Abstract
The paradigm of activation via ordered recruitment has evolved into a complicated picture as the influence of coactivators and chromatin structures on gene regulation becomes understood. We present here a comprehensive study of many elements of activation of ADH2 and FBP1, two glucose-regulated genes. We identify SWI/SNF as the major chromatin-remodeling complex at these genes, whereas SAGA (Spt-Ada-Gcn5-acetyltransferase complex) is required for stable recruitment of other coactivators. Mediator plays a crucial role in expression of both genes but does not affect chromatin remodeling. We found that Adr1 bound unaided by coactivators to ADH2, but Cat8 binding depended on coactivators at FBP1. Taken together, our results suggest that commonly regulated genes share many aspects of activation, but that gene-specific regulators or elements of promoter architecture may account for small differences in the mechanism of activation. Finally, we found that activator overexpression can compensate for the loss of SWI/SNF but not for the loss of SAGA.
The paradigm of eukaryotic gene activation centers on activators recognizing and binding to unique sequences of DNA and then recruiting coactivators and the transcription machinery (1). The order of recruitment events has been determined for several yeast genes, including HO (2), PHO5 (3–6), and the GAL genes (7–9). Based on these and other studies, the idea of activation has evolved beyond simple ordered recruitment.
One of the reasons for this evolving picture of activation is the finding that many coactivator complexes can play multiple roles. For example SAGA4 functions as a histone-acetyltransferase (HAT) at the HO promoter (10), but it is required for a non-HAT function at the GAL1 promoter (7). In cases such as these, knowing the order in which SAGA arrives at a promoter is insufficient for understanding its role in activation. The dual nature of coactivators emphasizes the need to look not only at their recruitment to the promoter, but also at the functional consequences of this recruitment. Glucose-regulated genes, under the control of Snf1, the homolog of the mammalian AMP-activated kinase, provide a model for such comprehensive studies. In response to glucose starvation, ∼200 genes are activated by Snf1, many of which depend on the transcription factors Adr1 and Cat8 (11). Previous work in our laboratory has established that SAGA, SWI/SNF, and Mediator are recruited by these two activators upon derepression (12). However, the specific roles of these coactivators and their interactions at these genes remain undetermined.
This subset of genes allowed us to address several factors that may influence the mechanisms of activation. First, we focused on two genes, ADH2 and FBP1, which have differential dependences on both the activators Adr1 and Cat8 (13), and on the repressor Mig1 (14), allowing us to determine if trends were regulator-specific. Second, these promoters have established chromatin structures providing a basis for examining the impact of promoter architecture on the events leading up to activation (12, 15, 16). Finally, because these genes are both regulated by Snf1, we could ask whether or not genes that share a common upstream regulator also share a common mechanism of activation.
We sought to delineate the steps leading up to activation, including the roles of the coactivators and the order in which they appear at the promoter, using a combination of expression, chromatin immunoprecipitation (ChIP), and chromatin-remodeling assays. The results from these combined studies did not always support the paradigm of activated transcription; they suggest that coactivator recruitment is possible without associated changes in chromatin remodeling or gene expression, and furthermore that chromatin remodeling does not necessarily lead to gene activation. In addition we clearly demonstrate that one role for coactivators is to stabilize Cat8 binding. We provide evidence that SAGA is required for forming a scaffold at the promoter that is important for downstream recruitment events, and that SWI/SNF is required for chromatin remodeling. Our results suggest that there are both shared and unique aspects in the mechanism of activation for co-regulated genes and that there are also redundancies in this mechanism.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth of Cultures—All strains used in this study were derived from W303 and are described in supplemental Table S1. Deletions of activators and epitope tagging of all protein components were introduced according to previously published work (17, 18). The med17ts strain was constructed by first transforming with YCpsrb4–138(leu2::URA3), which was created from RY2882 (srb4–138 LEU2 ars cen), a gift from S. Hahn, followed by deletion of chromosomal MED17 according to a previous study (17). For the med17 ts experiments, cultures were grown overnight at room temperature and then shifted to 37 °C for 30 min before taking the repressed sample; cultures were then spun down in a pre-warmed centrifuge and resuspended in 37 °C low glucose media and grown at 37 °C. Wild-type strains were grown in the same manner for comparison to eliminate indirect effects of the high temperature. Strains overexpressing Adr1 were created by introducing a plasmid (pNKA1-U) based on pKD17, which expresses Adr1 from the ADH1 promoter, with only a minor modification from its original form (19), in the various coactivator mutant backgrounds. These strains were grown in synthetic media lacking the appropriate amino acid for plasmid selection with either 5% glucose (repressed) or 0.05% glucose (derepressed). In all experiments, repressed samples (R) were isolated just before cells were switched to low glucose media.
ChIP and Real-time PCR (QPCR)—ChIP was performed as previously described, using both dimethyl adipimate (Pierce Chemicals) and formaldehyde (FMA) as cross-linking agents (except where noted) and using QPCR instead of standard PCR (13). Additional ChIP for Cat8 was performed with two variations: 1) FMA was added directly to the culture at a final concentration of 1% for 15 min and then treated as above and 2) EGS (Pierce Chemicals) was used in place of dimethyl adipimate at a final concentration of 1.5 mm, with all other steps the same as above. Monoclonal antibodies against c-Myc (9E10, Santa Cruz Biotechnology sc-40), HA (F-7, Santa Cruz Biotechnology sc-7293), and FLAG (Sigma F3165) epitopes were used for most immunoprecipitations except for pol II, which was immunoprecipitated using the 8WG16 antibody (Abcam ab817). To carry out ChIP for five proteins in the same cell lysates for the time-course experiment (Fig. 6), an antibody against Snf2 (gift from J. Reese) was used, as well as IgG-Sepharose to bind tandem affinity purification epitopes. Sequences of primers used for QPCR are available upon request. QPCR was performed on an MJ Research DNA Engine using SYBR Green SensiMix (Quantance Ltd., London, UK). Experiments were performed in biological duplicate or triplicate at two time points of derepression, and all results were averaged together. Values were calculated as the ratio of ChIP to input at the specific locus divided by the ratio at the telomere region. The associated error results from the standard deviation of the biological replicates.
FIGURE 6.
