Abstract
Large conductance voltage and Ca2+-activated K+ (MaxiK) channels couple intracellular Ca2+ with cellular excitability. They are composed of a pore-forming α subunit and modulatory β subunits. The pore blockers charybdotoxin (CTx) and iberiotoxin (IbTx), at nanomolar concentrations, have been invaluable in unraveling MaxiK channel physiological role in vertebrates. However in mammalian brain, CTx-insensitive MaxiK channels have been described [Reinhart, P. H., Chung, S. & Levitan, I. B. (1989) Neuron 2, 1031–1041], but their molecular basis is unknown. Here we report a human MaxiK channel β-subunit (β4), highly expressed in brain, which renders the MaxiK channel α-subunit resistant to nanomolar concentrations of CTx and IbTx. The resistance of MaxiK channel to toxin block, a phenotype conferred by the β4 extracellular loop, results from a dramatic (≈1,000 fold) slowdown of the toxin association. However once bound, the toxin block is apparently irreversible. Thus, unusually high toxin concentrations and long exposure times are necessary to determine the role of “CTx/IbTx-insensitive” MaxiK channels formed by α + β4 subunits.
MaxiK (large conductance, voltage- and Ca2+-activated K+) channels are composed of a pore-forming α subunit (1) and modulatory transmembrane β subunits (2–4). Association with tissue-specific modulatory β subunits may account for much of the phenotypic MaxiK channel variability observed in native tissues (5). β subunits increase the apparent Ca2+/voltage sensitivity of the α subunit (3, 6, 7), modify channel kinetics (8–10), and alter its pharmacological properties (3, 11). MaxiK channel β1 and β2 subunits can alter the charybdotoxin (CTx) and iberiotoxin (IbTx) interaction in electrophysiological (9–11) and biochemical (12) studies; however, none of the reported β subunits can account for the toxin-insensitive MaxiK channels found in brain (13).
MaxiK channels are important regulators of neuronal function. In presynaptic terminals, MaxiK channels are found colocalized with voltage-dependent Ca2+ channels and, therefore, are thought to play a critical role in the regulation of neurotransmitter release (14–17). However, because of the lack of IbTx effects in striatal brain slices (18, 19), the general involvement of MaxiK channels in neurotransmitter release remains uncertain.
IbTx is considered to be a specific MaxiK channel blocker, whereas CTx also blocks other potassium channels (20). Because of its specificity, IbTx is used as a pharmacological agent to dissect the role of MaxiK channels in physiological processes, and generally it is used at a concentration of 100 nM. In contrast to smooth and skeletal muscles, where MaxiK channels are normally blocked by 100 nM IbTx or CTx, in neurons both CTx-sensitive and -insensitive MaxiK channels are found. CTx-insensitive MaxiK channels were first described in rat brain membrane preparations (13) and later in embryonic telencephalic neuroepithelium (21). Interestingly, in rat supraoptic magnocellular neurons a CTx-sensitive isoform is localized on the cell body (22), whereas a CTx-insensitive isoform is present at the nerve endings (23). In addition, MaxiK currents in other neuronal preparations are only partially blocked by CTx and IbTx (15, 24).
Herein, we report that coexpression of a neuronal MaxiK channel β subunit (β4) leads to a MaxiK channel phenotype that displays a low apparent IbTx and CTx sensitivity due to dramatically decreased toxin association rates, by ≈250–1,000 fold. Exchange of the extracellular loop between the smooth muscle β1 and neuronal β4 subunits shows that this domain is responsible for toxin resistance. We propose that the assembly of the neuronal MaxiK channel β4 subunit with its pore forming α subunit in the brain forms the molecular basis for toxin-insensitive MaxiK channels found in neurons.
Experimental Procedures
Cloning and Expression.
The MaxiK β3 and β4 subunits were identified in the expressed sequence tag (EST) database. β3 (accession no. AF160968) and β4 (accession no. AF160967) cDNA clones were isolated from an arrayed, normalized human cDNA library (β3, L12-D12; β4, F23F16; HUCL-Array library, Stratagene). The β3 clone contains several in-frame upstream stop codons before the start methionine. The sequence upstream of the Kozak consensus sequence (25) of β4 clone F23F16 is unusually GC-rich (82.5% G + C in 337 bases), a feature frequently found in 5′-untranslated regions, and lacks an upstream in-frame stop codon. For functional expression and in vitro translation experiments, the untranslated (UT) regions were removed and replaced with 182 bp 5′-UT region from the Shaker potassium channel. The β4 5′-GC-rich region was confirmed by genomic sequencing (P1 clone, Genome Systems, St. Louis). The genomic sequence is identical with the β4 cDNA sequence from position 1 to 673 (ends at CCAAG). After position 673 of the cDNA sequence, the genomic clone contains an intron.
The β3ΔN38-ball construct contained the N-terminal 19-aa “ball” from the β2 subunit (9), a 7-aa linker (AMGRHEG) followed by β3 (from amino acid 39). Chimeric “β1–β4 loop exchange” constructs β1Lβ4 and β4Lβ1 were made using the overlap extension method (26). The predicted protein sequence can be deduced from the sequence alignments (Fig. 1) by swapping the sequence between transmembrane regions TM1 and TM2 between β1 and β4. DNA was sequenced on both strands. Northern and Dot blots (CLONTECH) were hybridized at high stringency (9). Internal standards were β1, β2, and β4 cRNA transcripts (in 200 ng of human placenta total RNA as carrier) spotted on Nytran Plus (Schleicher & Schuell) membranes. The internal standards were processed together with the dot blot membranes, and signals were quantified with a phosphorimager.
