Abstract
Protein kinase D (PKD) is a novel family of serine/threonine kinases targeted by the second messenger diacylglycerol. It has been implicated in many important cellular processes and pathological conditions. However, further analysis of PKD in these processes is severely hampered by the lack of a PKD-specific inhibitor that can be readily applied to cells and in animal models. We now report the discovery of the first potent and selective cell-active small molecule inhibitor for PKD, benzoxoloazepinolone (CID755673). This inhibitor was identified from the National Institutes of Health small molecule repository library of 196,173 compounds using a human PKD1 (PKCμ)-based fluorescence polarization high throughput screening assay. CID755673 suppressed half of the PKD1 enzyme activity at 182 nm and exhibited selective PKD1 inhibition when compared with AKT, polo-like kinase 1 (PLK1), CDK activating kinase (CAK), CAMKIIα, and three different PKC isoforms. Moreover, it was not competitive with ATP for enzyme inhibition. In cell-based assays, CID755673 blocked phorbol ester-induced endogenous PKD1 activation in LNCaP cells in a concentration-dependent manner. Functionally, CID755673 inhibited the known biological actions of PKD1 including phorbol ester-induced class IIa histone deacetylase 5 nuclear exclusion, vesicular stomatitis virus glycoprotein transport from the Golgi to the plasma membrane, and the ilimaquinone-induced Golgi fragmentation. Moreover, CID755673 inhibited prostate cancer cell proliferation, cell migration, and invasion. In summary, our findings indicate that CID755673 is a potent and selective PKD1 inhibitor with valuable pharmacological and cell biological potential.
Protein kinase D (PKD)3 belongs to a subfamily of the Ca2+/calmodulin-dependent kinases (CAMKs) (1). PKD is a novel target of the second messenger diacylglycerol and phorbol esters, the natural products from plants and potent tumor promoters in mouse skin (2). Three isoforms of PKD (PKD1, -2, and -3) have been identified, which share high sequence homology (3-6). The regulatory domain of PKD contains a C1 domain that binds diacylglycerol/phorbol esters with high affinity and a PH domain that mediates protein-protein interactions. The entire regulatory domain appears to exert a negative effect on catalytic activity, possibly serving as an autoinhibitory domain for PKD (7). The activity of PKD is controlled through a protein kinase C (PKC)-dependent mechanism (8). PKC is the primary target of diacylglycerol/phorbol esters and it activates PKD by directly binding and phosphorylating PKD on two serine residues in the activation loop. In most cellular systems examined, PKD is an effector of selective PKC isoforms, acting in a canonical PKC/PKD pathway that leads to a unique set of biological responses including cell proliferation, survival, protein transport, and immune responses (2, 9).
PKD regulates many fundamental cellular functions and has been implicated in the pathogenesis of several diseases. PKD is a key regulator of protein transport from the Golgi to the plasma membrane (10-12). It plays a major role in the epigenetic control of gene expression through regulating class IIa histone deacetylases (HDAC4, -5, -7, and -9), which coincides with its crucial role in pathological cardiac remodeling (13, 14). PKD also promotes cell proliferation and modulates apoptotic responses. These effects of PKD have been demonstrated in various normal and tumor cell lines (15-17). PKD is activated by oxidative stress and triggers a cell survival response through activating NF-κB signaling (18). Furthermore, PKD modulates cell migration and tumor cell invasion in normal and tumor cells (19-22). Thus, PKD is a key regulator of basic biological processes and is a potential druggable target for cardiovascular diseases and cancer.
Despite these important discoveries, a more detailed analysis of the regulation and biology of PKD has been greatly hampered by the lack of a potent and PKD-specific inhibitor. Since the discovery of the first PKD isoform (PKD1) in 1994 (4, 6), no PKD-specific inhibitors have been reported. The most widely used PKD inhibitor in many studies is Gö6976, which inhibits purified PKD at an IC50 of 20 nm (23). However, Gö6976 is foremost known as a PKC inhibitor that preferentially inhibits cPKC isoforms at single digit nanomolar concentrations (24). When paired with Gö6983 (a pan-PKC inhibitor that inhibits PKD poorly), Gö6976 has been shown to be useful in assessing the involvement of PKD in cellular processes. This combination is far from ideal for therapeutic purposes due to the apparent lack of specificity. For similar reasons, other PKD inhibitors such as the PKA inhibitor H-89, which was reported to inhibit PKD at 0.5 μm have not been actively pursued (25). In addition, other compounds such as trans-3,4′,5-trihydroxystilbene (resveratrol), an antioxidant and effective chemopreventive agent, have been reported to inhibit PKD, however, resveratrol lacks the requisite specificity to be a suitable PKD inhibitor (2, 26).
In this study, we report the discovery of the first potent and selective, cell-active, PKD inhibitor benzoxoloazepinolone (CID755673). This compound was discovered through an immobilized metal affinity for phosphochemical (IMAP)-based fluorescence polarization (FP) high throughput screening (HTS) assay (27). It was highly potent and selective for PKD1. When applied to cultured cells, CID755673 impeded various PKD-mediated cellular responses. Moreover, the cancer-associated phenotypic properties of proliferation, migration, and invasion were blocked by CID755673, underscoring the therapeutic potential of targeting PKD in cancer therapy.
EXPERIMENTAL PROCEDURES
Chemicals, Compound Libraries, and Reagents—Enzymatically active recombinant human protein kinase D1 (PKD1) and CDK7/cyclin H/Mat1 (CDK activating kinase (CAK)) were obtained from Millipore (Billerica, MA). Purified enzymatically active recombinant human PKD2, PKD3, PLK1, and AKT1 were purchased from Cell Signaling Technology (Danvers, MA), and PKD3 was purchased from Biomol International (Plymouth Meeting, PA). 5-FAM-KKLNRTLSVA (PKD), 5-FAM-KKRNRRLSVA-OH (PLK1, CAK), and 5-FAM-GRPRTSSFAEG (AKT) substrate peptides were purchased from Molecular Devices (Sunnyvale, CA). The IMAP™ progressive binding reagent, binding buffer (pH ∼ 5.5), bovine serum albumin, and Tween 20-based kinase reaction buffers (10 mm Tris-HCl, pH 7.2, 10 mm MgCl2, 0.05% NaN3, 1 mm dithiothreitol, and 0.01% Tween 20 or bovine serum albumin) were purchased from Molecular Devices. Black and white opaque small volume microtiter plates were purchased from Greiner (Monroe, NC) and used for all IMAP-based experiments. ATP was purchased from GE Healthcare. The 196,173 compound library screened for PKD1 small molecule inhibitors was made available by the Pittsburgh Molecular Library Screening Center (PMLSC, Pittsburgh, PA) as part of the National Institutes of Health MLSCN Roadmap Initiative (NIH-SMR). Cherry-picked compounds from the PMLSC library were supplied by Biofocus DPI (A Galapagos Company, San Francisco, CA). DMSO was purchased from Sigma. CID755673 and CID797718 were synthesized from commercially available starting materials by the PMLSC Chemistry Core (University of Pittsburgh, Pittsburgh, PA).