Recruitment of coactivators is nearly simultaneous. Occupancy of transcription factors and coactivators in a wild-type strain (RBY143) at different times of derepression at ADH2 (A) and FBP1 (B). Values, based on QPCR, were normalized to the highest value for each protein over the time course.
mRNA Isolation and QPCR—mRNA was isolated from strains grown in either repressing (YPD (yeast extract/peptone/dextrose) with 5% glucose) or derepressing (YPD with 0.05% glucose) media for the time indicated and processed as detailed in a previous study (12). Samples were prepared from biological triplicates and quantitated in duplicate.
Chromatin Remodeling Assays—NuSA was performed as in Ref. 12 using samples from cells grown in either repressing medium or derepression medium for 4 h (see above). Supercoiling assays were performed as in a previous study (16) using the pLLTY1 plasmid, carrying the –640 to +135 region of the ADH2 gene and the pLLTY3 plasmid in which the ADH2 TATAA box was changed to GAGAA. The Southern blots were probed using either a 32P-labeled probe as in Tachibana (16) or using the AP direct labeling and detection system from GE, following the manufacturer's instructions.
RESULTS
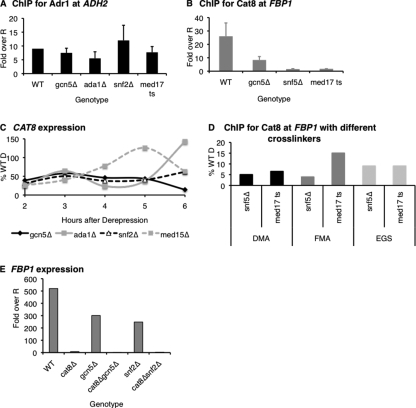
Coactivators Are Required for Stable Cat8 Binding—We previously established that SAGA, SWI/SNF, and Mediator occupy the promoters of ADH2 and FBP1 and contribute to their activation (12). To determine the roles of these coactivators, we first asked whether these complexes are required for the transcription factors Adr1 and Cat8 to bind to the promoters. ChIP assays for Adr1 and Cat8 in coactivator deletion strains showed that Adr1 binding at ADH2 was unaffected by these mutations, but Cat8 binding at FBP1 was reduced (Fig. 1, A and B). This result was surprising in light of the fact that the Cat8 binding site is in a nucleosome-free region at FBP1 (20). Several activators whose binding sites are similarly accessible bind without the aid of coactivators, such as Pho2/Pho4 at PHO8, Swi5 at HO, and Gal4 at GAL1/GAL10 (21–23).
FIGURE 1.
Adr1 and Cat8 show differential dependences on coactivators for stable binding. A, ChIP for Adr1 at ADH2 in coactivator mutants (RBY3, RBY110, RBY111, RBY117, and RBY119). Values, based on QPCR, were first normalized to a negative control locus (TEL) and to an input sample, and then expressed as the -fold over the WT repressed (R) value. B, ChIP for Cat8 at FBP1 in coactivator mutants (CTYTY18, RBY126, RBY127, and RBY123). Values as in A. C, mRNA analysis of CAT8 in WT (W303a) and coactivator mutants (LLTY72, LLTY73, KKTY3, and RBY93). Values, based on QPCR, were first normalized to ACT1 and then expressed as a percent of the maximum WT Derepressed (D) value. D, ChIP for Cat8 with different cross-linkers was performed as described under “Experimental Procedures” with either dimethyl adipimate (DMA) plus formaldehyde (FMA), just FMA, or EGS plus FMA at FBP1 in med17 ts (RBY123) and snf5Δ (RBY127). ChIP/Input values (obtained as in A) were expressed as the percent of ChIP/Input value of the WT strain (RBY135). E, mRNA analysis of FBP1 in WT (W303a), cat8Δ (RBY19), gcn5Δ (EAY12), gcn5Δcat8Δ (EAY16), snf2Δ (KKTY3), and snf2Δcat8Δ (RBY160). Values, based on QPCR, were first normalized to ACT1, and then expressed as the -fold over repressed (R).
To eliminate the possibility that the weak Cat8 signal was due to an indirect effect of deleting coactivators, we monitored CAT8 expression in these strains. It was moderately reduced compared with wild type (Fig. 1C), but ADR1 expression was similarly reduced (supplemental Table S2) with no observable defect in Adr1 binding. Thus it does not seem likely that the lack of binding was due to compromised CAT8 transcription. We also measured Cat8 protein levels by immunoprecipitating tagged Cat8 from equal amounts of cell extract isolated from either wild type or SAGA, SWI/SNF, and Mediator mutant strains and found comparable levels after 4 h of derepression (supplemental Fig. S1). Taken together, this demonstrates that the ChIP results were not due simply to reduced levels of Cat8.
We next asked if Cat8 was still at the promoter but unable to be detected by ChIP due to transient or unstable Cat8 binding in the coactivator mutants. To test this, we repeated the ChIP with two variations: adding formaldehyde directly to the cultures in an attempt to rapidly “trap” any transient binding events, or using EGS, a 16.1-Å-long cross-linker that uses N-hydroxysuccinimide ester chemistry, which has been used to improve weak ChIP signals (24). Both of these methods gave marginally improved results; occupancy of Cat8 in snf5Δ increased from roughly 5% of the wild-type level with the standard assay to ∼10% when EGS was used, and in med17 ts, occupancy increased modestly when EGS was used and nearly doubled when FMA was used (Fig. 1D). This increase suggested that some Cat8 was at the promoter in these mutants, but probably less stably bound than in wild type.
To further confirm our hypothesis that Cat8 was still at the promoter in these mutants despite the low ChIP signal, we asked whether the FBP1 expression in the coactivator mutant strains was Cat8-dependent. When we combined either GCN5 or SNF2 and CAT8 deletions, expression of FBP1 decreased to background (Fig. 1E). The fact that FBP1 expression was still strongly Cat8-dependent in the coactivator mutant backgrounds proves that Cat8 was at the promoters, but with less frequency or stability than in a wild-type strain.
The Role of SAGA—To extend our analysis of the functions of coactivators at glucose-repressed genes we looked in depth at each complex using a three-pronged approach: gene expression, ChIP, and chromatin remodeling assays. We began with the deletion of the HAT component of SAGA, Gcn5. Gcn5 is required for changes in nucleosome position that accompany derepression and full activation of ADH2 (25). We confirmed a role for Gcn5 in activation by measuring gene expression over 6 h of derepression (Fig. 2). Loss of Gcn5 resulted in >65% reduction in mRNA levels of both ADH2 and FBP1.