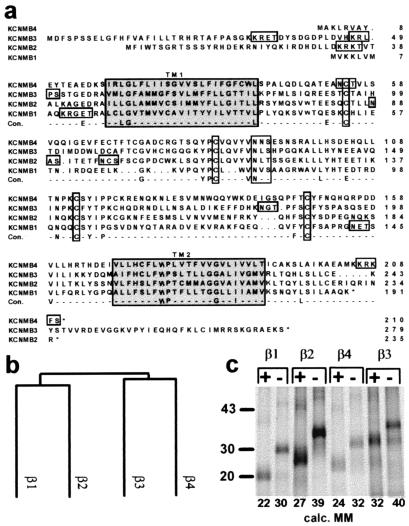
Figure 1.
Molecular properties of the MaxiK channel β3 and β4 subunits. (a) Multiple sequence alignment of four human MaxiK channel β subunits: β1 (KCNMB1) (8), β2 (KCNMB2) (9), β3 (KCNMB3) (28), and β4 (KCNMB4) (GenBank accession nos. β1, U25138; β2, AF099137; β3, AF160968; and β4, AF160967). Con, consensus sequences; TM1 and TM2, transmembrane regions (shaded boxes). Boxes show consensus sequences for protein kinase A phosphorylation (K/R,K/R,X1–2,S/T), N-linked glycosylation (N,X,S/T), and conserved cysteines. (b) Dendrogram of human β subunits. Length of vertical branches is inversely proportional to the similarity among sequences. The sequence similarity among the four human β subunits ranges from 36 to 54%. (c) In vitro translation and deglycosylation of β1–β4 mRNA. Transcripts were in vitro translated in a reticulocyte lysate in presence of microsomal membranes (Promega) and [35S]methionine. Microsomal proteins were treated with (+) or without N-glycosidase F (NEB) (−) and resolved by SDS/PAGE. Molecular weights were calculated with the gcg program; for each N-linked glycosylation site, 4 kDa was added to the molecular mass (calc. MM). Standards are in kilodaltons (kDa).
Chromosomal Localization.
The bacteriophage P1 clone carrying the human genomic β4 gene hybridized to the long arm of chromosome 12 using fluorescent in situ hybridization (Genome Systems). This localization was confirmed by cohybridization of the digoxigenin-labeled β4 genomic clone with a biotin-labeled probe specific for the centromere of chromosome 12. Hybridization was visualized with a fluoresceinated antidigoxigenin antibody (β4-probe) and Texas red avidin for the chromosome 12 centromere-specific probe.
Electrophysiology.
Xenopus laevis oocytes were injected with 2 ng of human MaxiK α subunit (hslo; GenBank accession no. U11058) and 4 ng of β subunit cRNAs, which corresponded to a ≈6 fold molar excess of β cRNA. Based on our previous studies with other β subunits, it is likely that β3 and β4 subunits are at saturating concentrations. HEK293T cells were transfected with β4 in pcDNA3. Pipette and bath solutions (mM): 110 K+-methanesulfonate, 5 KCl, 10 Hepes, 5 N-(2-hydroxyethyl)ethylenediamine triacetic acid, pH 7.0; patch pipettes had resistances of 1–2 MΩ. Free Ca2+ concentrations were calculated with chelator (27) and measured with a Ca2+ electrode. Data were analyzed as described (8). Values are mean ± SD. Student's t test was used. IbTx and CTx were a kind gift from M. L. Garcia or from commercial sources. BSA (0.001%) was present in all toxin and washing solutions.
Curve Fitting.
Association (kon) and dissociation (koff) rates, and equilibrium dissociation constants (Kd) were obtained by fitting the data to a first order bimolecular reaction scheme. The unblocking rate koff [s−1] was determined according to I(t) = (I0 − (I0 − Ir) * exp [−Δtw*koff] with I0, current before drug application; Δtw, time elapsed from the beginning of washout; and Ir, residual current before washout. The blocking rate kon [M−1s−1] was determined by a fit of the blocking reaction according to I(t) = Iss + (I0 − Iss) exp {−Δtb*([Tx]*kon + koff)} with Δtb, time elapsed from the beginning of drug application; and Iss, steady-state current at a given toxin concentration, Iss = 1/(1 + [Tx]/Kd) * I0. Kd = koff/kon.
Results
Cloning and Molecular Characteristics of the MaxiK β3 and β4 Subunits.
Screening public data bases reveals at least four human β subunit genes: (i) the smooth muscle β1 subunit (KCNMB1); (ii) the inactivating β2 subunit (KCNMB2), (iii) a β3 subunit (KCNMB3; Fig. 1) that is duplicated in the dup(3q) Syndrome region (28), and (iv) the neuronal β4 subunit characterized here (KCNMB4) (Fig. 1).
The β4 and β3 cDNAs were isolated from an arrayed, normalized human cDNA library as the available EST clones were incomplete at the N terminus. The β3 subunit (Fig. 1a) differs from the previously reported β3 protein (28) at the N terminus. Its functional analysis (Ca2+/V and IbTx/CTx sensitivities) did not yield any distinct phenotype when compared to the α subunit alone. However, addition of the β2 subunit inactivating ball to the N terminus of truncated β3 (β3ΔN38-ball) yielded inactivating currents suggesting that it interacts with the α subunit (not shown).
