SUMMARY
G12 class heterotrimeric G proteins stimulate RhoA activation by RGS-RhoGEFs. However, p115RhoGEF is a GTPase Activating Protein (GAP) towards Gα13 while PDZRhoGEF is not. We have characterized interaction between the PDZRhoGEF rgRGS domain (PRG-rgRGS) and the alpha subunit of G13, and determined crystal structures of their complexes in the inactive state bound to GDP, and in the active states bound to GDP·AlF (transition state) and GTPγS (Michaelis complex). PRG-rgRGS interacts extensively with the helical domain and the effector binding sites on Gα13 through contacts that are largely conserved in all three nucleotide-bound states, although PRG-rgRGS has highest affinity to the Michaelis complex. An acidic motif in the N-terminus of PRG-rgRGS occupies the GAP binding site of Gα13 and is flexible in the GDP·AlF complex, but well ordered in the GTPγS complex. Replacement of key residues in this motif with their counterparts in p115RhoGEF confers GAP activity.
INRODUCTION
Heterotrimeric G proteins (Gα) and the monomeric G proteins of the Ras superfamily are characterized by a GTPase cycle in which signaling activity is achieved by the exchange of GDP for GTP at the G protein catalytic site and inactivation is accomplished by hydrolysis of the bound GTP (Bokoch and Der, 1993; Gilman, 1987; Sprang, 1997). The modulated rates of these two processes regulate the GTPase cycle and determine the amount of activated G protein available at any given time. Exchange of guanine nucleotides is largely limited by the rate of dissociation of GDP; proteins that stimulate this rate of exchange are called Guanine nucleotide Exchange Factors (GEFs). The intrinsic GTPase activity of G proteins can be stimulated by GAPs (GTPase activating proteins) (Donovan et al., 2002; Geyer and Wittinghofer, 1997). Many proteins that contain RGS (Regulator of G protein Signaling) domains are GAPs for heterotrimeric G proteins (Berman and Gilman, 1998; Ross and Wilkie, 2000). Several G protein effectors also function as GAPs for their activating G proteins, including PLCβ for Gq (Biddlecome et al., 1996) and members of the RGS domain-containing RhoGEFs for G12 and G13 (Kozasa et al., 1998).
RGS-containing RhoGEFs (RGS-RhoGEFs), including p115RhoGEF, LARG, PDZRhoGEF and its rat splice variant GTRAP48, function as potential regulatory links between G protein coupled receptors that activate the G12 class of heterotrimeric G proteins and Rho-mediated pathways that lead to cytokinesis and transformation (Sternweis et al., 2007). The C-terminal halves of these RGS-RhoGEFs contain tandemly arranged Dbl homology (DH) and pleckstrin homology (PH) domains that constitute the functional unit of GEF activity in most Rho-directed GEFs (Cerione and Zheng, 1996). The N-terminal portions of RGS-RhoGEFs possess RhoGEF-RGS (rgRGS) domains. These domains share remote sequence similarity to the RGS family of protein domains, which are characterized by a conserved ~120 residue helical fold (the “RGS-box”) (Tesmer et al., 1997). In contrast to the canonical RGS domains, rgRGS domains require elements outside of the conserved RGS-box for structural integrity (Chen et al., 2001) and to interact with activated Gα12 or Gα13 (Wells et al., 2002).
Crystal structures of the rgRGS domain of p115RhoGEF (p115-rgRGS), a GAP for Gα13, have been solved by itself (Chen et al., 2001) and as a complex with a chimeric Gα13/i1 subunit (Chen et al., 2005). The latter structure revealed a mode of interaction between the rgRGS and Gα13 that was very different from those observed in other RGS·Gα complexes (Slep et al., 2001; Tesmer et al., 1997). Like other RGS domains with GAP activity, p115-rgRGS stabilizes the GDP·AlF4−·Mg2+-bound conformation of Gα13. GDP·AlF4−·Mg2+ (GDP·AlF) appears to be an analog of the transition state for GTP hydrolysis (Coleman et al., 1994; Sondek et al., 1994). Hence, the conformation of Gα that was observed in this complex represents a catalytically activated, or "transition state" form of the enzyme, in contrast to that of the Michaelis complex with GTP and Mg2+ (Coleman and Sprang, 1999; Noel et al., 1993). In contrast to GAP proteins that contain canonical RGS domains, the N-terminal subdomain of rgRGS that lies outside of the RGS-box mediates GAP activity of p115RhoGEF, and the interface between the RGS-box and Gα resembles that of a Gα·effector complex (Chen et al., 2005).
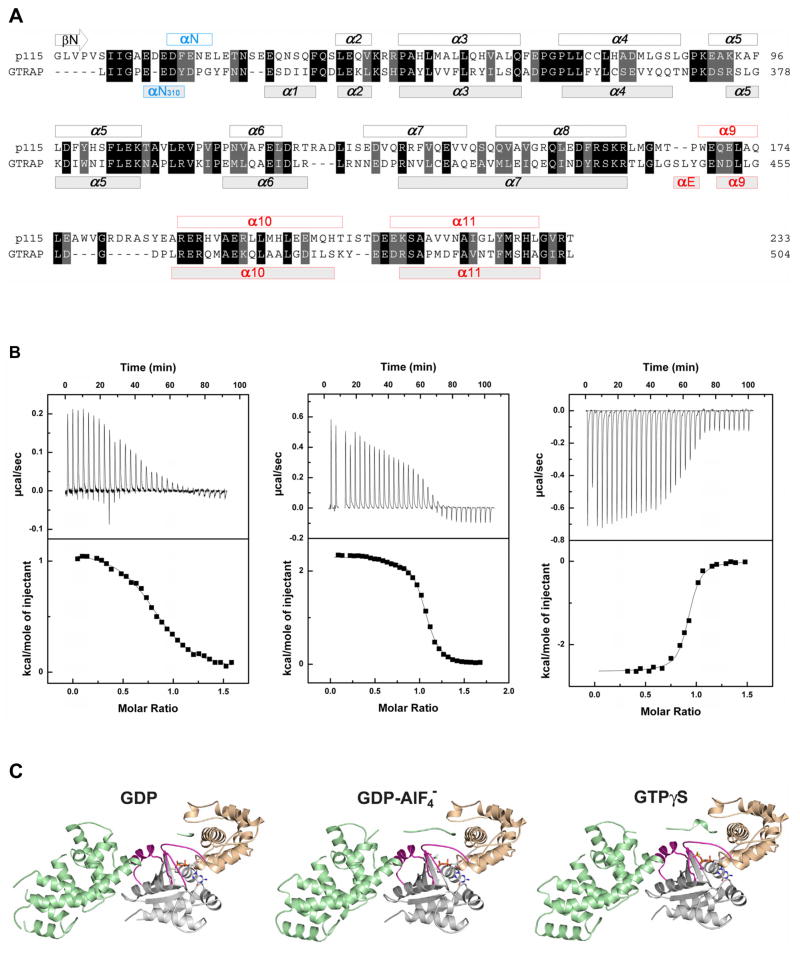
Gα13 binds PDZRhoGEF and GTRAP48 (Jackson et al., 2001) and stimulates the GEF activity of these RhoGEFs in vivo (Fukuhara et al., 1999). However, neither PDZRhoGEF nor GTRAP48 has GAP activity towards Gα13 (Wells et al., 2002). The N-terminal subdomain of p115RhoGEF possesses a series of acidic amino acid residues followed by an aromatic residue (Figure 1A), 27EDEDF, which stably interacts with the GTPase active site of Gα13 in the transition state (Chen et al., 2005). Substitution of the phenylalanine residue in the EDEDF motif, or of certain acidic residues, is sufficient to abolish GAP activity (Chen, et al, 2003). The rgRGS domain from PDZRhoGEF/GTRAP48 (PRG-rgRGS) possesses a similar N-terminal motif, 312EEDY, bordered by two proline residues (Figure 1A). The absence of GAP activity in PDZRhoGEF was attributed to this shortened acidic sequence, and substitution of phenylalanine for tyrosine in the corresponding EEDY motif (Chen et al., 2005). PRG-rgRGS is otherwise structurally similar to p115-rgRGS (Longenecker et al., 2001). Both have an RGS-box-like helical domain followed by a layer of additional 3–4 helices packed against the RGS-box, which is the locus of sequence and structural divergence between the two rgRGS domains (Figure 1A).
Figure 1.
