our recent data indicate that Atrophin proteins bring about transcriptional repression by drawing on both histone deacetylation and histone H3-K9 methylation. As described above, RERE, via its ELM2 domain, recruits HDAC1 and HDAC2 [Wang et al., 2006] and, through its SANT domain, recruits the histone H3-K9 methyltransferase G9a [Wang et al., 2008]. In studying the properties of the ELM2 and SANT domains of RERE, we found that, by coordinating the action of HDAC1/2 and G9a, these two domains of RERE catalyze sequential molecular events that first cause the deacetylation of H3-K9 and then allow this deacetylated residue to be methylated by G9a [Wang et al., 2008]. As a result of these modifications of H3-K9, chromatin structures at the regions where RERE binds become compact and favor gene silencing (see the model shown in Figure 2). This mode of action may also apply to other transcriptional regulators containing the ELM2 and SANT domains, since our data additionally show that all three MTA proteins and MIER1 not only interact with HDAC1/2, but also bind G9a [Wang et al., 2008].
Connections with EGFR signaling
The Atrophin-HDAC1/2-G9a complex is also linked to the EGFR signaling pathway in Drosophila [Wang et al., 2008]. As mentioned earlier, wing vein formation is initiated by the activated EGFR in Drosophila [Martin-Blanco et al., 1999]. In keeping with the report that Atro acts as a negative factor in the EGFR signaling pathway [Charroux et al., 2006], mutation of Atro or reduced expression of Atro (using the double-stranded RNA method) results in ectopic vein formation in the intervein regions [Charroux et al., 2006; Kankel et al., 2004; Wang et al., 2008]. This observed mutant Atro-mediated ectopic wing vein phenotype is enhanced when G9a or Rpd3 (the fly HDAC1/2) is also mutated [Wang et al., 2008]. These genetic data indicate that Atro, dG9a and Rpd3 act together to repress wing vein formation, perhaps by antagonizing the activities of EGFR. Strong genetic interaction among Atro, dG9a and Rpd3 also takes place in other tissues. For example, melanotic masses appear in the Drosophila head when both Atro and dG9a or Rpd3 are removed or mutated. We surmise that the formation of these melanotic masses is a result of accumulated hemocytes. Whether this phenotype involves a hyperactive EGFR signaling pathway remains to be investigated.
Atrophin appears to be involved in growth-factor signaling pathways in other species as well. Julie Ahringer’s group (University of Cambridge, UK) has shown that EGL-27, the worm Atrophin-like protein, acts to inhibit vulval development in C. elegans by antagonizing Ras signaling [Solari and Ahringer, 2000], which takes place downstream of growth-factor signaling pathways. Therefore, negative regulation of the EGFR-Ras pathway may be a conserved property shared by Atrophin proteins and their associating factors. Since hyperactive EGFR and Ras are associated with various cancers, their oncogenic effects may result from their interference with the transcriptional repressive properties of the Atrophin-HDAC1/2-G9a protein complex.
Janus-faced transcriptional properties, and beyond transcription
Owing to their relationship with transcriptional repressors and HDACs, Atrophin proteins have been viewed primarily as dedicated corepressors. However, considerable evidence suggests that Atro can regulate gene transcription in a positive manner as well. For example, Ian Duncan’s group (Washington University, St. Louis) reported that ftz expression is reduced in Drosophila embryos mutant of Atro [Kankel et al., 2004]. In the same report, they provided additional evidence suggesting that Atro is a member of the trithorax group (trxG) proteins, which are known to regulate the expression of homeotic genes positively. Their evidence includes: (1) mutations of Atro enhance the loss-of-function phenotypes of antennapedia complex and bithorax complex genes; (2) the phenotypes caused by the mutations of two trxG genes, brahma and trithorax, are enhanced by mutations of Atro; and (3) the leg phenotype caused by mutations in Polycomb-like, a polycomb group (PcG) gene, is suppressed by mutations of Atro. (Note that PcG genes have opposite effects from trxG genes on regulating the expression of homeotic genes.)
In many respects, the observed behavior of Atro resembles that of Osa, as also suggested by Ian Duncan’s group. Osa is a trxG group protein known to associate with the Brahma chromatin remodeling complex [Collins et al., 1999; Vazquez et al., 1999]. While the functions of Osa have been associated primarily with transcriptional activation, it has been linked with transcriptional repression as well. For example, Jessica Treisman’s group (Skirball Institute, New York University) showed that several wingless target genes are repressed by Osa [Collins and Treisman, 2000]. Therefore, both Atro and Osa may be bifunctional transcriptional regulators whose transcriptional properties (either positive or negative) can be influenced by other factors or by posttranslational modifications. This idea is further supported by the finding that the C-terminal region of RERE contains a domain that can activate gene transcription through its association with the transcriptional coactivator p300 [Shen et al., 2007]. Since our recent data indicate that RERE is subject to protein cleavage [Wang et al., 2008], the cleavage event—which allows the C-terminal transcriptional activating module to be separated from the N-terminal transcriptional repressive module—could be responsible for converting RERE from a transcriptional repressive form to a transcriptional activating form. My laboratory is currently investigating this possibility.