The similarity of the β4 and β3 proteins with the other members of the MaxiK channel β subunit family suggests a common structure with two membrane spanning regions (TM1 and TM2) separated by a large extracellular loop (Fig. 1a and Fig. 3b, Inset). The β4 protein has a consensus sequence for protein kinase A (PKA) phosphorylation at the C terminus; whereas β1, β2, and β3 carry PKA sites at the N terminus. The β4 and β3 extracellular loops contain two consensus sequences for N-linked glycosylation, one of these sites is conserved in all four human β subunit homologs, and the other is unique (Fig. 1a). Remarkable is the conservation of four cysteine residues among the β subunit family (12). A dendrogram illustrating the sequence similarities among β-subunit proteins shows that β4 is closer to the β3 subunit than to the β1 and β2 subunits (Fig. 1b).
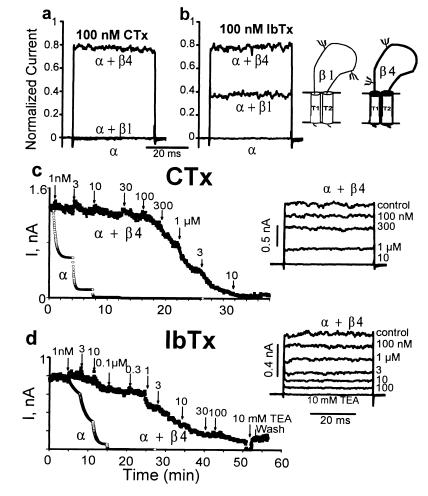
Figure 3.
CTx and IbTx resistant MaxiK channel formed by α + β4 subunits. Outside-out currents in 110 mM symmetrical K+ (100 μM Ca2+ in the pipette) from ooyctes expressing α + β4, α + β1 and α subunits. The holding potential was 0 mV. Pulses were to + 40 mV for 40 ms with 2-s interpulse interval. Channels with an open probability near 1 were examined. Currents were measured 3–5 min after toxin application. (a and b) Traces are normalized (to control currents before toxin application) currents after application of 100 nM CTx (a) or 100 nM IbTx (b). (Inset) Topology of β1 and β4 subunits. (c and d) Time course for CTx (c) and IbTx (d) block of α + β4 subunits. Arrows mark the time of toxin addition. (Insets) Original current traces. To illustrate the dramatic difference between α and α + β4 toxin blockade, we calculated the time course for the α subunit blockade using the CTx and IbTx average kon and koff values (see Fig. 4).
To confirm the proposed structural features of the β4 and β3 subunits, we performed in vitro translation in the presence of microsomes and deglycosylation with N-glycosidase F. Electrophoretic size separation of in vitro translated β-subunit proteins leads to bands of the expected molecular mass (Fig. 1c). Treatment with N-glycosidase (Fig. 1c, +) reduces the apparent molecular weight by ≈8 kDa (β1, β3, and β4) and ≈12 kDa (β2), consistent with the number of consensus sequences for N-linked glycosylation. Therefore, it is likely that all N-linked glycosylation sites in these MaxiK β subunits are available for N-glycosylation in the native protein.
The MaxiK Channel β4 Subunit mRNA Is Abundant in Brain.
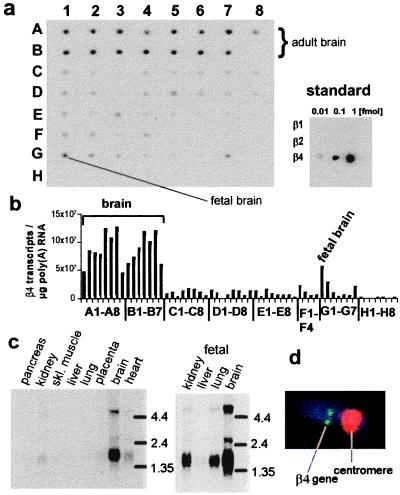
Dot blot and Northern blot analysis show strong β4 signals in all brain regions (Fig. 2a, lanes A and B, G1) with lower signals in many other tissues. Dot blot quantification (Fig. 2b) using an internal standard with known amounts of β-subunit transcripts (Fig. 2a, Inset) shows that β4 mRNA is abundant in brain. Assuming an average transcript length of 2,000 nucleotides, β4-subunit mRNA constitutes ≈0.01% of the total mRNA, which is comparable to the levels of the MaxiK β1-subunit transcripts in smooth muscle tissues (29).
Figure 2.