Structures of PRG-rgRGS:Gα13 complexes. (A) Structure based sequence alignment of p115-rgRGS and PRG-rgRGS domains. Helices are represented with bars on top (for p115-rgRGS) and at bottom (for PRG-rgRGS) of the amino acid sequences. Additional elements N- and C-terminal to the consensus RGS-box are colored blue and red, respectively. (B) ITC profile for the binding of PRG-rgRGS to Gα13·GDP (left), Gα13·GDP·AlF4− (middle), and Gα13·GTPγS (right). Nonlinear least squares fit using a "one set of sites" model resulted in the fit shown (solid line). (C) Ribbon diagrams depicting tertiary structures of PRG-rgRGS in a complex with Gα13·GDP (left), Gα13·GDP·AlF4− (middle), and Gα13·GTPγS (right). The Ras-like domain of Gα13 is colored gray, with the switch regions colored purple. The helical domain of Gα13 is colored wheat. PRG-rgRGS is colored cyan. GDP, AlF4−, Mg2+ and GTPγS are depicted as ball-and-stick models and colored as follows: Oxygen, nitrogen, carbon and phosphorus atoms are colored red, blue, gray and yellow, respectively. Magnesium is colored green. AlF4− is colored light blue. Water molecules are colored red.
Here, we describe the results of an investigation aimed at understanding why p115-rgRGS is a GAP towards Gα13, whereas PRG-rgRGS is not. We show by thermodynamic analysis that PRG-rgRGS has nearly equal sub-micromolar affinity for the GTPγS- and GDP·AlF-bound states of Gα13, and micromolar affinity for the GDP-bound form of the G protein. Crystal structures of all three complexes show Gα13 in the activated state characteristic of GTP- and GDP·AlF-bound Gα proteins. PRG-rgRGS forms essentially identical contacts with the effector-binding sites of Gα13 in all three nucleotide-bound states. In contrast, the N-terminal segment of PRG-rgRGS, which occupies the GAP-binding site, adopts different conformations in the GTPγS and GDP·AlF-bound states of Gα13. Biochemical studies with site-directed mutants of PRG-rgRGS reveal the residues required to convert PRG-rgRGS into a GAP for Gα13. The results of these studies demonstrate that members of the RGS-RhoGEF family have evolved distinct recognition mechanisms for the transition-state conformation of Gα versus its Michaelis complex with GTP.
RESULTS
PRG-rgRGS Preferentially Binds the GTPγS Complex of Gα13
RGS domains for the most part bind with highest affinity to the transition state conformation (GDP·AlF) of Gα proteins (Berman et al., 1996; Chen et al., 1996; Watson et al., 1996). Thus, the dissociation constants for binding of RGS4 with the GDP·AlF, GppNHp·Mg2+ and GDP-bound forms of Gαi1 were measured by isothermal titration calorimetry (ITC) to be 0.1, 5, and 400 μM, respectively (Thomas et al., 2004). Similarly, p115-rgRGS binds to the GDP·AlF complex of Gα13 with 10-fold higher affinity than to GTPγS-bound Gα13 (Chen et al., 2005). In contrast, PRG-rgRGS was found by ITC to bind with highest affinity (Kd ~ 300 nM) to the GTPγS-bound form of Gα13 and with somewhat lower affinity (Kd ~500 nM) to the GDP·AlF complex (Figure 1B, Table 1). PRG-rgRGS has considerably lower, but substantial, affinity for Gα13·GDP (Kd ~ 4–5 μM). Thus, PRG-rgRGS is unusual among RGS proteins in that it discriminates among the three nucleotide-bound forms of Gα with less than 2 kcal/mol of binding free energy, and binds with nearly equal affinity to Gα13 stabilized by GTP or GDP·AlF in the active conformation. Association of PRG-rgRGS with GTPγS-bound Gα13 is exothermic in contrast to endothermic binding to GDP·AlF and GDP, a difference that may be attributed to the divergent binding mechanisms revealed by the crystal structures of the complexes.
Table 1.
Thermodynamic parameters of the binding of PRG-rgRGS and Gα13
| N | Kd | ΔH | ΔG | ΔS | |
|---|---|---|---|---|---|
| nM | kcal mol−1 | kcal mol−1 | cal mol−1 K−1 | ||
| GDP | 0.9 | 4800 | 1.1 | −6.8 | 28 |
| GDP·AlF4− | 1.1 | 520 | 2.3 | −8.0 | 37 |
| GTPγS | 0.9 | 340 | −2.6 | −8.4 | 20 |
Thermodynamic parameters for the binding of PRG-rgRGS to different forms of Gα13.
Determinations were made using ITC as described in experimental procedures. Kd, ΔH, ΔG, ΔS, and N represent the dissociation constant, enthalpy, free energy, entropy, and stoichiometry, respectively. Results shown are for a single experiment. Error in ΔH is ~10% based on similar experiments using the same ITC equipment.
Structures of PRG-rgRGS:Gα13 Complexes
Complexes of PRG-rgRGS were prepared with N-terminally truncated Gα13 bound to either GDP·AlF, GTPγS or GDP (see Experimental procedures). Although binding of GDP to Gα does not require Mg2+ (Gilman, 1987), the metal ion is required for formation of the PRG-rgRGS:Gα13·GDP complex. Crystal structures of the PRG-rgRGS:Gα13·GDP, PRG-rgRGS:Gα13·GDP·AlF, and PRG-rgRGS:Gα13·GTPγS complexes (Figure 1C) were determined at 2.5 Å, 2.25 Å and 2.0 Å resolution, respectively (Table 2). The N-terminal subdomain (residues 307–318) of PRG-rgRGS, which is disordered in the structure of PRG-rgRGS alone (Longenecker et al., 2001), is partially ordered in the Gα13·GDP complex, and well ordered in complexes with the activated (GDP·AlF- and GTPγS-bound) forms of Gα13 (Figure 1C). Structural differences between the free and Gα13-bound RGS-box subdomain (residues 322–502, root mean square difference for all atoms is 1.09Å) are largely confined to residues with high temperature (B) factors, and lowest in segments that contact Gα13. The interface between PRG-rgRGS and activated Gα13, which buries a solvent accessible surface area of over 3,000 Å2, is similar to that observed in the p115-rgRGS:Gα13/i1 complex (Chen et al., 2005). However, in contrast to the latter, the PRG-rgRGS:Gα13 complexes do not form symmetric dimers.
Table 2.
Data collection and refinement statistics
| GDP | GDP·AlF | GTPγS | |
|---|---|---|---|
| Data Collection | |||
| Source | APS SBC 19ID | APS GM/CA 23ID | APS SBC 19ID |
| Wavelength (Å) | 0.97921 | 0.97939 | 0.97934 |
| Space group | P212121 | P212121 | P212121 |
| Unit cell (Å) | |||
| a | 58.04 | 57.95 | 58.11 |
| b | 66.87 | 66.37 | 66.12 |
| c | 150.90 | 151.19 | 152.52 |
| Dmin (Å) | 2.50 | 2.25 | 2.00 |
| Unique reflections | 20,508 | 26,607 | 39,804 |
| Redundancy 1 | 6.4 | 5.9 | 5.2 |
| Rsym (%) 1,2 | 8.6 (57.4) | 8.6 (41.1) | 9.2 (46.7) |
| Completeness (%)1 | 98.0 (83.3) | 93.6 (60.2) | 98.1 (90.7) |
| <I>/σ <I>1 | 17.0 (1.5) | 25.6 (2.0) | 22.9 (2.3) |
| Mosaicity (°) | 0.33 | 0.27 | 0.31 |
| Wilson B-factor (Å2) | 49.8 | 37.2 | 26.4 |
| Refinement | |||
| Resolution (Å) | 75 - 2.5 | 45 - 2.25 | 40 - 2.0 |
| Number of nonhydrogen atoms | 4,303 | 4,331 | 4,513 |
| Protein | 4,129 | 4,182 | 4,182 |
| Water | 145 | 114 | 298 |
| Heterogen | 29 | 34 | 33 |
| Rwork (%) 3 | 22.6 | 22.8 | 22.2 |
| Rfree (%) 4 | 28.6 | 26.9 | 25.5 |
| rms deviations | |||
| Bond lengths(Å) | 0.008 | 0.008 | 0.007 |
| Bond angles (°) | 1.296 | 1.305 | 1.293 |
| Average B-factor (Å2) 5 | 40.5 | 42.3 | 32.5 |
| Included in the final model 6 | |||
| Gα13 residues | 47–336, 340–369 | 47–336, 341–369 | 47–336, 341–369 |
| PRG-rgRGS residues | 307–313, 323–372, 374–407, 409–502 | 307–318, 322–502 | 307–318, 322–502 |
Numbers in parentheses correspond to the last resolution shell.
Rsym = ΣhΣi|Ii(h) − <I(h)>|/ΣhΣi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h, respectively.
Rwork = Σh ||Fo(h)| − |Fc(h)||/Σh |Fo(h)|, where Fo(h) and Fc(h) are the observed and calculated structure factors, respectively. No I/σ cutoff was used in the final calculations of R-factors.
