Abstract
The intrinsic electrical properties and the synaptic input-output relationships of neurons are governed by the action of voltage-dependent ion channels. The localization of specific population of ion channels with distinct functional properties at discrete sites in neurons dramatically impacts excitability and synaptic transmission. Molecular cloning studies have revealed a large family of genes encoding voltage-dependent ion channel principal and auxiliary subunits, most of which are expressed in mammalian central neurons. Much recent effort has focused on determining which of these subunits co-assemble into native neuronal channel complexes, and the cellular and subcellular distributions of these complexes, as a crucial step in understanding the contribution of these channels to specific aspects of neuronal function. Here we review progress made on recent studies aimed at determining the cellular and subcellular distribution of specific ion channel subunits in mammalian brain neurons using in situ hybridization, and immunohistochemistry. We also discuss the repertoire of ion channel subunits in specific neuronal compartments and implications for neuronal physiology. Finally, we discuss the emerging mechanisms for determining the discrete subcellular distributions observed for many neuronal ion channels.
I. OVERVIEW OF MAMMALIAN BRAIN VOLTAGE-DEPENDENT ION CHANNELS
A. Introduction
Mammalian central neurons express a large repertoire of voltage-dependent ion channels (VDICs) that form selective pores in the neuronal membrane and confer diverse properties of intrinsic neuronal excitability. This allows mammalian neurons to display a richness of firing behaviors over a wide range of stimuli and firing frequencies. The complex electrical behavior of mammalian neurons is due to a huge array of VDICs with distinct ion flux rates and selectivity, although the major VDICs underlying neuronal excitability and electrical signaling are those selective for Na+, K+ and Ca2+ ions. Neuronal VDICs also exhibit widely differing properties of how sensitive their gating, or the opening or closing of the channels pore, is to changes in membrane potential. Different VDICs also differ in the kinetics of these gating events. Importantly in the terms of mammalian brain, different VDICs differ widely in their cellular expression and subcellular localization, impacting their relative contribution to brain function. This functional diversity is based on expression of dozens of VDIC subunits that can assemble into complicated multisubunit protein complexes with distinct properties, and their subsequent targeting to and retention at specific sites in the neuronal membrane. Molecular cloning and genomic analyses have revealed a diversity of ion channel subunits that was arguably unanticipated from previous physiological and pharmacological studies. The molecular definition of the mammalian VDIC family has led to the development of molecular tools that has allowed for studies aiming to link expression and function of specific VDIC subunits with neuronal excitability and electrical signaling in specific classes of mammalian brain neurons and neuronal networks. Such efforts find justification in leading towards a better fundamental understanding of the molecular processes that shape neuronal function, but also in identifying and validating novel targets for discovery research aimed at developing new therapeutics for CNS disorders. Here we review the findings from these studies and the implications for these goals.
B. General Structural Features of the Principal Subunits of Voltage-Dependent Ion Channels
VDICs selective for Na+, K+ and Ca2+ are referred to as Nav, Kv and Cav channels, respectively. The macromolecular proteins complexes that form these channels comprise numerous subunits with distinct structural and functional features. All mammalian VDICs contain one (Nav, Cav) or four (Kv) transmembrane pore forming and voltage-sensing subunits termed α (for Nav and Kv) or α1 (for Cav). These polypeptides exist in two general forms: individual Kv channel α subunits (Fig. 1) with six transmembrane segments (termed S1-S6) that assemble posttranslationally to form tetrameric complexes, and Nav channel α (Fig. 3) and Cav channel α1 (Fig. 5) subunits that resemble four tandemly concatenated Kv α subunits and contain four internally repeated “pseudosubunit” S1-S6 domains and comprise a single 24 transmembrane segment subunit (376). These principal VDIC subunits form the major structural and functional unit of the channel and have been the focus of some of the most exciting biophysical and structural studies in all of biology in the past two decades. Much of this work has focused on the Kv channel α subunits, which from a molecular standpoint are more amenable to structure-function analyses than are the larger Nav α and Cav α1 subunits (187, 207).
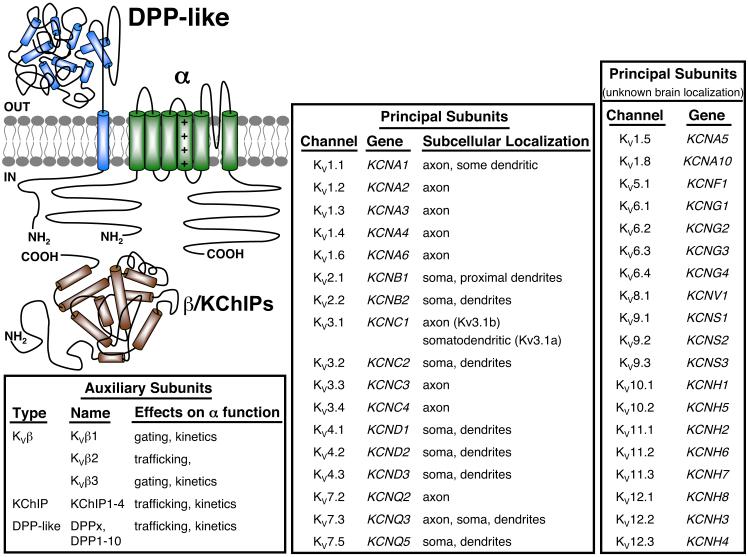
FIG. 1.
Subunit composition and subcellular localization of Kv channel principal and auxiliary subunits in mammalian central neurons. Schematic representation of a single Kv α subunit, four of which (within the same subfamily) assemble to form functional Kv channels. Native channels also comprise auxiliary Kvβ (Kv1 subfamily), or KChIP and DPP-like (Kv4 family) subunits. Top middle box shows the classification, genetic nomenclature, and subcellular localization of Kv channel α subunits with well-characterized subcellular localization in mammalian central neurons. Top right box, the classification and genetic nomenclature of Kv channel principal subunits with unknown subcellular localization in mammalian brain. Bottom right box, classification of Kv channel auxiliary subunits expressed in mammalian central neurons, and their functional effects on Kv α subunits.
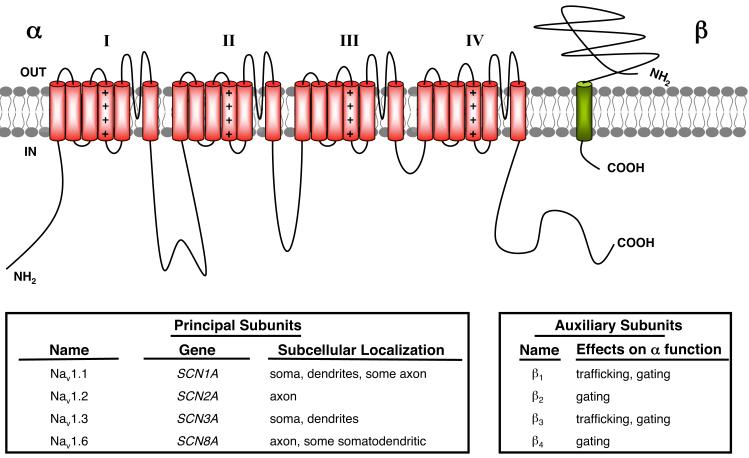
FIG. 3.
Subunit composition and subcellular localization of Nav channel principal and auxiliary subunits in mammalian central neurons. Schematic representation of a single Nav α subunit that forms macromolecular complexes with auxiliary Navβ subunits. Bottom left box, the classification, genetic nomenclature, and subcellular localization of mammalian brain Nav channel principal α subunits. Bottom right box, classification of Navβ auxiliary subunits expressed in mammalian central neurons, and their functional effects on coexpressed Nav α subunits.
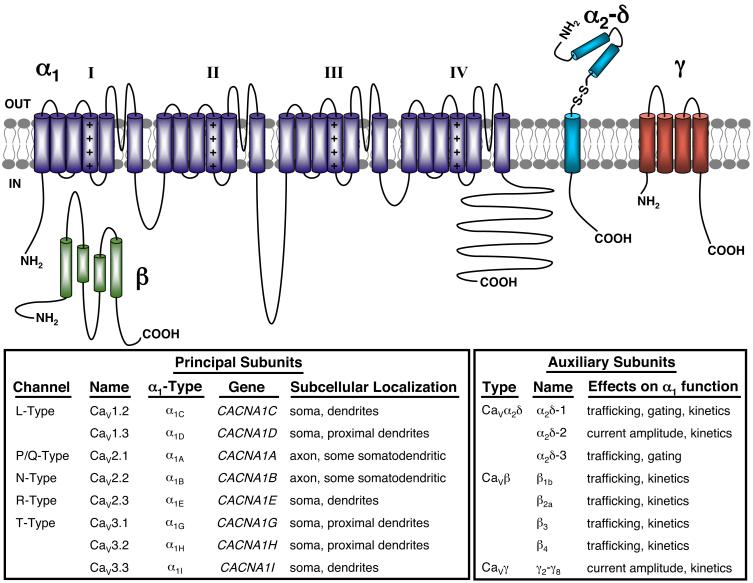
FIG. 5.
Subunit composition and subcellular localization of Cav channel principal and auxiliary subunits in mammalian central neurons. Schematic representation of a single Cav α1 subunit that forms macromolecular complexes with auxiliary Cavβ, Cavα2δ, and Cavγ subunits. Bottom left box, the classification, genetic nomenclature, and subcellular localization of mammalian brain Cav channel principal α1 subunits. Bottom right box, classification of Cavβ, Cavα2δ, and Cavγ auxiliary subunits expressed in mammalian central neurons, and their functional effects on coexpressed Nav α subunits.
The picture that emerges is that the core region, containing the six transmembrane segments of the Kv α subunit (or Nav α and Cav α1 pseudosubunit), is divided into two distinct modular domains, the voltage-sensing module bounded by transmembrane segments S1-S4, and the pore module bounded by transmembrane segments S5-S6. The fourth transmembrane segment, or S4, of each subunit acts as the main voltage-sensing component of the voltage sensor module, which responds to changes in the transmembrane electrical potential and undergoes conformational changes that lead to the voltage-dependent gating of the channel (141, 317). The ionic conductance pathway or pore, which is responsible for rapid and selective potassium ion flux, is formed by the close association of the last two transmembrane segments (S5 and S6) from each of the four Kv α subunits (or Nav α and Cav α1 pseudosubunits) around a central water filled Cavity (79, 180). Allosteric conformational changes link voltage-dependent movement of the voltage-sensor module to the opening of the channel pore, although the precise details of these movements are not yet clear (141, 308). Molecular differences in structures of the voltage-sensor and pore modules account for the bulk of the functional differences between different VDICs in terms of their ion flux rate and selectivity, voltage-dependence of gating, and kinetics (376). The S1-S6 core region also contains all of the extracellular domains of the principal subunit and as such contains all of the binding sites for externally acting drugs and neurotoxins (376).
The principal subunits of VDICs also have extensive cytoplasmic domains that can profoundly affect the functional characteristics described above (268). These domains can influence the coupling between the movement of the voltage-sensor module and the opening of the channel pore, and confer rapid inactivation. The cytoplasmic domains also mediate the interaction of the channel with the rest of the cell, acting as the target site for interacting proteins and posttranslational modifications that influence channel function (233). As discussed in detail below, the cytoplasmic domains of the principal VDIC subunits also play a major role in the intracellular trafficking, targeting and retention events that shape channel expression and localization.
C. Molecular Diversity of the Principal Subunits of Voltage-Dependent Ion Channels
The transmembrane, pore-forming and voltage-sensing α (for Nav and Kv) or α1 (for Cav) subunits of mammalian VDICs are encoded by a large and diverse family of homologous genes (374). Genes encoding the Nav and Cav α/α1 subunits arose from gene duplication and fusion of the coding regions of genes encoding six transmembrane polypeptides resembling Kv α subunits (6, 97, 315). Sequence homology analyses reveal that these gene-duplication and fusion events occurred after the divergence of eukaryotes and prokaryotes (6). The pore-forming and voltage-sensing functionalities of Kv channels are formed as tetrameric assemblies of four independent α subunits, while Nav and Cav channels are formed by a single α/α1 subunit (40). Diversity within the families then came about from subsequent additional gene duplication events (376). Principal α/α1 subunits are typically associated with smaller auxiliary subunits, as detailed below, which can either be cytoplasmic, or single-, double- or tetra-span transmembrane polypeptides (109). The auxiliary subunit composition of the native channel complexes is diverse, and can dramatically influence channel function, intracellular trafficking, post-translational modification, stability and localization (333).
The complexity of the ion channel gene family, combined with a dizzying array of names given to the genes and/or cDNAs as they were isolated, have resulted in a fair degree of confusion in the molecular nomenclature of VDICs. As a response to the growing number of homegrown appellations for cloned Kv channel α subunits, Chandy (45) proposed a systematic nomenclature system that is now widely accepted. This system was later adopted for Cav (87) and Nav (98) channel α subunits by consortia of involved investigators. Together, this has resulted in a clear, systematic and unambiguous nomenclature for nearly all VDICs (374). The nomenclature system is based on the chemical symbol for the principal physiologically permeant ion (K for potassium, Ca for calcium and Na for sodium), followed by the abbreviation of the ligand, which, in the case of this review, is always voltage (v). Thus we will be focusing this review on voltage-dependent potassium (Kv), calcium (Cav) and sodium (Nav) channels. The nomenclature and molecular relationships within each of these families of voltage-gated channels has recently been updated in detailed compendiums; we refer the reader to these as excellent and comprehensive sources for information on Kv (107), Nav (42) and Cav (43) channels.
The remainder of the nomenclature relates to the gene subfamilies within these broad ion channel families. For example, the prototypical Kv channels have been divided into twelve subfamilies (Kv1-Kv12) based on relative sequence homology (107). Kv1-4 channels are in one cluster, Kv7 in another, Kv5, 6, 8, and 9 in another, and Kv10-12 in the last (374, 376). Nav channels are primarily within the Nav1 family, which contains all of the classical voltage-dependent sodium channels (42). The outlying second family, both in terms of sequence homology and function (termed Nax), contains channels whose sequences share some similarity to Nav1 sequences but have never been functionally expressed to verify their ability to function as Nav channels (42), although genetic evidence suggests a role in sodium homeostasis in mouse brain (120). Cav channels have been divided into three groups based on both sequence homology and function: Cav1, which comprise L-type high voltage activated channels, Cav2, which form P/Q-, N- and R-type high-voltage-activated channels, and Cav3, which are the T-type low-voltage activated channels (43).
A parallel nomenclature for ion channel genes has been developed by mammalian geneticists, who assigned official HUGO Human Gene Nomenclature symbols in conjunction with the human genome project (30). The Kv α subunit genes are named KCN*, with the original four gene families assigned the letters A-D (i.e. Kv1-Kv4=KCNA-KCND). Families Kv5-12 have other designations as detailed below. The specific gene number is then derived from the subunit nomenclature, such that Kv1.1 = KCNA1, Kv1.4 = KCNA4, Kv2. 1 = KCNB1, Kv4.2 = KCND2, etc. Similarly, Nav1.1-Nav1.5 α subunit genes are simply assigned the name SCN, and the numerical designation used in the Nav1 nomenclature followed by an A for α subunit (e.g. Nav1.1=SCN1A, Nav1.2=SCN2A, Nav1.3=SCN3A, etc.). For Nav1.6-Kav1.9 α subunits, the HUGO gene name is offset by two relative to the accepted α subunit nomenclature, as Nax subunits account for SCN6A and SCN7A. As such Nav1.6=SCN8A, Nav1.7=SCN9A, etc. The nomenclature for Cav genes is even more complex, as the names are based on the classification system that was in place before a more systematic nomenclature was adopted, such that the gene for Cav1.1 (nee α1S) is CACNA1S, Cav3.1 (nee α1G), is CACNA1G, etc. Thus, some knowledge of the physiological and pharmacological characteristics of the channel, as well as the history of its identification and characterization, is needed to accurately sort through the HUGO nomenclature for Cav genes. Throughout this review, we will predominantly use the nomenclature system developed by ion channel researchers (i.e. Kv, Cav and Nav), but will refer to the HUGO system for clarification or when referring to the gene itself.
Voltage-dependent ion channel α/α1-subunit genes are dispersed throughout the genome, although certain highly related channel genes are found clustered in multigene complexes. Examples of ion channel gene clustering in the mammalian genome include Nav1.1, Nav1.2, Nav1.3 and Nav1.7 (the SCN1A, SCN2A, SCN3A and SCN9A loci, respectively) on 2q23-24 (14, 193), Nav1.5, Nav1.8 and Nav1.9 (the SCN5A, SCN10A, and SCN11A loci, respectively) on 3p21-24, and Kv1.1, Kv1.5 and Kv1.6 (KCNA1, KCNA5 and KCNA6) on 12p13 (4). These clusters presumably represent the products of gene duplication events that led to the diversity of Nav and Kv genes observed in higher mammals (6, 42, 97). The genes themselves can range from the quite simple to the staggeringly complex. On the simple end of the spectrum, all six of the Shaker-related Kv1 α subunit genes expressed in mammalian brain (Kv1.1-Kv1.6) are encoded by intronless open reading frames (47, 107). At the opposite end of the spectrum is the CACNA1E gene (encoding the Cav2.3 channel α1 subunit) that encodes a polypeptide of ≈2300 amino acids in 49 exons within a gene that encompasses >388 kB of genomic sequence on human chromosome 1 (http://www.genecards.org/cgi-bin/carddisp.pl?gene=CACNA1E). Even genes encoding small, cytoplasmic auxiliary subunits can be quite complex. The KCNAB1 gene is over 250 kB in length, and 21 exons are used to encode the relatively small (400 amino acid, 40 kD) gene product (169) (http://www.genecards.org/cgi-bin/carddisp.pl?gene=KCNAB1).