IMAP-based High-throughput and Secondary Screening Assays—An IMAP-based PKD HTS FP assay (Molecular Devices) was used for primary HTS of the PMLSC library to identify hit compounds and subsequent secondary hit confirmation studies. For primary HTS activities and concentration response studies, PKD1 kinase reactions were assembled as previously described (28). Briefly, PKD1 kinase reactions were generated by the stepwise addition of 3 times concentrations of substrate/ATP (300 nm/60 μm), test compound, and PKD1 enzyme (0.18 milliunits/ml). PKD1 kinase reactions were incubated for 90 min at room temperature and stopped with the addition IMAP binding reagent. Assay plates were then incubated for 2 h. For IMAP-based PLK1 timed-resolved fluorescence energy transfer (TR-FRET) dose-response specificity assays, PLK1 kinase reactions were assembled and processed as previously described (28). AKT IMAP FP kinase specificity assays were performed using 3 times concentrations of substrate/ATP (900 nm/30 μm), test compound, and AKT enzyme (0.15 milliunits/ml). AKT kinase reactions were incubated for 90 min at room temperature, stopped with the addition IMAP binding buffer, and incubated for 45 min prior to data collection. CAK IMAP TR-FRET specificity assays were implemented using 3 times concentrations of substrate/ATP (1500 nm/75 μm), test compound, and CAK enzyme (280 milliunits/ml). CAK kinase reactions were incubated for 3 h at room temperature, stopped with the addition of terbium-supplemented binding reagent, and incubated for 2 h. Kinase reactions for each IMAP-based screening assays were performed in miniaturized reaction volumes (i.e. 6 μl) and all IMAP-based FP and TR-FRET data were captured on a Molecular Devices Spectra-Max M5 (excitation A485, emission A520, and excitation A330; emission A490 and A520, respectively). Kinase reaction and binding buffers for each IMAP enzyme-substrate pair were used according to the manufacturer's instructions. The IC50 determinations for each compound in each IMAP (PKD1, PLK1, CAK, and AKT) assay were conducted within the linear range of the captured signal readout.
HTS Data Analysis and Visualization—HTS data analysis and subsequent compound IC50 determinations were performed using ActivityBase (IDBS, Guildford, UK), CytoMiner (UPDDI, Pittsburgh, PA), and Spotfire (Somerville, MA) software. The Pubchem data base (pubchem.ncbi.nim.nih.gov) was used to determine whether test compounds displayed inhibitory effects on additional kinases.
In Vitro Radiometric PKD or CAMK Kinase Assay—The radiometric kinase assay was carried out by coincubating 0.5 μCi of [γ-32P]ATP (PerkinElmer Life Sciences), 20 μm ATP, 50 ng of purified recombinant human PKD (PKD1, PKD2, and PKD3) or CAMKIIα (Cell Signaling Technology) proteins, and 2.5 μg of Syntide-2 (Sigma) in 50 μl of kinase buffer that contains 50 mm Tris-HCl, pH 7.5, 4 mm MgCl2, 10 mm β-mercaptoethanol. The reaction was carried out under conditions that the initial rate was within the linear kinetic range. For CAMK assay, 0.5 mm Ca2+ and 30 ng/μl calmodulin was added to each reaction mixture. The reaction was incubated at 30 °C for 10 min, followed by spotting 25 μl of the mixture onto a Whatman P81 filter paper (Whatman Inc., Clifton, NJ). The filter papers were then washed three times in 0.5% phosphoric acid, air-dried, and counted using a Beckman LS6500 multipurpose scintillation counter (Beckman).
In Vitro Radiometric PKC Kinase Assay—The kinase assay was carried out by coincubating 1 μCi of [γ-32P]ATP (PerkinElmer), 20 μm ATP, 100 ng of purified rat PKCδ protein (a kind gift from Dr. Peter M. Blumberg, NCI, National Institutes of Health) or 50 ng of recombinant human PKCα or PKCβI (Cell Signaling Technology), and 5 μg of myelin basic protein 4-14, 0.25 mg/ml bovine serum albumin, 0.1 mg/ml phosphatidylcholine/phosphatidylserine (80/20%) (1 μm), 1 μm phorbol dibutyrate in 50 μl of kinase buffer that contains 50 mm Tris-HCl, pH 7.5, 4 mm MgCl2, 10 mm β-mercaptoethanol. The reaction was incubated at 30 °C for 10 min, followed by spotting 25 μl of the reaction mixture onto a Whatman P81 filter paper. The filter papers were then washed three times in 0.5% phosphoric acid, air-dried, and counted using a Beckman LS6500 multipurpose scintillation counter.
Cell Lines and Western Blot Analysis—HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemental with 10% fetal bovine serum, 1000 units/liter penicillin, and 1 mg/ml streptomycin in an atmosphere of 5% CO2 at 37 °C. Prostate cancer cells (LNCaP, PC3, and DU145) were cultured as described previously (29). Western blot analysis was carried out as reported (30). Briefly, cells were lysed in a buffer containing 200 mm Tris-HCl, pH 7.4, 100 μm 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 mm EGTA, and 1% Triton X-100. Equal amounts of protein were subjected to SDS-PAGE and electrotransferred to nitrocellulose membranes. After blocking with 5% nonfat milk and 2% bovine serum albumin in Tris-buffered saline, membranes were probed with a primary antibody, followed by a secondary anti-mouse or anti-rabbit secondary antibody conjugated to horseradish peroxidase (1:1000, Bio-Rad). Bands were visualized by the enhanced chemiluminescence (ECL) Western blotting detection system (Amersham Biosciences). The primary antibodies included p-S742-PKCμ/PKD antibody (Invitrogen), p-S916-PKD1 antibody (Millipore), PKD1 (Cell Signaling Technology), PKD2 (Bethyl Laboratories, Montgomery, TX), and a previously custom-made polyclonal anti-PKD3 antibody (31).
Nuclear Localization of YFP-HDAC5—HeLa cells were transiently transfected with YFP-HDAC5 (a kind gift from Dr. Ronald M. Evans, the Salk Institute for Biological Studies). Two days after transfection, the cells were pretreated with the PKD inhibitor for 45 min followed by phorbol 12-myristate 13-acetate (PMA) stimulation at 1 μm for 4 h. The cells were then washed with phosphate-buffered saline and fixed in 4% formaldehyde for 30 min at room temperature. Images were captured under a ×63 objective, using a confocal fluorescence microscope (Olympus Fluoview). The nuclear localization of YFP-HDAC5 was scored visually. Cells with predominant nuclear YFP-HDAC5 were considered positive.
Vesicular Stomatitis Virus Glycoprotein (VSVG) Transport Assay—The plasmid encoding the temperature-sensitive (ts045) VSVG with a green fluorescent protein (GFP) tag was kindly provided by Dr. Meir Aridor (University of Pittsburgh). HeLa cells were grown on round glass coverslides in 35-mm culture dishes and transfected with plasmid encoding VSVG-GFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Two days after transfection, cells were transferred to a 40 °C incubator for 2 h, and then transferred to 20 °C for 30 min to collect VSVG-GFP protein in the Golgi. CID755673 was applied to cells for 30 min at 20 °C, followed by a 2-h incubation at 20 °C in medium containing 30 μm ilimaquinone to induce Golgi fragmentation or 1 h incubation at 32 °C to allow protein transport to the cell surface. Cells were then fixed in 4% formaldehyde and imaged under a confocal fluorescence microscope.
Wound Healing Assay—DU145 cells were seeded in 6-well plates in growth medium containing 10% fetal bovine serum. The cells were allowed to grow to confluent monolayer. The wound-induced migration was triggered by scraping the cells with a plastic pipette tip, and the wound was imaged immediately. The cells were then were treated with or without CID755673 at different concentrations. The wound was imaged immediately (0 h) and at different intervals as indicated with an inverted phase-contrast microscope with a ×10 objective. At the end of the assay, cells were fixed with methanol and stained with crystal violet for a final image. The wound gap was measured on the images and the % wound healing = (wound gap at 0 h - wound gap at x h)/wound gap at 0 h × 100, and the average % wound healing was calculated based on at least 9 determinations of the wound.