Rfree is the R-factor obtained for a test set of reflections consisting of a randomly selected 10 % of the data.
B-factors are calculated using the Temperature Factor Analysis in CCP4i Suite.
Remaining residues of Gα13 and PRG-rgRGS are disordered.
rgRGS and Gα13 interact at two distinct sites. The first involves the N-terminal subdomain of rgRGS that binds to the helical domain and switch regions of Gα13. The second contact region comprises the RGS-box subdomain of PRG-rgRGS, which binds to a surface on the Ras-like domain of Gα13 that includes the characteristic Gα effector binding site (Sprang, et al. 2007). Interactions with these subdomains of PRG-rgRGS involve distinct, largely non-overlapping surfaces of Gα13, yet are mutually reinforcing. The N-terminal subdomain includes a conserved Ile-Ile-Gly sequence that binds in identical fashion to the three nucleotide-bound states of Gα13, whereas interactions with the succeeding, predominantly acidic, sequence differ among the three states. Interactions with the RGS-box are also largely invariant with respect to the nucleotide bound to the G protein. The switch regions of the Gα13·GTPγS and Gα13·GDP·AlF complexes with PRG-rgRGS adopt activated conformations characteristic of free Gα subunits in the corresponding nucleotide-bound states. The Gα13·GDP complex also adopts an active conformation similar to the GTPγS-bound state. Participation of Mg2+, which engages in most of the coordinating interactions observed in the GTP-bound state with the nucleotide, switch I and switch II, is required to form this complex. Inspection of symmetry-related molecules shows that the activated switch conformations of the GDP complex cannot be explained by crystal packing constraints. Thus, interactions with PRG-rgRGS tend to constrain Gα13 in the active conformation. Superposition of the Gα13 subunits in the three complexes yields an average root mean square deviation (rmsd) of 0.31 Å at all Cα positions, and 0.28 Å for the 22 Cα atoms in switch I and switch II.
The Conserved IIG Motif Forms Gα12/13-Specific Interactions
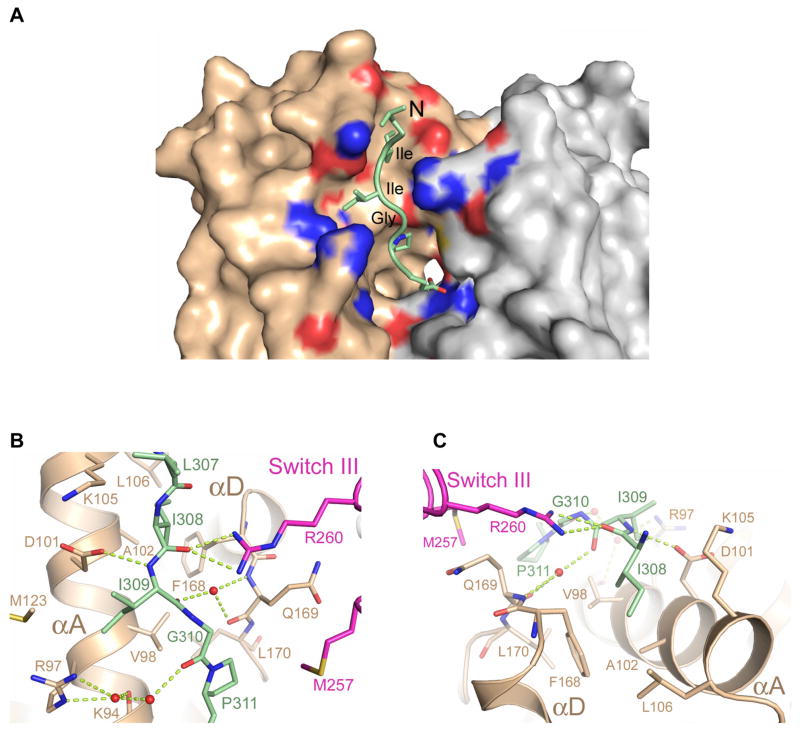
The N-terminal 308Ile-Ile-Gly (IIG) motif of PRG-rgRGS is conserved among all RGS- containing RhoGEFs (Figure 1A) and forms one of the anchor points between rgRGS and the Gα subunit. The IIG motif threads through a narrow trough formed by the αA and αD helices in the helical domain of Gα13 (Figure 2). The carboxylate of Asp-101 from the αA helix forms a hydrogen bond with the main chain amide group of r-Ile-309 (for clarity, rgRGS residue names are preceded by "r-") and the guanidinium group of Arg-260 from switch III forms hydrogen bonds with the main chain carbonyl group of r-Ile-308 (Figure 2B). This interaction, which occurs also in the complex with Gα·GDP, may help to constrain the protein in an active conformation, by pulling the helical and Ras domains together (Supplemental Figure 1). In the structure of uncomplexed Gα13·GDP (Kreutz et al., 2006), these domains are rotated 8° further apart, and the distance between Asp-101 and Arg-260 increases by 5 Å relative to the complex with PRG-rgRGS. This set of contacts provides specific recognition of Gα12/13. In Gαi1, for example, the polarity of the Asp-101/Arg-260 charge pair is reversed, by substitution with Arg and Glu, respectively. The side chain of r-Ile-308 docks into a hydrophobic pocket in the helical domain of Gα13 (Figure 2C) that is conserved among all Gα subunits, and may constitute a binding site for other effectors or regulators of Gα subunits. The interaction between Arg-260 and the main chain of the IIG motif positions r-Ile-309 into a cavity formed predominantly by the αA helix of Gα13 helical domain (Figure 2A, B). Finally, the backbone carbonyl groups of rIle-309 and r-Gly-310 form water-mediated interactions with Gα13 (Figure 2B).
Figure 2.
Interface between the conserved IIG motif from PRG-rgRGS and Gα13. (A) The N-terminus of PRG-rgRGS is depicted as a ribbon diagram with side chains drawn as ball and stick models. The solvent accessible surface of domains of Gα13 is colored as in Figure 1C. Residues contacting the IIG motif are colored according to electrostatic potential in the range of −10kT (red) to +10kT (blue), where k is the Boltzmann’s constant, and T is temperature (K). (B) Ribbon diagram showing electrostatic interactions between main chain atoms of the IIG motif and Gα13. Hydrogen bonds are drawn as dotted lines; waters are shown as orange spheres. Note that Ile-309 of the rgRGS is accommodated by residues Arg-97, Val-98, Asp-101 and Met-123 from the helical domain of Gα13. (C) Ribbon diagram showing the Ile-308 binding pocket on the helical domain of Gα13. Ile-308 of the rgRGS is accommodated in a hydrophobic pocket formed by Val-98, Ala-102, Leu-106 and Phe-168 from the helical domain of Gα13.
The EEDY Motif is Partly Disordered in the Gα13 Transition State Complex
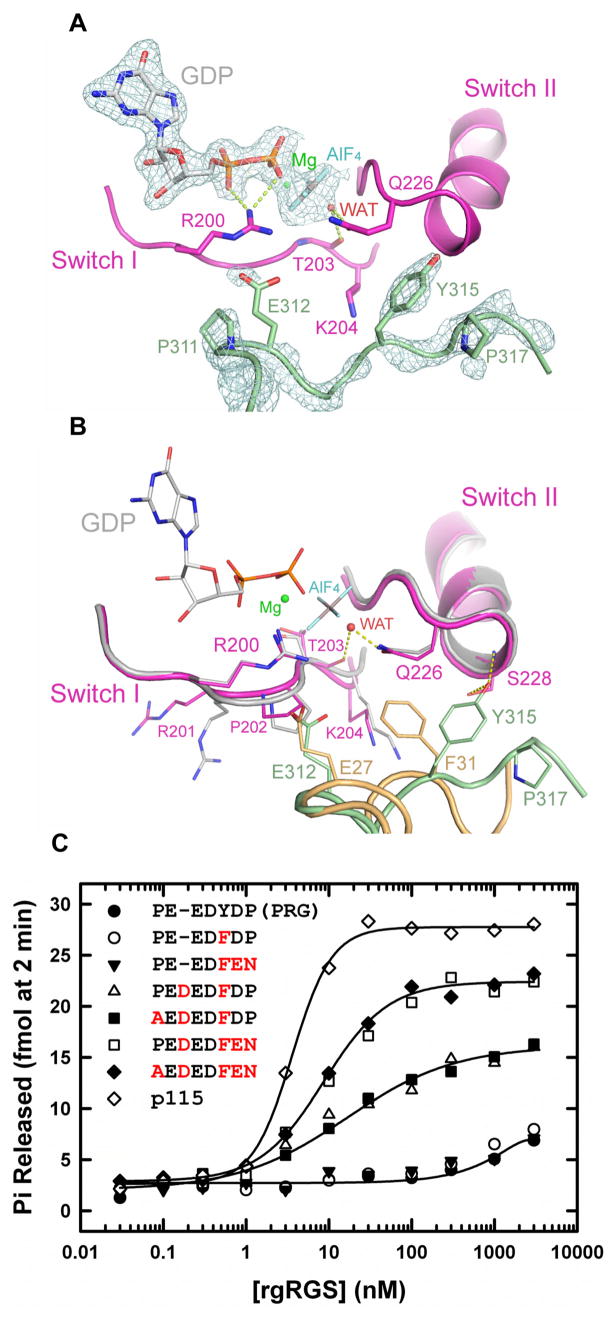
The overall structure of the switch regions of Gα13 in the PRG-rgRGS:Gα13·GDP·AlF complex is very similar to that in the p115-rgRGS:Gα13/i1·GDP·AlF complex. The conformations of the catalytic Arg-200 from switch I and Gln-226 from switch II are identical in these structures (Figure 3). However, the interaction of PRG-rgRGS with the Gα catalytic site is quite different than that of p115-rgRGS. In the latter complex, the 27EDEDF sequence adopts an α-helical conformation, wherein r-Glu-27 and r-Phe-31 directly contact key residues in the switch regions of Gα13 (Chen et al., 2005). Although shorter by one residue, the 312EEDY sequence in PRG-rgRGS traverses the same path between the switch segments of Gα13 by adopting a more extended secondary structure that contains a turn of 310 helix (Figure 3B). If the two transition state complexes are superimposed, the Cα atoms of r-Glu-312 and r-Tyr-315 in PRG-rgRGS are displaced by 0.8 Å and 0.5 Å from those of r-Glu-27 and r-Phe-31 in p115-rgRGS, respectively. However, the 310 helix is poorly ordered in the transition state complex (Figure 3A); the B-factors for the EEDY motif exceed 60 Å2, in contrast to values less than 40 Å2 for the IIG motif.