D. Auxiliary Subunits of Voltage-Dependent Ion Channels
Native neuronal VDIC protein complexes also contain a variety of auxiliary subunits that are stable components of the channel complex. The subunits profoundly influence the functional properties of associated principal subunits, as well as acting as determinants of expression and localization.
Native neuronal Kv channel complexes contain both cytoplasmic and transmembrane auxiliary subunits. The best characterized of these are the cytoplasmic Kvβ subunits (Fig. 1) associated with Kv1 family members (240). The bulk of Kv1 channel complexes in mammalian brain have associated Kvβ subunits (259). Four Kvβ subunit genes exist in the human genome, and alternative splicing can generate a number of functionally distinct isoforms (240). Inclusion of the Kvβ1.1 subunit in Kv channel complexes containing Kv1.1, or Kv1.2 dramatically alters the channel gating properties, converting the channels from sustained, or delayed-rectifier type, to rapidly inactivating, or A-type (254). Moreover, the specific α and β subunit composition of native complexes can dramatically impact both the expression level and function of Kv1 channels in mammalian neurons (173, 331, 333). Kvβ subunits exhibit weak overall sequence similarity (198) but striking structural similarity (104) to aldo-keto reductase enzymes. Enzymatic activity of Kvβ subunits against artificial substrates has recently been demonstrated (354). As discussed in detail below, Kvβ subunits are also major determinants of Kv1 channel localization (101).
Accessory subunits for Kv4 channels have also been identified recently and are encoded by two distinct sets of proteins (139). One set is a family of cytoplasmic calcium binding proteins, called KChIPs (Fig. 1), that are members of the neuronal calcium sensor gene family (5). At least four KChIP genes have been reported to exist in mammals (5, 122), and multiple alternatively spliced isoforms of each KChIP gene product have been reported (26, 122, 322, 341). With the exception of the KChIP4a splice variant, in heterologous expression systems all KChIP isoforms increase the surface density (302) and slow the inactivation gating and speed the kinetics of recovery from inactivation (5) of coexpressed Kv4 channels. More recently, Rudy and colleagues (217) reported the identification of a transmembrane dipeptidyl-peptidase-like protein (DPPX) as an accessory subunit of native mammalian brain Kv4 channel complexes (Fig. 1). Co-expression of different combinations of DPPX, KChIPs and Kv4 subunits gives rise to A-type currents whose diverse biophysical properties match very closely the distinct properties of native somatodendritic A-type currents in different mammalian neurons (139).
In addition to these stereotypical Kv channel auxiliary subunits, there exist in the genome genes encoding a number of “electrically silent” α subunit-like polypeptides (80). These make up the Kv5, Kv6, Kv8 and Kv9 subfamilies of principal or α subunits, but are to some extent analogous to auxiliary subunits in that in themselves they cannot form functional channels, but in heterologous expression systems can co-assemble with and functionally modify channels formed from bona fide Kv α subunits (234). The contribution of these subunits to native mammalian brain Kv channels has not yet been established.
Native neuronal Nav channel complexes contain transmembrane auxiliary or Navβ subunits (336), of which there are four different isoforms (Navβ1-Navβ4; Fig. 3). The extensive extracellular domains of these subunits are similar to those of cell adhesion molecules, suggesting a role in cell-cell interactions (252). These subunits have dramatic effects on channel function and pharmacology, and mutations in Navβ subunits can lead to generalized epilepsy (204).
Native neuronal Cav channels have a wide array of auxiliary subunits (Fig. 5). Cavβ subunits resemble Kvβ subunits in that they are cytoplasmic and have similar effects on intracellular trafficking (263), although a number of additional functions have recently been identified, due to the presence of MAGUK-like guanylate kinase and SH3 domains that mediate diverse protein-protein interactions (118). There exist four Cavβ subunits Cavβ1-4. Cav α2 and δ subunits are the product of a single gene whose polypeptide product is posttranslationally cleaved and then subsequently covalently linked by disulfide bonding to yield the mature transmembrane auxiliary α2/δ subunit complex (67). Skeletal muscle Cav channels contain tetraspan Cavγ1 subunits (27, 136). Related proteins of the stargazin family are found in brain and interact with diverse membrane proteins (49) including AMPA-type glutamate receptors, to regulate their trafficking and function (226). A role for stargazins as auxiliary subunits of brain Cav channels has not been firmly established.
E. Techniques for Mapping Voltage-Dependent Ion Channel Expression in Mammalian Brain
The initial isolation of cDNA clones encoding Nav (220), Cav (325), and Kv (12, 144, 232) α subunits initiated the molecular definition of the VDIC family that became complete with the successful completion of sequencing of the human, mouse and rat genomes. These analyses have allowed for tremendous insights into the sequences of VDIC genes, mRNAs, and the encoded polypeptides. This allowed for the generation of subtype-specific molecular probes for measuring expression levels of VDIC mRNAs in extracts of brain tissue by Northern blots, RNAse protection assays, and more recently by RT-PCR. Such probes can also be used to determine cellular patterns of VDIC mRNA expression by in situ hybridization. These nucleotide probes can be designed with a high level of confidence as to their specificity, although the use of multiple probes targeting different regions of the target mRNA is justified (335). Detailed analyses of expression patterns have provided invaluable information as to the relative expression patterns of different VDIC mRNAs in adult brain, at different stages of brain development, in different brain regions, and in specific neuronal populations.
The isolation of cDNAs encoding VDIC subunits, and the subsequent deduction of the primary sequence of the encoded polypeptides, allowed for the efforts to generate specific antibodies targeting VDIC subunits. While this effort still continues today, there now exist extensive libraries of polyclonal and monoclonal antibodies from academic and commercial sources that can be used to identify, isolate and map cellular and subcellular expression patterns of VDIC principal and auxiliary subunits in mammalian brain. This has led to an explosion of information as to the expression and localization of specific VDICs, which combined with results from detailed physiological and pharmacological studies, can begin to yield a picture of the role of specific VDICs of defined subunit composition in mammalian central neurons. While there are numerous issues that have arisen from the use of insufficiently characterized antibodies in such studies, many of these have been addressed in a number of recent articles (262, 278, 279, 335). While these will not be addressed in detail here, we will try throughout to provide insights into the apparent level of characterization and validation of antibodies used in studies described below, to aid in critical evaluation of any subsequent results.
II. VOLTAGE-DEPENDENT POTASSIUM CHANNEL LOCALIZATION IN MAMMALIAN BRAIN
A. Overview of Mammalian Brain Voltage-Gated Potassium Channels
Kv channels are members of a diverse gene family. The nomenclature system for Kv channel α subunits, originally proposed by Chandy and colleagues (45, 107) and now widely accepted is based primarily on the relatedness of amino acid sequences between the different Kv α subunits. The remainder of the nomenclature relates to the gene families within these ion channel groups (Fig. 1). The prototypical Kv channels have been divided into four families (Kv1-Kv4) based on the relative similarity of their amino acid sequences, and on their relatedness to their single gene orthologues in Drosophila: Kv1 (Shaker), Kv2 (Shab), Kv3 (Shaw), and Kv4 (Shal). More recently discovered α subunits make up the Kv5-Kv12 subfamilies. Note that among these subfamilies, the Kv5, Kv6, Kv8 and Kv9 α subunits have not been shown to yield functional expression, but in heterologous cells are able to modulate the expression level and/or gating of channels comprising coexpressed bona fide Kv channel α subunits (107). Note that while the vast majority of publications unambiguously adhere to this nomenclature, there are still examples of ambiguity. These are noted below when appropriate to assist in accurate evaluation of the primary literature relative to the standard nomenclature, which is used exclusively here.
A parallel nomenclature for Kv channel α subunit genes has also been established by the HUGO Gene Nomenclature Committee (Fig. 1). Genes encoding the four original Kv1-Kv4 subfamilies of α subunits are named KCN*, and assigned the letters A-D (i.e., Kv1-Kv4 = KCNA-KCND) and the specific gene numbers following the Kv nomenclature (e.g. Kv1.1 = KCNA1, Kv1.4 = KCNA4, Kv2.1 = KCNB1, etc). Kv5-Kv12 subfamilies have been assigned KCNF (Kv5), KCNG (Kv6), KCNQ (Kv7), KCNV (Kv8), KCNS (Kv9), and KCNH (Kv10-12) gene names by HUGO.
Molecular characterization of Kv channels originally lagged behind that of Nav and Cav channels. Due to the diversity of Kv channel α subunit genes, the potential for oligomerization and the complex repertoire of Kv channels in any given excitable cell, the biochemical approach of purifying Nav channels from eel electroplax (1) and rat brain (113), and Cav channels from skeletal muscle (63), and that ultimately yielded success in cloning Nav (220) and Cav (325) channel α subunits did not yield amounts of Kv channel subunits amenable for protein sequencing. The breakthrough for Kv channel molecular characterization came from concerted genetic and molecular analyses of potassium channel mutants in the fruit fly Drosophila melanogaster (133). These efforts resulted in the isolation of cDNAs encoding the Kv channel α subunit encoded at the Shaker gene locus (12, 144, 232). It was immediately clear from the deduced Shaker amino acid sequence that this Kv channel subunit strongly resembled one of the four internally repeated homologous pseudosubunit domains of an Nav or Cav channel. This led to the proposal, initially substantiated by biophysical analyses (186) and more recently by direct visualization of cryo-electron microscopic (155, 156, 224, 307) and crystallographic (180) structures, that Kv channels are comprised of functional tetramers of individual α subunits. In Drosophila, in addition to Shaker, four other Kv channel genes were present, named Shab, Shal and Shaw (2 members) (275). Subsequent genetic analyses revealed the existence of Kv7 (KCNQ), Kv10 (Eag), Kv11 (Erg) and Kv12 (Elk) genes in Drosophila (177).
cDNAs encoding multiple members of each of the corresponding mammalian gene families (Shab=Kv2 or KCNB; Shaw=Kv3 or KCNC; Shal=Kv4 or KCND; KCNQ=Kv7 or KCNQ; Eag=Kv10=KCNH1, 5; Erg=Kv11=KCNH2, 6, 7; and Elk=Kv12=KCNH3, 4, 8) have now been isolated and expressed. Using the Shaker cDNAs as probes, Tempel and coworkers isolated the first mammalian Kv channel cDNA, Kv1.1 (326). In rapid succession, cDNAs encoding other Kv1 (i.e. Shaker-related) family members (Kv1.2-Kv1.7, the products of the KCNA1-7 genes) were isolated [reviewed in (46)]. These different mammalian Kv1 family members had distinct functional properties when expressed alone (i.e. as homotetramers) in heterologous cells (316). Different Kv1 family members could also co-assemble into channels with mixed subunit composition, and such heterotetrameric channels exhibited functional properties intermediate between those of channels formed from homotetramers of the constituent subunits (123, 129, 271). Certain Kv1 channels contain strong trafficking determinants, while others do not (195). Heteromeric assembly of different subunits yields channels with intermediate trafficking characteristics (194). Thus, the subunit composition of Kv1 channels not only determines their gating and kinetic properties, but also dramatically affects their expression and localization (340).
Coincident with the identification of mammalian Kv1 α subunits, a cDNA encoding the rat brain Kv2.1 α subunit was isolated by expression cloning in Xenopus oocytes (91). The manner of cloning Kv2.1 is noteworthy as it reflects the high level expression of Kv2.1 in mammalian brain. Low stringency hybridization screening of mammalian brain cDNA libraries led to the isolation of cDNAs encoding Kv3 (184, 199, 270, 373), Kv4 (10, 266) Kv5 and Kv6 (80), and Kv10, Kv11 and Kv12 (182, 351) α subunits. The Kv8 family was identified using RT-PCR performed with degenerate primers (125), and Kv9 by in silico analyses of EST databases (274). The Kv7 subfamily was initially identified in human genetic analyses of genes associated with a congenital heart disorder called long QT syndrome (350).
The human genome contains a total of forty genes encoding Kv channel α subunits. Some of these genes generate messages that are subject to alternative splicing. In mammalian brain, the expression of many of these Kv channel α subunits is restricted to neurons, although glial cells may express a subset of the neuronal repertoire. In general, Kv channels exhibit subfamily-specific patterns of subcellular localization (Fig. 1), as detailed below. Kv1 channels are found predominantly on axons and nerve terminals, Kv2 channels on the soma and dendrites, Kv3 channels can be found in dendritic or axonal domains, depending on the subunit and cell type, and Kv4 channel are concentrated in somatodendritic membranes (335). Kv7 family members are predominantly on axons, although evidence also exists for somatodendritic Kv7 channels (110). Little information is available for the subcellular localization of Kv5-6 and Kv8-12 α subunits.
B. Kv1 Subfamily and Associated Auxiliary Subunits
When expressed in heterologous systems, the neuronal Kv1 α subunits can generate either transient (Kv1.4) or sustained (Kv1.1-Kv1.3, Kv1.5-Kv1.6) Kv currents. Moreover, heteromeric assembly with one another, and co-assembly with auxiliary Kvβ subunits can generate a diversity of function from the resultant α4β4 channel complexes (340). In neurons Kv1 channels are found predominantly on axons and nerve terminals, although dendritic expression is also found in certain neurons (335). Kv1 family members exhibit extensive coassembly to generate heteromeric channels with distinct characteristics (239). In addition, assembly with auxiliary subunits can dramatically impact expression, localization and function of the resultant channel complexes (173). For example, inclusion of the Kvβ1.1 subunit in Kv1 channel complexes containing Kv1.1 or Kv1.2 dramatically alters channel gating properties, converting the channels from sustained, or delayed-rectifier type, to rapidly inactivating, or A-type (254). The specific subunit composition of native complexes can also dramatically impact the expression level, localization and function of Kv1 channels in mammalian neurons (339).
The predominant Kv1 cellular staining pattern throughout the brain is neuronal, and subcellularly, axonal. The three most abundant Kv1 subunits expressed in mammalian brain, Kv1.1, Kv1.2 and Kv1.4, are found predominantly localized to axons and nerve terminals (335). In many cases, these subunits are components of heteromeric channel complexes, as Kv1.1, Kv1.2 and Kv1.4 exhibit precise patterns of colocalization (260), and extensive association as shown by co-purification (57, 260, 293). However, the subunit composition of channels containing these subunits varies across brain regions (288). The Kv1.1 α subunit in mammalian brain appears to be segregated into two major subpopulations: one associated with Kv1.2, and one associated with Kv1.4. Kv1.1 and Kv1.2 are found in the absence of Kv1.4 in cerebellar basket cell terminals (167, 202, 203, 258, 260, 346, 347), the juxtaparanodal membrane adjacent to axonal nodes of Ranvier (Fig. 4D) (243, 245, 248, 249, 258, 260, 338, 346, 347), and in the terminal segments of axons (73). Kv1.1 and Kv1.2 are also present at axon initial segments (72, 128, 342), sometimes in association with Kv1.4, where they control axonal action potential waveform and synaptic efficacy (160). Kv1.1/Kv1.2 channels also play a role in μ-opioid receptor-mediated modulation of GABAergic inputs into basolateral amygdala neurons (90) and in serotonin-modulated glutamate release from thalamocortical nerve terminals (165). Low-threshold, slowly inactivating axonal Kv1.2-containing channels, presumably containing either Kv1.4 α or Kvβ1 subunits to confer inactivation, are involved in the flexible properties of intracortical axons of layer 5 pyramidal neurons and may contribute significantly to intracortical processing (304). Kv1-containing channels are also important in setting the firing rate of layer II/III pyramidal neurons (103). Kv1.1 and Kv1.4 are found robustly expressed in the relative absence of Kv1.2 within the striatal efferents in globus pallidus and pars reticulata of substantia nigra (260, 297).
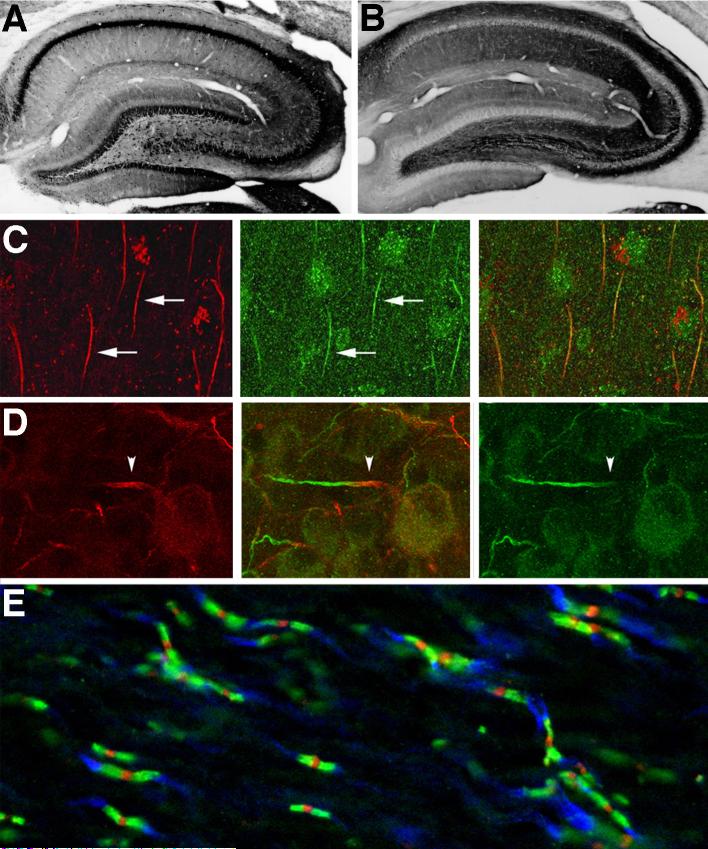
FIG. 4.