β4 mRNA is abundant in brain. (a) PolyA+ RNA Dot blot (CLONTECH) of 50 human tissues were hybridized at high stringency with a 32P-labeled β4 (681 bp, AccI–AccI fragment) probe. Tissues are: whole brain (1A), amygdala (2A), caudate nucleus (3A), cerebellum (4A), cerebral cortex (5A), frontal lobe (6A), hippocampus (7A), medulla oblongata (8A), occipital lobe (1B), putamen (2B), substantia nigra (3B), temporal lobe (4B), thalamus (5B), nucleus accumbens (6B), spinal cord (7B), heart (1C), aorta (2C), skeletal muscle (3C), colon (4C), bladder (5C), uterus (6C), prostate (7C), stomach (8C), testis (1D), ovary (2D), pancreas (3D), pituitary gland (4D), adrenal gland (5D), thyroid gland (6D), salivary gland (7D), mammary gland (8D), kidney (1E), liver (2E), small intestine (3E), spleen (4E), thymus (5E), peripheral leukocyte (6E), lymph node (7E), bone marrow (8E), appendix (1F), lung (2F), trachea (3F), placenta (4F), no sample (5F-8F, G8), fetal brain (G1), fetal heart (G2), fetal kidney (G3), fetal liver (G4), fetal spleen (G5), fetal thymus (G6), fetal lung (G7), 100 ng yeast total RNA (H1), 100 ng of yeast tRNA (H2), 100 ng of E. coli rRNA (H3), 100 ng of E. coli DNA (H4), 100 ng of poly r(A) (H5), 100 ng of human Cot 1 DNA (H6), 100 ng of human DNA (H7), and 500 ng of human DNA (H8). The amount of RNA in each dot is between 89 and 514 ng, due to a normalization with respect to ubiquitously expressed mRNAs. (Inset) Internal dot blot standard of β1, β2, and β4 transcripts. The internal standard shows a detection limit of 0.01 fmol or 6 × 106 transcripts. (b) Quantification of Dot blot signals using phosphorimaging. The signals were background subtracted and normalized to equal amounts of RNA. Absolute transcript numbers were obtained using the signals from the internal standard assuming similar hybridization efficiencies. (c) Northern Blots were hybridized and visualized as in a. The signals on the blot with fetal tissues are stronger than for the blot with adult tissues (c); however, for a comparison of expression levels, compare signals on the same blot (a and b). (d) β4 gene is localized in chromosome 12q15–21. Double labeling of chromosome 12 with a chromosome 12 centromere-specific probe (red signal) and genomic β4 probe (green signal) (n = 10 measurements).
Northern blot hybridization confirms the high levels of expression in the brain and shows that the predominant transcript size is ≈1.5 kb with weaker bands at ≈2.6 kb and 5 kb (Fig. 2c). Analysis of 25 3′-end β4 sequences in the EST database suggests that at least part of the transcript size heterogeneity may be explained by the use of different polyadenylation sites. The Dot blots and Northern blots (Figs. 2a–c) show that β4 RNA is present at lower levels in many smooth muscles, secretory tissues, heart, lung, and kidney but is not detectable in skeletal muscle, pancreas, liver, and bone marrow. Low levels of the β4-subunit mRNA are present in smooth muscles where β1 is abundant; thus, it is possible that more than one β subunit exists in a functional channel complex resulting in heteromultimeric channels composed of α/β1/β4 subunits with intermediate functional properties. Transcripts of the smooth muscle β1 subunit (29), the inactivating β2 subunit (9) and the β3 (our unpublished results; cf. ref. 28) subunit are virtually absent or scarce in adult brain (9). Thus, the β4 subunit seems to be the predominant β subunit in the brain. Fig. 2d shows that the β4 gene is located on chromosome 12 at position 12q15–21.
β4 Subunit Makes MaxiK Channels Resistant to Nanomolar Concentrations of CTx and IbTx.
Because β4 subunit transcripts are abundant in brain (Fig. 2a) and IbTx/CTx resistant MaxiK channel isoforms have been reported in neuronal preparations (13, 15, 22, 24), we decided to investigate if the β4 subunit influences CTx and IbTx block of the MaxiK channel pore-forming α subunit. Toxin block was assessed in outside-out patches excised from oocytes and HEK293T cells expressing α, α + β1, and α + β4 subunits. Application of 100 nM CTx, a concentration that completely blocks the channel formed by the α subunit, and α + β1 subunits, blocks only ≈20% of the α + β4 current within 5 min (Figs. 3a and c). The effect of CTx over a large concentration range (1 nM–10 μM) was investigated in cumulative dose-response experiments (Fig. 3c). Currents were evoked every 2 s by pulses from 0 to + 40 mV in 100 μM Ca2+ (open probability ≈1). Due to the slow nature of the toxin equilibration process (see below), cumulative toxin additions were made at ≈5 min intervals. During ≈5 min exposure per toxin concentration, the β4 subunit lead to a diminution of the apparent half-maximal inhibitory concentration (IC50) of the α subunit from 1 ± 0.5 nM (n = 3) to 365 ± 17 nM (n = 3). In contrast, in presence of the β1 subunit the change in the α subunit apparent toxin sensitivity (2.8 ± 1 nM, n = 5) was not significant (P > 0.05).
The same experimental conditions were used to evaluate the effect of the selective MaxiK channel blocker, IbTx. As with CTx, MaxiK channels associated with the β4 subunit are much less sensitive to the blocking effects of IbTx (Fig. 3 b and d). The IC50 was 2.6 ± 0.3 μM (n = 3) in α + β4 channels as compared with 3.6 ± 0.6 nM (n = 3) in α subunit alone. However, in contrast to CTx, which shows the same potency on α and α + β1 channels (Fig. 3a), the apparent IbTx sensitivity is clearly reduced in channels formed by α + β1 subunits (IC50 = 65 ± 7 nM, n = 3; Fig. 3b). Note that within 3–5 min, toxin blockade of α + β4 channels does not reach steady-state (Fig. 3c and d; see Fig. 4). In the experiment shown in Fig. 3d, IbTx up to 100 μM did not fully block α + β4 channels, whereas 10 mM tetraethylammonium fully abolished the remaining current in a reversible manner as expected for a MaxiK current. Results of IbTx and CTx blockade obtained in HEK293T cells for α+ β4 were similar to those found in oocytes (not shown).