Matrigel Invasion Assay and Cell Migration Assay—A fixed number of LNCaP or DU145 cells (1.0 × 105 cells/ml) in RPMI 1640 media containing 0.1% fetal bovine serum were seeded into the upper chamber of BioCoat control inserts that contain filters with pore size of 8 μm or BioCoat Matrigel invasion inserts that contain Matrigel-coated filters (BD Pharmingen), and the lower chamber contained 20% serum-containing medium. The cells were incubated for 22 h with or without inhibitors. Following incubation, cells that did not migrate through the filters of the control inserts, or did not invade through the Matrigel of the invasion inserts were removed with a cotton swab. Cells that traversed or invaded through the filter/Matrigel were detected by fixing in 100% methanol, followed by staining the cells in 1% crystal violet and visualized under a microscope. After staining, cells in both control inserts and invasion chambers were counted under a microscope in 10 random fields (magnification ×200). Cell invasion was determined as the percent invasion through the Matrigel matrix compared with the number of cells migrated through the control inserts. Cell migration was assessed by counting cells that migrated through the control inserts in 10 random fields.
Cell Proliferation Assay and Cell Cycle Analysis—Cell proliferation was determined by counting the number of viable cells upon trypan blue staining as described (29). Cell proliferation was also measured by CellTiter-Glo Luminescent Cell Viability Assay according to the manufacturer's instructions (Promega, Madison, WI). Cell cycle analysis was conducted as described previously (29).
Statistical Analysis—All statistical analysis was done using GraphPad Prism IV software. A p value <0.05 was considered statistically significant.
RESULTS
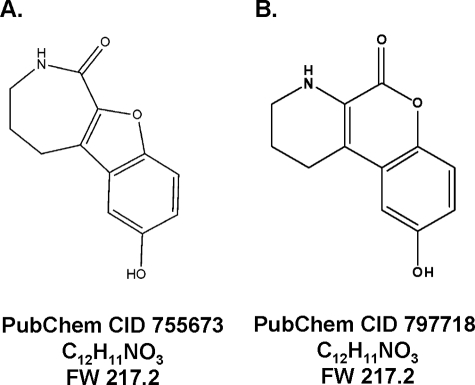
An Unbiased HTS Identified CID755673 as an Inhibitor of PKD1 in Vitro Kinase Activity—The Pubchem compound CID755673 (Fig. 1) was identified after interrogating the PMLSC 196,173 member library as a small molecule that at 10 μm produced >50% inhibition in a screen of PKD1 activity (Pubchem AID 797). CID755673 and CID797718, a structural analog of CID755673, were synthesized by the PMLSC Chemistry Core. Concentration-response studies demonstrated a 50% inhibitory concentration (IC50) for CID755673 of 0.50 ± 0.03 μm (Table 1) for PKD1. CID755673 was also 30-100-fold more selective for PKD1 inhibition compared with PLK1, CAK, or protein kinase B (AKT/PKB) (Table 1). The structural CID755673 analogue, CID797718, was a less potent PKD1 inhibitor with an IC50 of 7.0 ± 0.8 μm (Table 1). The exquisite specificity of CID755673 as a PKD1 inhibitor was further supported by an interrogation of the current Pubchem data base, which indicated that CID755673 was not biologically active in more than 200 other assays including about 14 kinase assays that target Rho kinase 2 (ROCK2), focal adhesion kinase, AKT, PLK1-protein binding domain, JNK3, Wee1, PKA, pyruvate kinase, mitogen-activated kinase 1, phosphomevalonate kinase, mevalonate kinase, Her kinase, Ephrin type B receptor, and Ephrin type A receptor.
FIGURE 1.
Chemical structures of CID755673 and CID797718. A, chemical structure of CID755673, a compound identified and confirmed as a PKD1 inhibitor after interrogation of the PMLSC library. B, chemical structure of CID797718, an analogue of CID755673 obtained during the synthesis of the latter structure.
TABLE 1.
Initial IC50 determinations of CID755673 and CID797718 against PKD1 and various kinases using IMAP-based FP or TR-FRET kinase assays
|
Kinase
|
IC50 (μm)
|
|
|---|---|---|
| CID755673 | CID797718 | |
| PKD1 | 0.5 ± 0.03 | 7.0 ± 0.8 |
| PLK1 | 20.3 ± 10.9 | 21.9 ± 6.5 |
| CAK | 15.3 ± 1.8 | 8.4 ± 1.6 |
| AKT | >50 | >50 |
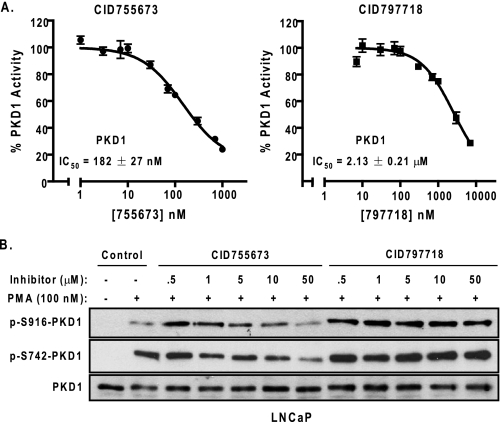
CID755675 Was a Potent Small Molecule Inhibitor of PKD1—The PKD1 inhibitory activity of CID755673 was evaluated in an orthogonal radiometric PKD kinase assay: recombinant human PKD1 was incubated with the substrate syntide-2 and 10 different concentrations of CID755673 or CID797718 (1-10,000 nm). Confirming the FP results, we found CID755673 was a potent inhibitor of recombinant PKD1 with an even lower IC50 value of 182 ± 27 nm (n = 5), whereas CID797718 was 10-fold less potent than CID755673 (IC50 = 2.13 ± 0.21 μm, n = 3) (Fig. 2A). We next assessed the ability of CID755673 to inhibit the activity of endogenous PKD1 in LNCaP prostate cancer cells in which PKD1 was shown to be the predominant isoform (8). PKD1 is activated by PMA through PKC-mediated phosphorylation on serine 738 and 742 (Ser738/Ser742) in the activation loop (32, 33). Thus, levels of phospho-Ser738 and/or phospho-Ser742 are indicative of PKD1 activity. PKD1, once activated, autophosphorylates serine 916 (Ser916) in the C terminus of PKD1, and the status of p-Ser916-PKD correlates well with the catalytic activity of PKD1 (34). Therefore, we used both the p-Ser916- and p-Ser742-PKD1 antibodies to monitor the activity of PKD1 in LNCaP cells. As shown in Fig. 2B (lane 2), PMA alone induced phosphorylation of Ser742 and Ser916 of PKD1. The addition of CID755673 caused a concentration-dependent inhibition of PMA-induced PKD1 phosphorylation at both Ser742 and Ser916. The inhibitory effect was more apparent on phospho-Ser916 levels with near complete blockade of the phosphorylation at 50 μm CID755673. In contrast, the less potent PKD1 antagonist CID797718 did not significantly reduce phospho-Ser742 or phospho-Ser916 levels of PKD1, or slightly reduced the phospho signals at the highest 50 μm concentration. As controls, the compounds did not alter the endogenous expression of PKD1 in LNCaP cells (Fig. 2B). Taken together, these results suggested that CID755673 directly inhibited PKD1 activity both in vitro and in cells.
FIGURE 2.
The inhibitory activities of CID755673 and CID797718. A, inhibition of recombinant human PKD1 in vitro. The kinase activity of PKD1 was assayed in the presence of 10 different concentrations of CID755673 by a radiometric PKD kinase assay. The IC50 values were the mean ± S.E. of at least three independent experiments with triplicate determinations at each drug concentration in each experiment. The data were plotted as a function of drug concentration and a representative graph is shown. B, inhibition of endogenous PKD1 activity by CID755673 in cells. LNCaP cells were pretreated with different doses of CID755673 or CID797718 for 45 min, followed by PMA stimulation at 100 nm for 20 min. Cell lysates were subjected to immunoblotting for p-S742-PKD1 and p-S916-PKD1. PKD1 was blotted as loading control. Note that the low p-S916-PKD1 signal in lane 2 (PMA alone) was likely caused by uneven loading. The experiment was repeated five times and a representative blot is shown.