Cellular and subcellular distribution of Nav channels in mammalian brain and retina. A, B: Immunoperoxidase staining of adult rat hippocampus. Reproduced with permission from reference (100). A, Nav1.1. Note staining in somata and proximal dendrites of dentate granule cells, CA3-CA1 pyramidal cells and interneurons. B, Nav 1.2. Note staining in the mossy fiber pathway within CA3 s. lucidum and in s. radiatum of CA1. C, Double immunofluorescence staining for Nav1 channels (red) and spectrin IVβ (green) in the axonal initial segments (arrows) of pyramidal cells in human temporal neocortex. Reproduced with permission from reference (128). D, Double immunofluorescence staining for Nav1.1 (red) and Nav1.6 (green) in the axonal initial segments of retinal ganglion cells. Arrowhead denotes proximal portion of initial segment positive for Nav1.1 Reproduced with permission from reference (342). E, Triple immunofluorescence staining for nodal Nav1 channels (red), paranodal Caspr (green) and juxtaparanodal Kv1.2 (blue) in adult rat optic nerve. Image courtesy of Dr. Matthew N. Rasband.
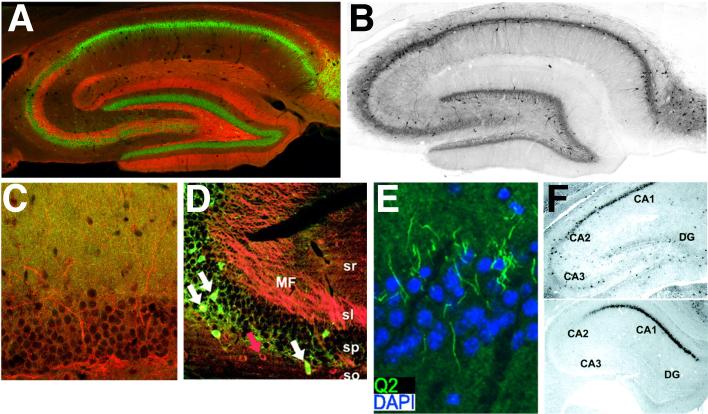
Within the excitatory circuitry of the hippocampus, a number of patterns for expression for these three Kv1 subunits emerge, providing a striking example of the complex heterogeneity of subunit association (260). Kv1.1, Kv1.2 and Kv1.4 are highly expressed in the middle third of the molecular layer of the dentate gyrus (Fig. 2A) where they are associated with axons and terminals of the medial perforant path (212, 258, 260, 296, 297, 344, 346, 347). Kv1.1, Kv1.2 and Kv1.4 are also found in Schaffer collateral axons, while Kv1.1 and Kv1.4 colocalize, in the absence of Kv1.2, in mossy fiber axons (Fig. 2A) (61, 296, 297, 346, 347), where they regulate Ca2+ influx and transmitter release (96).
FIG. 2.
Cellular and subcellular distribution of Kv channels in adult hippocampus. A-C, F: rat, D-E: mouse. A, Double immunofluorescence staining for Kv1.4 (red) and Kv2.1 (green). Note Kv1.4 staining in terminals fields of the medial perforant path in the middle molecular layer of the dentate gyrus, and mossy fiber axons and terminals in s. lucidum of CA3. B, Immunoperoxidase staining for Kv3.1b. C, Double immunofluorescence staining for Kv4.2 (green) and Kv4.3 (red) in dentate gyrus. Note uniform staining for both Kv4 α subunits in granule cell dendrites in molecular layer, and Kv4.3 staining in scattered interneurons. D, Double immunofluorescence staining for Kv7.2 (red) and parvalbumin (green) in CA3. White arrows depict Kv7.2-negative, and red arrow Kv7.2-positive neurons. MF: mossy fibers, sr: s. radiatum, sl: s. lucidum, sp: s. pyramidale, so: s. oriens. Reproduced with permission from reference (59). E, Double immunofluorescence staining for Kv7.2 (green) and DNA (DAPI, blue) in CA1. Reproduced with permission from reference (229). F, In situ hybridization of Kv11.1 (top panel) and Kv11.3 (bottom panel). DG: dentate gyrus. Reproduced with permission from reference (272).
However, in spite of their colocalization, it is not clear that heteromeric channel complexes containing co-associated Kv1.1, Kv1.2 and Kv1.4 are present on perforant path and Schaffer collateral axons. Lesions in entorhinal cortex have distinct effects on the distribution of Kv1.2 and Kv1.4 in the middle third of the dentate molecular layer, suggesting that while these subunits may colocalize at the light microscope level, they may be expressed, along with Kv1.1, on different components of the perforant path (212). Similar results are obtained with lesions placed in other subfields. For example, in CA3 the predominant Kv1 channels appear to be composed of Kv1.1 with Kv1.4, while in the Schaffer collateral pathway, Kv1.1 is likely to be associated with Kv1.2 and Kv1.4 (212).
Electron microscopic immunohistochemical studies have demonstrated that Kv1.1, Kv1.2 and Kv1.4 are concentrated in the axonal membrane immediately preceding or within axon terminals (61, 346, 347). The immunoreactivity for Kv1.1 and Kv1.2 has been localized to the pre-terminal axonal membrane in stratum (s.) radiatum (346, 347), while immunoreactivity for Kv1.4 has been localized to the pre-terminal extensions of mossy fiber axons (61). Activation of Kv1 channels at these sites can play a critical role in regulating nerve terminal excitability and thereby regulate neurotransmitter release, as shown by pharmacological and genetic knockdown of Kv1 function (74).
The other Kv1 α subunits appear to be expressed at lower levels in mammalian brain. Kv1.6 is found predominantly in interneurons, although some dendritic staining is seen throughout the brain on principal cell dendrites, which also express Kv1.1 and Kv1.2 (260), and presumably underlie the “D” current (313, 378). Kv1.3 is highly expressed in the cerebellar cortex. The bulk of this expression is in the parallel fiber axons of cerebellar granule cells, as strong in situ hybridization signal is present in the granule cell layer (163), while strong immunostaining (344) and 125I-margatoxin binding (specific for Kv1.2 and Kv1.3) (159) is found in the molecular layer. The molecular layer also contains high levels of staining for Kv1.1 (260, 344) suggesting that Kv1.1 and Kv1.3 have the opportunity to form heteromeric channels on parallel fibers. The expression of Kv1.5 in the brain is overall quite low (88). What Kv1.5 expression there is may be restricted to non-neuronal cells. For example, Kv1.5, and Kv1.3 are components of delayed rectifier currents in glia (52, 150, 230) and endothelial cells (206). In mammals, Kv1.7 is expressed in skeletal muscle, heart and pancreatic islets, but not brain (143).
In situ hybridization, immunoprecipitation and immunohistochemical analyses have also localized sites of expression of the Kvβ1 and Kvβ2 auxiliary subunits in mammalian brain (212, 254, 258-260). Kvβ2 appears to be a component of many if not all Kv1-containing channel complexes in mammalian brain, and immunoreactivity for Kvβ2 is present in almost every location where immunoreactivity for Kv1-family subunits is observed (259, 260). However, there is also extensive immunostaining for Kvβ2 in somata and dendrites that is not observed for Kv1 α subunits, and certain sites of intense Kv1 α subunit staining (e.g. hippocampal mossy fibers) lack robust Kvβ2 immunoreactivity (212, 259, 260). In the hippocampus, excitotoxic lesion of distinct components of the extrinsic and intrinsic circuitry reveal that the majority of Kvβ2 that is associated with Kv1 α subunits is associated with axons and presynaptic terminals of the perforant path and Schaffer collateral pathways (212). The Kvβ1 subunit, which exerts dramatic effects on the inactivation kinetics of Kv1 channels, appears to be included in Kv1 channel complexes more selectively than is Kvβ2. Interestingly, the pattern of immunoreactivity for Kvβ1 closely matches the expression pattern for Kv1.1 and Kv1.4, in that Kvβ1 is found to co-localize with Kv1.4 in the medial perforant path, mossy fiber pathway, and in striatal efferents to the globus pallidus (259, 260). Kvβ2 is found in the absence of Kvβ1 at many sites that exhibit colocalized Kv1.1 and Kv1.2, for example in cerebellar basket cell terminals and juxtaparanodes of nodes of Ranvier (248, 260). As such there exist distinct pairings of Kv1 α and Kvβ subunits in heteromeric channel complexes in mammalian brain.
C. Kv2 Subfamily
Kv2 family members form delayed rectifier Kv channels that are prominently expressed in mammalian brain, where they are localized in the somatodendritic domain of neurons. Kv2.1 was the first member of this family isolated by molecular cloning approaches, and is unique in that it was identified and isolated from an adult rat brain cDNA library by expression cloning in Xenopus oocytes (91), suggestive of an unusually high level expression of Kv2.1 mRNA in adult rat brain. Thus it was not surprising when immunostaining revealed that Kv2.1 was highly expressed, and has an extensive distribution throughout the mammalian brain (82, 127, 332). In fact, the cellular distribution in neurons is so broad that in many regions the cellular staining pattern of Kv2.1 resembles that of a Nissl stain. However, in spite of this broad neuronal expression, within individual neurons the staining for Kv2.1 is highly restricted, being present only on the somatic and proximal dendritic membrane (Fig. 2A), and absent from axons and nerve terminals (332). Immuno-electron microscopy (82) and excitotoxic lesion studies (212) have unambiguously confirmed the somatodendritic localization of Kv2.1. The striking subcellular distribution is accentuated by the fact that within these domains Kv2.1 is present in large clusters (82, 127, 190, 281, 332). These clusters are present on the cell surface membrane immediately facing astrocytic processes, and over subsurface cisterns underlying the plasma membrane facing astrocytes (82). The physiological basis for the highly clustered, discrete localization of Kv2.1 to these specialized membrane domains is not known.
In spite of its widespread cellular distribution, there are certain cells that stand out for having especially prominent Kv2.1 expression. In the cortex, pyramidal cells in layers II/III, and in layer V are especially striking for their high levels of Kv2.1 expression. Kv2.1 is also present in high levels throughout the hippocampus, although the levels in dentate granule cells and CA1 pyramidal cells exceed that in CA3 and CA2 pyramidal cells in both rat and mouse (Fig. 2A). However, it should be stressed that Kv2.1 is found on both principal cells and interneurons (Fig. 2A) throughout the hippocampus (82). Among interneurons, Kv2.1 is found in the majority of cortical and hippocampal parvalbumin, calbindin and somatostatin-containing inhibitory interneurons (82). Note that Kv2.1 localization in rat brain, especially the extent of clustering, is dramatically affected by neuronal activity (209) and ischemia (208) due to changes in phosphorylation state, such that state of the subject and preparation of the sample could impact details of Kv2.1 localization.
Kv2.2 is expressed in many of the same cells that express Kv2.1. However, unlike other Kv channels (Kv1, Kv3 and Kv4 family members), the two members of the mammalian Kv2 family apparently do not readily form heteromultimers in native neurons (although see (21)), as the subcellular localizations of Kv2.1 and Kv2.2 expressed in the same cells are distinct (126, 175). Kv2.2 is present on dendrites, but is present uniformly and along the entire length of the dendrite. The clustered, proximal dendritic localization of Kv2.1 is not observed for Kv2.2. Kv2.2 is present at high levels in olfactory bulb neurons, and in cortical pyramidal neurons.
D. Kv3 Subfamily
Kv3 family members have unique functional characteristics, including fast activation at voltages positive to -10 mV and very fast deactivation rates. These properties are thought to facilitate sustained high-frequency firing, and Kv3 subunits are highly expressed in fast-spiking neurons, such as neocortical and hippocampal interneurons as well as midbrain auditory neurons (269). Kv3 currents can either have sustained (Kv3.1, Kv3.2) or transient (Kv3.3, Kv3.4) characteristics, and can form hetero-oligomeric channels with intermediate gating characteristics (269). Kv3 mRNAs are somewhat unusual among Kv α subunits in that they are subjected to extensive alternative splicing to generate subunits that differ only at their cytoplasmic carboxyl termini (184). This complicates studies of localization of these subunits, as the specific nature of the in situ hybridization or antibody probes used can affect which Kv3 alternative splicing isoforms are detected.
Initial in situ hybridization analyses revealed that unlike many other Kv subunits, Kv3.1 and Kv3.2 transcripts were expressed in only a small subset of cells in the cerebral cortex and hippocampus (236, 353). Interestingly, the in situ hybridization patterns of Kv3.1 and Kv3.2 were distinct, suggesting a strict cellular specificity to expression of these highly related Kv channel α subunits (353). Initial immunolocalization studies were performed using antibodies raised against the major splice variant of Kv3.1, termed Kv3.1b, which has a longer carboxyl terminus than the less abundant Kv3.1a variant. These and subsequent studies revealed that Kv3.1b was highly expressed in interneurons (Fig. 2B), and that expression was very low/undetectable in principal cells, such as neocortical and hippocampal pyramidal cells, and dentate granule cells (289, 352). Kv3.1b is also robustly expressed in fast-spiking cells in the cochlear nucleus (236). Double labeling experiments (289) revealed that the subset of cortical cells labeled with anti-Kv3.1b antibodies corresponded to GABAergic interneurons expressing the calcium-binding protein parvalbumin, which are distinguished by their fast-spiking properties. Interestingly, Kv3.2 α subunits were found in non-fast-spiking, somatostatin- and calbindin-containing interneurons (53). Thus, the expression patterns of Kv3.1 and Kv3.2 α subunits can distinguish different populations of interneurons, raising the possibility that interneuron firing patterns rely to some extent on the subtype of Kv3 channels expressed (166, 174, 269). Kv3.2 is also found in the cerebellar pinceau (22), in the terminals of basket cells that synapse onto Purkinje cell axon initial segments.
Another site of prominent expression of Kv3.1b subunits is in auditory neurons (83, 172, 352), especially in globular bushy cells in the anterior ventral cochlear nucleus (AVCN) and principal cells in the medial nucleus of the trapezoid body (MNTB). In AVCN neurons Kv3.1b is present in somatodendritic compartments, and in MNTB neurons in presynaptic terminals (83). The expression of Kv3.1b follows a tonotopic gradient and is present in cells transmitting high frequency tones (83, 172). In these cells Kv3.1b is found basally phosphorylated by protein kinase C, which suppresses Kv3.1b currents (185). Activity-dependent dephosphorylation enhances Kv3.1b currents and allows for high frequency firing at auditory synapses (309). Kv3.1b is also present at a subset of nodes of Ranvier in axons of central but peripheral neurons, and was not present at axon initial segments (70).
In the case of mammalian brain Kv3.1, alternative splicing does in fact lead to a difference in the polarized expression of Kv3.1 variants in mammalian brain (228). The initial studies of Kv3.1b localization in mammalian brain revealed staining that was present in the soma, proximal dendrites, unmyelinated axons, and axon terminals of the parvalbumin-positive interneurons (53, 289, 352) and neurons in the cochlear nucleus (235). In contrast, Kv3.1a proteins were prominently expressed in the axons of some of the same neuronal populations, but there was little or no Kv3.1a protein expression in somatodendritic membrane.
Studies on the exogenous expression of three different Kv3.2 splice variants (Kv3.2a, b and c) in polarized epithelial cells revealed that alternative splicing led to differences in subcellular localization. The Kv3.2a variant was localized to the basolateral membrane, while the Kv3.2b and Kv3.2c isoforms were found apically (238). The epithelial cell:neuron analogous membrane hypothesis (77) predicts that as such, in neurons, Kv3.2a would be localized to the somatodendritic domain, and Kv3.2b and Kv3.2c would be localized to the axon. Thus, as for the case of Cav2.3 discussed below, alternative splicing of ion channel transcripts can generate functionally similar variants of the same channel with altered subcellular distributions (228).
Kv3.3 α subunits are also widely expressed at the mRNA level in brain (269). In forebrain, Kv3.3 are coexpressed with Kv3.1b and Kv3.2 subunits in parvalbumin-positive interneurons (48). Robust Kv3.3 staining is also observed in mossy fiber axons of hippocampal dentate granule cells (48). Both Purkinje cells in cerebellar cortex, and deep cerebellar nuclei contain high levels of Kv3.3b message (99). Purkinje cells have Kv3.3 protein in axons, on somata, and in proximal and distal dendrites (48, 196), where a Kv3 channel complex of Kv3.3 and Kv3.4 may play a role in shaping depolarizing events. Most brainstem auditory neurons also express Kv3.3 mRNA (172) and protein (48), where it may co-assemble with Kv3.1 in a subset of cells.
Unlike Kv3.1 and Kv3.2, in neocortex and hippocampus Kv3.4 is present in principal cells (256, 353). Moreover, Kv3.4 appears to be localized to axons and nerve terminals of these cells, such that in a number of brain regions Kv3.4 is found co-localized with Kv1 family members. Combined in situ hybridization and immunohistochemistry yield a picture whereby Kv3.4 is found in terminals of the perforant path, as high levels of Kv3.4 mRNA, but not protein, are found in entorhinal cortex, and high levels of Kv3.4 immunostaining are present in the middle third of the molecular layer of the dentate gyrus (256, 344). Intense Kv3.4 staining is also observed in s. lucidum of CA3, and appears to be associated with mossy fiber axons and/or terminals (256, 344), where it may be present in heteromeric Kv3 complexes with Kv3.3 (48). In these regions Kv3.4 may be present in the same axons and terminals as Kv1.1 and Kv1.4. Kv3.4 is also found in cerebellar basket cell terminals, which also contain high levels of Kv1.1 and Kv1.2. Immuno-electron microscopy (167) revealed that although the localization of staining of Kv1.1, Kv1.2 and Kv3.4 overlaps at the light microscope level, these subunits have distinct ultrastructural localizations. Kv1.1 and Kv1.2 are present in septate-like junctions formed between basket cell terminals and Purkinje cell axons, while Kv3.4 is found in nonjunctional regions of the terminals (167). Kv3.2 is also found in basket cell terminals at sites distinct from those that contain Kv3.4, perhaps corresponding to active zones of the presynaptic terminals (22). These findings highlight the extent to which different highly related ion channel subunits can be precisely localized in neuronal membrane domains. It should be noted that recent data suggest that in certain fast-spiking cells Kv3.4 may also associate with Kv3.1 and/or Kv3.2 α subunits to generate a fast delayed rectifier current (11). However, examples of neurons in which Kv3.4 was found colocalized with Kv3.1 and Kv3.2 were not provided. Studies of Kv3.1b (65) and Kv3.4 (29) localization in certain brainstem nuclei suggest that these subunits may both localize to presynaptic terminals, for example those in nucleus tractus solitarius. As noted above, Purkinje cell dendrites may contain a Kv3.3/Kv3.4 heteromeric channel that is important in shaping responses to certain, relatively strong depolarizing events (196).