Figure 3.
GTPase active site in the PRG-rgRGS:Gα13·GDP·AlF4− complex. (A) Electron density (cages) at the active site from a 2.25 Å σA-weighted 2Fo−Fc difference map (Read, 1986) is contoured at 1.6 standard deviations above the mean. Only densities of the PRG-rgRGS N-terminus, GDP·Mg2+·AlF4− and the axial water molecule bound to AlF4− are shown. Hydrogen bonds are drawn as dotted lines. (B) Structural comparison of active sites from PRG-rgRGS:Gα13·GDP·AlF4− complex and p115-rgRGS:Gα13/i1·GDP·AlF4− complex. Elements from Gα13/i1 are colored gray and the N-terminus of p115-rgRGS is colored brown. (C) Stimulation of GTPase activity of Gα13 by increasing concentrations of wild type and mutated PRG-rgRGS. Amino acids mutated in PRG-rgRGS are colored red.
The ionic interaction between r-Glu-27 of p115-rgRGS and the switch I Arg-200, which is presumed to stabilize the transition state conformation of Gα13·GDP·AlF, is not present in the corresponding complex with PRG-rgRGS and Gα13 in which the corresponding r-Glu-312 (Figure 3A) is disordered. p115-rgRGS also stabilizes the transition state by inserting the aromatic ring of r-Phe-31 between switch I Lys-204 and switch II Gln-226 of Gα13 (Chen et al., 2005). This sterically constrains the catalytic Gln-226 for interaction with the γ phosphorus non-bridging oxygen atoms, as mimicked by the fluorine atoms of AlF, and the water nucleophile (Figure 3B). In contrast, the corresponding r-Tyr-315 in PRG-rgRGS is directed away from the catalytic site, stabilized by hydrogen bonds with Ser-228 from switch II (Figure 3B). In the absence of this bulky rgRGS residue, the side chain of Lys-204 is 0.6 Å closer to Gln-226 in switch II (Figure 3B). The transition state conformation of Gln-226 is stabilized, not by r-Tyr-315 from rgRGS, but rather by the transition state analog, GDP·AlF, as seen in other Gα·GDP·AlF structures (Coleman et al., 1994; Kreutz et al., 2006).
Mutagenic Conversion of PRG-rgRGS to a GAP
The structural similarity between the two transition state Gα complexes with p115-rgRGS and PRG-rgRGS, especially with respect to the positions of key residues critical for GAP activity (Figure 3B), suggests that it might be possible to convert PRG-rgRGS to a GAP. Earlier mutagenesis experiments demonstrated that mutation of the EDEDF motif phenylalanine residue in p115RhoGEF was sufficient to abolish GAP activity (Chen et al., 2003). To test the possibility that the phenylalanine is sufficient for GAP activity, we mutated r-Tyr-315 to a phenylalanine, in the context of the EEDY motif of PRG-rgRGS, to attempt restoration of the Glu-Phe pair that is required for GAP activity in p115-rgRGS (Chen et al., 2005). However, this single mutation fails to confer GAP activity upon PRG-rgRGS (Figure 3C). Because the 312EEDF sequence likely adopts a 310 helical conformation, rather then the α helix formed by the longer acidic sequence in p115-rgRGS, we next replaced the EEDY sequence in PRG-rgRGS with EDEDF. This mutation did generate some GAP activity, though at much lower potency and efficacy compared with p115-rgRGS. We then tested the possible role of the proline residues flanking the EDEDF sequence in destabilizing the α helical structure observed in p115-rgRGS, which may be required to position GAP residues at the Gα active site. Whereas mutation of the N-terminal r-Pro-311 had little effect, mutation of the C-terminal r-Pro-317 to asparagine greatly increased the efficacy of the EDEDF mutant of PRG-rgRGS for GAP activity to a level similar to p115-rgRGS, although the potency of the EDEDFEN mutant is still less than that of p115-rgRGS (Figure 3C). It is possible that GAP activity is influenced by the length and composition of the sequence that links the N-terminal acidic residues to the RGS-box subdomain, although this sequence is disordered in the crystal structures of both p115-rgRGS and PRG-rgRGS.
PRG-rgRGS Forms Well-ordered Interactions with Gα13·GTPγS
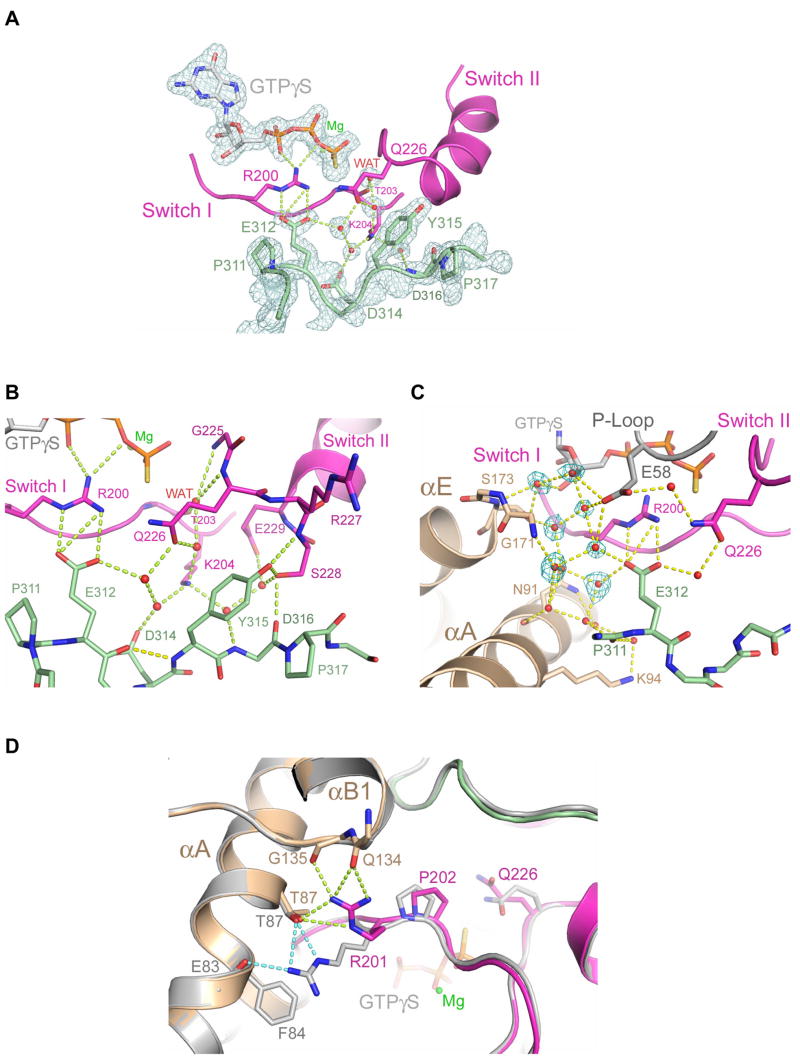
Except for the switch regions of Gα13 and the rgRGS 312EEDY sequence, the overall structure of the PRG-rgRGS:Gα13·GTPγS complex is almost identical to that of PRG-rgRGS:Gα13·GDP·AlF (0.35 Å rmsd for all Cα atoms) (Figure 1C). The 312EEDY sequence (310 helix), which is partly disordered in the transition state complex, is well ordered in the complex of PRG-rgRGS with Gα13·GTPγS (Figure 4A). The average B-factor for the 310 helix are similar to that of the IIG motif and well below the average B-factor for the entire rgRGS domain. The side chain of r-Tyr-315 is inserted into a cavity formed by Lys-204 from switch I, the side chains of residues 226–229 in switch II, Glu-258 from switch III, and r-Pro-317 (Figure 4B). The hydroxyl group of r-Tyr-315 forms hydrogen bonds with the backbone amide group and the side chain hydroxyl group of Ser-228, which itself is stabilized by hydrogen bonds (Figure 4B). Thus, r-Tyr-315, which makes contact with elements from all three switch regions of Gα13, adopts essentially the same position in the Gα13·GTPγS complex as in that with Gα13·GDP·AlF, but is more highly ordered.