FIGURE 2.
SAGA, SWI/SNF, and Mediator are required for full expression of ADH2 and FBP1. mRNA analysis throughout 6 h of derepression of (A) ADH2 and (B) FBP1 from gcn5Δ (LLTY72), ada1Δ (LLTY73), snf2Δ (KKTY3), and med17 ts (RBY93). Values, based on QPCR, were normalized to ACT1 and then expressed as the percent of the wild-type (W303a) level after 4-h derepression. Error bars represent the standard deviation of two biological replicates.
There is precedent that the HAT activity of SAGA is required at promoters to loosen chromatin and allow successive recruitment events, notably at HO and PHO8 (10, 26). To see if Gcn5 was similarly required at ADH2 and FBP1 we assayed occupancy of other coactivators in a gcn5Δ strain after 4 and 6 h of derepression. Because expression in the coactivator mutants was very low at early points in derepression, we chose to look at recruitment of other coactivators at later times to ensure that if we observed little or no recruitment, it was due to loss of the coactivator rather than simply due to low levels of transcription. We did look at one earlier time in the gcn5Δ strain, however, but no significant recruitment was observed at this point (supplemental Fig. S5). Occupancy of Snf2, Med15, and Med17 was less than half the wild-type levels at both promoters (Fig. 3A). In contrast, binding of Sua7, a component of TFIIB, and pol II persisted in this mutant, indicating that HAT activity was not required for stable recruitment of the transcription machinery. We did not observe an increase in recruitment between 4 and 6 h of derepression, and so, for simplicity sake, these values have been averaged together (as was done for all other ChIP experiments). Additionally, we assayed for two other SAGA components, Ada1 and Spt8, both of which occupied the promoters under derepressed conditions in a gcn5Δ strain, verifying that loss of Gcn5 resulted only in loss of HAT activity, and not total loss of the complex (supplemental Fig. S2, A and B).
FIGURE 3.
Coactivators binding is interdependent at ADH2 and FBP1. ChIP for coactivators in gcn5Δ (RBY110, RBY115, RBY116, and RBY155) (A); ada1Δ (RBY106, RBY108, RBY111, RBY113, and RBY154) (B); snf2Δ (RBY109, RBY112, RBY117) (C); and med17 ts (RBY119, RBY120, RBY121) at ADH2 (light bars) and FBP1 (black bars) (D). Data for coactivator occupancy in a WT strain was obtained using RBY3, RBY5, RBY8, RBY9, and RBY11. Values are expressed as the -fold over the WT repressed (R) value, as in Fig. 1A (see “Experimental Procedures” for details). WT data is shown for 4-h derepression (no error bar) or an average of 4- and 6-h derepression (with error bars representing the standard deviation) and data for all mutant strains is based on the average for 4- and 6-h derepression. Error bars of the mutant values indicate the standard deviation of at least three independent biological replicates.
Previous work in our laboratory demonstrated that activation of ADH2 is associated largely with changes in nucleosome density and not nucleosome position (12). Therefore, to determine the role of Gcn5 in chromatin remodeling, we employed a plasmid-based supercoiling assay that allows us to monitor the nucleosome density at the ADH2 promoter (16). Under the conditions used, a downward shift in the topoisomer distribution represents a decrease in nucleosome density on the plasmid. In a wild-type strain, there was a clear shift in the density of topoisomers upon derepression, indicating the loss of approximately one nucleosome (Fig. 4, A (lanes 1 and 2) and B).
FIGURE 4.
Coactivators differentially contribute to chromatin remodeling at the ADH2 promoter. A, supercoiling analysis on strains containing a plasmid with the ADH2 promoter was performed as described under “Experimental Procedures.” Samples from WT (W303a) stain containing the WT plasmid, pLLTY1 or from WT strain containing a plasmid in which the ADH2 TATAA box was mutated to GAGAA, pLLTY3, were repressed (R) or 4-h derepressed (DR). The locations of nicked DNA and Band 1 are indicated. Quantitation of supercoiling assays in WT (W303a) (B), gcn5Δ (LLTY72) (C), ada1Δ (LLTY73) (D), snf2Δ (KKTY3) (E), and med15Δ (LLTY120) (F), all carrying pLLTY1. Samples were from cells either repressed (dashed), or derepressed for 2.5 h (gray) or 5 h (black) and subjected to supercoiling analysis. The topoisomer distributions were quantified using phosphorimaging. The data are presented as relative intensity for each of eight bands. Error bars represent the standard deviation of three independent experiments. A shift to the right represents a decrease in overall nucleosome density.
We first established that this loss was not simply a result of transcription per se, but rather due to the concerted action of chromatin-remodeling complexes by performing the assay using a version of the plasmid in which the TATA box had been mutated, abolishing transcription. We still saw the shift with this mutant (Fig. 4A, lanes 3 and 4), confirming that the loss of nucleosomes occurs in the steps leading up to initiation. Importantly, the transcription from the plasmid driven by the ADH2 promoter was glucose-repressed and both Adr1- and Cat8-dependent as was the decrease in nucleosome density upon derepression (data not shown). This agrees with published studies (12, 27) and demonstrates that our plasmid-based system is an accurate reflection of the events on the chromosome. The shift indicative of decreased nucleosome density that accompanied derepression in a wild-type strain was only partially observed in the gcn5Δ strain (Fig. 4C). This is consistent with previously published data showing a role for Gcn5 in ADH2 chromatin remodeling (28).
Previous work establishing the role of SAGA in ADH2 regulation was limited to examining the effects of deleting Gcn5. SAGA has functions other than HAT activity, however, such as recruitment of TATAA-binding protein, deubiquitination, and stabilization of pre-initiation complexes (PICs) (29) that are differentially required for expression. For example, it is known that the GAL genes require SAGA for activation, but not Gcn5 (30). To investigate whether SAGA has a function in the regulation of ADH2 and FBP1 beyond its HAT activity, we used a strain lacking Ada1, an adaptor protein required for the structural integrity of SAGA (31). Expression of both ADH2 and FBP1 was significantly reduced by loss of Ada1 (Fig. 2).