E. Kv4 Subfamily and Associated Auxiliary Subunits
The Kv4 α subunits Kv4.1, Kv4.2 and Kv4.3 form transient or A-type Kv channels (139). Experimental knockdown of Kv4 α subunit expression in mammalian neurons results in suppression of A-type Kv channels (154, 168, 191, 192, 379). Kv4.1 is expressed at very low levels in mammalian brain (290), and what expression that can be detected in neurons does not correlate with A current density (114). In contrast, Kv4.2 and Kv4.3 are expressed at relatively high levels (290) and the expression of these subunits correlates well with neuronal A-type current density in a number of neuronal types (114, 176, 303, 310, 330). In situ hybridization analyses show that the expression of Kv4.2 and Kv4.3 is widespread throughout the brain, and that while in many brain regions the cellular expression of these two Kv4 genes is reciprocal or complementary, there are numerous cell populations in which Kv4.2 and Kv4.3 are co-expressed (290).
Immunoreactivity for Kv4 subunits is concentrated primarily in the dendrites of central neurons (335). Kv4.2 is expressed at high levels in many principal cells, while Kv4.3 is found in a subset of principal cells and in many interneurons (335). In the hippocampus, dentate granule cells express high levels of Kv4.2 and Kv4.3 mRNA (290), and robust staining for both of these subunits (Fig. 2C) is present in the molecular layer (190, 205, 257, 297). That this immunostaining is in the distal granule cell dendrites that receive input from the lateral perforant path is supported by elimination of such staining upon excitotoxic lesion of the dentate gyrus (257).
A similar colocalization of Kv4.2 (190, 205, 257, 297) and Kv4.3 (205, 257) is seen in the distal basal (in s. oriens) and apical dendrites (in s. lucidum and s. radiatum) of CA3 pyramidal neurons. That the staining for Kv4.2 and Kv4.3 in CA3 s. lucidum is to CA3 pyramidal cell dendrites, as opposed to dentate granule cell axons that form mossy fibers, is supported by the lack of an effect of excitotoxic lesions within the dentate gyrus on this staining (257). Interestingly, CA2 and CA1 pyramidal cells express Kv4.2 mRNA (290, 297) and protein (190, 205, 257, 297) in the absence of Kv4.3 mRNA (290) and protein (205, 257), suggesting a unique role for Kv4.2 homotetramers in the distal dendrites of these cells relative to the situation in CA3. Conversely, throughout the hippocampus, Kv4.3 mRNA (290) and protein (205, 257) is found in the absence of Kv4.2 in many interneurons, such as those located adjacent to the granule cell layer in dentate gyrus (Fig. 2C). Thus, different hippocampal cell types appear to have different requirements for homotetrameric and heterotetrameric Kv4.2 and Kv4.3 channels. The specific targeting of Kv4 channels to dendrites of principal cells in the hippocampus has been confirmed by excitotoxic lesion studies (257). In all cases the staining of Kv4 channels on dendrites tends to be quite uniform, with little evidence of local concentrations of clustered immunoreactivity associated with dendritic spines, postsynaptic densities or other subcellular domains.
Principal cells and interneurons in neocortex also exhibit specific patterns of Kv4.2 and Kv4.3 staining (32, 257). Pyramidal cells in layer II of rat parietotemporal cortex exhibit high levels of Kv4.3 staining, while Kv4.2 staining predominates in those in layer V (257). The expression of Kv4.2 and Kv4.3 in pyramidal cell dendrites was further supported by studies in transgenic mice expressing fluorescent protein in a subset of pyramidal neurons in visual cortex (32). Clear staining for both Kv4.2 and Kv4.3 was observed in fluorescent dendrites (32). Immuno-electron microscopic studies reveal a high density of Kv4.2 and Kv4.3 immunoreactivity in distal dendrites of pyramidal neurons, and in apical tufts found in layer I (32). Interestingly, the majority of this immunostaining is associated with GABAergic and not excitatory synapses (32).
Kv4.3 is also found in interneurons scattered throughout layers II-VI in rat neocortex (257). Kv4.3 in also found in numerous GABAergic interneurons in mouse visual cortex (32). Kv4.2 was also found in GABAergic interneurons in mouse visual cortex, although the cellular populations are not identical to those expressing Kv4.3 suggesting different patterns of expression in different interneuron populations (32). Kv4 channels are also highly expressed in cerebellum. Cerebellar granule cells express both Kv4.2 and Kv4.3 mRNA (290, 297) and protein (205, 257, 297, 314). In the cerebellar granule cell layer, reciprocal gradients of mRNA and protein expression are observed for Kv4.2 (anterior > posterior) versus Kv4.3 (posterior > anterior) (290, 314).
The expression and localization of Kv4-associated accessory cytoplasmic KChIP subunits has also been studied in some detail (205, 257). Specific KChIPs exhibit distinct patterns of localization, and of colocalization, with Kv4 α subunits. The circuitry of the hippocampus is again exemplary for these patterns. For example, in hippocampus, KChIP2, KChIP3 and KChIP4 exhibit precise colocalization with Kv4.2 and Kv4.3 in dendrites of dentate granule cells, and CA3 pyramidal neurons (205, 257). Interestingly, in the dendrites of CA1 pyramidal cells, which lack Kv4.3, KChIP3 is also not expressed, such that CA1 dendrites express Kv4.2, KChIP2 and KChIP4 (205, 257). The distribution of KChIP1 in hippocampus is restricted to interneurons, where it is coexpressed and colocalized with Kv4.3 in the same subset of GABAergic cells. KChIP1 expression in neocortical and striatal interneurons also closely matches the cellular and subcellular distribution of Kv4.3 (257). These stereotypical Kv4:KChIP combinations (i.e. Kv4.2 with KChIP2 and KChIP4, and Kv4.3 with KChIP1) that are found in most brain regions are not present in cerebellum. KChIP1 is more highly expressed in granule cells in anterior cerebellar cortex, which also express high levels of Kv4.2, than in granule cells in posterior cerebellum, which where higher levels of Kv4.3 are found (314). KChIP3 and KChIP4 are expressed somewhat uniformly in granule cells across the anterior-posterior gradient axis (314). In each case the expression of KChIPs in cerebellar granule cells precisely colocalizes with that of Kv4 α subunits, in the somatic plasma membrane, and in the distinct glomerular synapses that form between granule cell dendrites and afferent inputs (205, 257, 314).
A recent immunohistochemical study of the expression of KChIPs in the Kv4.2 knockout mouse provided a dramatic example of the intimate relationship between Kv4 α subunit and KChIP auxiliary subunit expression (205). Remarkably, genetic ablation of Kv4.2 expression precisely altered the expression of the KChIPs that exhibited colocalization with Kv4.2, and not other KChIPs nor Kv4.3. For example, in hippocampus, KChIP2 staining was virtually eliminated in the dendrites of CA1 pyramidal neurons, which normally express only Kv4.2. However, in dendrites of CA3 pyramids and dentate granule cells, KChIP2 staining was reduced but still present, perhaps due to the persistent expression of Kv4.3 in these neurons. Similar effects were observed in cerebellum, where KChIP1 expression in the Kv4.2 knockout mouse was dramatically diminished in granule cells in the anterior cerebellar cortex, which would normally express high levels of Kv4.2, versus posterior, which expresses Kv4.3 (205). Overall, the impact of the elimination of Kv4.2 expression on KChIPs was stereotypical of the extent of their colocalization with Kv4.2, suggesting that the expression of these subunits is coregulated, by an as yet unknown mechanism, in multiple brain regions.
While not studied in detail, the cellular expression patterns of Kv4-associated DPP family auxiliary subunits also colocalize with those of Kv4 α subunits in mammalian brain neurons. The initial report that the DPPX isoform could be copurified with Kv4 channels from mammalian brain also provided examples of DPPX immunoreactivity precisely colocalizing with that of Kv4.2 in cerebellar granule cells, and specifically in somatic membranes and glomerular synapses (217). DPPX immunoreactivity in hippocampus did not provide compelling evidence for a similar colocalization with Kv4 α subunits (217). More recently, another member of the DPP family, DPP10 was found to exhibit auxiliary subunit effects on Kv4 channels in heterologous cells (140, 382). In situ hybridization analyses revealed distinct patterns of cellular expression, and overlap with that of Kv4 α subunits, for DPPX and DPP10 (382). DPPX expression was robust in the principal cells of the hippocampus, including dentate granule cells and CA1-CA3 pyramidal cells. In contrast, DPP10 was expressed at high levels in GABAergic interneurons. This indicates that DPPX may participate in heteromeric Kv4.2/Kv4.3 complexes (in dentate granule cells and CA3 pyramids) and homomeric Kv4.2 complexes (in CA1 pyramids) while DPP10 is associated with homomeric Kv4.3 complexes in interneurons. Thus, as observed previously for KChIPs, specific DPP family auxiliary subunits have distinct patterns of coexpression (and presumably subcellular colocalization) with specific Kv4 α subunits in hippocampus. Other regions of brain also exhibit distinct patterns of DPPX and DPP10 mRNA expression (382). For example, as noted above, DPPX is highly expressed in cerebellar granule cells, which have low levels of DPP10 expression, while the converse is seen in cerebellar Purkinje cells, which have high levels of DPP10 (and Kv4.3) expression, and low levels of DPPX (and Kv4.2). Together with the analyses of Kv4 a subunit and KChIP auxiliary subunit expression described above, a picture is emerging wherein different brain neurons express different combination of Kv4 α, and KChIP and DPP auxiliary subunits to form dendritic A-type with distinct functional properties (139).
F. Kv7 Subfamily
Neuronal Kv7 or KCNQ channels are the principal subunits underlying the slowly activating and non-inactivating M current that suppresses neuronal firing in many types of brain neurons (349), and suppression of M current by muscarinic modulation enhances neuronal excitability (69). Mutations in neuronal M channel principal KCNQ subunits lead to neurological diseases, the most prominent being epilepsy (60, 138). The major Kv7 KCNQ subunits expressed in mammalian brain are Kv7.2 (KCNQ2), Kv7.3 (KCNQ3) and Kv7.5 (KCNQ5). For simplicity and consistency with the remainder of this section, here we shall refer to these as Kv7 subunits, although the bulk of the literature employs the KCNQ system.
Kv7.2, Kv7.3 and Kv7.5 mRNAs exhibit distinct patterns of cellular expression in mammalian brain. Initial in situ hybridization analyses revealed robust expression and extensive overlap of Kv7.2 and Kv7.3 mRNAs in principal cells of the hippocampus and neocortex (272, 286, 328). Kv7.3 had additional expression in amygdala and thalamus (272, 286). Kv7.5 mRNA is present at high levels in hippocampus, caudate putamen, and in neocortex, especially in piriform and entorhinal cortex (285). In hippocampus, expression is especially high in CA3 pyramidal neurons relative to other principal cell types (285). Northern blot analyses of regional expression patterns in human brain yield similar results, with the exception that human cerebellum, in contrast to the situation in rat, expresses high levels of Kv7.5 mRNA (285).
Immunohistochemical localization of the cellular and subcellular localization of Kv7.2 and Kv7.3 subunits in brain has revealed a predominant axonal localization. Initial reports of staining in mouse (59) and rat (267) hippocampus revealed intense staining in mossy fiber axons and their terminals (Fig. 2D), and very little or no labeling in dentate granule cells themselves. Low levels of labeling were observed in somata of principal cells in CA3-CA1 in both mouse and rat, although moderate levels of staining in neuropil of s. oriens, and s. radiatum in these regions was observed. A subpopulation of GABAergic interneurons in s. oriens of CA3 (Fig. 2D), and in s. oriens, s. pyramidale, and s. radiatum of CA1 also exhibited robust immunoreactivity for Kv7.2 (59, 267), which in mouse comprised constituents of both parvalbumin-positive and -negative populations (59). In the mouse basal ganglia, strong somatic Kv7.2 staining was also observed in parvalbumin-positive neurons of the striatum, globus pallidus, and the reticular nucleus of the thalamus, dopaminergic neurons of the substantia nigra compacta and ventral tegmental area, and cholinergic neurons of the striatum and near the globus pallidus (59). In cerebellum, low levels of staining were present in granule cells, although intense labeling of a population of neurons scattered throughout the granule cell layer that presumably correspond to Golgi cells was observed (59). Staining was also seen in Purkinje cell bodies, and in the molecular layer of the cerebellar cortex, perhaps in Purkinje cell dendrites or granule cell axons (59). Kv7.2 is also present at axon initial segments of hippocampal CA3 and CA1 (Fig. 2E) pyramidal neurons and pyramidal neurons in cerebral cortex, and at nodes of Ranvier in spinal cord and optic nerve (71), in a pattern very similar to that of Nav channels and ankyrin-G. The localization at axon initial segments of adult rat hippocampal CA1 pyramidal neurons has been observed in independent studies using independently generated antibodies (250)
Immunohistochemical localization of Kv7.3 reveals many similarities with Kv7.2, consistent with the model whereby native M channel complexes are heteromers of Kv7.2 and Kv7.3 subunits (349). Initial immunohistochemical studies of Kv7.3 localization in rat brain yielded labeling that was cellular/nuclear and mainly present in interneurons and astrocytes (267), and did not match the cellular expression patterns predicted from previous in situ hybridization studies (272, 286), in spite of the fact that a standard control of antibody specificity, the inclusion of immunizing peptide, eliminated all detectable staining. Subsequent reports using independently generated antibodies revealed Kv7.3 immunostaining at nodes of Ranvier (71, 229) and axon initial segments (71, 229, 250), consistent with localization of Kv7.2 at these sites.
The immunohistochemical analysis of the more recently identified Kv7.5 has only been performed in human brain (381). The immunostaining presented in neocortex and hippocampus was cellular in nature and closely matched the cytoarchitecture of the regions examined. This staining, while eliminated in the presence of excess immunizing peptide, did not match that observed for Kv7.3 in rodent, in spite of the fact that co-immunoprecipitation experiments revealed extensive interaction between Kv7.3 and Kv7.5 in rat brain (381). This suggests that either differences in localization of Kv7 subunits between species, or problems with tissue preservation and or antibody specificity, confound comparisons of Kv7.5 localization in human brain. A recent study of Kv7.5 localization in rat auditory system presented clear immunostaining in presynaptic terminals of the auditory nerve (34). Additional staining was observed in dendrites (34). A more comprehensive view of Kv7.5 localization in mammalian brain awaits further study.
G. Kv5, Kv6, Kv8-12 Subfamilies
The cellular and subcellular localization of members of Kv subfamilies Kv5, Kv6, Kv8-Kv12 have not been as extensively characterized as have the members of the Kv1-Kv4 and Kv7/KCNQ subfamily. For most of these Kv channel principal subunits, some information on expression in brain is available, mainly based on analyses of mRNA isolated from whole brain or from different brain regions, or on in situ hybridization analyses of mRNA levels in brain sections. However, immunohistochemical analyses of the subcellular localization of these subunit proteins have for the most part not been accomplished.
The single member of the Kv5 family, Kv5.1, was originally isolated from an adult rat brain cDNA library, and in situ hybridization analyses show strongest signals in neocortex, especially in deeper layers (80, 345). Kv6.1, the product of the KCNG1 gene, is highly expressed in brain (80, 345). Kv6.1 mRNA is found in each of the principal cell populations of the hippocampus, and in the granule and Purkinje cell layers in the cerebellum (80, 345). Expression in neocortex is in more superficial layers than is Kv5.1 (80, 345). Kv6.2 has been detected in fetal rat (387) and human (371) (387) brain, as well as other tissues. Kv6.3 is prominently expressed in adult human brain. RT-PCR analyses showed expression in adult human brain (227), although note that in this report what is now termed Kv6.3 (107) is referred to as Kv10.1. A more detailed regional analysis of human brain Kv6.3 mRNA levels by RT-PCR (277) revealed prominent expression in all brain regions examined except cerebellum (277). Kv6.4 is also present in adult human brain (227), although note that in this report Kv6.4 is referred to as Kv6.3.
Kv8.1 was originally cloned from a hamster insulinoma cell line but is also expressed in hamster brain (125). In situ hybridization analyses of adult hamster brain reveal strong labeling of all principal cell populations of the hippocampus, of cerebellar granule and Purkinje cells, and throughout cerebral cortex (125). Similar findings for expression of Kv8.1 (termed Kv2.3rc in this report) were obtained from Northern blot and in situ hybridization analyses of rat brain (38). In a single study on Kv8.2 expression (although note that is termed Kv11.1 in this report) no detectable expression in adult human brain was found by RT-PCR (227). Kv8.2 is expressed in human retina, and specifically in photoreceptors, where mutations lead to lifelong visual loss due to cone dystrophy with supernormal rod electroretinogram (367). Kv8.2 mRNA was also found in rat retina by RT-PCR analyses, and in human and rat photoreceptors by in situ hybridization (64).