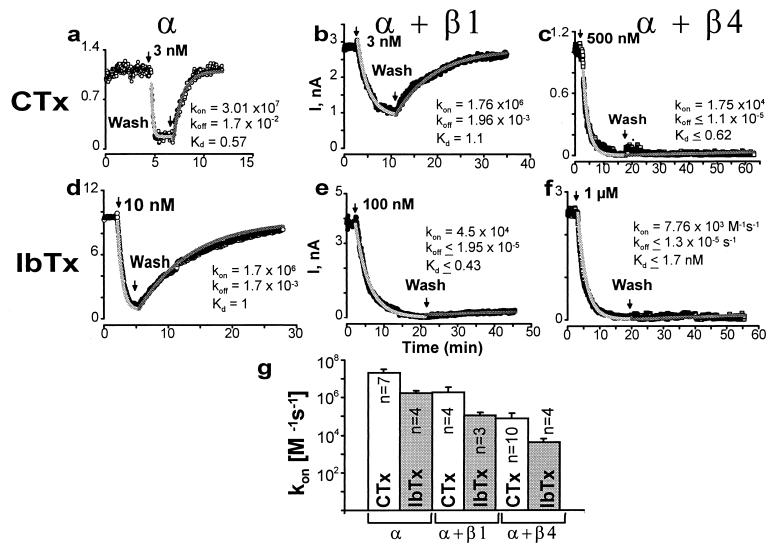
Figure 4.
β4 subunit dramatically slows down CTx and IbTx association and dissociation. Currents in outside-out patches were evoked by repetitive pulses as in Fig. 3. Currents were measured at the end of the pulse and plotted as a function of time; arrows mark the time of toxin application and washout. To attain toxin block in a reasonable time, CTx concentrations were 3 nM for α (a) and α + β1 (b), and 500 nM for α + β4 (c). IbTx concentrations were 10 nM for α (d), 100 nM for α + β1 (e), and 1 μM for α + β4 (f). kon, koff, and Kd values are given in M−1s−1, s−1, and nM, respectively, and were obtained as described in Experimental Procedures. Gray lines are fits to data. (g) Mean kon values (M−1s−1) ± SD (error bars) for CTx and IbTx block in α, α + β1, and α + β4 channels. CTx: kon-α = 2 ± 1 × 107 (n = 7), kon-α+β1 = 1.9 ± 1.8 × 106 (n = 4), and kon-α+β4 = 8 ± 7 × 104 (n = 10); IbTx: kon-α = 2 ± 0.7 × 106 (n = 4), kon-α+β1 = 1.1 ± 0.5 × 105 (n = 3), and kon-α+β4 = 4 ± 2 × 103 (n = 4). n = number of experiments. For both toxins, values for α + β1 and α + β4 were significantly different from those of α subunit (P < 0.005). In two out of seven cases, IbTx block of α + β4 channels had even slower on-rates (not included in mean value).
Coexpression of the β4 Leads to a Dramatic Slowdown of IbTx and CTx Channel Blockade.
We noticed that the blocking kinetics for both CTx and IbTx are slower in the presence of the β4 subunit (Fig. 3 c and d), when compared with the toxin blockade of the α subunit alone. To directly address the effect of β4 subunit on toxin blocking kinetics, we performed toxin wash-in and wash-out experiments on channels formed by α + β4, α + β1, and the α subunit alone. Currents were evoked as described in the legend for Fig. 3 and a single dose of CTx or IbTx was applied to the extracellular surface of the channel. For blocking effects to occur in a reasonable time period, the toxin concentration was increased from 3 nM (α and α + β1) to 500 nM (α + β4) for CTx (Fig. 4a-c), and for IbTx, the concentration was raised from 10 nM (α) to 100 nM (α + β1) to 1 μM (α + β4) (Fig. 4 d–f). After a substantial block, the toxin was washed-out. The association and dissociation rates were determined by fitting the data to a simple bimolecular blocking reaction. The association of the toxins to the channel was ≈250–1,000 fold slower in presence of β4 subunit; whereas it was only ≈10–20 fold slower in the presence of β1 subunit (mean values ± SD in Fig. 4g). β1 subunit led to a proportional decrease of both CTx association and dissociation (koff) rates, resulting in a similar Kd value as that of the α subunit alone (≈1 nM) (Fig. 4 a and b). Because toxin block is essentially irreversible in the presence of the β4 subunit (up to 50 min wash; Fig. 4 c and f), we can only estimate an upper limit for the koff and Kd values under the assumption that a bimolecular blocking reaction is still applicable. Therefore, the affinity for toxins at equilibrium may even be higher in the presence of the β4 subunit. The same is true for IbTx block of α + β1 channels (Fig. 4e).
The kinetic experiments (Fig. 4) indicate that the apparent IbTx and CTx insensitivity of α + β4 channels under nonequilibrium conditions, as shown in Fig. 3, is the result of the slow toxin association rate, even although they probably have a higher affinity under equilibrium conditions. Thus, the apparent values for half-maximal blockade (IC50) in cumulative dose-response experiments are much higher than the actual equilibrium dissociation constants (Kd values).