Without Ada1, recruitment of coactivators to ADH2 was very low (Fig. 3B, light bars), despite full Adr1 binding (Fig. 1A), which we previously showed was sufficient for coactivator recruitment in a wild-type strain (12). Furthermore, Sua7 and pol II occupancy were very low, consistent with the reduced transcription in this mutant. Recruitment of coactivators and the general transcription machinery to FBP1 in the ada1Δ strain was also very low, in agreement with the weak gene activation we observed (Fig. 3B, dark bars).
Chromatin remodeling of the ADH2 promoter in this mutant closely resembled wild type (Fig. 4D). Deletion of Ada1 destabilizes SAGA, including Gcn5 (supplemental Fig. S2C), and yet the chromatin remodeling defect in this mutant was milder than the gcn5Δ defect (Fig. 4C). This discrepancy could be explained by compensation by another chromatin-remodeling complex (such as NuA4) that gains access to the promoter in the absence of the whole SAGA complex, but not when only Gcn5 is missing.
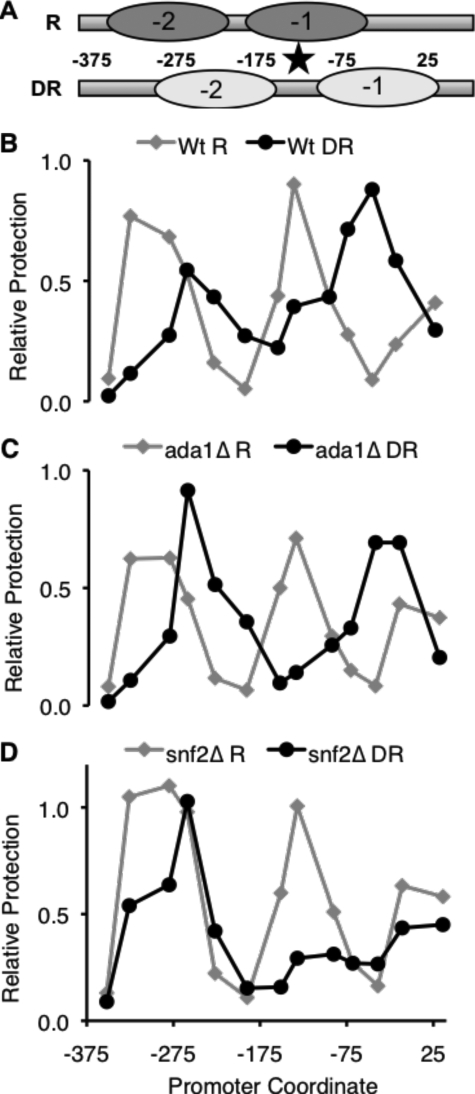
We also wanted to assay chromatin remodeling at the FBP1 promoter in the ada1Δ strain to determine if SAGA was functioning in the same manner as at ADH2. In contrast to the ADH2 promoter, the chromatin remodeling at FBP1 includes nucleosome repositioning (12), a change that is not detectable by supercoiling assays. We instead used NuSA (nucleosome scanning assay), which maps the positions of nucleosomes at the chromosomal locus by determining the relative protection of DNA during micrococcal nuclease digestion (12). (To assess the comparability of these two methods, we performed NuSA on the supercoiling plasmid from wild-type cells and observed similar results on the plasmid and for the chromosomal locus for the ADH2 promoter (Ref 12 and data not shown)). When we performed NuSA on the FBP1 promoter with either the wild type or the ada1Δ strain, we saw the same shift in nucleosome position as previously reported: both N-2 and N-1 nucleosomes shifted 3′, increasing accessibility of TATAA box (Fig. 5, A–C). As with ADH2, the fact that loss of Ada1 did not affect chromatin remodeling at FBP1 suggests that an open chromatin structure is not sufficient for full activation, because expression of FBP1 in this mutant was <15% of wild type. Previous studies showing that chromatin remodeling did not lead to activation were done under repressing conditions (16, 25), and our finding here expands on the idea that the sole purpose of coactivators is not simply to provide access to the DNA, but they may also be important transducers of upstream regulatory signals (for example, a regulatory kinase may phosphorylate a coactivator at the promoter, causing a change that leads to activation). It also implies that if there is a compensatory complex (see above), it is limited in its function to chromatin remodeling and cannot overcome the rest of the requirement on SAGA.
FIGURE 5.
Chromatin remodeling at the FBP1 promoter requires SWI/SNF, but not SAGA. A, schematic of the nucleosome positions in the FBP1 promoter in a WT strain under repressed (R) and derepressed (DR) conditions (12). The star indicates the location of the FBP1 TATAA box. NuSA was used to determine the nucleosome positioning at the FBP1 promoter in wild type (W303a) (B), ada1Δ (LLTY73) (C), and snf2Δ (KKTY3) (D). Cells were either repressed (gray lines) or derepressed for 4 h (black lines). Each point shows the relative protection from micrococcal nuclease digestion for each amplicon.
The Role of SWI/SNF—The limited effects on chromatin remodeling in the SAGA mutants suggested another complex may be the primary remodeling complex at glucose-repressed genes. Such a candidate is the ATP-dependent chromatin-remodeling complex SWI/SNF, which is recruited to these promoters upon activation (12). Loss of Snf2, the catalytic subunit, led to roughly 50% reductions in both ADH2 and FBP1 mRNA levels (Fig. 2). Even greater reductions in expression were observed when a second component, Snf5, was deleted (supplemental Table S2).
Recruitment of Gcn5 to ADH2 and FBP1 was only reduced a small amount by deletion of Snf2 (Fig. 3C). Med17 was recruited with normal efficiency in this strain. These data suggest that SWI/SNF plays a role downstream of the recruitment of these two coactivators. ChIP for Sua7 showed strong reductions in the amount of occupancy at these promoters, but pol II levels, whereas down at FBP1, were relatively high at ADH2. Again, this is not the only example where pol II occupancy does not correlate with expression, both in our hands (see above and Ref. 16) and more broadly (32).