Kv9 family members Kv9.1 and Kv9.2 were originally cloned from adult mouse brain, where Northern blots revealed high levels of expression in mRNA from adult mouse brain and no detectable expression in any other adult mouse tissues examined (274). Similar brain-specific expression of Kv9.1 was seen in Northern blots of rat tissues (312). In situ hybridization of adult mouse brain revealed strikingly similar patterns of cellular expression for Kv9.1 and Kv9.2 (274). High levels of expression were observed in hippocampal dentate granule cells, and in CA3-CA1 pyramidal neurons, as well as presumptive interneurons scattered through s. oriens and s. radiatum of CA3-CA1, and the hilar region of the dentate gyrus. In cerebellum, hybridization signal was observed in Purkinje cells, and in the granule cell layer, and in cerebral cortex intense signal was observed throughout all cortical layers (274). Kv9.3 was also found expressed in adult rat brain by RT-PCR analyses (234) and by Northern blot analyses (58, 234, 312), although expression was also observed in other adult rat tissues.
The original cloning of rat Kv10.1 revealed expression in brain by Northern blot analysis, and in situ hybridization showing strong labeling in cerebellar granule cells, in hippocampal dentate granule cells and CA3 but not CA1 pyramidal neurons, and in neocortex (182). Northern blot (183, 273) and RT-PCR (273) analyses of Kv10.2 mRNA levels showed expression in adult brain, with olfactory bulb and brainstem having much higher levels that forebrain and midbrain structures. Initial in situ hybridization analyses revealed strong and laminar expression in cerebral cortex, in the granule cell layer of the olfactory bulb and olfactory cortex (273). More comprehensive in situ hybridization analyses of the two Kv10 or Eag family members in adult rat brain (272) revealed that Kv10.1 (Eag1) and Kv10.2 (Eag2) transcripts exhibit pronounced colocalization in specific neuronal populations, although in general the expression of Kv10.2 was more restricted such that many neurons express only Kv10.1. In neocortex, prominent coexpression of both Kv10 family members was observed in layers II-VI, with most robust expression of both transcripts in layer IV. Strong signal for Kv10.1 but not Kv10.2 was observed in each of the principal cell populations of the hippocampus, with more intense labeling of CA2 and CA3 pyramids than in CA1 and dentate gyrus, and no obvious labeling of interneurons. Kv10.1 signal in cerebellum was present in the granule cell layer, which lacked signal for Kv10.2.
Two independent in situ hybridization analyses of the three Kv11 (Erg) family members in adult rat brain revealed primarily consistent results (231, 272), although Kv11.2 signal was not observed in the initial study (272). The cellular patterns of expression suggested the possibility of different combinations of subunits in heteromeric channels, based on studies showing heteromeric Kv11 channel formation in heterologous cells coexpressing multiple Kv11 subunits (363, 364). Kv11.2 (Erg2) and Kv11.3 (Erg3) mRNAs exhibit precise cellular colocalization in adult rat hippocampus, with expression limited to principal cells in dentate gyrus and in pyramidal layers CA3-CA1, although Saganich, Rudy and colleagues (272) observed principal cell labeling for Kv11.1 and Kv11.3 only in CA1 (Fig. 2F). In addition to labeling in principal cells, Erg1 also exhibits robust labeling of interneurons scattered in the hilar region of the dentate gyrus, and in s. oriens and s. radiatum of CA3-CA1 (Fig. 2F), some of which are also positive for parvalbumin. Labeling in s. pyramidale of CA3-CA1 was also more limited than observed for Kv11.2 and Kv11.3. Each of the Erg subunits displayed extensive expression in neocortex, with especially robust expression in layer V pyramidal neurons. In the striatum, Kv11.1 mRNA predominated, with strong in situ hybridization signals in lateral and medio-dorsal compartments of the caudate-putamen, where expression was found predominantly in interneurons (272). Kv11.1 also dominated in the thalamus, with strong signals in anterodorsal and anteromedial nuclei, especially in reticular thalamic nucleus. All three Kv11 family members were found expressed in cerebellar Purkinje neurons. Kv11.1 was also expressed in cerebellar granule cells, with an especially high signal in isolated neurons scattered throughout the granule cell layer, perhaps corresponding to Golgi cells.
Immunohistochemical analyses of the cellular and subcellular localization of Kv11.1 in adult rat brain yielded staining that was mostly cellular in nature (231), While all observed staining was eliminated by preincubation of antibody with immunizing peptide, in most cases the staining was intracellular suggesting that the channel population relevant to control of neuronal membrane excitability was not the predominant target of the antibody. A number of consistent cellular targets of in situ hybridization signal (e.g. neocortical and hippocampal interneurons, cerebellar Purkinje neurons) also exhibited robust immunostaining.
In situ hybridization analyses (272) of Kv12 (Elk) subfamily members revealed little expression of Kv12.1 (Elk1) in adult rat brain. This was consistent with previous reports presenting either RNAse protection assays that yielded positive signals for Kv12.1 in mRNA prepared from sympathetic ganglia but little detectable signal in that prepared from whole adult rat brain (300), and RT-PCR analyses showing low levels of Kv12.1 mRNA in adult brain, although levels were much higher in mRNA prepared from the brains of embryonic day 18 rats (86). Kv12.2 (Elk2) and Kv12.3 (Elk3) mRNAs were found robustly expressed in adult rat brain in both studies (86, 300), as well as in a later study reporting independent isolation of Kv12.2 (Bec1) and Kv12.3 (Bec2) cDNAs from rat and human brain (210). Note that in the study of Engeland, Pongs and colleagues (86), what was later named Kv12.3 was referred to as Elk1, and Kv12.1 as Elk3. In situ hybridization analyses revealed expression of Kv12.2 in cerebellum in granule cells but not Purkinje cells, whereas detectable Kv12.3 signal was not found in cerebellum (272). Kv12.3 was expressed at high levels in the caudate putamen, consistent with earlier studies of regional expression (86). Most cells in caudate exhibited Kv12.3 hybridization signal, suggesting expression in principal cells (272). In neocortex, upper layers (II-III) had highest levels of both Kv12.2 and Kv12.3 mRNA. In hippocampus, hybridization signal for both Kv12.2 and Kv12.3 was prominent in CA1 pyramidal neurons and dentate granule neurons. Similar regional expression of Kv12.2 and Kv12.3 was found in human brain in studies using Northern blots (210) and quantitative RT-PCR (389).
III. VOLTAGE-DEPENDENT SODIUM CHANNEL LOCALIZATION IN MAMMALIAN BRAIN
A. Overview of Mammalian Brain Voltage-Dependent Sodium Channels
The molecular characterization of Nav channels, and in fact of all VDICs, began with the cloning of a cDNA encoding the electric eel electroplax Nav channel α subunit (220). The α subunit of the electroplax Nav channel was the first subunit of a VDIC to be cloned, and early analyses of the deduced amino acid sequence led to a number of important insights into how ion channel primary structure might relate to function. Moreover, this eel Nav channel cDNA was used to isolate, in rapid succession, cDNAs encoding rat brain Nav channel α subunits (218). The nine mammalian Nav channel isoforms that have been identified and functionally expressed exhibit ≥75% pairwise amino acid identify in the transmembrane and extracellular domains. These will be referred to using the established nomenclature (42) that consists of the prefix Nav to indicate the principal permeating ion and the principal physiological regulator, followed by a number that indicates the gene subfamily. Currently Nav1 is the only subfamily within the Nav family (Fig. 3). The number following the decimal point identifies the specific channel isoform (e.g. Nav1.1) and generally reflects the order in which the subunit was cloned (42). A tenth isoform exists, termed Nax, that exhibits approximately 50 % sequence identity to mammalian Nav channel α subunits (42). Because the Nax α subunits have not been functionally expressed, it is possible that this gene does not encode a functional Nav α subunit, although genetic ablation yields altered sodium homeostasis in neurons (120). Representatives of all nine isoforms and Nax have been identified and characterized in human, mouse and rat, and representatives of some isoforms in other species.
Nav channels (Fig. 3) consist of a highly posttranslationally modified α subunit, which is approximately 260 kD, associated with smaller auxiliary Navβ subunits (39). Nav channels in the adult central nervous system and heart contain Navβ1-Navβ4 subunits, whereas Nav channels in adult skeletal muscle have only the Navβ1 subunit (130). The pore-forming α subunit is sufficient for functional expression (219, 334), but the kinetics and voltage dependence of channel gating are modified by Navβ subunits (130). Navβ subunits are also involved in channel localization and interaction with cell adhesion molecules, extracellular matrix, and intracellular cytoskeleton (130-132). Nav α subunits are organized in four internally repeated homologous domains (I-IV), each of which contains six transmembrane segments (S1-S6) and an additional pore loop located between the S5 and S6 segments (Fig. 3). The pore loops line the outer, narrow entry to the pore, whereas the S5 and S6 segments line the inner, wider exit from the pore. The S4 segments in each domain contain positively charged amino acid residues at every third position. These residues serve as gating charges and move across the membrane to initiate channel activation in response to depolarization of the membrane. The short intracellular loop connecting homologous domains III and IV serves as the inactivation gate, folding into the channel structure and blocking the pore from the inside during sustained depolarization of the membrane (39).
At least nine of the ten Nav1 channel α subunit genes present in the mammalian genome are expressed in the nervous system, the exception being the muscle-specific Nav1.4 (334). When expressed in heterologous systems, Nav1 α subunits generate Nav currents whose functional properties are highly conserved (97), compared to the heterogeneity of function observed among the different Cav and Kv family members. Certain subtypes, such as Nav1.7, Nav1.8, Nav1.9 and Nax, are expressed predominantly in the peripheral nervous system and will not be discussed here. Nav1.4 is the predominant Nav channel in adult skeletal muscle, and Nav1.5 in adult cardiac and embryonic skeletal muscle. The remaining four α subunits (Nav1.1, Nav1.2, Nav1.3 and Nav1.6), are expressed at high levels in brain, although in rodents Nav1.3 is predominantly found in embryonic and neonatal, but not adult brain (16). In addition, Nav1 channels are found in both neurons and glia. A number of groups have used antibodies raised against subtype-specific sequences in the intracellular inter-domain I-II linker region or the cytoplasmic C-terminus to define the localization of Nav1.1, Nav1.2, Nav1.3 and Nav1.6 in rat, mouse and human brain.
Nav1 channels initiate and propagate action potentials in virtually all mammalian central neurons. The different subtypes exhibit different subcellular localization, as detailed below. Nav1.1 and Nav1.3 are predominantly localized to the neuronal soma and to proximal dendrites, where they control neuronal excitability through integration of synaptic impulses to set the threshold for action potential initiation and propagation to the dendritic and axonal compartments. Nav1.2 is predominantly expressed in unmyelinated axons, where it conducts action potentials. Nav1.6 is prominently found at nodes of Ranvier, the gaps in the myelin sheaths of myelinated axons, where it propagates action potentials, and at axon initial segments where action potentials initiate. Modulation of Nav1 currents is undoubtedly important in vivo (37), and mutations that subtly alter Nav1 channel function can lead to human diseases of hyperexcitability such as epilepsy (161).
B. Nav1 subfamily
The availability of Nav1 α subunit cDNAs allowed for the generation of subtype-specific molecular probes and led to the first studies of the regional and developmental patterns of Nav1 gene expression in mammalian brain (15, 16, 19, 31). The subsequent generation of isoform-specific antibodies allowed for initial analyses of the cellular and subcellular distribution of the corresponding Nav1 α subunits in rat brain (33, 100, 357). These studies and many that followed are especially relevant for understanding how each isoform contributes to cellular excitability.
1. Nav1.1
Initial studies of Nav1.1 mRNA distribution in rat brain showed high expression in cerebellum, lower levels in the striatum, hippocampus and thalamus, and no detectable signal in cortex (19, 31). Immunostaining revealed Nav1.1 polypeptide predominantly in the cell bodies and dendrites of expressing cells (100, 357). In the adult rat (Fig. 4A) and human hippocampus, strong Nav1.1 staining was present on the soma of dentate granule cells, and of interneurons dispersed throughout the dentate hilus (100, 357). Intense staining was also seen in s. pyramidale of all hippocampal subfields (CA1, CA2 and CA3) with staining restricted to the pyramidal cell somata and proximal dendrites (Fig. 4A). Recently, an Nav1.1-dense subregion was observed within the axon initial segment of a subset of initial segments adjacent to or within the pyramidal cell layer of hippocampal area CA3 (342). Nav1.1 was located at the proximal end of the axon initial segment, nearest to the soma, in this subset of CA3 neurons. Additional Nav1.1 staining was also observed in a subpopulation of interneurons dispersed throughout s. radiatum and s. oriens (Fig. 4A). The identity of this subpopulation of interneurons has not been experimentally determined. Robust Nav1.1 staining was also observed on the soma and apical dendrites of pyramidal cells in layer V of the cerebral cortex. In the cerebellum, studies of Nav1.1 expression in the cerebellum are quite inconsistent. Some studies showed low level expression of Nav1.1 in granule cells (56, 343), while in other studies no Nav1.1 expression was detected (89, 225). However, cerebellar Purkinje cells have strong somatic Nav1.1 staining, and clear staining is also seen on Purkinje cell dendrites. It should be noted that strikingly similar patterns of Nav1.1 staining were observed in human brain (361), suggesting that this distinct pattern of localization is evolutionarily conserved and that somatodendritic targeting of Nav1.1 is essential for normal neuronal function across species. Nav1.1 channels are presumably involved in propagating synaptic signals from dendrites to soma, and in integration of electrical signals within the soma prior to the initiation of axonal action potentials.
2. Nav1.2
While Nav1.2 mRNA has a somewhat similar pattern of cellular expression to that observed for Nav1.1 (31), the Nav1.2 polypeptide was found specifically localized in axons and terminals (100, 357). These early studies revealed that the highly related rat brain Nav1.1 and Nav1.2 polypeptides (≈88% amino acid identity) could be targeted to distinct subcellular domains (somatodendritic versus axonal) in rat central neurons. This localization also suggested that in central neurons, Nav1.2 is a major component of the axonal action potential conductance mechanism, as well as a regulator of neurotransmitter release in presynaptic terminals.
High levels of Nav1.1 and Nav1.2 mRNA are expressed in many of the same cells. However, the subcellular staining pattern of Nav1.2 is in many instances a reciprocal image of that for Nav1.1. The contrasting subcellular localizations of Nav1.1 and Nav1.2 are especially clear in the hippocampus (Fig. 4), where the exquisitely laminar cytoarchitecture lends itself to such comparisons. In many cells expressing robust somatodendritic Nav1.1 staining (Fig. 4A), very little if any Nav1.2 staining is observed on the somatic or dendritic membrane (Fig. 4B). Thus the principal cell layers of the hippocampus are virtually lacking Nav1.2 staining, while areas rich in axons and nerve terminals exhibit robust staining. In the dentate gyrus, Nav1.2 is present at high levels in the middle third of the molecular layer, site of termination efferents from the entorhinal cortex to the hippocampus (the so-called medial perforant path), and absent from dentate granule cell bodies (Fig. 4B). Additional strong staining is seen in s. lucidum of CA3, the site of densely packed mossy fiber axons of dentate granule cells, while CA3 pyramidal cell bodies are not stained (Fig. 4B). Nav1.2 is also concentrated in s. radiatum and s. oriens of CA1 and appears to be associated with axons and terminals of the Schaffer collaterals (s. radiatum), and the commissural/associational pathway (s. oriens). As in other pyramidal cell layers, Nav1.2 is not present on CA1 pyramidal cell bodies. In general, the staining pattern of Nav1.2 (260, 297) is quite similar to that of Kv1.4 (compare Fig. 2A and Fig. 4B), for which experimental lesions (212) and immuno-electron microscopy (61) studies have strongly supported an axonal localization. However, neither lesion studies nor immuno-electron microscopy have been utilized to verify that the Nav1.2 staining is in fact localized to axons and terminals. It should be noted that in the large pyramidal cells of hippocampal CA1, and of layer V of the neocortex, some apical dendritic Nav1.2 staining is apparent (100). However, this staining appears to be intracellular and presumably represents a pool of newly synthesized Nav1.2 α subunits transiting through the rough endoplasmic reticulum and/or Golgi apparatus prior to subsequent trafficking to axons.
Similar contrasting localization of Nav1.1 and Nav1.2 is observed in the cerebellar cortex, where strong Nav1.1 staining is seen in Purkinje cells, and strong Nav1.2 staining on the axons of the granule cells and interneurons of the molecular layer (100, 357). There are conflicting reports concerning Nav1.2 expression in Purkinje cells. Some in situ hybridization studies detect Nav1.2 mRNA (19, 89) but other in situ hybridization and immunolocalization studies fail to detect this isoform (31, 134, 282). One interesting finding of a recent study is the graduated intensity of Nav1.2 staining in the cerebellum, with the highest intensity found in the posterior lobes and much lower intensity in the anterior lobes (134), similar to the gradient of Kv4 channels detailed above.
3. Nav1.3 and Nav1.5
In contrast to Nav1.1 and Nav1.2 transcripts, in rodents Nav1.3 mRNA was found at highest levels in embryonic and early postnatal brain. A detailed immunohistochemical study of Nav1.3 localization in brain has not been performed in the rat (although see (358)). However, recent studies of Nav1.3 mRNA (360) and protein (361) in human brain have revealed that the distribution of Nav1.3 in adult human brain is quite extensive. Certain human brain regions express moderate to high levels of Nav1.3 staining, which in all cases, as in the rat, (358) is associated with neuronal somata and dendrites. Especially notable is staining in neocortex, where robust staining of layer III pyramidal cells is apparent (361). This strong staining extends some distance along the large apical dendrites along these layer III pyramidal cells, while in other neocortical laminae staining is less intense and restricted to somata. In rat, both layer III and layer V pyramidal cells exhibit strong Nav1.3 staining extending along the length of the apical dendrites (358). In the human hippocampus, robust staining is observed to a subpopulation of interneurons scattered throughout s. oriens of CA1 and CA2, s. lucidum of CA3, and the polymorphic layer of the dentate gyrus. Less intense somatodendritic staining is also observed in pyramidal cells, but little or no staining is seen in dentate granule cells. In cerebellar cortex, moderate staining is observed in granule cells and in the molecular layer. Thus in human brain Nav1.3 may act together with Nav1.1 in determining the active properties of neuronal somata and dendrites. Moreover, a recent study showed a upregulation and misexpression of the Nav1.3 sodium channel in neurons of the ventral posterolateral nucleus of the thalamus after a peripheral nerve injury (385). Their data document abnormal expression of the Nav1.3 mRNA in the ventral posterolateral nucleus during this time of neuronal hyperresponsiveness and reduced nociceptive threshold, with no changes in the expression of other Nav channels (e.g. Nav1.1, Nav1.2, or Nav1.6) in ventral posterolateral neurons.