Although Atro has been investigated predominantly as a nuclear protein, Helen McNeill’s group (Samuel Lunenfeld Research Institute, Mt. Sinai Hospital, Canada) has reported that Atro physically interacts with the cytoplasmic domain of the atypical cadherin Fat in vitro [Fanto et al., 2003], and that Atro collaborates with Fat to control planar polarity in the Drosophila eye. These results suggest that Atro might have non-nuclear functions as well. Furthermore, ATN1 localizes predominantly to the cytoplasm of neuronal cells [Knight et al., 1997], and can interact with several proteins with functions outside the realm of transcriptional regulation. Several ATN1-interacting proteins (AIPs) isolated from yeast two hybrid screens [Wood et al., 1998] are known to function in the cytoplasm. For example, AIP1 is a membrane-associated guanylate kinase family protein that interacts with the tumor suppressor PTEN and enhances its ability to suppress Akt activation [Wu et al., 2000]. AIP4 is an ubiquitin E3 ligase, which regulates the degradation of the human enhancer of filamentation 1 (HEF1) in TGF-β (transforming growth factor β) signaling pathways [Feng et al., 2004]. ATN1 may also function in an insulin-related signaling pathway, since ATN1 can bind the SH3 (Src homology 3) domain of a human homologue of the insulin receptor tyrosine kinase substrate protein of 53kD (IRSp53) in the cytoplasm [Okamura-Oho et al., 1999].
A comparison between the Atrophin- and SMRT-family proteins
In many respects, the functional properties discovered for Atrophin family proteins resemble those found for SMRT family proteins, which are, by far, the best characterized nuclear receptor corepressors. Aside from their functional resemblance, these two groups of nuclear receptor corepressors also exhibit their own unique features. A comparison of their known properties is described below and summarized in Figure 3.
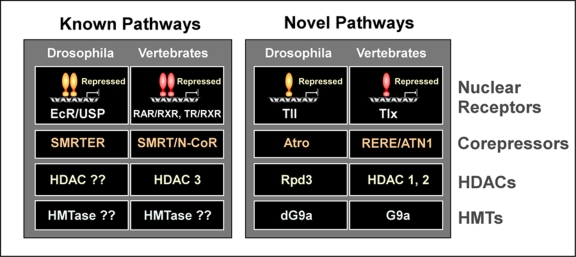
Figure 3. A comparison of the known properties of SMRT- and Atrophin- proteins.
The well characterized SMRT class of corepressors, including SMRT (silencing mediator of retinoid and thyroid hormone receptors), N-CoR (nuclear receptor corepressor), and their Drosophila cognate SMRTER (SMRT-related ecdysone receptor interacting factor), preferentially bind members of subgroup 1 of the nuclear receptor superfamily, such as TR (thyroid hormone receptor), RAR (retinoic acid receptor), or EcR (ecdysone receptor), and recruits HDAC3. The newly-identified Atrophin class of corepressors, including RERE, ATN1, and their Drosophila cognate Atro, in contrast, selectively bind members of the subgroup 2 of nuclear receptors, such as Tll and Tlx, and recruits HDAC1/2. (Note: Rpd3 is the fly HDAC1/2). Whereas Atrophin proteins, except for ATN1, bind G9a both in mammalian cells and in fly, none of the members of the SMRT family proteins have so far been shown to bind HMTases. RXR (retinoid-X receptor) and USP (Ultraspiracle, the fly RXR), are the common binding partners of RAR and TR, and EcR, respectively.
(1) Both families' proteins are conserved. Each includes a Drosophila protein (Atro and SMRTER), and two vertebrate paralogues (ATN1, RERE and SMRT, N-CoR) [Chen and Evans, 1995; Erkner et al., 2002; Horlein et al., 1995; Koide et al., 1994; Nagafuchi et al., 1994; Tsai et al., 1999; Yanagisawa et al., 2000; Zhang et al., 2002]. All of these proteins contain RE-repeats and all, except ATN1, also contain one or two SANT domains. Moreover, all proteins in both groups, except RERE, contain at least one glutamine-tract.
(2) Both families' proteins interact with nuclear receptors in a selective way. SMRT, N-CoR and SMRTER preferentially bind nuclear receptors, such as retinoic acid receptor (RAR), thyroid hormone receptor (TR), peroxisome proliferator-activated receptor (PPAR) and ecdysone receptor (EcR) [Jepsen and Rosenfeld, 2002; Lazar, 2003; Privalsky, 2001; Tsai and Fondell, 2004], but not Tll and Tlx [Wang et al., 2006]. Atrophin proteins, in contrast, interact strongly with Tll and Tlx, but barely with RAR, TR, PPAR or EcR [Wang et al., 2006]. Their differential affinity towards different groups of nuclear receptors may result from the different structural motifs these two groups of proteins use to interact with nuclear receptors. For example, Mitch Lazar’s group (University of Pennsylvania) identified a CoRNR box (or L/I-X-X-I/V-I motif) in both SMRT and N-CoR that enables both proteins to interact with nuclear receptors [Hu and Lazar, 1999]. We identified a stretch of highly conserved amino acids, termed the Atro-box (Figure 1), in all Atrophin proteins that are essential for them to bind Tll and Tlx [Wang et al., 2006].
(3) Both families' proteins interact with HDACs, also in a selective way. SMRT and N-CoR bind HDAC3 strongly [Guenther et al., 2000; Li et al., 2000], while their associations with HDAC1 and HDAC2 are relatively weak [Wang et al., 2006]. Atrophin proteins show the opposite preferences, with Atro and RERE strongly interacting with HDAC1 and HDAC2, but barely with HDAC3 [Wang et al., 2006]. Considering that SMRT proteins can also bind to type II HDACs, such as HDAC 4, 5, and 7 [Huang et al., 2000; Kao et al., 2000], it is possible that Atrophin proteins may associate selectively with type II HDACs as well.
(4) Atrophin proteins, except ATN1, associate with HMTases such as G9a [Wang et al., 2008]. Although direct interactions between SMRT family proteins and HMTase have not been reported, data from our laboratory indicate that transcriptional repression mediated by SMRT family proteins also involves HMTases. This finding, together with the results described above, suggests that recruiting both HDACs and HMTases is an important and shared strategy used by nuclear receptor corepressors to repress gene transcription.