Resistance to Toxin Block Is Determined by the Extracellular Loop of the β4 subunit.
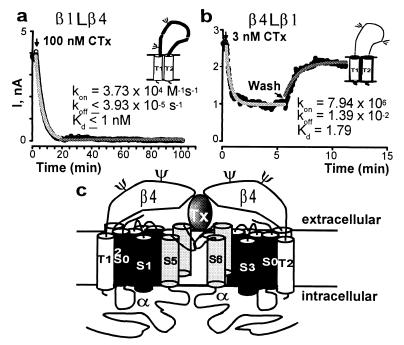
MaxiK channel β subunits have large extracellular loops (Fig. 3A, Inset), which may interfere with the IbTx and CTx blocking and unblocking reactions. To test this hypothesis, we exchanged the extracellular loops between the smooth muscle β1 subunit and the neuronal β4 subunit, coexpressed the chimeras together with the α subunit, and tested for toxin block in wash-in, wash-out experiments. Channels coexpressed with the β1 subunit carrying the extracellular loop of the β4 subunit (β1Lβ4) show toxin blocking characteristics indistinguishable from those coexpressed with the wild-type β4 subunit, both in terms of toxin association and irreversibility (Fig. 5a). These results show that the β4 extracellular loop is responsible for the dramatic effects on CTx and IbTx blocking and unblocking kinetics. Furthermore, when the β4 subunit carrying the extracellular loop of β1 subunit (β4Lβ1) was coexpressed, channels were highly sensitive to toxin block, as expected for the β1 phenotype; although, their on and off rates were slightly faster (Fig. 5b). Thus, the extracellular loops of β1 and β4 subunits determine the kinetics of toxin block of the MaxiK channel α subunit (Fig. 5c).
Figure 5.
The extracellular loops of β4 and β1 subunit determine the kinetics of toxin block. Experiments were performed as in Fig. 4. (a) 100 nM CTx was applied to α + β1Lβ4 channels (β1 carrying the loop of β4) and 3 nM CTx (b) to α + β4Lβ1 channels (β4 carrying the loop of β1). Mean kon values (M−1s−1) ± SD were: for α + β1Lβ4, 4 ± 2 × 104 (n = 3); for α + β4Lβ1, 7 ± 2 × 106 (n = 4). Mean koff values (s−1) were for α + β4Lβ1, 1 ± 0.2 × 10−2. (c) Model of channels formed by α + β subunits where the β-subunit extracellular loop may contribute to the toxin (Tx) receptor. Two of the four subunits are shown.
Discussion
The receptor for IbTx and CTx binding in MaxiK channels resides in the pore of its α subunit (30–32). Our results show that the neuronal MaxiK channel β4 subunit and the smooth muscle β1 subunit influence the toxin-channel interaction to different degrees. Remarkably, in the presence of the neuronal β4 subunit the toxin association is so slow that α + β4 channels become apparently toxin resistant, suggesting that α + β4 channels form the molecular basis for “CTx and/or IbTx insensitive” MaxiK channels found in neurons (13, 22, 23).
The dramatic slowing of the IbTx and CTx association in the presence of MaxiK channel β subunits has important implications for the usage of IbTx in dissecting the physiological role of MaxiK channels in native cells and tissues. For MaxiK channels formed by human α + β1 (smooth muscle), it would take ≈20 min for 100 nM IbTx to completely block channel activity (Fig. 4e). In channels formed by human α + β4 (neuronal) at least 1 μM IbTx is required for a complete block within 20 min. In tissue preparations, additional limitations in toxin diffusion may demand even longer toxin applications and/or higher toxin concentrations.
Besides toxin insensitive MaxiK channels, neurons also express IbTx and CTx sensitive MaxiK channels (13, 33). It is therefore likely that despite the high mRNA level of the β4 subunit in brain, not all neuronal MaxiK channel α subunits are associated with this neuronal β4 subunit. It is tempting to speculate that the GC-rich 5′-UT region in the β4 mRNA inhibits protein expression and that neuronal MaxiK channel β4 subunit expression is dependent on locally regulated translation. In accordance, toxin sensitive and insensitive MaxiK channels are differentially expressed in the cell bodies and nerve endings in supraoptic magnocellular neurons (22, 23).
Our kinetic analysis shows that besides slowing down the toxin association (≈250- to 1,000-fold), β4 causes an even greater effect on toxin dissociation kinetics resulting in an apparently irreversible block. This leads to the paradox that despite the apparent toxin resistance of α + β4 MaxiK channels, the actual toxin affinity at equilibrium is higher for α + β4 channels than for channels formed by the α subunit alone. We cannot exclude, that channels formed by α + β4 subunit become truly irreversibly blocked, in which case the concept of equilibrium would not be applicable.
When α + β4 currents are partially blocked by IbTx or CTx (not at steady-state), we always observe that a fraction of blocked channels recover upon toxin washout (not shown). One possible explanation for this observation may be that there are at least two sequential toxin-blocked states, a superficial reversible blocked state, and a deeper, apparently irreversible blocked state. Thus, the model of a bimolecular blocking reaction used to fit our data is only an approximation and the actual blocking process of β subunit associated MaxiK channels may be more complicated.