Interestingly, Nav1.5, which is the predominant cardiac sodium channel, has recently been shown to be expressed at both the mRNA (112) and protein (368) level in mammalian brain. In mouse brain, Nav1.5 is localized to axons (368).
4. Nav1.6
The cDNA encoding Nav1.6 was originally isolated from rat brain (284), and Nav1.6 mRNA is highly expressed in cerebellar granule cells and in pyramidal and granule cells of the hippocampus (282). In many central neurons, immunoreactivity for Nav1.6 is found in the soma and proximal dendrites of cells expressing the corresponding mRNA (33, 162). In adult neurons with myelinated axons, Nav1.6 is robustly expressed at high densities at nodes of Ranvier (24, 33), and axon initial segments (25), where Nav1 channels are clustered at high densities (Fig. 4C, E). When nodes of Ranvier initially form in response to developmental myelination, Nav1.2 channels are found in high densities at most if not all nodes, and Nav1.6 is not found (24). However, with further maturation of the node Nav1.6 becomes the predominant Nav isoform such that Nav1.2 is seen in only a small subpopulation of nodes, where it is always found in conjunction with Nav1.6. In neurons such as retinal ganglion cells, where the myelinated portion of the axon within the optic nerve lies distal to extensive non-myelinated portions within the retina, it is clear that Nav1.2 is highly expressed in the non-myelinated portions in the absence of Nav1.6 (24). However, in the axon initial segment, Nav1.6 is again found at high levels in conjunction with Nav1.1 and Nav1.2 (25). These findings point to the fact that a number of distinct membrane domains can exist along the length of mammalian axons. Some of these domains are defined by highly-specific cell-cell interactions, such as that which occurs between neurons and glia at the node of Ranvier of myelinated axons (as discussed below). However, intrinsic neuronal machinery must also exist to restrict the expression of Nav isoforms to different locations in the non-myelinated portions of axons, such as axon initial segments.
Nav1.6 is also robustly expressed in Purkinje cells, where it is present in the somata and throughout the dendritic arbor in rat (162) and human (361). This robust Purkinje cell staining is consistent with the finding that mutant mice lacking Nav1.6 expression are missing a Purkinje cell resurgent Nav current (242). In situ hybridization and immunolocalization studies demonstrated that Nav1.6 is more highly expressed in the granule cells of lobules posterior to the boundary at lobules V and VI (282, 283), similar to Nav1.2 and Kv4 channels. Other somatodendritic Nav1.6 staining is less impressive, with neurons in many brain regions, such as cortex and hippocampus, exhibiting somatodendritic staining that is somewhat intracellular in appearance (33, 162). This may simply represent newly synthesized Nav1.6 prior to its targeting to initial segments and/or nodes of Ranvier of myelinated axons, as opposed to a functional somatodendritic pool of Nav1.6.
C. Nav β Auxiliary Subunits
Unlike the Kvβ and Cavβ auxiliary subunits, the Navβ subunits are transmembrane glycoproteins. Like other VDIC auxiliary subunits, Navβ subunits are multifunctional. They modulate channel gating and regulate the level of channel expression in the plasma membrane (130, 131, 241), and saxitoxin binding studies show that Navβ1 mediates a four-fold increase in surface expression of coexpressed Nav1.2 channels (132). More recently, Navβ subunits have been shown to function as cell adhesion molecules that mediate interaction with extracellular matrix, regulation of cell migration, cellular aggregation, and interaction with the cytoskeleton (200, 311). Moreover, mutations in Navβ subunit cause effects on channel function and subcellular distribution that bias neurons toward hyperexcitability and epileptogenesis (204, 370). All four Navβ subunits, Navβ1, Navβ2, Navβ3 and Navβ4, are expressed in brain (36, 171, 223, 292, 375). However, Navβ3 is expressed primarily in embryonic brain, with mRNA levels decreasing around postnatal day 3, except in the hippocampus and striatum (292). Navβ3 mRNA is highly expressed throughout the prenatal brain with a hybridization signal temporally similar to that of both Nav1.2 and Nav1.3 subunits. It is possible that Navβ3 may couple with Nav1.2 and/or Nav1.3 subunits during development to form a functional Nav channel complex. In contrast, Navβ1 and Navβ2, while expressed in embryonic brain, only reach maximum expression levels at postnatal day 21 and expression remains elevated in adults (292). In the postnatal brain, Navβ1 and Navβ2 mRNA show some synchrony in their regional and temporal expression in cortex, hippocampus and cerebellum (292). Navβ1 mRNA first appears around P3 (280) with expression increasing rapidly in most brain areas to reach a maximum by P14. Navβ2 mRNA expression was also found to increase throughout the CNS between P3. The analyses of Navβ association during development, by immunoblot under non-reducing conditions, show that the association of both Nav1.1 and Nav1.2 with Navβ changes during the development of the brain (100). Navβ3 mRNA is expressed at very high levels throughout the CNS at P1 with levels decreasing in most areas after this stage (292). Navβ3 mRNA has a complementary distribution with Navβ1 after P9, with distinct differences appearing by adulthood (213, 292). The subcellular distribution of these Navβ subunits has not been extensively characterized.
The Navβ4 mRNA is expressed in a restricted pattern throughout the cerebral cortex and present at high levels cerebellar Purkinje cells. In the hippocampal region, Navβ4 is expressed at very low levels (375). In general, Navβ4 protein expression was in accordance with the in situ hybridization data. Both Navβ2 and Navβ4 are coexpressed in many areas of the brain, but Navβ2 exhibited a more widespread distribution. For example, in the hippocampus, Navβ4 protein is found in isolated pyramidal neurons but is absent from the dentate gyrus apart from occasional hilar neurons. In contrast, Navβ2 protein is highly expressed in hippocampal pyramidal cells and in the hilus and granule cells of the dentate gyrus (251). This complementary distribution may indicate selective association with distinct Nav1 α subunits in those regions that may differentially influence Nav channel properties or dictate differences in protein-protein interactions.
IV. VOLTAGE-DEPENDENT CALCIUM CHANNEL LOCALIZATION
A. Overview of Voltage-Dependent Calcium Channels
Cav channels mediate calcium influx in neuronal cells in response to membrane depolarization. Calcium entry through Cav channels mediates a wide range of intracellular processes such as activation of calcium-dependent enzymes, gene transcription, and neurotransmitter exocytosis/secretion (41). Their activity is an essential requirement for the coupling of electric signals in the neuronal plasma membrane to physiological events within the cells. Biochemical characterization of native brain Cav channels revealed that, in addition to the large principal α1 subunit, the existence of numerous auxiliary subunits (Fig. 5), each encoded by multiple genes (41). The α1 subunit is the largest and principal subunit, and incorporates the ion conduction pore, the membrane voltage-sensor and gating apparatus, a number of different types of which have been identified and characterized in the mammalian nervous system (Fig. 5), each with specific physiological functions and electrophysiological and pharmacological properties (41). These are classified into three families: the Cav1 (L-type), Cav2 (P/Q-, N-, and R-type), and Cav3 (T-type) family (43). These principal α1 subunits, like the α subunits of Nav channels, consist of four internally repeated homologous domains (I-IV), each with six transmembrane segments (S1-S6), wherein each S4 segment serves as the voltage sensor (Fig. 5). In addition to these principal pore-forming α1 subunits, native functional Cav channels consist of auxiliary α2δ, β, and γ subunits (Fig. 5), with exemplary subunit composition of many native Cav channels being α1α2δβγ in a 1:1:1:1 ratio (43). Diversity among these Cav auxiliary subunits leads to a variety of isoform-specific effects that are crucial in determining the expression level, localization and function of the associated Cav α1 subunits (121). These different Cav channel subunits have distinct cellular and subcellular distributions in the mammalian brain supporting their individual physiological roles (335).
B. Cav1 Subfamily
The Cav1 family or L-type Cav channels play a critical role in somatodendritic calcium influx in many mammalian central neurons (43). Cav1 channels are involved in dendritic calcium signaling resulting from back-propagating action potentials, synaptic plasticity, and excitatory activity-dependent modulation of gene transcription in mammalian brain neurons (142). Among the four Cav1 family members, only Cav1.2 (α1C), and Cav1.3 (α1D) are expressed in mammalian central neurons. These underlie prominent somatodendritic high-voltage activated L-type Cav currents, as determined by specific pharmacological inhibition of channel currents by dihydropyridine inhibitors (305, 335). In situ hybridization studies in rat brain first suggested the expression of Cav1.2 mRNA in olfactory bulb, hippocampus, cerebellum, and amygdala; Cav1.3 mRNA was also detected here as well as in cortex and habenula (181). Immunolocalization studies on brain L-type Cav1 channels suggested the expression of Cav1.2 and Cav1.3 in the cortex, cerebellum, and hippocampus, with distinct subcellular localization in the neuronal soma and proximal dendrites (2, 116, 355). In the cerebellum, initial studies with an α2-specific monoclonal antibody, suggested to reflect Cav1 expression, suggested expression and somatodendritic localization of L-type channels in the Purkinje cells and neurons in the granular and molecular layers (355). In the hippocampus Cav1 channels were found expressed and localized in the soma and proximal dendrites of pyramidal neurons in CA1-CA3 region, granule cells in the dentate gyrus, neurons in the subiculum and entorhinal cortex, and interneurons scattered throughout these region (85, 116, 355).
Subsequent Cav1 isoform-specific antibody staining suggested that Cav1.2 and Cav1.3 α subunits have distinct subcellular localizations in the soma and dendrites of principal cells in the hippocampus (85, 116). Specific immunoreactivities of Cav1.2 and Cav1.3 were found to make up ∼75% and ∼20% of the total dihydropyridine binding activity in rat brain membranes respectively. Cav1.2 staining was consistently found associated with the soma and proximal dendrites, with a clustered localization pattern, in hippocampal pyramidal cells, cerebral cortical neurons, and cerebellar neurons (116), and closely resembled the staining pattern previously obtained with the α2-specific monoclonal antibody. These observations suggest that Cav1.2 is the major constituent of the brain L-type Cav channel population, whose clustering may be critical to proximal dendritic calcium entry in response to synaptic activity (85, 189). Cav1.2 is also found on distal dendrites, especially in the molecular layer of the dentate gyrus and CA3/CA2 s. radiatum (116). These clusters are shown localizing at or near axosomatic and axodendritic postsynaptic regions, and are often associated with β2-adrenergic receptor signaling complexes (66). Subsequent immunolocalization studies on both endogenous and exogenous Cav1.2 in cultured mouse hippocampal neurons indicated plasma membrane expression of these channels in small clusters in the soma and apical dendrites with some expression in the axon initial segments (222). Higher resolution microscopic analyses revealed that these channel clusters are localized in both extrasynaptic as well as synaptic regions in the dendrites, the latter associated with glutamatergic synapses in dendritic spines (222). A recent ultrastructural study in hippocampus provided evidence for Cav1.2 localization on both dendrites and axon terminals in all hippocampal subfields (329).
Cav1.3 exhibited a staining pattern similar to Cav1.2 (Fig. 6D-F), and was found mainly in the soma and proximal dendrites but without the clustering seen for Cav1.2 (116). A more recent report on exogenous Cav1.2 and Cav1.3 channels in cultured rat hippocampal neurons revealed subtype-specific localization patterns, with Cav1.3 surface clusters less abundant, but larger in size and brighter than Cav1.2 clusters (383). Most of the Cav1.2 clusters (∼80%) are located at/near synapses at the postsynaptic region, as opposed to Cav1.3 clusters, which are less abundant in these regions. However, co-expression of the postsynaptic adapter protein, Shank-1B, promoted synaptic localization of dendritic Cav1.3 clusters (383). In developing neurons Cav1.2 is robustly expressed in the growth cone, but upon neuronal maturation are excluded from the distal axons and nerve terminals, suggesting that the specific targeting machinery in different neuronal compartments changes during differentiation (222).
FIG. 6.
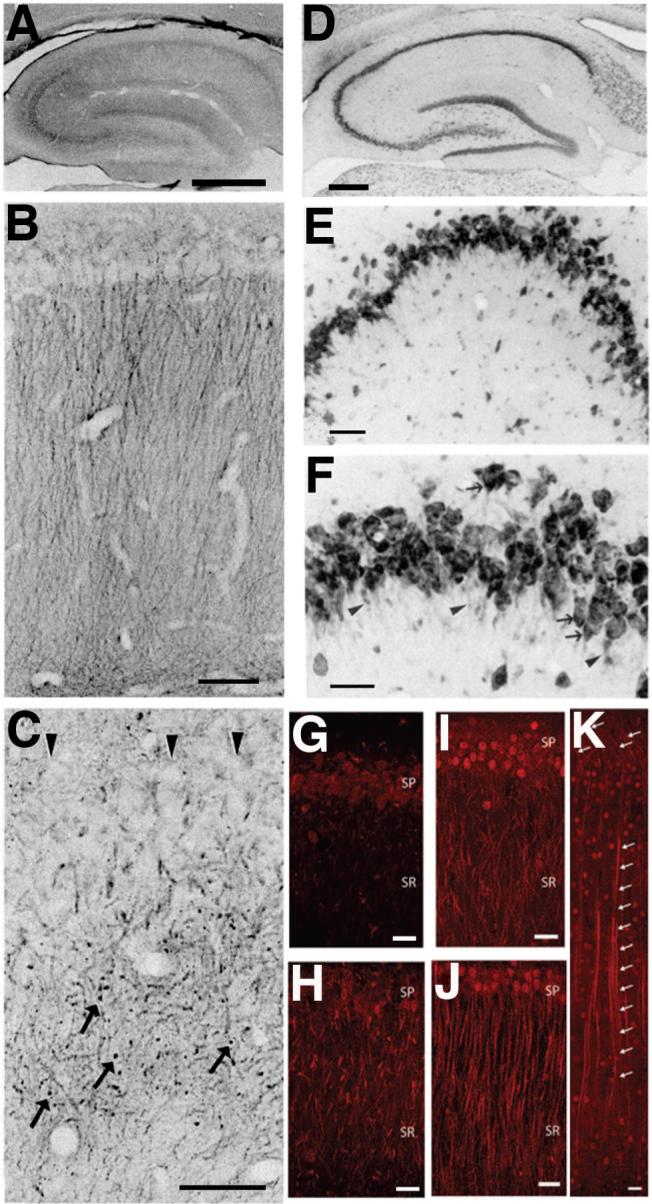
Cellular and subcellular distribution of Cav channels in adult rat brain. A-F: Immunoperoxidase staining. G-K, Immunofluorescence staining. A, D-J: Hippocampus. B, C, K: Neocortex. A, Cav2.1. Note staining in terminals fields of the medial perforant path in the middle molecular layer of the dentate gyrus, and mossy fiber axons and terminals in s. lucidum of CA3. B, Higher magnification view of Cav2.1 staining in CA1. C, Higher magnification view of Cav2.1 staining in CA3, arrows indicate mossy fiber terminals, and arrowheads CA3 pyramidal cell somata. D, Cav1.3. Note staining in somata and proximal dendrites of dentate granule cells, CA3-CA1 pyramidal cells and interneurons. E, Higher magnification view of Cav1.3 staining in CA3-CA2. F, Much higher magnification view of Cav1.3 staining in CA3-CA2. Arrows: base of apical dendrites. Arrowheads: proximal dendrites. G-I: Cav3.1 (G), Cav3.2 (H), and Cav3.3 (I) staining in CA1. J, Cav3.3 staining in subiculum. K, Cav3.3 staining in neocortex. SP, s. pyramidale. SR, s. radiatum. Reproduced with permission from reference: A-C: (359); D-F: (116); G-I: (201).
C. Cav2 Subfamily
The Cav2 family of P/Q-, N- and R-type Cav channels plays critical roles in somatodendritic calcium influx, and regulates neurotransmitter release from presynaptic terminals in many mammalian central neurons. All three members of Cav2 α1 subunits (Fig. 5), the P/Q-type or Cav2.1 (α1A), the N-type or Cav2.2 (α1B), and the Cav2.3 (α1E) are expressed in mammalian central neurons, as determined by specific pharmacological inhibition of these isoform-specific Cav currents by the funnel web spider toxin ω-agatoxin-IVA, the cone snail toxin ω-GVIA-conotoxin, and the tarantula toxin SNX-482, respectively. Pharmacological studies using these toxins suggested that Cav2.1 and Cav2.2 channels constitute most of the presynaptic voltage-dependent calcium influx, which triggers neurotransmitter release from nerve terminals. In contrast to specific subcellular localization of Cav1 channels in the soma and dendrites, Cav2 family channels have both axonal as well as somatodendritic localization in mammalian brain neurons.