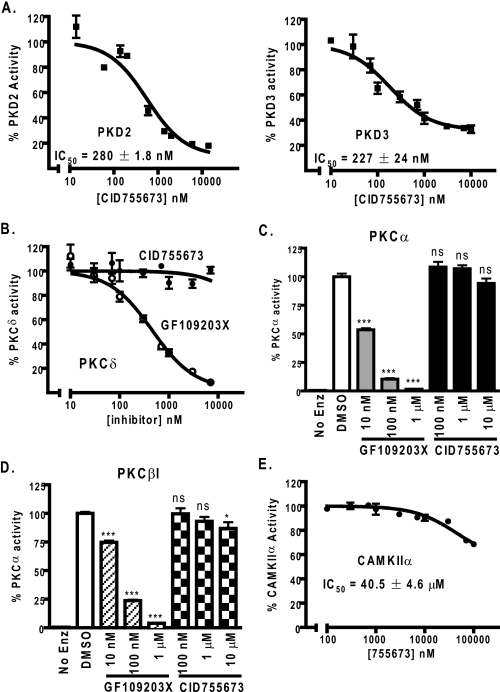
CID755673 Was a Selective PKD Inhibitor—The high homology of the PKD family members stimulated our interest in examining the effects of CID755673 on two other closely related kinases: PKD2 and PKD3. In the radiometric kinase assays, CID755673 inhibited full-length human PKD2 and PKD3 with IC50 values of 280 ± 1.8 (n = 2) and 227 ± 24 nm (n = 3), respectively (Fig. 3A). Thus, CID755673 inhibited all three PKD isoforms with similar potency.
FIGURE 3.
Selectivity of CID755673. A, PKD isoform selectivity of CID755673. Inhibition of human recombinant PKD2 and PKD3 by CID755673 was assayed by the radiometric PKD kinase assay. The experiment was repeated three times and a representative graph is shown. B, CID755673 did not inhibit PKCδ. Inhibitory activity of CID755673 for PKCδ was measured using a radiometric PKC kinase assay. Inhibition of PKCδ by GF109203X was assayed as control. Data are from one of three representative experiments. C and D, inhibition of PKCα (C) or PKCβI (D) was determined at 100 nm, 1 μm, and 10 μm CID755673. As controls, the PKC inhibitor GF109203X potently inhibited PKCα or PKCβI activity. Data are the mean ± S.E. of two independent experiments. ns, not statistically significant; *, p < 0.05; ***, p < 0.001. E, inhibition of CAMKIIα was measured by the radiometric CAMK kinase assay. The experiment was repeated twice and a representative curve is shown.
Because of the commonality of the signaling pathways of PKD with the classical and novel PKC isoforms (for example, PKCα, PKCβI, and PKCδ), the creation of pharmacological tools that would enable the dissection of the role of PKD in biological processes would be valuable. The absence of any evidence for kinase inhibition in the Pubchem data base for CID755673 was encouraging but we are unaware of any attempts to determine its activity against any PKC isoforms. Therefore, we examined the inhibitory activity of CID755673 on several recombinant full-length PKC proteins in the radiometric PKC kinase assay. No inhibitory activity was detected for PKCδ at any concentrations examined (0.007-7 μm) (n = 3), whereas the PKC inhibitor GF109203X inhibited PKCδ with an IC50 of 459 nm (Fig. 3B). Similarly, the compound did not significantly inhibit PKCα activity up to 10 μm (Fig. 3C) and only caused a slight reduction of PKCβI activity (<15%) at the highest inhibitor concentration (10 μm). As controls, GF109203X potently inhibited both PKC isoforms in a concentration-dependent manner (Fig. 3, C and D). These data indicated that CID755673 was a poor inhibitor for PKC isoforms.
PKD is classified as a subfamily of CAMKs. The high homology, especially at the catalytic domain, between PKD and CAMKs led us to examine the inhibitory activity of CID755673 on CAMKs. As shown in Fig. 3E, CID755673 inhibited full-length human CAMKIIα with an IC50 of 40.5 ± 4.6 μm (n = 2), an over 200-fold weaker inhibitory activity for CAMKIIα as compared with PKD1 (Fig. 3A). Thus, the compound was likely a poor inhibitor of CAMKs. Overall, our data thus far supported the conclusion that CID755673 was a selective PKD inhibitor.
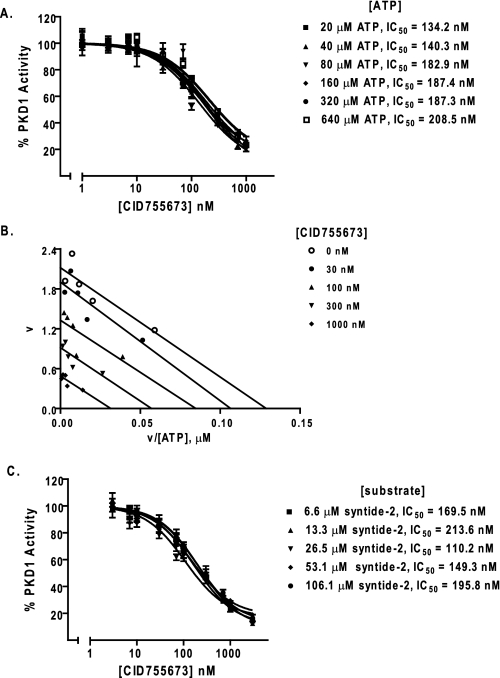
Analysis on the Mode of Action of CID755673—To provide insight on how CID755673 inhibited PKD1, we examined the effects of increasing concentrations of ATP or substrate had on compound inhibition. As indicated in Fig. 4A, the IC50 value for CID755673 was not significantly affected by ATP concentrations ranging from 20 to 640 μm. Next, Eadie-Hofstee plots were generated by plotting the reaction velocities (v) as a function of the velocity versus ATP concentration ratio (v/[ATP]) at different concentrations of the compound. The points were fitted by linear regression. As shown Fig. 4B, the lines generated at different inhibitor concentrations were parallel, indicating that the Km (the slope of the line is -Km) of substrate phosphorylation remained constant, whereas Vmax (the intercept with the y axis) decreased with increased inhibitor concentrations. These results indicated that CID75573 was not an ATP-competitive inhibitor.
FIGURE 4.
CID755673 was not competitive with ATP or substrate for PKD1 inhibition. A, ATP competition analysis. PKD1 kinase activity was measured as a function of increasing doses of CID755673 in the presence of varying concentrations of ATP. B, Eadie-Hofstee plot analysis. The reaction velocity (v) was plotted as a function of the velocity versus ATP concentration ratio (v/[ATP]) for each concentration of CID755673. The points were fitted by linear regression analysis. C, substrate competition analysis. Inhibitory activity of CID755673 was determined in the presence of varying concentrations of substrate peptide syntide-2. Each point of the curves represent the average of triplicate determinations.
Next, we examined whether increasing concentrations of the minimal substrate syntide-2 altered the CID755673 inhibitory activity. As indicated in Fig. 4C, the IC50 values of the inhibitor obtained over a range of substrate concentrations (6.6-106.1 μm) were similar. Further analysis of these data using Michaelis-Menten and visualization of these data using a Eadie-Hofstee plot indicated complexity with respect to substrate competition (data not shown). Overall, our data clearly indicated that the inhibition was non-competitive with respect to ATP, whereas it was complex with respect to the substrate syntide-2.