Mechanistically, the changes in toxin binding induced by MaxiK β subunits may be explained by (i) an allosteric mechanism, (ii) a direct contribution of parts of the β subunit to the toxin binding site, (iii) steric hindrance for toxin binding and unbinding, or (iv) a combination of all of these factors. Our results showing that the loops of β1 and β4 subunits determine the toxin association and dissociation characteristics of MaxiK channels formed by α + β subunits (Fig. 5), favor a model where the β subunit loops form part of the toxin receptor and a structural barrier for the travel of the rather bulky toxin molecules into and from their binding site.
Structurally, it is conceivable that the extracellular loops of the MaxiK channel β subunits may form an extended external vestibule, covering the otherwise rather flat entrance of the MaxiK channel α subunit pore (34), analogous to the extracellular pore-flanking regions that form the external vestibule in Na+ and Ca2+ channels.
Electrophysiological and binding experiments show discrepancies in the Kd values obtained for CTx block of α and α + β1 channels. The Kd for CTx in binding studies decreases from ≈50 pM to ≈1 pM in the presence of β1 subunit (12, 35) whereas in our electrophysiological measurements the Kd value is not altered (≈1 nM) (Fig. 4 a and b). This apparent discrepancy is likely due to different experimental conditions especially ionic strength (10 mM Na+ vs. 110 mM K+) because it was shown that the affinity for CTx is much higher at low ionic strength (36, 37). Similar, although less pronounced, ionic strength effects have been reported for IbTx blockade on skeletal muscle MaxiK channels (30).
Conserved cysteine residues and Leu-90, Tyr-91, Thr-93, and Glu-94 (see Fig. 1) in the smooth muscle β1 subunit are critical for the enhanced binding of CTx to α + β1 channels in 10 mM sodium (12). It will be interesting to study the effect of mutations in the equivalent residues in β4 on CTx and IbTx block under physiological ionic strength.
Recently, the biochemical purification of a glycosylated neuronal MaxiK channel subunit with an apparent molecular weight of 25 kDa has been reported (38). The difference in molecular weight of ≈10 kDa with the β4 subunit reported here (32 kDa; Fig. 1c) makes it unlikely that these two subunits are identical.
In summary, we have characterized a human MaxiK channel β subunit from brain, which leads to channels with an apparently low sensitivity to IbTx and CTx caused by a very slow toxin association rate. The β4 subunit extracellular loop is the domain that confers the resistance to toxin block. The high levels of mRNA in the brain suggest that β4 subunits form the molecular basis of toxin-insensitive MaxiK channels reported in neurons.
Acknowledgments
We thank Drs. Adrian Gross, Enrico Stefani, and Francisco Bezanilla for critically reading the manuscript. This work was supported by an American Heart Association National Center Grant-in-Aid 9750745N (P.M.), American Heart Association Western States Affiliate Beginning Grant in-Aid 9960028Y (M.W.), National Institutes of Health Grant HL54970, and Human Frontier Science Program grants (L.T.). L.T. is an Established Investigator of the American Heart Association.
Abbreviations
- MaxiK channel
large conductance, voltage- and Ca2+-activated K+ channel
- CTx
charybdotoxin
- IbTx
iberiotoxin
Note Added in Proof
A biophysical characterization of the same KCNMB3 and KCNMB4 was recently reported by Brenner et al. (39).
Footnotes
Data deposition: The sequences reported in the paper have been deposited in the GenBank database [accession nos. AF160968 (KCNMB3) and AF160967 (KCNMB4)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100118597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100118597
References
- 1.Adelman J P, Shen K Z, Kavanaugh M P, Warren R A, Wu Y N, Lagrutta A, Bond C T, North R A. Neuron. 1992;9:209–216. doi: 10.1016/0896-6273(92)90160-f. [DOI] [PubMed] [Google Scholar]
- 2.Knaus H G, Folander K, Garcia-Calvo M, Garcia M L, Kaczorowski G J, Smith M, Swanson R. J Biol Chem. 1994;269:17274–17278. [PubMed] [Google Scholar]
- 3.McManus O B, Helms L M, Pallanck L, Ganetzky B, Swanson R, Leonard R J. Neuron. 1995;14:645–650. doi: 10.1016/0896-6273(95)90321-6. [DOI] [PubMed] [Google Scholar]
- 4.Meera P, Wallner M, Toro L. In: Channels in Cardiovascular Biology. Archer S L, Rosch N, editors. New York: Plenum; 1999. [Google Scholar]
- 5.Latorre R, Oberhauser A, Labarca P, Alvarez O. Ann Rev Physiol. 1989;51:385–399. doi: 10.1146/annurev.ph.51.030189.002125. [DOI] [PubMed] [Google Scholar]
- 6.Wallner M, Meera P, Ottolia M, Kaczorowski G, Latorre R, Garcia M L, Stefani E, Toro L. Recept Channels. 1995;3:185–199. [PubMed] [Google Scholar]