Figure 4.
GTPase active site in the PRG-rgRGS:Gα13·GTPγS complex. (A) Electron density (cages) at the active site from a 2.0 Å σA-weighted 2Fo−Fc difference map is contoured at 1.6 standard deviations above the mean. Only densities of the PRG-rgRGS N-terminus, GTPγS, Mg2+ and key water molecules are shown. Hydrogen bonds are drawn as dotted lines. (B) Ribbon diagram depicting extensive contact between the N-terminus of PRG-rgRGS and the switch regions of Gα13. Hydrogen bonds are drawn as dotted lines. The hydrogen bond between E312 and Y315 as a result of the 310 helical fold is colored yellow. (C) Ribbon diagram depicting water-mediated contact between the N-terminus of PRG-rgRGS and the helical domain and P-loop of Gα13. Electron density (cages) for the seven additional water molecules, which are only present in the GTPγS complex, is shown. Hydrogen bonds are drawn as dotted lines. (D) Structural comparison of active sites from the PRG-rgRGS:Gα13·GTPγS complex and PRG-rgRGS:Gα13·GDP·AlF4− complex. The PRG-rgRGS:Gα13·GDP·AlF4− complex is colored gray. Hydrogen bonds between Arg-201 and the helical domain are drawn as dotted lines and colored lime in the GTPγS complex and light blue in the GDP·AlF4− complex. The distance between the Cα atom positions of Arg-201 in these two structures when superimposed is about 0.8 Å.
Certain interactions between PRG-rgRGS and Gα13·GTPγS may be pro-catalytic while others may have a negative impact on catalytic activity. The ion pair interaction between r-Glu-312 and Arg-200 in switch I, occurs also in the p115-rgRGS:Gα13/i1·GDP·AlF complex (Chen et al., 2005). This possibly pro-catalytic interaction supports ionic contacts between Arg-200 and the β and γ phosphate oxygen atoms of GTP. In contrast, the catalytic Glu-226 forms no interactions with either the water nucleophile or the γ phosphate as occurs in GDP·AlF complexes (Figure 3A, B). Instead, Gln-226 is oriented away from the γ phosphate by a network of hydrogen bonds to ordered water molecules that are themselves stabilized by r-Glu-312 and switch I Lys-204 (Figure 4B). Nevertheless, Glu-226 is not tightly constrained in a non-catalytic conformation by direct interactions with protein atoms. Therefore, even though PRG-rgRGS forms extensive and well ordered interactions with Gα·GTPγS, it does not inhibit Gα13-catalyzed hydrolysis of GTP by preferentially stabilizing the Michaelis complex.
The putative water nucleophile in the PRG-rgRGS complex with Gα13·GTPγS is located ~4 Å from the γ-phosphorus as in other Michaelis complexes, for example that of Gαi1 with GTPγS (Coleman et al., 1994). In contrast, as an axial ligand of AlF, the water is 1.3 Å closer to switch II in the GDP·AlF complex (Figure 4B). The nucleophile forms hydrogen bonds with the main-chain amide groups of Gly-225 and Gln-226 in switch II, and with the main-chain carbonyl oxygen of Thr-203 in switch I as observed in the Michaelis and GDP·AlF complexes of free Gα subunits (Sprang, 1997).
Other interactions appear to contribute to the higher binding affinity between PRG-rgRGS and Gα13· GTPγS relative to the transition state conformation of the G protein. For example, Lys-204 forms water-mediated contacts with the carboxyl group of r-Asp-314 and the backbone amide group of r-Asp-316 in PRG-rgRGS (Figure 4B). These stabilizing interactions are absent in the PRG-rgRGS:Gα13·GDP·AlF complex. Ordered active site water molecules participate in a web of hydrogen bond interactions in which the acidic motif of the rgRGS domain, the Gα13 P-loop (via Glu-58), the catalytic arginine and glutamine of switch I and II, are knit together with the main chain and side chain groups of the αA and αE helices from the helical domain of Gα13 (Figure 4C). This network, which includes the guanine nucleotide by virtue of P-loop interactions (Sprang, 1997) appears to provide overall structural integrity to the complex. Potential perturbation of local water structure, due to γ-thiol moiety of GTPγS does not appear to be the cause of the water-mediated hydrogen bonded network that supports the catalytic residues, but rather, is permissive of it. The extensive involvement of water molecules in the complex might rationalize the relatively low entropy, and high negative enthalpy of complex formation relative to that of the GDP and GDP·AlF complexes.
In addition to changes in the switch II region, a ~1 Å rearrangement of switch I in Gα13 is also observed in the GTPγS complex relative to the transition state complex (Figure 4D). Most noticeable is the shift of Arg-201 in switch I towards αB1 (switch IV) in the helical domain. Arg-201 exchanges hydrogen partners in αA for new contacts with the C terminus of αB1 (Figure 4D). The same switch I–switch IV interactions are observed in the p115-rgRGS:Gα13/i1·GDP·AlF complex, and might reinforce interactions with the rgRGS N-terminal subdomain (Supplemental Figure 2).
Structural Adaptation of Gα13 for Recognition of Divergent rgRGS Proteins
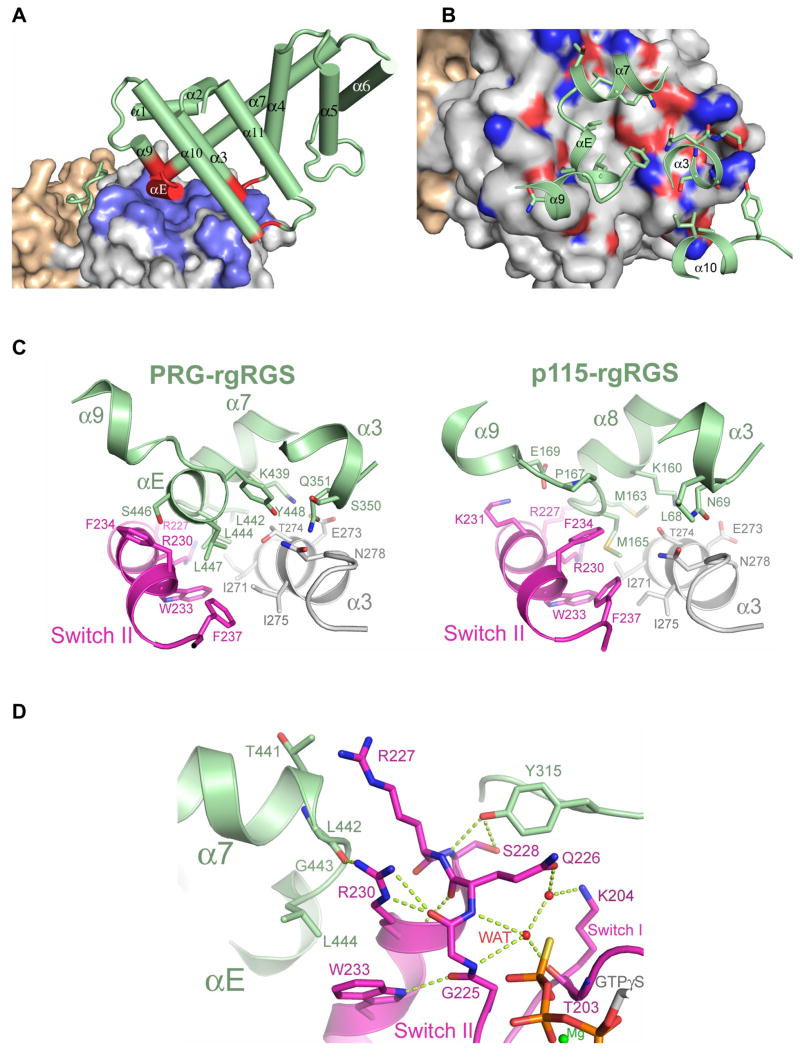
RGS-boxes of rgRGS domains possess C-terminal extensions (α9–α11, Figure 1A) not present in other members of the RGS superfamily (Chen et al., 2001), and these form part of the effector interaction surface with switch II and the α3 helix in Gα13 (Figure 1C) (Sprang et al., 2007). The RGS-boxes of PDZRhoGEF and p115RhoGEF are about 30% identical in amino acid sequence and the C-terminal halves of the domains differ in the lengths of several of the inter-helical connecting segments (Figure 1A). Structural adaptations in Gα13 permit binding of both RGS-boxes, despite their substantial divergence. The RGS-box of PRG-rgRGS projects a larger footprint (1,500 Å2 of solvent accessible surface) on Gα13 in all three complexes than does p115-rgRGS onto Gα13/i1 (1,200 Å2) (Chen et al., 2005). We note that all of the rgRGS-contacting residues of Gα13 are conserved in Gα13/i1. A short α-helix (αE), which is unique to PRG-rgRGS (Figure 1A), lies at the center of the interface, and forms a loose three-helix bundle with switch II and the α3 helix from Gα13 (Figure 5). Three leucine residues of the rgRGS from the α7–αE loop (r-Leu-442 and 444) and the αE helix (r-Leu-447), project into the highly conserved hydrophobic trough between switch II and the α3 helix (Figure 5C). r-Leu-442 and r-Leu-444 correspond to r-Met-163 and r-Met-165 in p115-rgRGS (Figure 1A), which project into the same cavity on Gα13/i1(Chen et al., 2005). However, there is no equivalent αE helix in p115-rgRGS. In order to accommodate this additional element from PRG-rgRGS, the effector binding cleft expands by ~3 Å at the αE contact surface between Phe-237 in switch II and Val-280 in α3 (Figure 5C). The side chain of Phe-234 from switch II also rotates outward from the center of the cleft to provide access to the site for r-Leu-447 from αE (Figure 5C). Thus, the conformational plasticity of switch II is expressed not only in the transition between active and inactive states for signaling, but in binding to the divergent surfaces of different effectors as well.