The contribution by SWI/SNF to chromatin remodeling was also assessed. Of all the coactivators in this study, SWI/SNF appeared to be most important for full nucleosome remodeling. There was virtually no change upon derepression at ADH2 in the supercoiling assay (Fig. 4E). The NuSA at FBP1 showed some remodeling upon derepression, but it was incomplete (Fig. 5D). The N-2 nucleosome did not change position to the extent seen with the wild-type strain, and the N-1 nucleosome diminished in occupancy rather than shifting in position. This NuSA profile is very similar to the one we reported previously in a cat8Δ strain, in which there was neither activation nor chromatin remodeling (12). Taken together, this suggests that SWI/SNF is essential for creating the proper chromatin structure for wild-type levels of activation at both ADH2 and FBP1. We note that full remodeling is not essential for all gene activation, because there was still about a 100-fold increase in both ADH2 and FBP1 expression over repressed conditions in this mutant (Fig. 2). This increase, however, is only ∼20% of the full wild-type level of expression, indicating that chromatin remodeling is very important for activation.
The Role of Mediator—To clarify the role of Mediator at ADH2 and FBP1, we used a med17 ts mutant (med17–138) (33). Mediator is a large complex divided into three sub-modules, head, middle, and tail, that serves as an adaptor between activators and general transcription factors (34). Med17 is the largest subunit of the head sub-module. Expression of ADH2 and FBP1 was <5% of the expression in a wild-type strain at all times assayed, attesting to the importance of this coactivator (Fig. 2). Several recent studies have reached different conclusions about the effects on the rest of Mediator in the med17 ts mutant: some found that the tail sub-module of Mediator is still recruited to promoters at the restrictive temperature (35, 36), whereas others found the entire complex had dissociated (37). For this reason, we also looked at gene expression in a tail mutant, med15Δ. The expression defects in this strain were also significant, although not as severe as in the med17 ts strain. (supplemental Table S2).
Occupancy of coactivators in the med17 ts strain was varied (Fig. 3D). ChIP for Gcn5 at ADH2 gave a reduced signal compared with the wild type; at FBP1 its occupancy was not significantly different than the wild-type level. Levels of Snf2 occupancy, however, were reduced to background at both promoters. Given the proposed role of Mediator, it was unsurprising that levels of both Sua7 and pol II were diminished in the med17 ts strain. Because of differing reports about the affect of the med17–138 allele in terms of its effect on the structure of the Mediator complex (see above), we also looked at the binding of several other Mediator components in this mutant background (supplemental Fig. S4). We confirmed that the head sub-module was dissociated from all promoters tested in the med17 ts strain, as was the middle sub-module, based on occupancy of Med14. In contrast, we found that the tail component Med15 occupied some promoters tested, but not others. We suggest that the contradiction in the literature may be a result of the phenomenon we observed within this small set of genes: effects of this mutation may be promoter-specific. We also note, however, the continued presence of the tail sub-module at FBP1 did not contribute to gene activation (supplemental Table S2), and if it did improve the recruitment of other coactivators, it was limited to a marginal increase in Gcn5 recruitment (Fig. 3D).
Although Mediator itself is not reported to have chromatin-remodeling activities, we wanted to know how loss of this complex would indirectly affect promoter chromatin structure. Due to the inherent changes in chromatin structure at high temperatures, we chose to use the med15Δ strain rather than the med17 ts strain for our chromatin remodeling assays. Supercoiling assays of the ADH2 promoter in a med15Δ strain showed that the reduction in nucleosome density associated with derepression still occurred (Fig. 4F). Interestingly, compared with wild type, there was a shift toward lower nucleosome density even under repressed conditions, suggesting a role for the tail of Mediator in gene repression. The tail of Mediator has long been implicated in negative regulation, such as at the HO promoter (38). No increase in transcription was detected in repressing conditions in the med15Δ mutant, however, although other studies have reported a significant increase in Adr1-dependent constitutive activation of an ADH2/lacZ reporter gene in the same background (39). It was also observed in these studies that loss of Med15 was accompanied by significant Adr1 occupancy of the ADH2 promoter in repressing conditions. Together these results suggest that in the absence of Med15 Adr1 may bind and recruit chromatin remodeling factors but not pol II in repressing conditions.
An Order of Events?—The combined data from the ChIP experiments would be consistent with a definite order of recruiting events. To test this, we assayed recruitment of Adr1, Cat8, Gcn5, Med15, Snf2, and pol II in a wild-type strain at ADH2 and FBP1 over a time course of derepression. Initially, we looked at very early time points (5, 10, 15, and 30 min after removal of glucose), but we were unable to detect any binding events prior to 30 min derepression (data not shown). Despite the slow activation, we did not observe distinct recruitment events, as all proteins assayed gave the same binding patterns (Fig. 6). The inability to resolve binding events may be a reflection of the highly interdependent nature of the recruitment of these complexes (Fig. 3). The early peak of recruitment at 1 h was observed in several experiments, with a corresponding spike in expression (∼100-fold over repressed, less than a tenth of the full activation, data not shown) at this time. This may be due to metabolic synchronization and cycling that is known to occur in aerobically grown yeast cultures (40). There is some prior evidence, however, that would support the notion that there are two distinct phases of induction based on studies of the SUC2 gene (41).
Overexpression of Adr1 Can Compensate for Loss of Snf2—Thus far, we have shown generally that coactivators contribute to ADH2 and FBP1 activation, but no single coactivator appeared to be absolutely required, even loss of Mediator, which was very important for expression, still resulted in chromatin remodeling and some recruitment. This observation led us finally to ask if coactivators were merely “optimization widgets,” helpful in achieving full, rapid gene activation, either by stabilizing activators and the general transcription machinery at the promoter via protein-protein contacts, or by creating enhanced access to promoter elements through chromatin remodeling. We previously demonstrated that overexpression of Adr1 can compensate for loss of Cat8 at ADH2 (but not at FBP1) (12). We now asked whether or not overexpression of Adr1 could also compensate for loss of coactivators in a similar fashion. ADH2 gene expression in strains carrying a high copy plasmid with ADR1 under control of the strong ADH1 promoter (pNKA-1U) and deleted for various coactivators are shown in Table 1. At ADH2 clear compensation for loss of Snf2 was observed, but not for loss of either Gcn5 or Ada1. This suggests that SAGA at least is more than just an optimization widget, as the function(s) it performs cannot be compensated for by overexpression of Adr1.
TABLE 1.