In situ hybridization studies in rat brain first suggested the expression of Cav2.1 mRNA in most regions of the brain except in habenula (181). Immunostaining of adult rat brain with a Cav2.1-specific antibody (Fig. 6A-C) revealed specific staining in the hippocampus, brain stem, habenula, frontal cortex, substantia nigra, molecular layer of cerebellar cortex, Purkinje cell dendrites, periglomerular cell soma and dendrites in the olfactory bulb, and deep layers of entorhinal cortex, neocortex and striatum (119). Subsequently, immunolocalization studies in rat brain using another Cav2.1 isoform-specific antibody suggested expression of these channels in the synaptic terminals of cerebellar molecular layer, and the mossy fiber zone of hippocampal CA3 (178, 253, 359). This presynaptic localization of Cav2.1 channels is consistent with the role of P/Q-type channels in the regulation of neurotransmitter release through interaction with release machinery (44, 255). However, in cerebellar Purkinje cells, prominent somatodendritic staining of Cav2.1 was also observed (178, 253, 337, 359), where staining colocalizes with that of metabotropic glutamate receptor subtype 1 receptor through a direct physical interaction (157). Immuno-electron microscopic analysis in globus pallidus neurons also suggested that in addition to prominent presynaptic expression, the Cav2.1 channels are also localized in the dendrites (111). Subsequently, several reports have shown dendritic localization of Cav2.1 channels, which are subjected to neurotransmitter regulation (85, 147, 179, 188, 214). Given that Cav2.1 transcripts are extensively processed by alternative splicing, it is attractive to speculate that different Cav2.1 splice variants are targeted to different subcellular compartments in mammalian brain neurons.
In situ hybridization studies in rat brain revealed the expression of Cav2.2 mRNA in most regions of the brain except in habenula (181). Localization studies on Cav2.2 channels using autoradiography of the Cv2.2-specific 125I-labelled-GIVA-conotoxin (149) in mammalian brain revealed high-density expression in neuronal synaptic terminals in the striatum, hippocampus, cortex, and cerebellum (3, 9, 148, 320, 321). Higher resolution analyses in the hippocampus revealed intense 125I-ω-GIVA-conotoxin-binding signals in regions rich in axons and presynaptic terminals, mainly in the CA3 s. lucidum where mossy fiber axons and terminals are located (9). Strong 125I-ω-GIVA-conotoxin-binding signals were also detected in the infragranular polymorphic region of the dentate gyrus, and s. radiatum and s. lacunosum moleculare of CA1, which contains the Schaffer collateral, and lateral perforant path, axons and nerve terminals (9). Immunolocalization studies using a Cav2.2-specific antibody yielded similar results with strong immunoreactivity in punctate/synaptic structures in rat brain (356). Although robust Cav2.2 immunoreactivity was seen in the mossy fiber expansions / presynaptic terminals in the s. lucidum of hippocampal CA3 relative to the immunoreactivity in the preceding axon shafts, some Cav2.2 immunoreactivity were also seen on the dendrites of pyramidal cells in the layers II/III and V of neocortex, and of layers CA1-CA3 of hippocampus (85, 356). Studies in organotypic cultures of rat hippocampal CA3 revealed stronger Cav2.2 immunoreactivity associated in CA3 pyramidal cell dendrites than in mossy fiber terminals (85). Analyses of Cav2.2 localization in human hippocampus with a distinct antibody revealed less immunoreactivity in mossy fiber terminals in comparison to that in rats, although the labeling corresponded well with that for calbindin, a standard marker for CA3 mossy fiber terminals (68).
While the basis for the differences in the localization of Cav2.2 is not yet apparent, it is known that alternative splicing of Cav2.2 mRNA at the C-terminus leads to the generation of long (Cav2.2a) and short (Cav2.2b) channel isoforms (362). The long Cav2.2a variant/isoform is specifically targeted to presynaptic terminals when expressed exogenously in cultured hippocampal neurons, whereas the short Cav2.2b isoform is localized uniformly in the soma and dendrites (197). Such a difference in the targeting of exogenously expressed Cav2.2 splice-variants may underlie the earlier observation of nonpolarized expression of the total neuronal pool of Cav2.2 channels in mammalian brain (356). However, thorough immunolocalization studies using splice-variant-specific Cav2.2 alpha-subunit antibodies has not yet been accomplished to demonstrate the specific subcellular localization Cav2.2 alternative splice products.
In situ hybridization studies in rat brain showed the expression of Cav2.3 mRNA in almost all regions of the brain including habenula, where Cav2.1 and Cav2.2 mRNAs were not detected (181). Immunolocalization studies on Cav2.3 suggested a more prominent localization in the midbrain and hindbrain region than in forebrain structures, where Cav2.1 and Cav2.2 are more prominently localized (372). Strong immunoreactivity for Cav2.3 was observed in the soma of most cell types, however, close inspection of somatic immunoreactivity in the principal cells in neocortex and hippocampus at higher magnifications suggested perinuclear/intracellular localization of these channels, perhaps representing biosynthetic pool of inefficiently trafficked channels (372). Neurons in the globus pallidus, thalamus, anterior amygdala, subthalamic nuclei, and hypothalamus exhibited strong plasma membrane immunoreactivity in the soma and dendrites. Prominent Cav2.3 immunoreactivity was also observed in the soma and dendrites of cerebellar Purkinje neurons (372). However, subsequent immuno-electron microscopic analyses revealed that in rat globus pallidus neurons, Cav2.3 immunoreactivity was clearly present in dendritic structures as well as in a subpopulation of presynaptic boutons (111).
D. Cav3 Subfamily
The Cav3 family or T-type Cav channels underlie transient currents that activate at subthreshold membrane potentials, and that are crucial for the regulation of plasma membrane calcium permeability near resting membrane potentials and during action potentials. Cav3 family members (Fig. 5) have diverse expression patterns in the mammalian brain (323, 380). Immunolocalization studies revealed that Cav3.1 channels are prominently expressed in hindbrain (cerebellum and medulla) with low-intensity immunoreactivity in forebrain and midbrain regions in human, rat, and mouse (62, 323, 380). In the cerebellar cortex, strong and specific Cav3.1 immunoreactivity was detected in the thick processes in the molecular layer, with or without additional immunoreactivity in Purkinje cell bodies (62, 380). This immunoreactivity pattern is presumably associated with the soma and dendrites of Purkinje cells, given the high levels of Cav3.1 mRNA in rat cerebellar Purkinje cell somata (62, 323), and relatively low mRNA levels in granule cells and basket cells. In the medulla, strong and specific Cav3.1 immunoreactivity was observed in the hypoglossal nucleus, the inferior olive, and the lateral reticular nucleus. Although in some neurons Cav3.1 immunoreactivity exhibited intracellular localization, in others it was present uniformly on the plasma membranes of somatic and proximal dendritic membranes (62). A recent study on immunolocalization of Cav3.1 channels in rat brain using isoform-specific antibody revealed expression of these channels in the neocortex, thalamus, hippocampus, and cerebellum (201). Specific Cav3.1 immunostaining was observed in the soma and proximal dendrites of layers 1-4 and layer V cortical pyramidal neurons, and hippocampal CA1, CA3, and subicular pyramidal neurons (Fig. 6G). However, in hippocampal dentate granule cells, neurons in the thalamus, midline thalamic nuclei, and in cerebellar Purkinje cells Cav3.1-specific immunostaining was more restricted to the cell somata (201).
In situ hybridization studies showed that in rat hippocampus the Cav3.2 mRNA was found at high levels in the dentate granule cells and CA pyramidal cells, as well as in olfactory bulb neurons (323). Cav3.2 immunostaining is present in the soma and medial dendrites of specific neurons in the neocortex, thalamus, hippocampus, and cerebellum (201). Cav3.2 immunostaining was found in the soma and proximal dendrites of layers 1-4 and layer V cortical pyramidal neurons. However, in hippocampal CA1, CA3, and subicular pyramidal neurons, Cav3.2 immunostaining was extended to medial dendrites, in addition to the somatic and proximal dendritic staining (Fig. 6H). Cav3.2-specific immunostaining, like that of Cav3.1, was also restricted to neuronal somata in hippocampal dentate granule cells, neurons in the thalamus, midline thalamic nuclei, and in cerebellar Purkinje cells, (201).
Initial in situ hybridization and immunolocalization studies on Cav3.3 channels in mammalian brain suggested prominent localization in olfactory bulb and mid brain, distinct from that for Cav3.1 (323, 380). Intense Cav3.3 immunoreactivity was also observed in the olfactory nerve layer and glomerular layer in mouse olfactory bulb (380). In hippocampus, Cav3.3-specific immunoreactivity was observed in a subset of interneurons dispersed throughout s. lucidum of the dentate gyrus, CA3, and CA2, and subiculum, with the highest intensity in the CA2. In the dentate gyrus, strong immunoreactivity was seen in the inner and outer thirds of the molecular layer, which suggests localization of Cav3.3 on terminals of the associational and commissural pathways of the dentate gyrus and of the lateral perforant pathway of entorhinal cortex, respectively (380). These staining patterns might represent input-specific localization of Cav3.3 channels to specific subdomains of dentate granule cell dendrites. In rat cerebellar cortex, strong Cav3.3 immunoreactivity was seen in the molecular layer, which appeared diffuse and associated with the parallel fibers and/or their terminals (380), consistent with the observation of high levels of Cav3.3 mRNA in the granule cell layer, but not in the Purkinje cell or molecular layer (323). A recent report revealed the expression and somatodendritic localization of Cav3.3 channels in specific neuronal subtypes in the neocortex, thalamus, hippocampus, and cerebellum (201). The Cav3.3 immunostaining was restricted to the soma and proximal dendrites of layers 1-4 cortical pyramidal neurons. However, in layer V cortical pyramidal neurons, hippocampal CA1, CA3, and subicular pyramidal neurons as well as in cerebellar Purkinje cells, Cav3.3 immunostaining was seen in the soma as well as throughout the dendrites (Fig. 6I-K). Like Cav3.1 and Cav3.2, in hippocampal dentate granule cells, neurons in the thalamus and midline thalamic nuclei, Cav3.2-specific immunostaining was restricted to somata (201).
E. Auxiliary Subunits of Cav Channels
Functional Cav channels are composed of four subunits, including the pore-forming Cav α1 subunit, and auxiliary Cavα2δ, Cavβ, and Cavγ subunits (Fig. 5) as determined originally by biochemical purification of Cav channel complexes (41, 145). Classification of these three subunits as bona fide Cav channels auxiliary subunits is based on their presence in purified Cav channel complexes, their direct interaction with the pore-forming α1 subunit, and their ability to modulate the biophysical and/or trafficking properties of coexpressed Cav α1 subunits (8, 76).
The Cavα2δ subunit is composed of two polypeptides (Cavα2 and Cavδ) that are the products of the same gene and generated by the posttranslational cleavage of a single large precursor polypeptide into Cavα2, and Cavδ polypeptides and their subsequent covalent linkage by disulfide bonds (Fig. 5). Topologically, the Cavδ peptide has a single transmembrane segment, with the Cavα2 polypeptide being completely extracellular (Fig. 5). Association of Cavα2δ subunits enhances the membrane trafficking and current density of coexpressed Cav α1 subunits, and also modulates the channel’s voltage-dependent gating (8). Some of these effects can be observed in the absence of coexpressed Cavβ subunits, whereas others display an absolute Cavβ requirement. Four distinct Cavα2δ subunits, (1-4) that interact with the Cav α1 subunits have been reported, of which only Cavα2δ-1, Cavα2δ-2, and Cavα2β-3 have been found expressed in mammalian brain (8, 106, 158). However, information on the specific subcellular localization of individual Cavα2δ subunits (although see description of studies with a general anti-Cavα2δ subunit antibody described above), as well as their distribution relative to Cav α1 subunits in mammalian brain is lacking.
The Cavβ subunit is a product of a single gene, which subsequently gives rise to several alternatively spliced isoforms (8, 75, 117). Four distinct Cavβ subunit genes (Cavβ1 - Cavβ4) have been described. Topologically, Cavβ subunits are cytoplasmic polypeptides (Fig. 5), although Cavβ1b and Cavβ2a isoforms can associate with the plasma membrane independent of Cav α1 subunits (23, 51). Interaction of Cavβ subunits with Cav α1 subunits promotes channel trafficking to the plasma membrane, and modulates voltage-dependent gating (18, 306). In situ hybridization and immunolocalization studies in mammalian brain revealed that the Cavβ1b splice variant mRNA is expressed throughout the brain, with a diffuse cytoplasmic distribution (181). The Cavβ2 subunit is found expressed only in specific cell types in thalamus, cerebellum and hippocampus, including cerebellar Purkinje neurons and hippocampal pyramidal neurons (17, 181). Strong immunostaining of both Cavβ3 and Cavβ4 is seen in mammalian brain, with prominent Cavβ3 immunoreactivity in olfactory bulb, cortex, hippocampus, habenula, and cerebellum, and prominent Cavβ4 immunoreactivity in cerebellum, with moderate to low immunoreactivity in olfactory bulb, cortex, hippocampus, and brainstem (181). Comparative distribution studies of Cav α1 and Cavβ subunits indicate no exclusive association between particular pairs of these principal and auxiliary subunits. In particular, the Cavβ subtype associated with presynaptic Cav channels is not known. The only known examples of presynaptic Cavβ subunits are Cavβ2 and Cavβ4 subunits in cerebellum (181, 359). Based on the polarized expression of exogenous Cavβ subunits in polarized epithelial cells (28), it is predicted that in cerebellar Purkinje neurons, the somatodendritic Cav2.1 α1 subunits might be associated largely with the Cavβ2a subunits, whereas the presynaptic Cav2.1 α1 subunits might be associated with the Cavβ4 subunits (75). However, thorough information on the subcellular localization of specific Cavα1-β combinations in mammalian brain is lacking.
The Cavγ subunits are transmembrane proteins having four transmembrane segments and predicted intracellular amino and carboxyl termini (Fig. 5). To date eight Cavγ subunits (designated Cavγ1 - Cavγ8) have been identified (8, 20), although only Cavγ1 and Cavγ6 may act as auxiliary subunits of Cav channels (49), with all others acting as glutamate receptor auxiliary subunits (226). All, with the exception of Cavγ1, have been found expressed in mammalian brain, where Cavγ2, Cavγ3, and Cavγ4 have been found associated and colocalized with Cav2.1 and Cav2.2 α subunits (294). Interestingly, unlike Cavα2δ and Cavβ subunits, Cavγ subunits do not affect the trafficking and plasma membrane expression of coexpressed Cav α1 subunits, but do impact the biophysical properties of the resultant Cav channels (8, 20). Nonetheless, thorough information on the role of Cavγ subunits in the function of specific neuronal Cav channels is lacking.
V. VOLTAGE-DEPENDENT ION CHANNEL REPERTOIRE OF SPECIFIC NEURONAL COMPARTMENTS
In this section, we provide selected examples of the VDIC distributions detailed above, but here organized by neuronal compartment. As different neurons can exhibit different subcellular distributions of specific VDICs, an exhaustive recapitulation of the details of VDIC expression in specific compartments of every type of neuron is well beyond the scope of even this detailed review. However, a brief summary of hallmarks of a subset of channels in well-characterized neurons is informative as to the repertoire of VDICs involved in compartment-specific functions.
A. Axons
The generation, propagation and modulation of action potentials in neurons requires the precise subcellular localization of axonal VDICs (13). Among VDICs, a restricted subset has been found expressed in the axons of mammalian brain neurons. This set includes Kv1.1, Kv1.2, Kv1.4 and their auxiliary Kvβ1 and Kvβ2 subunits, Kv3.1b, Kv3.3 and Kv3.4, Kv7.2 and Kv7.3, Nav1.1, Nav1.2 and Nav1.6, and Cav1.2 (25, 48, 70, 72, 128, 167, 222, 259, 342). Two prominent examples of high density VDIC clustering in mammalian axons are the axon initial segment and the node of Ranvier. The initial segment, an axonal subdomain essential for action potential initiation, expresses high density of Nav1.6, or Nav1.2 at younger ages (24), and occasionally Nav1.1 (342), Kv1.1 and Kv1.2 (72, 128), and Kv7.2 and Kv7.3 (229, 250). The node of Ranvier, characterized by regularly spaced, short 1 μm gaps in the myelin sheath, expresses high densities of clustered Nav1.6, or Nav1.2 in immature nodes, (24), and Kv7.2 and Kv3.1b, which generate the slowly activating (Ks) and the fast (Kf2) nodal currents, respectively (70, 71). High density clustering of Kv1.1, Kv1.2 and occasionally Kv1.4, along with Kv 2, are found in the juxtaparanode, a 5-15 μm long axonal membrane domain adjacent to the node but peripheral to the axoglial junction of the paranode (247, 259, 260, 346).
VDICs can be also prominently localized to unmyelinated axons. Many axons expressed high levels of Kv1.1, Kv1.2 and Kv1.4, and Kvβ1 and Kvβ2 (260). In neocortex and hippocampus, Kv3.4 appear to be localized to axons, frequently colocalizing with these Kv1 family members (167). Unmyelinated axons of dentate granule cells and cerebellar granule cells express Nav1.2 and Nav1.6 (31, 100, 282, 284, 357). Cav1.2 channels exhibit localization in axons and growth cones when exogenously expressed in cultured hippocampal neurons, but with further maturation are found in the soma and dendrites, although some localization in the axon initial segment can be observed (222).