Implications for diseases
DRPLA
Polyglutamine expansion within human ATN1 causes DRPLA, which is also known as Haw River Syndrome and Naito-Oyanagi disease. DRPLA is one of the nine late-onset polyglutamine neurodegenerative diseases, which also include Huntington’s disease, spinobulbar muscular atrophy, and spinal cerebellar ataxias 1, 2, 3, 6, 7, and 17 [Zoghbi and Orr, 2000]. The clinical features of DRPLA include dementia, choreoathetosis, myoclonus epilepsy, and ataxia [Tsuji, 1999]. Like most polyglutamine diseases, the pathogenic mechanism underlying DRPLA remains elusive. It had been speculated that glutamine-repeat expansion in ATN1 compromises its own function. This view, however, has been challenged by a recent report that Atn1-/- mice show no neurological defects or other phenotype [Shen et al., 2007]. Therefore, the dominant negative effect caused by the glutamine-repeat expanded form of ATN1 is likely mediated through pathways other than a simple loss of function of ATN1.
Several models have been proposed to explain the neurotoxicity caused by mutant ATN1, including the formation of nuclear aggregates or inclusions, which is a hallmark of polyglutamine diseases [Ross and Poirier, 2004], protein cleavage of ATN1 [Ellerby et al., 1999; Nucifora et al., 2003; Schilling et al., 1999], and phosphorylation of ATN1 [Okamura-Oho et al., 2003]. Thus far, the exact roles of such modifications of ATN1 in the pathogenesis of DRPLA have not been firmly established. With regard to protein cleavage, a caspase-3 cleavage site was mapped to the Asp109 of ATN1; expressing this cleaved form of ATN1 causes cytotoxicity in cultured cells [Ellerby et al., 1999]. Although the in vivo relevance of this cleaved form of ATN1 to DRPLA development is still unclear, the fact that ATN1 is involved in transcriptional regulation suggests that this cleavage event might affect its transcriptional (and perhaps toxic) properties.
Transcriptional abnormality has recently emerged as a potential pathogenic mechanism responsible for many polyglutamine diseases [Riley and Orr, 2006]. The glutamine-repeat expanded form of ATN1 interacts strongly with TAFII130 and can suppress CREB-dependent transcriptional activation [Shimohata et al., 2000]. CBP is sequestered into nuclear inclusions found in DRPLA postmortem brain tissue, and the N-terminal truncation of the expanded ATN1 can inhibit CBP-mediated transcription [Nucifora et al., 2001]. Correlatively, a DRPLA transgenic mouse model expressing ATN1 with 118Q exhibits hypoacetylation of histone H3 in brain tissues, whereas the administration of sodium butyrate, an HDAC inhibitor, ameliorates the neurodegenerative phenotypes of such mice [Ying et al., 2006]. These observations strongly suggest that abnormal gene repression is a contributing factor for the development of DRPLA.
Although sequestration of CBP into ATN1-mediated nuclear inclusions may partly account for mutant ATN1-mediated changes in gene expression profile and neurotoxicity, other mechanisms may also be involved. With our recent identification of Atrophin proteins as nuclear receptor corepressors, one plausible explanation for the development of DRPLA is that nuclear receptor signaling pathways are perturbed. It is noteworthy that ATN1's interacting partners, such as Tlx and COUP-TF, are expressed and function in the nervous system [Miyawaki et al., 2004; Park et al., 2003; Roy et al., 2002; Shi et al., 2004; Tripodi et al., 2004; Yu et al., 1994]. The pathogenesis of DRPLA might also involve RERE, considering that RERE can dimerize with ATN1 and that RERE-ATN1 interaction is enhanced when ATN1’s glutamine-repeat is expanded [Yanagisawa et al., 2000]. In fact, the latter model is supported by the effects caused by ATN1 in Drosophila: expressing a pathological form of ATN1 in Drosophila generates a phenotype similar to that caused by mutations of Atro [Charroux et al., 2006].
We here propose several mechanisms that might explain how ATN1 affects Atro-dependent pathways in Drosophila: (1) The functions of Atro may be compromised upon its association with the insoluble form of mutant ATN1; (2) stabilized mutant ATN1 may compete with Atro to bind common transcriptional cofactors; and (3) a stabilized mutant ATN1, which lacks intrinsic HDAC and HMTase activities [Wang et al., 2008; Wang et al., 2006], might gain both chromatin-modifying activities upon its association with Atro. Any of these hypothesized mechanisms would affect the transcriptional properties of Atro in Drosophila. With the observed effects of ATN1 in Drosophila in mind, one tempting theory for the mechanism of DRPLA in vertebrates is that the functions of RERE are affected by mutant ATN1. If so, we predict that the phenotypes displayed by DRPLA mice will be enhanced when one allele of the Rere gene is removed or mutated.
Cancers
The gene for Rere is located in the distal region of chromosome 1p, where frequent structural rearrangements have been reported in association with several human malignancies [Schwab et al., 1996]. For example, the neuroblastoma cell line NGP contains a reciprocal chromosomal translocation/duplication t(1;15)(p36.2;q24), Dp(1)(p36.2-p36.3) [Casciano et al., 1996]. It has been reported that the Rere gene unit is disrupted in NGP [Amler et al., 2000]. Additionally, the expression of Rere transcripts is altered in many neuroblastoma cell lines [Waerner et al., 2001]. In cultured cells, transiently overexpressed RERE colocalizes with BAX, a proapoptotic protein, and enhances caspase-dependent apoptosis [Waerner et al., 2001]. The latter result implies that mutation of Rere may prevent damaged cells from undergoing cell death, thus leading to cancer development.