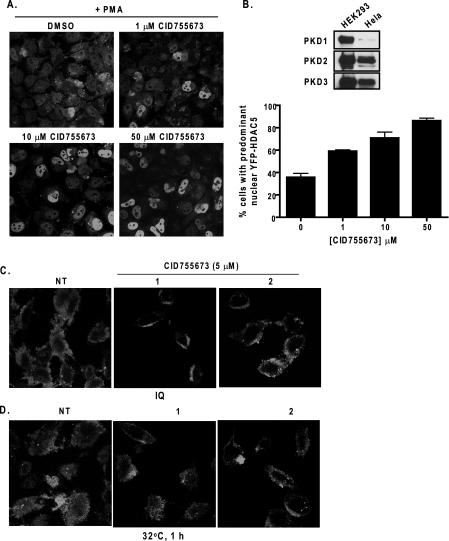
Effects of CID755673 on HDAC5 Localization and VSVG-GFP Trafficking—An effective PKD inhibitor should abrogate PKD-mediated cellular functions. PKD has been shown to directly regulate the class IIa HDACs (HDAC4, -5, -7, and -9), which control the activity of the myocyte enhancer factor-2 transcription factor that controls muscle-specific and stress-responsive gene expression (35). The activation of PKD results in the phosphorylation, nuclear export, and inactivation of the class IIa HDAC5, and this process has been implicated in pathological cardiac remodeling (13, 14). Phorbol esters trigger the nuclear export of HDAC5 in part through the activation of PKD (13). Here, the effect of CID755673 on PMA-induced nuclear export of HDAC5 was examined in HeLa cells. The presence of PKD (mainly PKD2 and PKD3) in HeLa cells was first confirmed by Western blot analysis (Fig. 5B, upper panels). Next, a yellow fluorescent protein (YFP)-tagged HDAC5 was transiently transfected in HeLa cells. At the basal state, YFP-HDAC5 was localized predominantly in the nucleus. PMA stimulation induced gradual nuclear export of HDAC5. After 4 h PMA treatment, the majority of transfected cells displayed even cytosolic and nuclear or predominant cytosolic distribution of HDAC5. Pretreatment with CID755673 significantly blocked PMA-induced nuclear export of HDAC5. The percentage of cells exhibiting predominant HDAC5 increased in a concentration-dependent manner in response to CID755673 (Fig. 5, A and B).
FIGURE 5.
Effects of CID755673 on HDAC5 localization and VSVG-GFP trafficking. A, effect of CID755673 on PMA-induced nuclear exclusion of HDAC5. HeLa cells were transiently transfected with YFP-HDAC5. Two days after transfection, cells were pretreated with or without different concentrations of CID755673 for 45 min, following by the addition of PMA at 1 μm for 4 h. Cells were fixed and imaged under fluorescence confocal microscope. B, quantitative analysis of YFP-HDAC5 nuclear localization. Percent cells with predominant nuclear YFP-HDAC5 were determined. Data were obtained from a total of 400 randomly captured cells in a given experimental condition. Upper panel, the expression of endogenous PKD isoforms in HeLa cells was evaluated by Western blot analysis. HEK293 lysates were probed as positive control. C and D, effect of CID755673 on ilimaquinone-induced Golgi fragmentation and on trafficking of VSVG-GFP from the Golgi to the plasma membrane. HeLa cells were transiently transfected with the VSVG-GFP plasmid. Two days after transfection, cells were incubated for 2 h at 40 °C, followed by incubation at 20 °C for 30 min to trap the VSVG-GFP protein to the Golgi. Cells were then pretreated with or without CID755673 (5 μm) at 20 °C, followed by a 2-h incubation at 20 °C with 30 μm ilimaquinone to induce Golgi fragmentation (C) or by a 1-h incubation at 32 °C to allow protein transport to the cell surface (D). Images of VSVG-GFP distribution were captured under a fluorescence confocal microscope. The experiments were repeated at least three times and representative images are shown. NT, no inhibitor treatment or DMSO only.
PKD activity is required for the maintenance of proper Golgi structure and protein transport from the Golgi to the plasma membrane (10, 11). The marine sponge metabolite ilimaquinone blocks protein transport by inducing reversible Golgi fragmentation, which is a PKD-dependent process (12). Thus, we verified cellular PKD inhibition by CID755673 using a GFP-tagged temperature-sensitive mutant of ts045 VSVG as a marker for Golgi fragmentation and vesicular trafficking from the Golgi. At the restrictive temperature (40 °C), ts045 VSVG is reversibly misfolded and retained in the endoplasmic reticulum. When the cells are shifted from the restrictive temperature to 20 °C, ts045 VSVG is transported out of the endoplasmic reticulum and trapped to the Golgi. The trafficking of VSVG from the endoplasmic reticulum or Golgi to the plasma member is then allowed by shifting the cells to the permissive temperature (32 °C). HeLa cells transiently transfected with ts045 VSVG-GFP were incubated at 40 °C for 2 h and then at 20 °C for 30 min. The VSVG-GFP trafficking was evaluated upon ilimaquinone treatment or 32 °C incubation. Consistent with a previous report (12, 36), treatment of cells with ilimaquinone at 20 °C caused vesiculation of the Golgi stacks, appearing as diffused small vesicles containing VSVG-GFP throughout the cytoplasm (Fig. 5C, left panel). The effect of ilimaquinone was substantially blocked by pretreatment with 5 μm CID755673 (Fig. 5C). Similarly, CID755673 significantly inhibited the transport of VSVG-GFP from the Golgi to the cell surface at the 32 °C permissive temperature (Fig. 5D). Taken together, these results indicated that CID755673 was effective at blocking PKD-mediated protein transport.
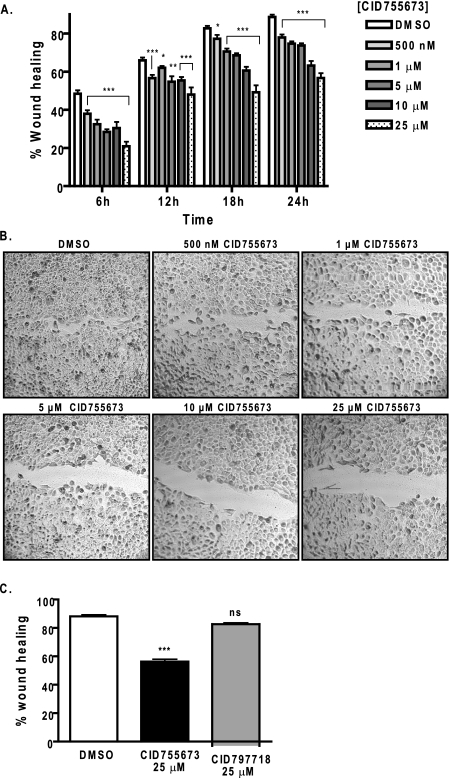
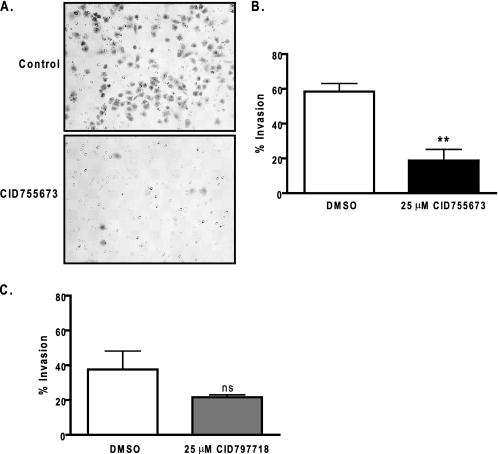
Effects of CID755673 on Tumor Cell Migration and Invasion—PKD has been shown to play important roles in cell motility, invasion, and adhesion (19-22). The effects of PKD on tumor cell migration and invasion were evaluated in prostate cancer cell lines. The expression of PKD in these cell lines has been demonstrated in our previous study with PKD1 and PKD2 predominantly expressed in LNCaP, whereas PKD2 and PKD3 were mainly expressed in DU145 and PC3 cells (29). To determine whether PKD regulates prostate cancer cell migration and invasion, two different studies were conducted. The effect of CID755673 on cell migration was examined by wound healing assays in DU145 prostate cancer cells. Confluent DU145 monolayers were wounded and then treated with varying concentrations of the PKD1 inhibitor. CID755673 inhibited wound closure in a concentration-dependent manner at all time points with ∼50% inhibition at 25 μm, approximately (Fig. 6, A and B). In contrast, the weaker analogue CID797718 did not significantly inhibit wound closure at 25 μm as compared with the vehicle control (DMSO) (Fig. 6C). The inhibitory effects of CID755673 on cell migration were further supported by a transwell cell migration assay, where CID755673 at 25 μm caused 2-5-fold reduction in migration of DU145 and LNCaP cells (supplemental Fig. 1S). CID755673 also significantly inhibited tumor cell invasion as examined by a Matrigel invasion assay. The inhibitor applied at 25 μm caused a 3-fold reduction in invasion of DU145 cells (invasion index = 0.32) (Fig. 7, A and B), whereas the weaker analogue CID797718 at 25 μm had no significant effects (Fig. 7C). Taken together, these results indicated that CID755673 potently blocked prostate cancer cell migration and invasion.