ADH2 expression in coactivator mutants with overexpressed Adr1
RNA was isolated from repressed cells or cells derepressed for 4 h (D) and quantitated by QPCR.
| Genotype (strain) | Relative expressiona | % WT D |
|---|---|---|
| WT (W303a) | 3438 | 100 |
| WT (W303a, pNKA-1U) | 5344 (456) | 155 |
| snf2Δ (KKTY3, pNKA-1U) | 9550 (306) | 277 |
| ada1Δ (LLTY73, pNKA-1U) | 1133 (629) | 33 |
| gcn5Δ (LLTY72, pNKA-1U) | 810 (200) | 28 |
Values were normalized to ACT1, and then the WT-repressed value was arbitrarily set to 1. The standard deviation for two independent experiments is shown in parenthesis.
DISCUSSION
We previously used glucose-repressed genes as a model for understanding the contributions to activation made by the two transcription factors, Adr1 and Cat8 (12). Our current work has expanded the analysis of the regulation of two of these genes, ADH2 and FBP1, to encompass the roles of coactivators. The comprehensive approach we used, integrating data from gene expression, ChIP, and chromatin remodeling experiments, provides one of the most complete pictures of gene activation to date. We have shown that coactivators stabilize Cat8 binding, contribute to full gene expression, and show interdependences for recruitment. Additionally, we identified SWI/SNF as the primary chromatin-remodeling complex at both ADH2 and FBP1. Our analysis provides evidence that genes like ADH2 and FBP1, which share a common activation signal (i.e. low glucose), share many other common aspects of activation, while some differences, perhaps due to different regulators and promoter architecture, do exist.
The Role of Coactivators—Using a series of coactivator mutants we showed that SAGA binding at ADH2 was independent of all other coactivators tested. This was also true at FBP1, although Cat8 and SAGA binding may be interdependent (see below). Binding of SWI/SNF and Mediator however, depended on SAGA, suggesting an important role for it in allowing other coactivators to be stably recruited. Because the chromatin remodeling assays showed partial remodeling in a gcn5Δ mutant and normal remodeling in an ada1Δ mutant (Figs. 4C, 4D, and 5C), we conclude that SAGA is not essential for chromatin remodeling. However, because loss of the HAT component of SAGA, GCN5, led to significant defects in activation, HAT activity must still play a role. Beyond chromatin remodeling, histone acetylation may be important for creation of binding sites for other coactivators, or even Cat8. Furthermore, histones are not the only acetylation target of HATs; for example, SAGA-dependent acetylation of Rsc4, a component of the chromatin structure remodeling complex, was recently reported (42). Thus SAGA could be marking transcription factors or coactivators at these promoters. The importance of acetylation for stable binding of activators and coactivators is suggested by the observation that a stable, complete “poised” PIC is formed in repressing growth conditions in the absence of the two major histone deacetylases, Rpd3 and Hda1, creating a hyperacetylated chromatin state (16).
The fact that SWI/SNF recruitment was observed only in the presence of all other coactivators (Fig. 3) was surprising in light of the finding that SWI/SNF was required for chromatin remodeling (Figs. 4E and 5D). This suggests that chromatin remodeling is not required for SAGA or Mediator to bind to the promoter. This conclusion is consistent with the observation that, when Mediator is recruited to the promoter artificially by tethering a tail component to the DNA binding domain of Adr1, neither SAGA nor SWI/SNF is required for ADH2 expression (39).
An alternative explanation is that SWI/SNF plays two different roles: an early role in chromatin remodeling during which it is only loosely associated with the promoter (and therefore undetectable by ChIP), and a second role after other coactivators are bound, at which point SWI/SNF stably occupies the promoter. Such a dual role was recently observed for SWI/SNF at the PHO5 promoter (3). It may be undetectable by ChIP in the first phase, because its interactions with nucleosomes preclude cross-linking to the DNA, or because it is in proximity to the DNA only transiently as it is evicting nucleosomes. A possible second role later in PIC formation is as a scaffold for TFIIB and TATAA-binding protein binding, or perhaps it functions even later in elongation, another putative role for SWI/SNF (43).
A recent report suggested that Mediator may be mostly important for gene activation during conditions of stress, including low glucose (44). Our results show that Mediator is essential for high levels of activation of glucose-repressed genes (Fig. 2 and supplemental Table S2). One reason for this strong dependence at these genes may be its central role in coactivator recruitment. Loss of Mediator resulted in reduced binding by all coactivators as well as the general transcription machinery (Fig. 3D). The presence of Mediator was not required for chromatin remodeling at ADH2 (Fig. 4F), ruling out even an indirect role for Mediator in this process. A similar study at the CHA1 promoter reached the same conclusion (35).
The order of recruitment may not be sufficient for describing the mechanism of activation given the complicated functional outcomes of coactivator recruitment, but nevertheless provides valuable information. Our time-course experiment revealed a tight association temporally in the recruitment events at both ADH2 and FBP1 (Fig. 6). This is in contrast to the GAL4 promoter, where distinct phases of recruitment were observed (8). The arrival of all coactivators together may again be reflective of the high level of interdependency between these complexes. Similar near-simultaneous recruitment of coactivators has been observed at the ARG1 promoter (45).
A Common Mechanism of Activation Despite Differences in Regulators and Promoter Architecture—A survey of the mechanisms of activation of other genes in Saccharomyces cerevisiae reveals that there is no single way to activate a gene; even genes regulated by the same activator, such as Gcn4-dependent genes, do not share the same set of required coactivators (46). One question has been to understand what, if anything, governs the mechanism of activation. ADH2 and FBP1 are both activated by low glucose via the upstream regulatory kinase Snf1 (11). Although both of these genes are bound and regulated by Adr1 and Cat8, ADH2 is equally co-dependent on these two transcription factors, whereas FBP1 is much more Cat8-dependent (12). Additionally, FBP1 is repressed by Mig1, whereas ADH2 has no known repressors (14, 47). The chromatin remodeling required at the two promoters is also distinctly different. Along with a decrease in nucleosome density, two of the nucleosomes in the FBP1 promoter significantly shift position upon derepression, whereas at the ADH2 promoter the positions of the nucleosomes remain the same but the overall density decreases (this report, Refs. 12 and 27, and unpublished data). Despite these differences, however, our findings here demonstrate that these two genes share many common aspects of activation. Not only did we observe the identical set of coactivators recruited to these genes upon activation, but they were recruited with the same patterns of interdependence (Fig. 3) (12). The role of each complex at these two promoters also appears to be the same. SWI/SNF was the primary chromatin-remodeling complex at both ADH2 and FBP1 (Figs. 4D and 5D), Mediator was the most important for gene expression (Fig. 2 and supplemental Table S2) and SAGA appeared to have dual HAT and non-HAT functions, based on the different phenotypes of the gcn5Δ and ada1Δ strains. We also obtained similar patterns when we looked at other Snf1-dependent glucose-regulated genes, including ADY2, JEN1, MLS1, and POT1 (supplemental Fig. S3, supplemental Table S2, and data not shown).