ATN1 has also been shown to interact with ETO/MTG8 [Wood et al., 2000], a transcriptional coregulator whose fusion with RUNX1 causes the most common form of t(8;21) acute myeloid leukemia (AML) [Kozu et al., 1993; Miyoshi et al., 1993; Nisson et al., 1992]. Interestingly, ETO/MTG8 has been shown previously to be an associating factor of SMRT, N-CoR, Sin3A and HDACs [Gelmetti et al., 1998; Lutterbach et al., 1998; Wang et al., 1998]. The association between ETO/MTG8 and various transcriptional corepressors suggests that aberrant transcriptional repression imposed on RUNX1-regulatory pathways by the ETO-RUNX1 fusion protein could be an underlying reason for the pathogenesis of AML1. If so, reducing the transcriptional repressive activities of either or both Atrophin-family and SMRT-family proteins could provide a feasible therapeutic strategy for treating AML1.
Therapeutic implications
Abnormal expression levels of MTAs and MIER1 have been linked to different forms of metastatic tumors or different carcinoma cell lines [Bowen et al., 2004; Ding et al., 2003; Kumar et al., 2003; Waerner et al., 2001]. Since our recent work shows that these proteins are associated with HDAC1/2 and G9a [Wang et al., 2008], we suspect that alteration of their expression levels may cause aberrant transcriptional repression and, consequently, may result in the development or progression of cancers. Recently, HDAC inhibitors [Marks et al., 2001] have been tested as promising agents in treating blood-borne cancers, and also in treating polyglutamine diseases [Carey and La Thangue, 2006; Hockly et al., 2003; McCampbell et al., 2001; Minamiyama et al., 2004; Steffan et al., 2001; Ying et al., 2006]. Since specific G9a inhibitors have also been recently reported by Thomas Jenuwein’s lab at the Vienna Biocenter, Austria [Kubicek et al., 2007], our study raises the possibility that these G9a inhibitors might also be used, alone or together with the current FDA-approved HDAC inhibitors, for treating cancers and neurological diseases involving Atrophin, MTAs, CoREST or MIER1 (Figure 2).
Concluding remarks
Rapid progress during the past few years has produced a much better understanding of the Atrophin protein family's molecular functions. The exciting discovery that Atrophin proteins are nuclear receptor corepressors enables us to look at these proteins from a nuclear receptor signaling perspective. Our efforts make it clear now that Atrophin proteins employ mechanisms, such as histone deacetylation and histone lysine methylation, to repress gene transcription. It is only a matter of time before more nuclear receptors, transcriptional factors, and histone/chromatin-modifying factors are identified that associate with Atrophin proteins. In-depth study of Atrophin proteins and their functions in connection with nuclear receptor, EGFR, and chromatin-modifying factors will not only lead to a better understanding of their roles in animal development and in human diseases, but will also pave the way for the identification of potential therapeutic targets for treating various human diseases.
Acknowledgments
We thank R. Head and H. Gui for commenting on this manuscript. CCT is a recipient of funding from NIH/NINDS (5R01NS050365), from the Leukemia Research Foundation, and from the New Jersey Commission on Cancer Research.
Abbreviations
- AIP
ATN1-interacting protein
- AML
acute myeloid leukemia
- ASH1
absent, small, or homeotic discs 1
- ATN1
Atrophin-1
- Atro
Atrophin
- BAH
bromo adjacent homology
- Bks
Brakeless
- CHD4
chromodomain helicase DNA binding protein 4
- CoREST
corepressor of REST
- COUP-TF
chicken ovalbumin upstream promoter-transcription factor
- DBD
DNA binding domain
- DRPLA
dentatorubral-phallidolusian atrophy
- EcR
ecdysone receptor
- EGFR
epidermal growth factor receptor
- ELM2
EGL-27 and MTA1 homology 2
- Eve
Even-skipped
- fgf8
fibroblast growth factor 8
- ftz
fushi-tarazu
- HDAC
histone deacetylase
- HEF1
human enhancer of filamentation 1
- HMTase
histone methyltransferase
- IRSp53
insulin receptor tyrosine kinase substrate protein of 53kD
- KD
lysine-aspartic acid
- KE
lysine-glutamic acid
- kni
knirps
- LBD
ligand binding domain
- LSD1
lysine specific demethylase 1
- MBD
methyl CpG binding protein
- MIER1
mesoderm induction early response 1
- MTA
metastasis-associated protein
- N-CoR
nuclear receptor corepressor
- NuRD
nucleosome remodeling and histone deacetylase
- Orc1
origin recognition complex 1
- PBAF
polybromo, BRG-1 associated factors
- PcG
polycomb group
- PPAR
peroxisome proliferator-activated receptor
- RAR
retinoic acid receptor
- RD
arginin-aspartic acid
- RERE
arginine glutamic acid repeats encoded protein
- RE-repeats
arginine-glutamic acid dipeptide repeats
- REST
RE1-silencing transcriptional factor
- RSC
remodeling the structure of chromatin
- RXR
retinoid-X receptor
- SANT
SWI3/ADA2/N-CoR/TFIII-B
- Sbb
Scribbler
- SH3
Src homology 3
- Sir
silent information regulator
- SMRT
silencing mediator of retinoid and thyroid hormone receptors
- SMRTER
SMRT-related ecdysone receptor interacting factor
- SVP
Seven-Up
- TGFβ
transforming growth factor β
- Tll
Tailless
- Tlx
vertebrate Tailless
- TR
thyroid hormone receptor
- trxG
trithorax group
- USP
Ultraspiracle
- UTR
untranslated region
- ZNF
zinc finger protein.