- 7.Ramanathan K, Michael T H, Jiang G J, Hiel H, Fuchs P A. Science. 1999;283:215–217. doi: 10.1126/science.283.5399.215. [DOI] [PubMed] [Google Scholar]
- 8.Meera P, Wallner M, Jiang Z, Toro L. FEBS Lett. 1996;382:84–88. doi: 10.1016/0014-5793(96)00151-2. [DOI] [PubMed] [Google Scholar]
- 9.Wallner M, Meera P, Toro L. Proc Natl Acad Sci USA. 1999;96:4137–4142. doi: 10.1073/pnas.96.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia X M, Ding J P, Lingle C J. J Neurosci. 1999;19:5255–5264. doi: 10.1523/JNEUROSCI.19-13-05255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dworetzky S I, Boissard C G, Lum-Ragan J T, McKay M C, Post-Munson D J, Trojnacki J T, Chang C P, Gribkoff V K. J Neurosci. 1996;16:4543–4550. doi: 10.1523/JNEUROSCI.16-15-04543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanner M, Vianna-Jorge R, Kamassah A, Schmalhofer W A, Knaus H G, Kaczorowski G J, Garcia M L. J Biol Chem. 1998;273:16289–16296. doi: 10.1074/jbc.273.26.16289. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart P H, Chung S, Levitan I B. Neuron. 1989;2:1031–1041. doi: 10.1016/0896-6273(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 14.Marrion N V, Tavalin S J. Nature (London) 1998;395:900–905. doi: 10.1038/27674. [DOI] [PubMed] [Google Scholar]
- 15.Yazejian B, DiGregorio D A, Vergara J L, Poage R E, Meriney S D, Grinnell A D. J Neurosci. 1997;17:2990–3001. doi: 10.1523/JNEUROSCI.17-09-02990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gola M, Crest M. Neuron. 1993;10:689–699. doi: 10.1016/0896-6273(93)90170-v. [DOI] [PubMed] [Google Scholar]
- 17.Meech R. Nature (London) 1989;340:594. doi: 10.1038/340594a0. [DOI] [PubMed] [Google Scholar]
- 18.Saria A, Seidl C V, Fischer H S, Koch R O, Telser S, Wanner S G, Humpel C, Garcia M L, Knaus H G. Eur J Pharmacol. 1998;343:193–200. doi: 10.1016/s0014-2999(97)01553-7. [DOI] [PubMed] [Google Scholar]
- 19.Fischer H S, Saria A. Neurosci Lett. 1999;263:208–210. doi: 10.1016/s0304-3940(99)00116-0. [DOI] [PubMed] [Google Scholar]
- 20.Kaczorowski G J, Garcia M L. Curr Opin Chem Biol. 1999;3:448–458. doi: 10.1016/S1367-5931(99)80066-0. [DOI] [PubMed] [Google Scholar]
- 21.Mienville J M, Barker J L. Pflügers Arch. 1996;431:763–770. doi: 10.1007/BF02253841. [DOI] [PubMed] [Google Scholar]
- 22.Dopico A M, Widmer H, Wang G, Lemos J R, Treistman S N. J Physiol (London) 1999;519:101–114. doi: 10.1111/j.1469-7793.1999.0101o.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, Thorn P, Lemos J R. J Physiol (London) 1992;457:47–74. doi: 10.1113/jphysiol.1992.sp019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meis S, Pape H C. J Neurophysiol. 1997;78:1256–1262. doi: 10.1152/jn.1997.78.3.1256. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton R M, Cai Z L, Ho S N, Pease L R. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 27.Schoenmakers T J, Visser G J, Flik G, Theuvenet A P. BioTechniques. 1992;12:870–879. [PubMed] [Google Scholar]
- 28.Riazi M A, Brinkman-Mills P, Johnson A, Naylor S L, Minoshima S, Shimizu N, Baldini A, McDermid H E. Genomics. 1999;62:90–94. doi: 10.1006/geno.1999.5975. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Z, Wallner M, Meera P, Toro L. Genomics. 1999;55:57–67. doi: 10.1006/geno.1998.5627. [DOI] [PubMed] [Google Scholar]
- 30.Candia S, Garcia M L, Latorre R. Biophys J. 1992;63:583–590. doi: 10.1016/S0006-3495(92)81630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKinnon R, Miller C. J Gen Physiol. 1988;91:335–349. doi: 10.1085/jgp.91.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meera P, Wallner M, Song M, Toro L. Proc Natl Acad Sci USA. 1997;94:14066–14071. doi: 10.1073/pnas.94.25.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholz A, Gruss M, Vogel W. J Physiol (London) 1998;513:55–69. doi: 10.1111/j.1469-7793.1998.055by.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stampe P, Kolmakova-Partensky L, Miller C. Biochemistry. 1994;33:443–450. doi: 10.1021/bi00168a008. [DOI] [PubMed] [Google Scholar]
- 35.Hanner M, Schmalhofer W A, Munujos P, Knaus H G, Kaczorowski G J, Garcia M L. Proc Natl Acad Sci USA. 1997;94:2853–2858. doi: 10.1073/pnas.94.7.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson C S, MacKinnon R, Smith C, Miller C. J Gen Physiol. 1988;91:317–333. doi: 10.1085/jgp.91.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacKinnon R, Latorre R, Miller C. Biochemistry. 1989;28:8092–8099. doi: 10.1021/bi00446a020. [DOI] [PubMed] [Google Scholar]
- 38.Wanner S G, Koch R O, Koschak A, Trieb M, Garcia M L, Kaczorowski G J, Knaus H G. Biochemistry. 1999;38:5392–5400. doi: 10.1021/bi983040c. [DOI] [PubMed] [Google Scholar]
- 39.Brenner R, Jegla T J, Wickenden A, Liu Y, Aldrich R W. J Biol Chem. 2000;275:6453–6461. doi: 10.1074/jbc.275.9.6453. [DOI] [PubMed] [Google Scholar]