FIGURE 6.
Effects of CID755673 on tumor cell migration. A, wound healing assay. DU145 cells were grown to confluence in 6-well plates. Monolayer was wounded and imaged immediately (0 h). Growth media containing a vehicle (DMSO) or varying concentrations of CID755673 was added. Wound closure was recorded every 6 h up to 24 h. The width of the wound is the average of 9 determinations per time point. Percent wound healing was calculated at each time point as described under “Experimental Procedures.” B, after 24 h, cells were stained with 0.25% crystal violet. Phase-contrast images of the final wounds were taken at ×100 magnification. The experiment was repeated three times and a representative experiment is shown. C, the weaker analogue CID797718 did not inhibit wound closure. DU145 monolayer was wounded and incubated in the presence of the vehicle (DMSO), 25 μm CID755673 or CID797718. Wound closure was measured after 24 h. Data represent one of two independent experiments. ns, not statistically significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 7.
Effects of CID755673 on tumor cell invasion. A, tumor cell invasion was evaluated by a Matrigel invasion assay. A fixed number of DU145 cells (1.0 × 105/ml) were seeded into the upper control or invasion chamber. After 22 h, non-invading cells were removed and cells that invaded through the Matrigel were fixed, stained, and photographed under a microscope. Magnification, ×200. B, percent invasion is expressed as the number of cells that invaded through the Matrigel matrix relative to the number of cells that migrated through the control insert. Cell number is determined by counting total cell number in 10 random fields. The experiment was repeated twice and a representative experiment is shown. C, the weaker analogue CID797718 did not significantly inhibit tumor cell invasion. DU145 cell invasion was measured using Matrigel invasion chambers after incubating with DMSO or 25 μm CID797718 for 22 h. ns, not statistically significant; **, p < 0.01.
Effects of CID755673 on Tumor Cell Proliferation and Cell Cycle Distribution—PKD has been shown to promote cell proliferation. The activation of the PKC/PKD pathway by various stimuli potentiates DNA synthesis and induces cell proliferation in normal and cancer cells (15, 17). Our previous study demonstrated that knockdown of PKD3 inhibits cell proliferation in prostate cancer cells (29). Therefore, the effect of the PKD inhibitor CID755673 was evaluated in prostate cancer cells. Cell proliferation was determined by counting the cell number for 6 consecutive days in the presence or absence of the inhibitor. CID755673 concentration dependently inhibited LNCaP and PC3 cell proliferation (Fig. 8, A and B). In contrast, the analogue CID797718 was much weaker at blocking PC3 cell proliferation as compared with the parental compound at 25 μm (Fig. 8B, right panel). Additionally, more potent growth-inhibitory effects of CID755673 were observed in PC3 cells as compared with LNCaP cells, accompanying significant morphological changes from small polygonal or spindle-like shapes to large, flat and round shapes of cells (Fig. 8C). The anti-proliferative effects of CID755673 were further confirmed by a CellTiter-Glo Luminescent Cell Viability Assay in LNCaP cells, as shown in supplemental Fig. 2S. The effects of CID755673 on cell cycle distribution were analyzed to provide insight into the anti-proliferative effects of the compound. As shown in Fig. 8D, CID755673 caused G2/M phase cell cycle arrest in a concentration-dependent manner in PC3 cells. Taken together, our data indicated that CID755673 was a potent inhibitor of prostate cancer cell proliferation.
FIGURE 8.
CID755673 inhibited cell proliferation in LNCaP and PC3 cells. A and B, LNCaP (A) or PC3 (B) cells were plated in triplicates in 24-well plates. Cells were allowed to attach overnight. A cell count at day 1 was made, and then either a vehicle (DMSO) or CID755673 at the indicated concentrations was added. Cells were counted daily for a total of 6 days. Fresh media and inhibitor were added every 2 days. The means of triplicate determinations were plotted over time. The experiment was repeated twice and results from one representative experiment are shown. Right panel (B), the effect of the weaker analogue CID797718 on PC3 cell proliferation. Results from one of two independent experiments are shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, CID755673 induced apparent morphological changes. Phase-contrast images of PC3 cells after 6 days of CID755673 treatment are shown. D, CID755673-induced G2/M phase cell cycle arrest. PC3 cells were incubated with a vehicle (DMSO) or varying concentrations of CID755673 for 6 days. Media with fresh inhibitors were replenished daily. Cell cycle distribution was evaluated by flow cytometry after propidium iodide labeling of fixed cells. One of two independent experiments is shown.
DISCUSSION
In this study, we report the discovery of the first PKD-specific inhibitor CID755673. This small heterocycle was identified by screening the full panel of the NIH small molecule repository (196,173 compounds) using an IMAP-based fluorescence polarization assay (27). The inhibitory activity of CID755673 was confirmed through a series of secondary screening assays including a radiometric PKD kinase assay. Of the two compounds examined in this study, CID755673 exhibited higher inhibitory activity for PKD1, whereas CID797718, a weaker analogue of CID755673, was 10 times less potent and significantly less selective. Data mining of Pubchem indicated that compound CID755673 had been tested in a total of 221 bioassays, however, was identified as a primary hit in only 3 assays. Besides the IMAP-based FP HTS assay for PKD1, this compound was also identified in a single concentration primary screening for an inhibitor of highly pathogenic avian influenza (H5N1) infection and for inhibition of Cdc25B catalytic domain protein-tyrosine phosphatase by us. However, in subsequent inhibitor validation assays for both targets the compound was not confirmed as an inhibitor. The compound is also a poor inhibitor for a number of protein kinases including AKT, PLK1, CAK, CAMKIIα, and several PKC isoforms. Thus, based on our current analysis, PKD appears to be a primary target of CID755673.