The difference between these genes is not the route to activation, but the coordination of those steps. At ADH2, Adr1 is the dominant activator in terms of coactivator recruitment, whereas at FBP1 Cat8 alone does most of the recruitment (12). Our finding here that Adr1 can bind to the ADH2 promoter unaided by coactivator complexes (Fig. 1A) agrees with the model wherein Adr1 coordinates the events leading up to initiation. This includes the binding of Cat8, whose binding site is adjacent to the Adr1 binding site, and which was previously shown to bind cooperatively with Adr1 at ADH2 (13). At the FBP1 promoter, their binding sites are separated by >200 bp, and the intervening region contains a well positioned nucleosome (12), and no cooperative binding was observed (13). Instead, we propose that Cat8 binding is stabilized at this promoter by coactivators. Loss of Cat8 reduced Gcn5 binding by ∼50% (12), supporting the idea of interdependent binding of these two proteins. A compelling alternative is suggested by the recent finding that Gcn4 requires Cyc8/Tup1 to bind to ARG1 and ARG4 (48). Cyc8/Tup1 appears to have dual roles, acting as both a corepressor and coactivating complex at some promoters (Ref. 48 and references therein). Interestingly, Mig1 represses FBP1 via the Cyc8/Tup1 complex (14), making this complex another candidate for stabilization of Cat8.
Finally, we attempted to answer a fundamental question about the necessity of coactivators. With the exception of Mediator, the effects on transcription in the coactivator mutants were surprisingly mild, implying that these complexes are not individually essential. Even in the Mediator mutants, there was still a significant increase in transcription when cells were derepressed, albeit much less than wild-type cells. We previously demonstrated that overexpression of Adr1 could compensate for loss of Cat8 at ADH2 (12). Here we found that Adr1 overexpression also compensated for loss of SWI/SNF (Table 1). It did not alleviate the defects caused by deletion of SAGA, though. Cat8 is required at ADH2 for chromatin remodeling (12). Overexpression of Adr1 may lead to a loosened chromatin structure, simply through increased binding to the promoter, and thus chromatin remodelers are no longer needed, explaining the compensation for both SWI/SNF and Cat8. SAGA, on the other hand, remained important for expression, supporting the idea that its main purpose at these promoters is not to remodel chromatin.
A Comprehensive Study of Activation—By combining three different experimental techniques we were able to investigate the role of coactivators in multiple aspects of gene activation. This approach uncovered several instances where recruitment or chromatin remodeling were not associated with expression and highlight relevant phenomena that deviate from the paradigm of activated transcription. Two cases in particular warrant illustration, the first of which is recruitment of coactivators and pol II without corresponding increases in expression. This was observed in the case of the snf2Δ mutant, where recruitment of most factors was normal (with the exception of Sua7), but expression was only 50% of the wild type. This shows that a promoter can be fully occupied by coactivators and even pol II without full activation. This result, similar to findings at the heat shock promoters, expands on the idea of a poised or stalled polymerase.
The second case is chromatin remodeling without gene activation, which we saw in the med15Δ mutant. Here, transcription levels at 4 h of derepression are very low (supplemental Table S2), and yet near wild-type levels of chromatin remodeling were observed (Fig. 4F). Additionally, loss of Mediator severely reduced occupancy by other coactivators (Fig. 3D), but did not affect chromatin remodeling (Fig. 4F). Taken together, these data illustrate that chromatin remodeling is not sufficient for stable coactivator binding or gene activation.
These specific cases demonstrate the complexity of activated transcription, a process that is not accurately described by ordered recruitment. Rather, it is a resilient mechanism with some redundancies, allowing cells to activate genes even in the absence of one or more coactivators. Such activation, however, is tightly regulated, as evidenced by the lack of expression even when chromatin is fully remodeled. One compelling idea that remains to be tested is whether or not coactivators serve as receptors for upstream regulatory signals, a hypothesis suggested by our findings with the Mediator mutants, where we observed chromatin remodeling, but very poor recruitment of other coactivators and weak activation. Such complexities can be easily masked when indirect assays, such as occupancy by pol II, are used as proxies for a measure of gene activation, a finding supported by recent genome-wide analysis of pol II occupancy (32) and demonstrate the strength of this type of comprehensive study.
Supplementary Material
Acknowledgments
We thank E. Chang for her contributions to the NuSA assays and C. Tachibana for helpful discussions and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM26079 (to E. T. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
Footnotes
The abbreviations used are: SAGA, Spt-Ada-Gcn5-acetyltransferase complex; SWI/SNF, SWI/SNF complex; ChIP, chromatin immunoprecipitation; QPCR, real-time quantitative PCR; FMA, formaldehyde; EGS, ethylene glycol bis(succinimidylsuccinate); pol II, RNA polymerase II; NuSA, nucleosome scanning assay; HA, hemagglutinin, HAT, histone acetyltransferase; PIC, pre-initiation complex; TFIIB, general transcription factor IIB; NuA4, nucleosome acetyltransferase of H4 complex.