Figure 5.
The interface between the RGS-box and C-terminal extension of PRG-rgRGS and Gα13. (A) The solvent accessible surface of Gα13 is colored as in Figure 1C, with residues contacting the rgRGS colored blue. The RGS-box and C-terminal extension is colored green, with elements contacting Gα13 colored red. (B) Residues of Gα13 that contact PRG-rgRGS are colored according to electrostatic potential as in Figure 2A. Side chains from PRG-rgRGS that directly contact Gα13 are represented as ball and stick models. In addition to the αE helix, the α3-α4 and α10-α11 loops of PRG-rgRGS also directly contact the effector binding site of Gα13. (C) Differences at the effector binding site on Gα13 upon binding to PRG-rgRGS (left) or p115-rgRGS (right). Residues directly involved in the rgRGS:Gα13 interface are represented as ball and stick models. (D) Ribbon diagram depicting the interaction interface between switch II of Gα13 and the N-terminal and the RGS-box subdomains of PRG-rgRGS. Main chain and side chain atoms are represented as ball and stick models. Hydrogen bonds are drawn as dotted lines.
The Two Subdomains of PRG-rgRGS Bind Switch II Cooperatively
Switch II forms a common interface at which the N-terminus and the RGS-box of PRG-rgRGS make mutual contact. These two subdomains of PRG-rgRGS are tethered by an unstructured peptide linker as in the case with p115-rgRGS, and otherwise make no direct contact with each other. Similarly, the GAP and effector binding surfaces on Gα13 with which the respective rgRGS subdomains interact are distinct and largely non-overlapping (Supplemental Figure 3). However, the two subdomains of PRG-rgRGS are cooperatively buttressed by switch II. A network of hydrogen bonds involving switch II Arg-230 with the α7–αE loop from the RGS-box, and the amide and hydroxyl groups of Ser-228 with the N-terminal r-Tyr-315 form a nexus of interaction that tie the three structural elements together (Figure 5D). These interactions may, in turn, reinforce the contacts between the Switch II amides of Gly-225 and Gln-226 and the putative water nucleophile in the GTPγS complex. Indeed, the interaction between the RGS-box and the effector binding site on Gα13 stabilizes the active conformation of switch II even in the GDP complex (Supplemental Figure 1). While largely disordered in the structure of Gα13·GDP alone (Kreutz et al., 2006), switch II in Gα13·GDP forms a structurally cooperative network of van der Waals interactions with the RGS-box in the complex with PRG-rgRGS.
DISCUSSION
The first known members of the RGS family were shown to have GAP activity towards Gα proteins (Abramow-Newerly et al., 2006; Hollinger and Hepler, 2002; Ross and Wilkie, 2000; Wilkie and Kinch, 2005). However, not all RGS domains function as GAPs, even those that bind Gα. The RGS-like domains of RGS-RhoGEFs (Kozasa et al., 1998) and GRK2 (Carman et al., 1999) belong to separate and distinct subgroups. The RGS domains of these proteins, which possess other protein-interaction, regulatory or catalytic domains, bind to Gα in the manner of effectors, in contrast to RGS GAPs, which bind to a different surface on Gα (Chen et al., 2005; Tesmer et al., 2005).
Comparison of the rgRGS complexes of two RGS-RhoGEFs with Gα13 is instructive of both the general features of the family and those that render them functionally divergent. First, dual sites for binding of the rgRGS domains to Gα appear to be characteristic of this RGS subfamily, regardless of their ability to function as GAPs. The guanine nucleotide-sensitive acidic motif at the N-terminus (312EEDY in PRG-rgRGS, and 27EDEDF in p115-rgRGS), which can mediate GAP activity, is bolstered by the preceding IIG sequence. The C-terminally extended RGS-box possesses a classical effector binding surface. Second, the lack of GAP activity of PRG-rgRGS is readily explained by the structures. Stepwise mutagenesis of its N-terminal subdomain demonstrates that GAP activity arises from the cumulative effect of mutations at multiple sites. Third, PRG-rgRGS, in contrast to p115-rgRGS, can interact effectively with both activated (GTP or GDP·AlF) and resting (GDP) states of the α subunit.
It is noteworthy that the mode by which Gα13 engages p115RhoGEF and PDZRhoGEF differs from that of the interaction of Gαq with its effector, p63RhoGEF (Lutz et al., 2007). The latter does not possess an RGS domain. Rather, Gαq relieves auto-inhibition of p63RhoGEF activity by interaction with an extensive contact surface: the PH domain occupies the canonical switch II/α3 effector binding site of Gαq, while the switch II-β4/α3-β5 elements bind to the DH-PH domain interface and perhaps alter interaction between the domains as a means of influencing activity. In contrast, RGS-RhoGEFs have significant basal GEF activity that is further stimulated by Gα13. The engagement of the switch II/α3 effector binding surface of Gα13 with the RGS-box region of these proteins, suggests a crucial but unknown mechanism for GEF stimulation. Additional interactions of Gα13 with the DH and PH domains of p115RhoGEF have been proposed to explain the in vitro regulation observed with this protein (Wells et al., 2001).
Even within the small family of RGS domain-containing RhoGEFs there has arisen functional and structural divergence. Although the RhoGEF activity of both is regulated by Gα13, p115RhoGEF expresses GAP activity towards Gα13 while PDZRhoGEF does not. Significant conformational differences between the respective transition state and the Michaelis complex are largely confined to switch I and the side chains of the catalytic switch I arginine and switch II glutamine residues. However, PRG-rgRGS binds both activated states with comparable affinity, whereas p115-rgRGS has higher affinity for the transition state conformation of Gα13. These alternative recognition modes can be ascribed to differences in the corresponding N-terminal acidic regions of the two rgRGS domains.
In PRG-rgRGS, this short peptide sequence, characterized by three to four acidic residues terminated by Phe or Tyr at the N-terminus of rgRGS, forms interactions with the transition state of Gα13 that are different from those with the Michaelis complex of the G protein. While the 312EEDY sequence of PRG-rgRGS is flexible in the active site of Gα13·GDP·AlF, it forms well-ordered interactions with the same residues of Gα13·GTPγS. These contacts do not appear to restrain active site residues from productive catalytic interaction with the nucleotide. This structural data agrees with biochemical measurements and indicates that interaction with the PRG-rgRGS allows normal intrinsic hydrolysis and termination of the GTP-induced activated state of Gα13.