References
- Aasland R., Stewart A. F., Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–8. [PubMed] [Google Scholar]
- Amler L. C., Bauer A., Corvi R., Dihlmann S., Praml C., Cavenee W. K., Schwab M., Hampton G. M. Identification and characterization of novel genes located at the t(1;15)(p36.2;q24) translocation breakpoint in the neuroblastoma cell line NGP. Genomics. 2000;64:195–202. doi: 10.1006/geno.1999.6097. [DOI] [PubMed] [Google Scholar]
- Andres M. E., Burger C., Peral-Rubio M. J., Battaglioli E., Anderson M. E., Grimes J., Dallman J., Ballas N., Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–8. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai Y., Chan D. K., Starr C. J., Kappler J. A., Kollmar R., Hudspeth A. J. Mutation of the atrophin2 gene in the zebrafish disrupts signaling by fibroblast growth factor during development of the inner ear. Proc Natl Acad Sci U S A. 2006;103:9069–74. doi: 10.1073/pnas.0603453103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P. J., Dowhan D. H., Franke K., Burke L. J., Downes M., Muscat G. E. Transcriptional repression by COUP-TF II is dependent on the C-terminal domain and involves the N-CoR variant, RIP13delta1. J Steroid Biochem Mol Biol. 1997;63:165–74. doi: 10.1016/s0960-0760(97)00079-4. [DOI] [PubMed] [Google Scholar]
- Bowen N. J., Fujita N., Kajita M., Wade P. A. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–7. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Langer M. R., Crowley K. A., Tan S., Denu J. M., Peterson C. L. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002;10:935–42. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Latek R. R., Peterson C. L. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–63. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Courvalin J. C., Mornon J. P. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–93. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- Carey N., La Thangue N. B. Histone deacetylase inhibitors: gathering pace. Curr Opin Pharmacol. 2006;6:369–75. doi: 10.1016/j.coph.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Casciano I., Marchi J. V., Muresu R., Volpi E. V., Rozzo C., Opdenakker G., Romani M. Molecular and genetic studies on the region of translocation and duplication in the neuroblastoma cell line NGP at the 1p36.13-p36.32 chromosomal site. Oncogene. 1996;12:2101–8. [PubMed] [Google Scholar]
- Charroux B., Freeman M., Kerridge S., Baonza A. Atrophin contributes to the negative regulation of epidermal growth factor receptor signaling in Drosophila. Dev Biol. 2006;291:278–90. doi: 10.1016/j.ydbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Evans R. M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Collins R. T., Furukawa T., Tanese N., Treisman J. E. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. Embo J. 1999;18:7029–40. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. T., Treisman J. E. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 2000;14:3140–52. doi: 10.1101/gad.854300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J. J., Yuan P., Hsu H. C., Li Z., Xu R. M., Sternglanz R. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol Cell Biol. 2006;26:3256–65. doi: 10.1128/MCB.26.8.3256-3265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz X., Lois S., Sanchez-Molina S., Martinez-Balbas M. A. Do protein motifs read the histone code? Bioessays. 2005;27:164–75. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- Ding Z., Gillespie L. L., Paterno G. D. Human MI-ER1 α and β function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol. 2003;23:250–8. doi: 10.1128/MCB.23.1.250-258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Gillespie L. L., Mercer F. C., Paterno G. D. The SANT domain of human MI-ER1 interacts with Sp1 to interfere with GC box recognition and repress transcription from its own promoter. J Biol Chem. 2004;279:28009–16. doi: 10.1074/jbc.M403793200. [DOI] [PubMed] [Google Scholar]
- Ellerby L. M., Andrusiak R. L., Wellington C. L., Hackam A. S., Propp S. S., Wood J. D., Sharp A. H., Margolis R. L., Ross C. A., Salvesen G. S., Hayden M. R., Bredesen D. E. Cleavage of atrophin-1 at caspase site aspartic acid 109 modulates cytotoxicity. J Biol Chem. 1999;274:8730–6. doi: 10.1074/jbc.274.13.8730. [DOI] [PubMed] [Google Scholar]
- Erkner A., Roure A., Charroux B., Delaage M., Holway N., Core N., Vola C., Angelats C., Pages F., Fasano L., Kerridge S. Grunge, related to human Atrophin-like proteins, has multiple functions in Drosophila development. Development. 2002;129:1119–29. doi: 10.1242/dev.129.5.1119. [DOI] [PubMed] [Google Scholar]
- Fanto M., Clayton L., Meredith J., Hardiman K., Charroux B., Kerridge S., McNeill H. The tumor-suppressor and cell adhesion molecule Fat controls planar polarity via physical interactions with Atrophin, a transcriptional co-repressor. Development. 2003;130:763–74. doi: 10.1242/dev.00304. [DOI] [PubMed] [Google Scholar]
- Feng L., Guedes S., Wang T. Atrophin-1-interacting protein 4/human Itch is a ubiquitin E3 ligase for human enhancer of filamentation 1 in transforming growth factor-β signaling pathways. J Biol Chem. 2004;279:29681–90. doi: 10.1074/jbc.M403221200. [DOI] [PubMed] [Google Scholar]
- Gelmetti V., Zhang J., Fanelli M., Minucci S., Pelicci P. G., Lazar M. A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–91. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin G. H., Nicolas R. H. The BAH domain, polybromo and the RSC chromatin remodelling complex. Gene. 2001;268:1–7. doi: 10.1016/s0378-1119(01)00428-0. [DOI] [PubMed] [Google Scholar]