To date several PKD1 inhibitors have been reported. The earliest include staurosporine (IC50 = 40 nm) and staurosporine-derived compounds such as K252a (IC50 = 7 nm) and Gö6976 (IC50 = 20 nm) (23). The indolocarbazole Gö6976, however, is more specific for PKC (24) and both K252a and staurosporine inhibit many other protein kinases. In addition, the isoquinoline sulfonamide H89 inhibits PKD1 at 0.5 μm, which is significantly higher than its activity for PKA (IC50 = 48 nm) and lower than that for PKCs (IC50 = 32 μm) (25). Although the potency of CID755673 did not supersede the previously known kinase inhibitors, its specificity was considerably superior and PKD appeared to be the only target that was inhibited by this compound at nanomolar concentration. We confirmed the selectivity of CID755673 for PKD against a number of serine/threonine kinases including a member of the CAMK family that is highly homologous to PKD. The lack of inhibitory activity for PKC is an important finding. Because PKD acts in a PKC/PKD pathway in most cellular systems, an inhibitor that targets PKD specifically will enable the dissection of PKD-controlled pathways and biological processes. We examined the inhibitory activities of the compound for two classical (PKCα and PKCβI) and one novel PKC (PKCδ) isoform, which have been implicated in activating PKD. CID755673 exhibited very weak or no inhibitory activity for these PKC isoforms, supporting the notion that CID755673 is capable of selectively blocking PKD-mediated cellular responses without altering the activities of the upstream broad-spectrum kinase PKC. Therapeutically, to treat diseases with deregulated diacylglycerol/PKC signaling, this may result in fewer side effects and a higher therapeutic index as compared with strategies, which target PKC.
Our data indicate that CID755673 is clearly not competitive with ATP for enzyme inhibition. The non-competitive nature of the inhibitor is supported by two findings: 1) varying ATP concentrations did not significantly alter the IC50 values of CID755673; 2) visualization of the data by the Eadie-Hofstee plot showed that increasing concentrations of the inhibitor did not alter Km but decreased Vmax, indicative of non-competitive inhibition. However, the inhibition was complex with respect to competition with the peptide substrate syntide-2 (data not shown). Our analysis of a known protein substrate of PKD1, myelin basic protein, was inconclusive due to the fact that myelin basic protein directly inhibited PKD1 activity (37).4 Analysis of other PKD protein substrates will be needed to confirm this finding regarding a potential mechanism of action. It is possible that the inhibitor works by modifying/blocking a unique site in PKD that is critical for catalysis but not for ATP or potentially substrate binding. Unique structural elements that regulate PKD enzyme activity have been reported, such as an acidic domain that may be involved in the activation and stabilization of an active state of PKD1 (37). Detailed structural analysis of inter- or intramolecular interactions in PKD will provide more insights. An interesting relevant observation is that the inhibition curves of CID755673 consistently leveled off at ∼15-20% PKD1/PKD2 or 37% PKD3 activity and increased inhibitor concentrations did not result in further inhibition. The plateaued inhibition at high inhibitor concentrations may indicate the formation of a non-competitive enzyme-inhibitor complex with low kinase activity. Finally, our key finding in this study is the lack of competition with respect to ATP. ATP-competitive inhibitors are often associated with low potency in cells or animals, lack of specificity, and development of drug resistance in patients. The fact that CID755673 is a non-ATP-competitive inhibitor implies that this compound may have many therapeutic advantages.
PKD is known to regulate several important biological processes. One of the best characterized functions is the regulation of protein transport. Both dominant negative PKD and H89 have been shown to inhibit protein transport from the Golgi to the plasma membrane (12). Using ts045 VSVG as a probe, we demonstrated that CID7555673 potently inhibited ilimaquinone-induced Golgi vesiculation as well as the transport of VSVG from the Golgi to the plasma membrane at the permissive temperature. We also examined the activity of the PKD inhibitor on nuclear export of HDAC5, a well characterized PKD substrate. The nuclear exclusion of HDAC5 activates myocyte enhancer factor-2 and leads to cardiac hypertrophy, thus the control of nuclear localization of HDAC5 is a key event in this biological response. Our data demonstrated a significant inhibitory effect of CID755673 in PMA-induced HDAC5 nuclear export, implying that the compound could be a valuable tool for establishing treatments of cardiac diseases resulted from pathological cardiac remodeling. Overall, our data demonstrated that CID755673 was capable of inhibiting PKD1-mediated cellular responses, providing the functional evidence for specific and effective targeting of PKD1 by this compound.
In addition to the biochemical and functional characterization of the inhibitor, application of the compound to cultured cells revealed novel biological actions of PKD in prostate cancer cells. Our data showed that CID755673 was a potent inhibitor of cell proliferation, cell migration, and invasion in several prostate tumor cell lines compared with its weaker analogue CID797718. The anti-proliferative effects of CID755673 coincided with our previous finding that the knockdown of PKD3 inhibits cell proliferation in PC3 cells (29). Interestingly, CID755673 induced G2/M phase cell cycle arrest, which differed from our previous results that depletion of PKD3 primarily resulted in accumulation of cells in the G0/G1 phase of the cell cycle. Because both PKD2 and PKD3 could be targets of CID755673 in PC3 cells, the G2/M arrest could be in part attributed to the inhibition of PKD2. Overall, these results provide strong support for targeting PKD for prostate cancer therapy and the compound CID755673 may be a valuable therapeutic agent.
CID755673 inhibited PKD1 at 182 nm in the in vitro assays. In cultured cells, it inhibited several biological responses (migration, invasion, and cell proliferation) with an IC50 between ∼10 and 30 μm. Up to 3-fold differences in IC50 values were noted between different cell lines (LNCaP versus PC3) for the same biological response (cell proliferation). The variations in IC50 between cell lines may be attributed to multiple factors including the relative levels and activities of endogenous PKDs. Our previous study suggests that PKD3, a major PKD isoform expressed in PC3 and DU145 cells, is hyperactive, which may explain the more potent anti-proliferative effects of CID755673 in these cells versus the non-PKD3-expressing LNCaP (29). Another contributing factor is that PKD inhibition by the compound was incomplete. Comparing the plateaued inhibitory activities of the inhibitor for PKD1, PKD2, and PKD3, we noted statistically significant differences between the average plateaued values of PKD1/PKD2 versus PKD3 at concentrations beyond 3 μm (***, p < 0.001 for PKD1 versus PKD3, and **, p < 0.01 for PKD2 versus PKD3 by an unpaired Student's t test), indicating the maximal extent of inhibition by CID755673 for PKD1/PKD2 is significantly greater than that for PKD3. This difference may have significant impact in its cellular effects when isoforms of PKD are differentially expressed. It is possible that cells expressing only PKD1 or PKD2 may be more sensitive to the compound than cells expressing only PKD3. Finally, it should be noted that our data do not exclude the possibility that CID755673 has other cellular targets, inhibition of which could be responsible for the variation in potency.
Analysis of CID755673 in intact cells indicates that the compound possesses minimal cytotoxicity and does not induce any measurable apoptosis. Percent cell death determined by trypan blue dye exclusion after 6-day incubation at 25 μm CID755673 showed no significant differences in % cell death for control (DMSO) versus inhibitor-treated cells (∼1.5% of total cell numbers) in LNCaP, PC3, and DU145 cells. Moreover, CID755673 has been assayed in at least 11 mammalian cell-based Pubchem AIDs including AID 430, 431, 463, 598, 620, 648, 719, 818, 827, 902, and 924 at concentrations ranging from 4 to 50 μm and was not found to be cytotoxic. Additionally, we did not observe significant apoptosis associated with the inhibitor treatment. In the cell cycle analysis by flow cytometry (Fig. 8D), the cell population generally associated with apoptosis, namely sub-G0/G1, remained marginal in cells treated with 1-10 μm CID755673 up to 6 days, and was only slightly increased with 25 μm. Overall, our data indicate that the compound is cytostatic and does not cause significant cell death.