PRG-rgRGS is evolved to recognize common features of the activated state of Gα13 stabilized by either GDP·AlF or GTPγS, by optimizing the bivalent interaction with Gα13. Within the N-terminal subdomain of PRG-rgRGS (and also observed in the p115-rgRGS:Gα13·GDP·AlF structure), the conserved Ile-Ile-Gly motif forms identical contacts with Gα13 regardless of its GTP hydrolysis state. This N-terminal anchor is Gα12/13-class specific, and represents one of the few examples, along with GoLoco-motif interactions (Kimple et al., 2002), in which the helical domains of Gα proteins play a distinct role in effector/regulator recognition. The second binding interface is the extensive contact surface between the RGS-box of PRG-rgRGS and the effector binding site of Gα13. Due to involvement of the αE extension in PRG-rgRGS, the contact area with Gα13 at this interface is several hundred square Angstroms greater than that observed with the RGS-box of p115-rgRGS. Hence, poor complementarity of the EEDY sequence with the GDP·AlF-bound conformation of switch I does not impose a significant cost in binding energy. In contrast, formation of a higher affinity p115-rgRGS:Gα13 complex requires productive contacts with the corresponding EDEDF motif that are realized in the transition state, and complement the lower affinity interaction with the RGS-box of p115RhoGEF. It seems unlikely that the GAP activity of p115RhoGEF has evolved to merely increase avidity of the protein for activated Gα13. It appears more likely that p115RhoGEF has evolved for rapid turnover while PDZRhoGEF may be capable of maintaining a more persistent signaling state. The latter may be necessary to effectively compete with binding of PDZRhoGEF to cellular constituents at other locals (Sternweis et al., 2007). Alternatively, the GAP activity of p115RhoGEF may contribute to kinetic coupling as a means to maintain a receptor signaling complex (Ross and Wilkie, 2000), whereas PDZRhoGEF uses its PDZ or other domains for sustained association with activated receptors (Sternweis et al., 2007; Yamada et al., 2005). It is intriguing that PDZRhoGEF stabilizes an activated conformation of Gα13 in the GDP-bound state. While the affinity of Gα13·GDP for the rgRGS is substantially lower than that for activated α subunits, the interaction is highly significant and suggests that the “inactive” αsubunit can functionally interact with PDZRhoGEF. This would be analogous to the ability of Gαs·GDP to stimulate the activity of adenylyl cyclase, albeit with lower affinity (Sunahara et al., 1997). This further suggests that hydrolysis of GTP by Gα13 is insufficient to fully terminate signaling. Rather, sequestration of the Gα13·GDP with βγ subunits or some other masking protein may play a key role. It will be of great interest to see whether this is a more general phenomenon among other Gα-effector pairs.
EXPERIMENTAL PROCEDURES
Expression Constructs
The coding region of the N-terminally truncated mouse Gα13 (residues 41–377) was ligated C-terminal to the coding region of rat Gαi1 (residues 1–28). The product was subcloned into the baculovirus transfer vector pFastBacHTb (Invitrogen, Carlsbad, CA). A Tobacco Etch Virus (TEV) protease recognition site was then inserted between the Gαi1 and Gα13 sequences. The expression construct for glutathione-S-transferase (GST)-PRG-rgRGS was generated by subcloning the coding region of GTRAP48 (residues 307–508) into a pGEX-KG vector, where a TEV protease recognition site was introduced before the start of the GTRAP48 sequence.
Expression and Purification of Proteins
Cells from 6L of High-5 (Invitrogen) culture expressing Gα were harvested by centrifugation 50–60 hours after infection with 10 ml/L of amplified baculovirus stock. Harvested cells were suspended with 600 ml of lysis buffer (50 mM NaHEPES, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, 10 μM GDP and protease inhibitors) and lysed by sonication. The lysate was clarified by centrifugation for 40 minutes at 30,000xg. Purification of hexahistidine-tagged Gα subunit from the supernatant utilized Ni-NTA resin according to the QIA-expressionist protocol (Qiagen). The Gαi1 element was cleaved by dialyzing overnight at 4 °C against Buffer A (25 mM NaHEPES, pH 8.0 and 5 mM β-mercaptoethanol) in the presence of 1 mg of TEV protease and 10 μM GDP. The dialyzed mixture was then applied to a Mono Q anion exchange column (Amersham Pharmacia Biotech) that had been pre-equilibrated with Buffer A. Elution was accomplished with a linear gradient of 0 to 0.5 M NaCl in Buffer A using an Acta-FPLC system (Amersham Pharmacia Biotech).
Expression and purification of GST-tagged PRG-rgRGS was carried out as described (Wells et al., 2002). The GST-tag was removed by TEV protease, and the rgRGS domain was further purified with Mono Q resin that had been pre-equilibrated with Buffer A and elution with a linear gradient of 0 to 0.5 M NaCl. The rgRGS domain of p115RhoGEF was produced in E. coli as a N-terminally 6His-tagged protein as described (Chen et al., 2001).
Binding of GTPγS to Gα13
Purified Gα13 was exchanged into binding buffer (20 mM NaHEPES, pH 8, 1 mM EDTA, 1 mM DTT, 50 mM NaCl, and 10 μM GDP) and concentrated to 100–250 μM. The concentrate was adjusted to 0.5 mM MgSO4, 0.005 % (v/v) polyoxyethylene 10 lauryl ether (Lubrol), and 1 mM GTPγS, and incubated at 25 oC for 48–72 hours.
Assay of GAP Activity of rgRGS
Single turnover GTPase activity assays with Gα13 and the rgRGS domains were carried out as described (Chen et al., 2003).
Formation of Gα13·rgRGS Complexes
All PRG-rgRGS:Gα13 complexes were purified by size-exclusion chromatography using Superdex 200/75 tandem gel filtration columns (Amersham Pharmacia Biotech) at 4°C with an Acta-FPLC. The complex with GDP-bound Gα13 was obtained from an equimolar (0.25 mM) mixture of Gα13(Q226L)·GDP and PRG-rgRGS which was subjected to gel filtration with Buffer A and 100 mM NaCl, 1 mM EDTA and 10 mM MgCl2. To form the complex in its transition state conformation, the mixture of Gα13·GDP and PRG-rgRGS was subjected to gel filtration with Buffer A and 100 mM NaCl, 10 μM GDP, 1 mM EDTA, 30 μM AlCl3, 10 mM NaF and 10 mM MgCl2. For the Michaelis complex, the mixture of GTPγS-loaded Gα13 and PRG-rgRGS was subjected to gel filtration with Buffer A and 100 mM NaCl, 1 mM EDTA and 1.25 mM MgCl2. Fractions that contained the Gα13·rgRGS complex (molecular weight of approximately 70 kDa as judged by elution volume) were pooled and concentrated using Amicon-Ultra 4 (10 kDa) concentrators (Millipore) to a final concentration of 10 mg/ml. Aliquots (50 μl) of the concentrated complex were flash frozen with liquid nitrogen and stored at −80 °C.
ITC Assays
Isothermal titration calorimetry (ITC) was performed at 6°C (279 K) or 12°C (285 K, for measurement of the GTPγS state) using a Microcal VP-ITC calorimeter (MicroCal, Northhampton, MA). Protein samples were dialyzed against Buffer A and 100 mM NaCl, 10 mM MgCl2, and 1 mM EDTA for measurement of the GDP and GTPγS states, or with an additional 10 μM GDP, 30 μM AlCl3, and 10 mM NaF for the GDP·AlF complex. Contents of the sample cell were stirred continuously at 300 rpm during the experiment. A typical titration of a Gα13 with the rgRGS domain involved 30–35 injections at 3 minute intervals of 8 μl of the rgRGS domain (0.4–0.6 mM) into a sample cell containing 1.5 ml of the Gα13 (60–80 μM). The heats of dilution of the titrants were subtracted from the titration data for baseline correction. The baseline-corrected data were analyzed with Microcal Origin* 5.0 software to determine the enthalpy (ΔH), association constant (Ka) and stoichiometry of binding (N). Thermal titration data were fit to the association model for 'single set of identical sites' available in the software (Microcal, 1998; Wiseman et al., 1989).
Crystallization and Data Collection
All crystallization procedures were carried out by vapor diffusion at 20°C. The PRG-rgRGS:Gα13·GDP complex was crystallized from 22–27 % polyethylene glycol 4000 and 100 mM NaHEPES, pH 7.2–8.1. The PRG-rgRGS:Gα13·GDP·AlF4−complex was crystallized from 18–23 % polyethylene glycol monomethyl ether 2000 and 100 mM Tris, pH 6.9–7.3. The PRG-rgRGS:Gα13·GTPγS complex was crystallized from 17–24 % polyethylene glycol 8000 and 100 mM NaHEPES, pH 7.3–7.8. All crystal forms were then cryoprotected with an additional 15 % (v/v) ethylene glycol. Native data were measured at 100 K at the Structural Biology Center (Beamline 19ID) or the GM/CA CAT (Beamline 23ID) at Argonne National Laboratory. Diffraction data were reduced using the HKL software package (Otwinowski and Minor, 1997).