- Grune T., Brzeski J., Eberharter A., Clapier C. R., Corona D. F., Becker P. B., Muller C. W. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–60. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–57. [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G., Barak O., Lazar M. A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A., Qi D., Lilja T., Moussian B., Andrioli L. P., Luschnig S., Mannervik M. Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos. PLoS Biol. 2007;5:e145. doi: 10.1371/journal.pbio.0050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M. A., Ch'ng Q., Hettenbach S. M., Ratliff T. M., Kenyon C., Herman R. K. EGL-27 is similar to a metastasis-associated factor and controls cell polarity and cell migration in C. elegans. Development. 1999;126:1055–64. doi: 10.1242/dev.126.5.1055. [DOI] [PubMed] [Google Scholar]
- Hoch M., Gerwin N., Taubert H., Jackle H. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel. Science. 1992;256:94–7. doi: 10.1126/science.1348871. [DOI] [PubMed] [Google Scholar]
- Hockly E., Richon V. M., Woodman B., Smith D. L., Zhou X., Rosa E., Sathasivam K., Ghazi-Noori S., Mahal A., Lowden P. A., Steffan J. S., Marsh J. L., Thompson L. M., Lewis C. M., Marks P. A., Bates G. P. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein A. J., Naar A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C. K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Huang E. Y., Zhang J., Miska E. A., Guenther M. G., Kouzarides T., Lazar M. A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Rosenfeld M. G. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–98. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Kanazawa I. Dentatorubral-pallidoluysian atrophy or Naito-Oyanagi disease. Neurogenetics. 1998;2:1–17. doi: 10.1007/s100480050046. [DOI] [PubMed] [Google Scholar]
- Kankel M. W., Duncan D. M., Duncan I. A screen for genes that interact with the Drosophila pair-rule segmentation gene fushi tarazu. Genetics. 2004;168:161–80. doi: 10.1534/genetics.104.027250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H. Y., Downes M., Ordentlich P., Evans R. M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Karres J. S., Hilgers V., Carrera I., Treisman J., Cohen S. M. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–45. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Knight S. P., Richardson M. M., Osmand A. P., Stakkestad A., Potter N. T. Expression and distribution of the dentatorubral-pallidoluysian atrophy gene product (atrophin-1/drplap) in neuronal and non-neuronal tissues. J Neurol Sci. 1997;146:19–26. doi: 10.1016/s0022-510x(96)00266-3. [DOI] [PubMed] [Google Scholar]
- Koide R., Ikeuchi T., Onodera O., Tanaka H., Igarashi S., Endo K., Takahashi H., Kondo R., Ishikawa A., Hayashi T. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- Kozu T., Miyoshi H., Shimizu K., Maseki N., Kaneko Y., Asou H., Kamada N., Ohki M. Junctions of the AML1/MTG8(ETO) fusion are constant in t(8;21) acute myeloid leukemia detected by reverse transcription polymerase chain reaction. Blood. 1993;82:1270–6. [PubMed] [Google Scholar]
- Kubicek S., O'Sullivan R. J., August E. M., Hickey E. R., Zhang Q., Teodoro M. L., Rea S., Mechtler K., Kowalski J. A., Homon C. A., Kelly T. A., Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Kumar R., Wang R. A., Bagheri-Yarmand R. Emerging roles of MTA family members in human cancers. Semin Oncol. 2003;30:30–7. doi: 10.1053/j.seminoncol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Lazar M. A. Nuclear receptor corepressors. Nucl Recept Signal. 2003;1:e001. doi: 10.1621/nrs.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. H., McInnis M. G., Margolis R. L., Antonarakis S. E., Ross C. A. Novel triplet repeat containing genes in human brain: cloning, expression, and length polymorphisms. Genomics. 1993;16:572–9. doi: 10.1006/geno.1993.1232. [DOI] [PubMed] [Google Scholar]
- Lutterbach B., Westendorf J. J., Linggi B., Patten A., Moniwa M., Davie J. R., Huynh K. D., Bardwell V. J., Lavinsky R. M., Rosenfeld M. G., Glass C., Seto E., Hiebert S. W. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–84. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney P. A., Lengyel J. A. The zygotic segmentation mutant tailless alters the blastoderm fate map of the Drosophila embryo. Dev Biol. 1987;122:464–70. doi: 10.1016/0012-1606(87)90310-1. [DOI] [PubMed] [Google Scholar]
- Manavathi B., Singh K., Kumar R. MTA family of coregulators in nuclear receptor biology and pathology. Nucl Recept Signal. 2007;5:e010. doi: 10.1621/nrs.05010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P. A., Richon V. M., Breslow R., Rifkind R. A. Histone deacetylase inhibitors as new cancer drugs. Curr Opin Oncol. 2001;13:477–83. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E., Roch F., Noll E., Baonza A., Duffy J. B., Perrimon N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development. 1999;126:5739–47. doi: 10.1242/dev.126.24.5739. [DOI] [PubMed] [Google Scholar]
- McCampbell A., Taye A. A., Whitty L., Penney E., Steffan J. S., Fischbeck K. H. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci U S A. 2001;98:15179–84. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamiyama M., Katsuno M., Adachi H., Waza M., Sang C., Kobayashi Y., Tanaka F., Doyu M., Inukai A., Sobue G. Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2004;13:1183–92. doi: 10.1093/hmg/ddh131. [DOI] [PubMed] [Google Scholar]
- Miyawaki T., Uemura A., Dezawa M., Yu R. T., Ide C., Nishikawa S., Honda Y., Tanabe Y., Tanabe T. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci. 2004;24:8124–34. doi: 10.1523/JNEUROSCI.2235-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Kozu T., Shimizu K., Enomoto K., Maseki N., Kaneko Y., Kamada N., Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. Embo J. 1993;12:2715–21. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran E., Jimenez G. The tailless nuclear receptor acts as a dedicated repressor in the early Drosophila embryo. Mol Cell Biol. 2006;26:3446–54. doi: 10.1128/MCB.26.9.3446-3454.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi S., Yanagisawa H., Ohsaki E., Shirayama T., Tadokoro K., Inoue T., Yamada M. Structure and expression of the gene responsible for the triplet repeat disorder, dentatorubral and pallidoluysian atrophy (DRPLA) Nat Genet. 1994;8:177–82. doi: 10.1038/ng1094-177. [DOI] [PubMed] [Google Scholar]