References
- 1.Ptashne, M. (2005) Trends Biochem. Sci. 30 275–279 [DOI] [PubMed] [Google Scholar]
- 2.Krebs, J. E., Kuo, M. H., Allis, C. D., and Peterson, C. L. (1999) Genes Dev. 13 1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins, M. W., Williams, S. K., Linger, J., and Tyler, J. K. (2007) Mol. Cell. Biol. 27 6372–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbaric, S., Luckenbach, T., Schmid, A., Blaschke, D., Horz, W., and Korber, P. (2007) J. Biol. Chem. 282 27610–27621 [DOI] [PubMed] [Google Scholar]
- 5.Nourani, A., Utley, R. T., Allard, S., and Cote, J. (2004) EMBO J. 23 2597–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R., and O'Shea, E. K. (2003) Science 299 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaumik, S. R., and Green, M. R. (2002) Mol. Cell. Biol. 22 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant, G. O., and Ptashne, M. (2003) Mol. Cell 11 1301–1309 [DOI] [PubMed] [Google Scholar]
- 9.Lemieux, K., and Gaudreau, L. (2004) EMBO J. 23 4040–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra, D., Parnell, E. J., Landon, J. W., Yu, Y., and Stillman, D. J. (2006) Mol. Cell. Biol. 26 4095–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young, E. T., Dombek, K. M., Tachibana, C., and Ideker, T. (2003) J. Biol. Chem. 278 26146–26158 [DOI] [PubMed] [Google Scholar]
- 12.Biddick, R. K., Law, G. L., and Young, E. T. (2008) PLoS ONE 3 e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana, C., Yoo, J. Y., Tagne, J. B., Kacherovsky, N., Lee, T. I., and Young, E. T. (2005) Mol. Cell. Biol. 25 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaragoza, O., Vincent, O., and Gancedo, J. M. (2001) Biochem. J. 359 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agricola, E., Verdone, L., Xella, B., Di Mauro, E., and Caserta, M. (2004) Biochemistry 43 8878–8884 [DOI] [PubMed] [Google Scholar]
- 16.Tachibana, C., Biddick, R., Law, G. L., and Young, E. T. (2007) J. Biol. Chem. 282 37308–37315 [DOI] [PubMed] [Google Scholar]
- 17.Guldener, U., Heck, S., Fielder, T., Beinhauer, J., and Hegemann, J. H. (1996) Nucleic Acids Res. 24 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999) Yeast 15 963–972 [DOI] [PubMed] [Google Scholar]
- 19.Dombek, K. M., and Young, E. T. (1997) Mol. Cell. Biol. 17 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Olmo, M. L., Sogo, J. M., Franco, L., and Perez-Ortin, J. E. (1993) Yeast 9 1229–1240 [DOI] [PubMed] [Google Scholar]
- 21.Burns, L. G., and Peterson, C. L. (1997) Mol. Cell. Biol. 17 4811–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosma, M. P., Tanaka, T., and Nasmyth, K. (1999) Cell 97 299–311 [DOI] [PubMed] [Google Scholar]
- 23.Gregory, P. D., Schmid, A., Zavari, M., Munsterkotter, M., and Horz, W. (1999) EMBO J. 18 6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowak, D. E., Tian, B., and Brasier, A. R. (2005) BioTechniques 39 715–725 [DOI] [PubMed] [Google Scholar]
- 25.Verdone, L., Cesari, F., Denis, C. L., Di Mauro, E., and Caserta, M. (1997) J. Biol. Chem. 272 30828–30834 [DOI] [PubMed] [Google Scholar]
- 26.Reinke, H., Gregory, P. D., and Horz, W. (2001) Mol Cell 7 529–538 [DOI] [PubMed] [Google Scholar]
- 27.Verdone, L., Camilloni, G., Di Mauro, E., and Caserta, M. (1996) Mol. Cell. Biol. 16 1978–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verdone, L., Wu, J., van Riper, K., Kacherovsky, N., Vogelauer, M., Young, E. T., Grunstein, M., Di Mauro, E., and Caserta, M. (2002) EMBO J. 21 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel, J. A., and Grant, P. A. (2007) Mutat. Res. 618 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhaumik, S. R., and Green, M. R. (2001) Genes Dev. 15 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sterner, D. E., Grant, P. A., Roberts, S. M., Duggan, L. J., Belotserkovskaya, R., Pacella, L. A., Winston, F., Workman, J. L., and Berger, S. L. (1999) Mol. Cell. Biol. 19 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muse, G. W., Gilchrist, D. A., Nechaev, S., Shah, R., Parker, J. S., Grissom, S. F., Zeitlinger, J., and Adelman, K. (2007) Nat. Genet. 39 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, C. M., and Young, R. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4587–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, B. A., and Reinberg, D. (2003) J. Cell Sci. 116 3667–3675 [DOI] [PubMed] [Google Scholar]
- 35.He, Q., Battistella, L., and Morse, R. H. (2008) J. Biol. Chem. 283 5276–5286 [DOI] [PubMed] [Google Scholar]
- 36.Linder, T., Zhu, X., Baraznenok, V., and Gustafsson, C. M. (2006) Biochem. Biophys. Res. Commun. 349 948–953 [DOI] [PubMed] [Google Scholar]
- 37.Takagi, Y., Calero, G., Komori, H., Brown, J. A., Ehrensberger, A. H., Hudmon, A., Asturias, F., and Kornberg, R. D. (2006) Mol Cell 23 355–364 [DOI] [PubMed] [Google Scholar]
- 38.Yu, Y., Eriksson, P., and Stillman, D. J. (2000) Mol. Cell. Biol. 20 2350–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young, E. T., Tachibana, C., Chang, H. W., Dombek, K. M., Arms, E. M., and Biddick, R. (2008) Mol. Cell. Biol. 28 2509–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu, B. P., Kudlicki, A., Rowicka, M., and McKnight, S. L. (2005) Science 310 1152–1158 [DOI] [PubMed] [Google Scholar]
- 41.Geng, F., and Laurent, B. C. (2004) EMBO J. 23 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.VanDemark, A. P., Kasten, M. M., Ferris, E., Heroux, A., Hill, C. P., and Cairns, B. R. (2007) Mol. Cell 27 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu, H., Hu, C., Yoon, S., Natarajan, K., Swanson, M. J., and Hinnebusch, A. G. (2004) Mol. Cell. Biol. 24 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan, X., Chou, D. M., and Struhl, K. (2006) Nat. Struct. Mol. Biol. 13 117–120 [DOI] [PubMed] [Google Scholar]
- 45.Govind, C. K., Yoon, S., Qiu, H., Govind, S., and Hinnebusch, A. G. (2005) Mol. Cell. Biol. 25 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson, M. J., Qiu, H., Sumibcay, L., Krueger, A., Kim, S. J., Natarajan, K., Yoon, S., and Hinnebusch, A. G. (2003) Mol. Cell. Biol. 23 2800–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irani, M., Taylor, W. E., and Young, E. T. (1987) Mol. Cell. Biol. 7 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, S. J., Swanson, M. J., Qiu, H., Govind, C. K., and Hinnebusch, A. G. (2005) Mol. Cell. Biol. 25 11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.