In summary, we report the discovery of a potent and selective PKD1 inhibitor, CID755673. Biochemical and functional analysis indicated that the compound inhibited all three PKD isoforms and was effective at blocking PKD-mediated cell functions. The inhibitor also revealed novel tumor-promoting functions of PKD isoforms in prostate cancer cells. It is conceivable that CID755673 will be a powerful small molecule tool for assessing the specific roles of PKD in biological processes, and most importantly, for therapeutic applications.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R03 MH082038-01, R01 DK066168-01, 1U54MH074411, and GM067082. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PKD, protein kinase D; CID755673, 7-hydroxy-2,3,4,5-tetrahydro-[1]benzoxolo[2,3-c]azepin-1-one; CID797718, 9-hydroxy-1,2,3,4-tetrahydrochromeno[3,4-b]pyridin-5-one; IMAP, immobilized metal affinity for phosphochemical; FP, fluorescence polarization; HTS, high throughput screening; TR-FRET, time-resolved fluorescence resonance energy transfer; PKC, protein kinase C; PMA, phorbol 12-myristate 13-acetate; HDAC, histone deacetylase; VSVG, vesicular stomatitis virus glycoprotein; PLK1, polo-like kinase 1; CAK, CDK activating kinase; AKT/PKB, protein kinase B; DMSO, dimethyl sulfoxide; GFP, green fluorescent protein; YFP, yellow fluorescent protein.
C. R. LaValle, E. R. Sharlow, P. Wipf, J. S. Lazo, and Q. J. Wang, unpublished data.
References
- 1.Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002) Science 298 1912-1934 [DOI] [PubMed] [Google Scholar]
- 2.Wang, Q. J. (2006) Trends Pharmacol. Sci. 27 317-323 [DOI] [PubMed] [Google Scholar]
- 3.Hayashi, A., Seki, N., Hattori, A., Kozuma, S., and Saito, T. (1999) Biochim. Biophys. Acta 1450 99-106 [DOI] [PubMed] [Google Scholar]
- 4.Johannes, F. J., Prestle, J., Eis, S., Oberhagemann, P., and Pfizenmaier, K. (1994) J. Biol. Chem. 269 6140-6148 [PubMed] [Google Scholar]
- 5.Sturany, S., Van Lint, J., Muller, F., Wilda, M., Hameister, H., Hocker, M., Brey, A., Gern, U., Vandenheede, J., Gress, T., Adler, G., and Seufferlein, T. (2001) J. Biol. Chem. 276 3310-3318 [DOI] [PubMed] [Google Scholar]
- 6.Valverde, A. M., Sinnett-Smith, J., Van Lint, J., and Rozengurt, E. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8572-8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldron, R. T., and Rozengurt, E. (2003) J. Biol. Chem. 278 154-163 [DOI] [PubMed] [Google Scholar]
- 8.Zugaza, J. L., Sinnett-Smith, J., Van Lint, J., and Rozengurt, E. (1996) EMBO J. 15 6220-6230 [PMC free article] [PubMed] [Google Scholar]
- 9.Rozengurt, E., Rey, O., and Waldron, R. T. (2005) J. Biol. Chem. 280 13205-13208 [DOI] [PubMed] [Google Scholar]
- 10.Liljedahl, M., Maeda, Y., Colanzi, A., Ayala, I., Van Lint, J., and Malhotra, V. (2001) Cell 104 409-420 [DOI] [PubMed] [Google Scholar]
- 11.Hausser, A., Storz, P., Martens, S., Link, G., Toker, A., and Pfizenmaier, K. (2005) Nat. Cell Biol. 7 880-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamora, C., Yamanouye, N., Van Lint, J., Laudenslager, J., Vandenheede, J. R., Faulkner, D. J., and Malhotra, V. (1999) Cell 98 59-68 [DOI] [PubMed] [Google Scholar]
- 13.Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N., and McKinsey, T. A. (2004) Mol. Cell. Biol. 24 8374-8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fielitz, J., Kim, M. S., Shelton, J. M., Qi, X., Hill, J. A., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3059-3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guha, S., Rey, O., and Rozengurt, E. (2002) Cancer Res. 62 1632-1640 [PubMed] [Google Scholar]
- 16.Sinnett-Smith, J., Zhukova, E., Hsieh, N., Jiang, X., and Rozengurt, E. (2004) J. Biol. Chem. 279 16883-16893 [DOI] [PubMed] [Google Scholar]
- 17.Wong, C., and Jin, Z. G. (2005) J. Biol. Chem. 280 33262-33269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storz, P., and Toker, A. (2003) EMBO J. 22 109-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods, A. J., White, D. P., Caswell, P. T., and Norman, J. C. (2004) EMBO J. 23 2531-2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prigozhina, N. L., and Waterman-Storer, C. M. (2004) Curr. Biol. 14 88-98 [DOI] [PubMed] [Google Scholar]
- 21.Bowden, E. T., Barth, M., Thomas, D., Glazer, R. I., and Mueller, S. C. (1999) Oncogene 18 4440-4449 [DOI] [PubMed] [Google Scholar]
- 22.Jaggi, M., Rao, P. S., Smith, D. J., Wheelock, M. J., Johnson, K. R., Hemstreet, G. P., and Balaji, K. C. (2005) Cancer Res. 65 483-492 [PubMed] [Google Scholar]
- 23.Gschwendt, M., Dieterich, S., Rennecke, J., Kittstein, W., Mueller, H. J., and Johannes, F. J. (1996) FEBS Lett. 392 77-80 [DOI] [PubMed] [Google Scholar]
- 24.Martiny-Baron, G., Kazanietz, M. G., Mischak, H., Blumberg, P. M., Kochs, G., Hug, H., Marme, D., and Schachtele, C. (1993) J. Biol. Chem. 268 9194-9197 [PubMed] [Google Scholar]
- 25.Johannes, F. J., Prestle, J., Dieterich, S., Oberhagemann, P., Link, G., and Pfizenmaier, K. (1995) Eur. J. Biochem. 227 303-307 [DOI] [PubMed] [Google Scholar]
- 26.Stewart, J. R., Christman, K. L., and O'Brian, C. A. (2000) Biochem. Pharmacol. 60 1355-1359 [DOI] [PubMed] [Google Scholar]
- 27.Sharlow, E. R., Leimgruber, S., Yellow-Duke, A., Barrett, R., Wang, Q. J., and Lazo, J. S. (2008) Nat. Protoc. 3 1350-1363 [DOI] [PubMed] [Google Scholar]
- 28.Sharlow, E. R., Leimgruber, S., Shun, T. Y., and Lazo, J. S. (2007) Assay Drug. Dev. Technol. 5 723-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, J., Deng, F., Singh, S. V., and Wang, Q. J. (2008) Cancer Res. 68 3844-3853 [DOI] [PubMed] [Google Scholar]
- 30.Lu, G., Chen, J., Espinoza, L. A., Garfield, S., Toshiyuki, S., Akiko, H., Huppler, A., and Wang, Q. J. (2007) Cell. Signal. 19 867-879 [DOI] [PubMed] [Google Scholar]
- 31.Chen, J., Lu, G., and Wang, Q. J. (2005) Mol. Pharmacol. 67 152-162 [DOI] [PubMed] [Google Scholar]
- 32.Waldron, R. T., Rey, O., Iglesias, T., Tugal, T., Cantrell, D., and Rozengurt, E. (2001) J. Biol. Chem. 276 32606-32615 [DOI] [PubMed] [Google Scholar]
- 33.Iglesias, T., Waldron, R. T., and Rozengurt, E. (1998) J. Biol. Chem. 273 27662-27667 [DOI] [PubMed] [Google Scholar]
- 34.Matthews, S. A., Rozengurt, E., and Cantrell, D. (1999) J. Biol. Chem. 274 26543-26549 [DOI] [PubMed] [Google Scholar]
- 35.McKinsey, T. A. (2007) Cardiovasc. Res. 73 667-677 [DOI] [PubMed] [Google Scholar]
- 36.Sonoda, H., Okada, T., Jahangeer, S., and Nakamura, S. (2007) J. Biol. Chem. 282 34085-34092 [DOI] [PubMed] [Google Scholar]
- 37.Gschwendt, M., Johannes, F. J., Kittstein, W., and Marks, F. (1997) J. Biol. Chem. 272 20742-20746 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.