- Nicolas R. H., Goodwin G. H. Molecular cloning of polybromo, a nuclear protein containing multiple domains including five bromodomains, a truncated HMG-box, and two repeats of a novel domain. Gene. 1996;175:233–40. doi: 10.1016/0378-1119(96)82845-9. [DOI] [PubMed] [Google Scholar]
- Nisson P. E., Watkins P. C., Sacchi N. Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells. Cancer Genet Cytogenet. 1992;63:81–8. doi: 10.1016/0165-4608(92)90384-k. [DOI] [PubMed] [Google Scholar]
- Nucifora F. C., Jr., Sasaki M., Peters M. F., Huang H., Cooper J. K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V. L., Dawson T. M., Ross C. A. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–8. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Nucifora F. C., Jr., Ellerby L. M., Wellington C. L., Wood J. D., Herring W. J., Sawa A., Hayden M. R., Dawson V. L., Dawson T. M., Ross C. A. Nuclear localization of a non-caspase truncation product of atrophin-1, with an expanded polyglutamine repeat, increases cellular toxicity. J Biol Chem. 2003;278:13047–55. doi: 10.1074/jbc.M211224200. [DOI] [PubMed] [Google Scholar]
- Okamura-Oho Y., Miyashita T., Ohmi K., Yamada M. Dentatorubral-pallidoluysian atrophy protein interacts through a proline-rich region near polyglutamine with the SH3 domain of an insulin receptor tyrosine kinase substrate. Hum Mol Genet. 1999;8:947–57. doi: 10.1093/hmg/8.6.947. [DOI] [PubMed] [Google Scholar]
- Okamura-Oho Y., Miyashita T., Nagao K., Shima S., Ogata Y., Katada T., Nishina H., Yamada M. Dentatorubral-pallidoluysian atrophy protein is phosphorylated by c-Jun NH2-terminal kinase. Hum Mol Genet. 2003;12:1535–42. doi: 10.1093/hmg/ddg168. [DOI] [PubMed] [Google Scholar]
- Onishi M., Liou G. G., Buchberger J. R., Walz T., Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–28. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Onodera O., Oyake M., Takano H., Ikeuchi T., Igarashi S., Tsuji S. Molecular cloning of a full-length cDNA for dentatorubral-pallidoluysian atrophy and regional expressions of the expanded alleles in the CNS. Am J Hum Genet. 1995;57:1050–60. [PMC free article] [PubMed] [Google Scholar]
- Pankratz M. J., Hoch M., Seifert E., Jackle H. Kruppel requirement for knirps enhancement reflects overlapping gap gene activities in the Drosophila embryo. Nature. 1989;341:337–40. doi: 10.1038/341337a0. [DOI] [PubMed] [Google Scholar]
- Pankratz M. J., Busch M., Hoch M., Seifert E., Jackle H. Spatial control of the gap gene knirps in the Drosophila embryo by posterior morphogen system. Science. 1992;255:986–9. doi: 10.1126/science.1546296. [DOI] [PubMed] [Google Scholar]
- Park J. I., Tsai S. Y., Tsai M. J. Molecular mechanism of chicken ovalbumin upstream promoter-transcription factor (COUP-TF) actions. Keio J Med. 2003;52:174–81. doi: 10.2302/kjm.52.174. [DOI] [PubMed] [Google Scholar]
- Paterno G. D., Ding Z., Lew Y. Y., Nash G. W., Mercer F. C., Gillespie L. L. Genomic organization of the human mi-er1 gene and characterization of alternatively spliced isoforms: regulated use of a facultative intron determines subcellular localization. Gene. 2002;295:79–88. doi: 10.1016/s0378-1119(02)00823-5. [DOI] [PubMed] [Google Scholar]
- Pignoni F., Baldarelli R. M., Steingrimsson E., Diaz R. J., Patapoutian A., Merriam J. R., Lengyel J. A. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990;62:151–63. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- Plaster N., Sonntag C., Schilling T. F., Hammerschmidt M. REREa/Atrophin-2 interacts with histone deacetylase and Fgf8 signaling to regulate multiple processes of zebrafish development. Dev Dyn. 2007;236:1891–904. doi: 10.1002/dvdy.21196. [DOI] [PubMed] [Google Scholar]
- Privalsky M. L. Regulation of SMRT and N-CoR corepressor function. Curr Top Microbiol Immunol. 2001;254:117–36. doi: 10.1007/978-3-662-10595-5_6. [DOI] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y., Allis C. D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–8. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Riley B. E., Orr H. T. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–92. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- Ross C. A., Poirier M. A. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 Suppl:S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Roy K., Thiels E., Monaghan A. P. Loss of the tailless gene affects forebrain development and emotional behavior. Physiol Behav. 2002;77:595–600. doi: 10.1016/s0031-9384(02)00902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling G., Wood J. D., Duan K., Slunt H. H., Gonzales V., Yamada M., Cooper J. K., Margolis R. L., Jenkins N. A., Copeland N. G., Takahashi H., Tsuji S., Price D. L., Borchelt D. R., Ross C. A. Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA. Neuron. 1999;24:275–86. doi: 10.1016/s0896-6273(00)80839-9. [DOI] [PubMed] [Google Scholar]
- Schwab M., Praml C., Amler L. C. Genomic instability in 1p and human malignancies. Genes Chromosomes Cancer. 1996;16:211–29. doi: 10.1002/(SICI)1098-2264(199608)16:4<211::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Shen Y., Lee G., Choe Y., Zoltewicz J. S., Peterson A. S. Functional architecture of atrophins. J Biol Chem. 2007;282:5037–44. doi: 10.1074/jbc.M610274200. [DOI] [PubMed] [Google Scholar]