Structure Determination and Model Refinement
Initial phases were generated by molecular replacement using the coordinates of Gα13/i1 (PDB entry 1ZCB) and the PDZRhoGEF rgRGS domain (PDB entry 1HTJ) as search models, using program PHASER (McCoy et al., 2007). Model building was performed using the program Coot (Emsley and Cowtan, 2004). The model was refined using CNS_SOLVE version 1.2 (Brunger et al., 1998), in alternate cycles of simulated annealing, energy minimization and individual B-factor refinement. Putative water molecules within hydrogen bonding distance of at least one protein atom or other water oxygen atoms and with refined B-factors <100 Å2 were included in the model. PROCHECK (Laskowski et al., 1993) indicates that over 90 % of the residues fall in the most favorable regions of φ, ψ conformational space (Ramachandran and Sassiekharan, 1968). Coordinates have been deposited in the Protein Data Bank (Berman et al., 2000) with accession code 3CX6 for the PRG-rgRGS:Gα13·GDP complex, 3CX7 for the PRG-rgRGS:Gα13·GDP·AlF4− complex, and 3CX8 for the PRG-rgRGS:Gα13·GTPγS complex. Atomic representations were created using Pymol (DeLano, 2002).
Supplementary Material
Acknowledgments
We thank Jana Hadas, Helen Aronovich, Celestine Thomas, and Stephen Gutowski for technical assistance, and Chad Brautigam and the staff of APS for assistance with data collection. This work was supported by National Institute of Health Grants GM31954 (to P. C. S.), and DK46371 (to S. R. S.), by the Robert A. Welch foundation (I-1262 to P. C. S. and I-1229 S. R. S.), the Alfred and Mabel Gilman Chair in Molecular Pharmacology (P. C. S.) and the John W. and Rhonda K. Pate Professorship in Biochemistry (to S. R. S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18:579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Berman DM, Gilman AG. Mammalian RGS proteins: barbarians at the gate. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- Berman DM, Kozasa T, Gilman AG. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlecome GH, Berstein G, Ross EM. Regulation of phospholipase C-β1 by Gq and m1 muscarinic cholinergic receptor Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J Biol Chem. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- Bokoch GM, Der CJ. Emerging concepts in the Ras superfamily of GTP-binding proteins. Faseb J. 1993;7:750–759. doi: 10.1096/fasebj.7.9.8330683. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carman CV, Parent JL, Day PW, Pronin AN, Sternweis PM, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. Selective regulation of Galpha(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J Biol Chem. 1999;274:34483–34492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Chen CK, Wieland T, Simon MI. RGS-r, a retinal specific RGS protein, binds an intermediate conformation of transducin and enhances recycling. Proceedings of the National Academy of Sciences USA. 1996;93:12885–12889. doi: 10.1073/pnas.93.23.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Singer WD, Sternweis PC, Sprang SR. Structure of the p115RhoGEF rgRGS domain-Galpha13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat Struct Mol Biol. 2005;12:191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- Chen Z, Singer WD, Wells CD, Sprang SR, Sternweis PC. Mapping the Galpha13 binding interface of the rgRGS domain of p115RhoGEF. J Biol Chem. 2003;278:9912–9919. doi: 10.1074/jbc.M212695200. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wells CD, Sternweis PC, Sprang SR. Structure of the rgRGS domain of p115RhoGEF. Nat Struct Biol. 2001;8:805–809. doi: 10.1038/nsb0901-805. [DOI] [PubMed] [Google Scholar]
- Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- Coleman DE, Sprang SR. Structure of Giα1·GppNHp, Autoinhibition in a Gα Protein- Substrate Complex. J Biol Chem. 1999;274:16669–16672. doi: 10.1074/jbc.274.24.16669. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS. A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. J Biol Chem. 1999;274:5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- Geyer M, Wittinghofer A. GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr Opin Struct Biol. 1997;7:786–792. doi: 10.1016/s0959-440x(97)80147-9. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G Proteins: transducers of receptor-generated signals. Ann Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- Jackson M, Song W, Liu MY, Jin L, Dykes-Hoberg M, Lin CI, Bowers WJ, Federoff HJ, Sternweis PC, Rothstein JD. Modulation of the neuronal glutamate transporter EAAT4 by two interacting proteins. Nature. 2001;410:89–93. doi: 10.1038/35065091. [DOI] [PubMed] [Google Scholar]
- Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Galpha subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. p115 RhoGEF, a GTPase activating protein for Gα12 and Gα13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- Kreutz B, Yau DM, Nance MR, Tanabe S, Tesmer JJ, Kozasa T. A New Approach to Producing Functional Galpha Subunits Yields the Activated and Deactivated Structures of Galpha(12/13) Proteins. Biochemistry. 2006;45:167–174. doi: 10.1021/bi051729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Longenecker KL, Lewis ME, Chikumi H, Gutkind JS, Derewenda ZS. Structure of the RGS-like domain from PDZ-RhoGEF: linking heterotrimeric g protein-coupled signaling to Rho GTPases. Structure. 2001;9:559–569. doi: 10.1016/s0969-2126(01)00620-7. [DOI] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJG. Structure of Gq-p63RhoGEF-RhoA Complex Reveals a Pathway for the Activation of RhoA by GPCRs. Science. 2007;318:1923–1927. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Microcal. ITC Data Analysis in Origin. Northampton, MA): 1998. [Google Scholar]
- Noel JP, Hamm HE, Sigler PB. The 2.2 A crystal structure of transducin-alpha complexed with GTPγS. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sassiekharan V. Conformation of polypeptides and proteins. Advances in Protein Chemistry. 1968;28:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallographica. 1986;A42:140–149. [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A. Nature. 2001;409:1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of G proteins from the 1.7 Å crystal structure of transducin α-GDP-AlF4−. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: Insights from structural analysis. Annual Review of Biochemistry. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Sprang SR, Chen Z, Du X. Structural basis of effector regulation and signal termination in heterotrimeric Ga proteins. In: Sprang SR, editor. Advances in Protein Chemistry. Elsevier Inc; 2007. pp. 1–65. [DOI] [PubMed] [Google Scholar]
- Sternweis PC, Carter AM, Chen Z, Danesh SM, Hsiung Y, Singer WD. Regulation of Rho Guanine Nucleotide Exchange Factors by G Proteins. In: Sprang SR, editor. Advances in Protein Chemistry. Elsevier Inc; 2007. pp. 189–228. [DOI] [PubMed] [Google Scholar]
- Sunahara RK, Dessauer CW, Whisnant RE, Kleuss C, Gilman AG. Interaction of Gsa with the cytosolic domains of mammalian adenylyl cyclase. J Biol Chem. 1997;272:22265–22271. doi: 10.1074/jbc.272.35.22265. [DOI] [PubMed] [Google Scholar]
- Tesmer JJG, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AIF4−-activated Giα1: Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Snapshot of activated G proteins at the membrane: the Galphaq-GRK2-Gbetagamma complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- Thomas CJ, Du X, Li P, Wang Y, Ross EM, Sprang SR. Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein α-subunit. Proc Natl Acad Sci U S A. 2004;101:7560–7565. doi: 10.1073/pnas.0304091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. RGS family members: GTPase-activating proteins for heterotrimeric G- protein α-subunits. Nature. 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- Wells CD, Gutowski S, Bollag G, Sternweis PC. Identification of potential mechanisms for regulation of p115 RhoGEF through analysis of endogenous and mutant forms of the exchange factor. J Biol Chem. 2001;276:28897–28905. doi: 10.1074/jbc.M102913200. [DOI] [PubMed] [Google Scholar]
- Wells CD, Liu MY, Jackson M, Gutowski S, Sternweis PM, Rothstein JD, Kozasa T, Sternweis PC. Mechanisms for Reversible Regulation between p115 RhoGEF and GTRAP48 and the G12 Class of Heterotrimeric G proteins. J Biol Chem. 2002;277:1174. doi: 10.1074/jbc.M105274200. [DOI] [PubMed] [Google Scholar]
- Wilkie TM, Kinch L. New roles for Galpha and RGS proteins: communication continues despite pulling sisters apart. Curr Biol. 2005;15:R843–854. doi: 10.1016/j.cub.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- Yamada T, Ohoka Y, Kogo M, Inagaki S. Physical and functional interactions of the lysophosphatidic acid receptors with PDZ domain-containing Rho guanine nucleotide exchange factors (RhoGEFs) J Biol Chem. 2005;280:19358–19363. doi: 10.1074/jbc.M414561200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.