- Shibata H., Nawaz Z., Tsai S. Y., O'Malley B. W., Tsai M. J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–24. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- Shi Y., Chichung Lie D., Taupin P., Nakashima K., Ray J., Yu R. T., Gage F. H., Evans R. M. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- Shimohata T., Nakajima T., Yamada M., Uchida C., Onodera O., Naruse S., Kimura T., Koide R., Nozaki K., Sano Y., Ishiguro H., Sakoe K., Ooshima T., Sato A., Ikeuchi T., Oyake M., Sato T., Aoyagi Y., Hozumi I., Nagatsu T., Takiyama Y., Nishizawa M., Goto J., Kanazawa I., Davidson I., Tanese N., Takahashi H., Tsuji S. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- Shi Y. J., Matson C., Lan F., Iwase S., Baba T., Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Solari F., Ahringer J. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol. 2000;10:223–6. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- Solari F., Bateman A., Ahringer J. The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development. 1999;126:2483–94. doi: 10.1242/dev.126.11.2483. [DOI] [PubMed] [Google Scholar]
- Steffan J. S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B. L., Kazantsev A., Schmidt E., Zhu Y. Z., Greenwald M., Kurokawa R., Housman D. E., Jackson G. R., Marsh J. L., Thompson L. M. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–43. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun G., Yu R. T., Evans R. M., Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci U S A. 2007;104:15282–7. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T., Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–3. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Tripodi M., Filosa A., Armentano M., Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–29. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Fondell J. D. Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam Horm. 2004;68:93–122. doi: 10.1016/S0083-6729(04)68003-4. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Kao H. Y., Yao T. P., McKeown M., Evans R. M. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–86. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Tsuji S. Dentatorubral-pallidoluysian atrophy (DRPLA): clinical features and molecular genetics. Adv Neurol. 1999;79:399–409. [PubMed] [Google Scholar]
- Vazquez M., Moore L., Kennison J. A. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–42. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- Waerner T., Gardellin P., Pfizenmaier K., Weith A., Kraut N. Human RERE is localized to nuclear promyelocytic leukemia oncogenic domains and enhances apoptosis. Cell Growth Differ. 2001;12:201–10. [PubMed] [Google Scholar]
- Wang L., Charroux B., Kerridge S., Tsai C. C. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Rep. 2008;9:555–62. doi: 10.1038/embor.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hoshino T., Redner R. L., Kajigaya S., Liu J. M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998;95:10860–5. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Rajan H., Pitman J. L., McKeown M., Tsai C. C. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 2006;20:525–30. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehn A., Campbell G. Genetic interactions among scribbler, Atrophin and groucho in Drosophila uncover links in transcriptional repression. Genetics. 2006;173:849–61. doi: 10.1534/genetics.105.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. D., Nucifora F. C., Jr., Duan K., Zhang C., Wang J., Kim Y., Schilling G., Sacchi N., Liu J. M., Ross C. A. Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J Cell Biol. 2000;150:939–48. doi: 10.1083/jcb.150.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. D., Yuan J., Margolis R. L., Colomer V., Duan K., Kushi J., Kaminsky Z., Kleiderlein J. J., Sharp A. H., Ross C. A. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11:149–60. doi: 10.1006/mcne.1998.0677. [DOI] [PubMed] [Google Scholar]
- Wu X., Hepner K., Castelino-Prabhu S., Do D., Kaye M. B., Yuan X. J., Wood J., Ross C., Sawyers C. L., Whang Y. E. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci U S A. 2000;97:4233–8. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G. T., Young M. K., Cote J., Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Xue Y., Canman J. C., Lee C. S., Nie Z., Yang D., Moreno G. T., Young M. K., Salmon E. D., Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–20. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H., Bundo M., Miyashita T., Okamura-Oho Y., Tadokoro K., Tokunaga K., Yamada M. Protein binding of a DRPLA family through arginine-glutamic acid dipeptide repeats is enhanced by extended polyglutamine. Hum Mol Genet. 2000;9:1433–42. doi: 10.1093/hmg/9.9.1433. [DOI] [PubMed] [Google Scholar]
- Ying M., Xu R., Wu X., Zhu H., Zhuang Y., Han M., Xu T. Sodium butyrate ameliorates histone hypoacetylation and neurodegenerative phenotypes in a mouse model for DRPLA. J Biol Chem. 2006;281:12580–6. doi: 10.1074/jbc.M511677200. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A., Green P., Liaw G. J., Rudolph K., Lengyel J., Hartenstein V. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol. 1997;182:270–83. doi: 10.1006/dbio.1996.8475. [DOI] [PubMed] [Google Scholar]
- Yu J., Li Y., Ishizuka T., Guenther M. G., Lazar M. A. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. Embo J. 2003;22:3403–10. doi: 10.1093/emboj/cdg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R. T., McKeown M., Evans R. M., Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370:375–9. doi: 10.1038/370375a0. [DOI] [PubMed] [Google Scholar]
- Zhang C. L., Zou Y., He W., Gage F. H., Evans R. M. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–7. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- Zhang S., Xu L., Lee J., Xu T. Drosophila atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell. 2002;108:45–56. doi: 10.1016/s0092-8674(01)00630-4. [DOI] [PubMed] [Google Scholar]
- Zhang C. L., Zou Y., Yu R. T., Gage F. H., Evans R. M. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006;20:1308–20. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zoghbi H. Y., Orr H. T. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–47. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- Zoltewicz J. S., Stewart N. J., Leung R., Peterson A. S. Atrophin 2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis. Development. 2004;131:3–14. doi: 10.1242/dev.00908. [DOI] [PubMed] [Google Scholar]