B. Presynaptic terminals
At the presynaptic terminal, VDICs play a critical role in regulating nerve terminal excitability and controlling calcium influx which consequently regulate neurotransmitter release. Kv1.1, Kv1.2 and Kv1.4 are concentrated in the axonal membrane immediately preceding or within the presynaptic terminal (61, 73, 347). Biochemical studies demonstrate that most Kv1 channels isolated from brain synaptic membranes contain Kv1.2 subunits, with Kv1.2 homomers being prevalent (293). Studies on calyx of Held presynaptic terminals show that Kv1.2 is located at the transition zone between the axon and terminal but are excluded from the terminal itself (73). Kv1.2 located here does not influence action potentials within the terminal per se, but instead suppress axonal excitability, ensuring that aberrant action potentials do not invade the nerve terminal and elicit transmitter release (73). In cerebellar basket cell terminals, Kv1.1, Kv1.2 and Kvβ2 and Kv3.4 colocalize at the level of the light microscope, but exhibit distinct ultrastructural localizations (167, 260). Kv1.1, Kv1.2 and Kvβ2 are present in the septate-like junctions formed between basket cell terminal and Purkinje cell axons, while Kv3.4 is found in nonjunctional region of basket cell terminals (167). Kv3.2 is also found in basket cell terminals at sites distinct from Kv3.4 sites (22). Kv3.2 is distributed to focal regions also distinct from the septate junctions (Kv1.1 and Kv1.2), and corresponding to vesicle clusters at the terminal active zone, such that basket cell terminals possess a mosaic of Kv channel subunits (22). Nav1.2 is also found concentrated near nerve terminals, such as in globus pallidus, presumably on nerve terminals of striatal afferents, and in CA3 s. lucidum, presumably on mossy fiber terminals (100). Specific immunolocalization of Cav2.1 and Cav2.2 has been reported in the presynaptic area of neurons in rat cerebellar molecular layer and the mossy fiber zone of hippocampal CA3 region (178, 359). The long splice variant of Cav2.2 (Cav2.2a), but not the short variant/isoform Cav2.2b, is specifically targeted to presynaptic terminals when expressed exogenously in cultured hippocampal neurons (197). In rat brain globus pallidus neurons, a subset of Cav2.3 is localized in a subpopulation of presynaptic boutons, as revealed by immuno-electron microscopy (111).
C. Soma and Proximal Dendrites
In neurons the initiation of dendritic action potentials in response to excitatory postsynaptic potentials, and their subsequent active propagation to the somata, requires somatodendritic VDICs, many of which also contribute to backpropagation of action potentials from soma to dendrites (13, 164). A large subset of VDICs have been found expressed in the soma and dendrites of mammalian central neurons, including Kv1.1, Kv1.2 Kv2.1, Kv2.2, Kv3.1a, and Kv3.2, Kv4.2, Kv4.3, and their auxiliary KChIP and DPP-like subunits, and Kv7.5 (127, 216, 257, 289, 296, 297, 332, 347, 381). Nav1.1, and to a lesser extent Nav1.6 are also found in dendrites (100, 282, 357), Cav1.2, Cav1.3, Cav2.1, Cav2.2, Cav2.3, Cav3.1, Cav3.2, and Cav3.3 and their auxiliary subunits (85, 116, 323, 372, 380). In terms of specific subcellular localization of VDICs on dendrites, there exists a distinction between those found in the soma and proximal dendrites, and those on distal dendrites. In the soma and proximal dendrites, there are high levels of Kv2.1, Kv2.2, Kv3.2a, Kv3.3, and in interneurons Kv3.1b, Kv3.3 and Kv4.3 (53, 77, 127, 297, 335). Among Nav channels Nav1.1 is predominantly expressed in the soma and proximal dendrites of many mammalian brain neurons, although Nav1.6 is also present in certain cell types (33, 357). Almost all the Cav channels have been found expressed in the soma and proximal dendrites, in varying degrees, and in different neurons (335). For example, although Cav2.1 and Cav2.2 are predominantly expressed in axon terminals of most central neurons, they are also expressed in the soma and proximal dendrites of cerebellar Purkinje neurons and hippocampal CA1-CA3 pyramidal neurons (85, 356). Cav3.1 and Cav3.2 are also restricted to the somata of certain cell types, such as hippocampal dentate granule neurons and cerebellar Purkinje neurons (201). It should be noted when evaluating immunostaining data showing localization of VDICs on the soma and proximal dendrites that these domains contain the bulk of the neuron’s rough endoplasmic reticulum and Golgi apparatus. As such, some of the immunostaining reported for VDICs may be intracellular biosynthetic pools destined for other compartments (e.g. axons), as opposed to functional plasma membrane populations. Moreover, these domains are also a major site of non-specific antibody staining (261). As such, the reported somatodendritic expression of certain VDICs may not be representative of channel pools regulating excitability of these domains.
D. Distal Dendrites
In distal dendrites of neurons, specialized expression of different VDICs is critical not only for the active propagation of dendritic action potentials from dendrites to soma, but also for controlling backpropagation of action potentials from the soma into dendrites. Kv4 channels play a crucial and dynamic role in the latter process (378), and principal and auxiliary subunits of Kv4 channels are prominently expressed in distal dendrites (205, 257, 297). Other Kv channels localized in the distal dendrites of mammalian brain neurons are Kv2.2, Kv7.2, Kv7.3, and Kv7.5, which are expressed in varying densities in different cell types (34, 127). A subpopulation of neuronal Kv1.1 channels is also found in the distal dendrites of certain mammalian brain neurons (50). Cav1.2 and Cav3.3 channels are expressed in the distal dendrites, in addition to their localization in the soma and proximal dendrites (85, 201). However, in certain types of mammalian brain neurons, Cav2.1, Cav2.2 and Cav2.3 are also expressed throughout the somatodendritic region (111, 119). Dynamic activity-dependent changes in the localization of Kv4.2 and Cav1.3 in distal dendrites, especially in relation to excitatory synapses, have recently been reported (153, 383).
VI. MECHANISMS UNDERLYING VOLTAGE-DEPENDENT ION CHANNEL LOCALIZATION
A. Kv Channels
As described in detail above, among VDICs expressed in mammalian central neurons, Kv channels have among the most diverse patterns of subcellular segregation. Kv1 channels are predominantly localized in axons. Recent studies suggest a prominent role for Kvβ subunits in determining this localization. Kv1.2 α subunits overexpressed in cultured hippocampal neurons exhibit a somatodendritic localization, whereas cotransfection of the Kvβ2 subunit, which in itself is preferentially localized to axons (35), yields a pronounced axonal localization of Kv1.2 (35, 101). Subsequent analyses of Kv1.2 deletion mutants and chimeric channels revealed that the amino terminal T1 domain, which comprises the Kvβ subunit binding site (105, 291, 377), is essential for axonal expression of Kv1.2 channels (101). Specific mutations to disrupt Kvβ2 binding also disrupted axonal expression of Kv1 α subunits in transfected neurons, further suggesting that endogenous Kvβ subunits were contributing to the axonal Kv1 localization (101). Remarkably, when appended to single-pass transmembrane reporter proteins, the Kv1.2 T1 domain, but not T1 domain mutants with altered Kvβ binding, was able to direct the axonal localization of the reporter proteins when co-expressed with Kvβ2 (101). Thus, cytoplasmic Kvβ subunits may affect not only early biosynthetic processing events and ER export (299), but also axonal localization (35, 101), of Kv1 channels.
Recent studies have shown that axonal targeting of Kv1 channels is dependent on the direct interaction of Kvβ2 with the microtubule plus-end tracking protein, EB1 (102). Analysis of Kvβ2 deletion mutants revealed that disruption of the association of Kvβ2 with EB1 dramatically decreases Kv1.2 axonal targeting (102). Suppression of endogenous EB1 expression using siRNA also impairs axonal targeting of endogenous Kv1 channels, but not of endogenous Nav channels (102), which appear to target to axons by distinct mechanisms and motifs (94). Another recent study shows that Kv1 channel axonal targeting is not only dependent on EB1 and Kvβ2, but also on the interaction of the Kv1 α subunit T1 domain with the kinesin KIF5B (264). The role of Kvβ2, which also binds at the T1 domain, was not addressed in this study, although endogenous Kvβ2 was presumably expressed in the neurons in the cultured cortical slices used in these studies (264). As such, the T1 domain of Kv1 α subunits mediates multiple protein-protein interactions, including tetrameric assembly of Kv1 α subunits, association with Kvβ subunits (and their EB-1 binding partner), and interaction with KIF5B.
Membrane-associated guanylate kinases (MAGUKs) such as PSD-95, Chapsyn-110, SAP102 and SAP97 function as scaffolding molecules to promote co-clustering of membrane receptors and ion channels (84). Mutational and structural analyses showed that the PDZ domains of PSD-95 and other MAGUKs bind to the PDZ-binding motif (S/TxV) at the carboxyl terminus of all Kv1 α subunits (78, 151, 152, 327). In most mammalian central neurons, MAGUKs are found at synapses, most prominently in the postsynaptic density (295), while Kv1 channels are predominantly found on axons (335). However, MAGUKs are found as prominent components of Kv1 complexes purified from mammalian brain (287). Examples where Kv1 channels and MAGUKs, in this case PSD-95, exhibit robust colocalize in neurons, are in the colocalization of Kv1.1, Kv1.2, and Kvβ2 with PSD-95 in cerebellar basket cell terminals (151, 167) and at the juxtaparanodal regions of nodes of Ranvier (247). Mutation of the Drosophila discs-large gene, the ortholog of PSD-95, results in failure to localize Shaker potassium channels to the neuromuscular junction of flies (388). However, mutation of PSD-95 in mice does not lead to altered Kv1 channel localization at juxtaparanodes and cerebellar basket cell terminals (245). At the level of the light microscope, the localization of Kv1.1 and Kv1.2 in basket cell terminals and juxtaparanodes is normal in transgenic mice expressing a truncated form of PSD-95 that is no longer expressed at these sites. Compensatory expression of other MAGUKs does not appear to be involved in maintaining Kv1 channel localization at these sites (245). However, genetic elimination of another PDZ binding protein found at the juxtaparanode, Caspr2, does lead to loss of Kv1 channels at these sites (237). While MAGUKs or other PDZ containing proteins remain attractive candidates for the interacting proteins that cluster Kv1 channels and Caspr2 at discrete plasma membrane subdomains such as juxtaparanodes of myelinated axons, axon initial segments, and near nerve terminals, the specific binding partners mediating the clustering and their regulation remain to be elucidated.
Initial studies of the polarized and clustered localization of Kv2.1 were performed in polarized epithelial MDCK cells. In these cells, Kv2.1 localizes in the basolateral membrane (281), consistent with the analogous membrane hypothesis whereby the apical and basolateral membranes of epithelial cells correspond to the axonal and somatodendritic membranes, respectively, of neurons (77). Interestingly, Kv2.1 is also present in large clusters in the basolateral domains of MDCK cells (281), as it is in neurons. Expression of truncation mutants that lacked relatively large portions of the cytoplasmic Kv2.1 carboxyl terminus revealed that an ≈ 130 amino acid segment approximately midway in the 440 amino acid carboxyl terminus of Kv2.1 was necessary for both polarized expression and clustering in MDCK cells (281). These findings were later extended by studies in primary cultures of hippocampal neurons (175), where further deletion analyses revealed an ≈ 25 amino acid segment that was necessary for polarized and clustered localization. This sequence is well conserved in Kv2.2, which has a localization distinct from Kv2.1, suggesting additional targeting determinants. An alanine scan through this region revealed that three of the four critical amino acids are serine residues, consistent with subsequent studies showing a role for phosphorylation in regulating Kv2.1 clustering (208, 209).
Electron microscopic analysis has shown that plasma membrane Kv2.1 clusters on the somata and proximal dendrites of pyramidal neurons lie over subsurface cisternae (82), and at F-type muscarinic synapses in spinal motor neurons (215). These specialized sites where intracellular Ca2+ stores come into close apposition with the plasma membrane represent a specialized neuronal signaling domain that may also contain elevated levels of voltage-dependent Ca2+ channels (355). A functional relationship between Kv2.1 and dendritic [Ca2+]i transients were revealed by antisense knock down of Kv2.1 in rat hippocampal neurons (81). Moreover, Kv2.1 clusters colocalize with ryanodine receptor intracellular Ca2+ release channels in cultured hippocampal neurons (7). These findings suggest that the clustered localization of Kv2.1 could affect dendritic Ca2+ signaling at these sites, and vice versa.
Some of the most exciting studies in the ion channel clustering field have recently revealed the dynamic nature of Kv2.1 clustering. These live cell imaging studies reveal that although Kv2.1 localization is restricted to clusters, within these clusters Kv2.1 is freely mobile (221). This has led to a perimeter fence model for Kv2.1 clustering, whereby Kv2.1 is not attached to scaffolding proteins per se, but is clustered through interaction with the actin-based cytoskeleton that is somehow involved in determining the perimeter of Kv2.1 clusters (324). It is not yet clear from this intriguing model for Kv2.1 clustering how point mutations in the PRC domain (175), and dephosphorylation of C-terminal Kv2.1 phosphorylation sites (208, 209), would eliminate clustering, and the relationship of these dynamic clusters to intracellular membranes.
The polarized expression of three different C-terminal Kv3.2 splice variants (Kv3.2a, b, and c) in polarized epithelial cells revealed that alternative splicing led to differences in subcellular localization. The Kv3.2a variant was localized to the basolateral membrane, whereas the Kv3.2b and Kv3.2c isoforms were found apically (238). A recent report suggests that the polarized expression of Kv3.1 channels may also be determined by C-terminal splicing that affects interaction with ankyrin-G (369). This may be consistent with the localization of Kv3.1b at nodes of Ranvier (70), which contain high levels of ankyrin-G. Is it intriguing that Kv3.1b is not found at the initial segments of the same axons, as many other nodal proteins are also found here, including ankyrin-G (229, 342).
As described above, Kv4 channels exhibit strict polarized somatodendritic localization in neurons (335). The determinants of dendritic targeting of Kv4 α subunits have recently been investigated using chimeras between Kv4.2 and axonally localized Kv1.3 (265). Analyses of these chimeras revealed a critical 16-amino acid dileucine-containing motif in the cytoplasmic carboxyl-terminal region of Kv4.2 that is conserved in all Kv4 family members from nematodes to mammals (265). Unlike the determinant for polarized localization and clustering of dendritic Kv2.1, the Kv4 targeting signal also targets a type 1 membrane protein reporter protein (CD8) to dendrites (265). It should be noted that these results were obtained in a neuronal background that presumably contains endogenous KChIPs and/or possibly DPPX, as such the contribution of these interacting proteins to mediating the effects of this targeting signal cannot be determined. Thus, it will be important to determine whether this trafficking signal directs polarized Kv4 targeting in an expression background lacking these proteins (e.g. MDCK cells), or whether Kv4.2 mutants that lack KChIP and DPP-like auxiliary subunits binding still localize to dendrites. Conversely, a direct determination of the role of auxiliary subunits in the polarized expression of Kv4 channels is needed. Dendritic targeting of Kv4 channels also appears to involve the action of the microtubule-based motor protein kinesin Kif17 (54). Knockdown of Kif17 inhibits dendritic localization of Kv4.2, and Kv4.2 and Kif17 can be copurified from brain and heterologous cells. Interestingly, Kif17 interacts with the Kv4.2 C-terminus outside of the motif previously defined as critical for Kv4.2 localization on dendrites, and knockdown of Kif17 does not affect dendritic localization of the CD8 reporter protein containing the Kv4 family dendritic localization signal (54). The authors suggest that the Kif17 motor may be the primary mechanism for Kv4.2 transport to dendrites, the dileucine-based motif is the primary determinant of whether this localization is maintained. The mechanism for this retention is not known.
Other proteins that interact with Kv4 α subunits in heterologous cells, such as PSD-95 and filamin, and enhance the steady-state cell surface expression level and clustering of Kv4.2 in non-neuronal cell lines may also play a role in the polarized localization of Kv4 channels on dendrites (366). However, it is interesting that in cultured hippocampal neurons, the obvious clusters of Kv4.2 and PSD-95 (as well as filamin) do not colocalize, but rather appear interdigitated along the dendrites (302). The roles of these proteins in the polarized expression of Kv4 channels remains to be elucidated.
Kv4.2 localization in cerebellar granule cells in organotypic culture is regulated by synapse formation and glutamatergic synaptic activity (301). Granule cells grown in the absence of synaptic contact from cerebellar mossy fiber axons have Kv4.2 aberrantly localized on somata. Upon mossy fiber:granule cell synapse formation Kv4.2 becomes clustered at/near these specialized synapses, in a process that requires ionotropic glutamate receptor activity. Stimulating glutamate receptors in the absence of synapse formation leads to translocation of Kv4.2 from the soma to the dendrites (301). A recent study reveals that the localization of Kv4.2 on dendrites of hippocampal neurons is also dynamically regulated in response to glutamatergic synaptic activity through clathrin-mediated endocytosis (153). Internalization can be triggered by either glutamate receptor stimulation or paradigms that induce long term potentiation (153). Internalization of Kv4.2 increases dendritic excitability and further potentiates excitatory signaling in dendrites (153). The molecular determinants of the dynamic regulation of Kv4 localization, and whether they are intrinsic to Kv4.2, or found on KChIP or DPPX auxiliary subunits, or other interacting proteins, are as yet undefined.
The localization of Kv7 channels at axon initial segments has been the focus of recent study (55, 229, 250). Kv7.2 and Kv7.3 have binding motifs on their C-terminal cytoplasmic tails for interaction with the cytoskeletal associated protein ankyrin-G (229) similar to those found on Nav channels (95, 170). Kv7.2 and Kv7.3 localization at initial segments is disrupted in ankyrin-G knockout mice, and the targeting of exogenous Kv7 C-terminal fragments to the initial segments requires the ankyrin-G binding motif (55, 229). Deletion of the ankyrin-G binding motif from full-length Kv7.2 and Kv7.3 also leads to loss of localization of the channel subunits at the axon initial segment (250). Interestingly, in heteromeric Kv7.2/Kv7.3 channels that form native M channels (349) the ankyrin-G binding motif of Kv7.3 appears to be dominant over that of Kv7.2 (250). This is consistent with the finding that C-terminal fragments from Kv7.3 were more effective at directing reporter proteins to initial segments than were those from Kv7.2 (55)
B. Nav Channels
As described above, in general, Nav channels have different subcellular distributions in mammalian brain: Nav1.1 and Nav1.3 are principally expressed on neuronal cell bodies, Nav1.2 is expressed on unmyelinated axons, and Nav1.6 is present on both dendrites and myelinated axons. The resulting compartmentalized, non-uniform distribution can dramatically impact neuronal excitability. These distinct localization patterns presumably reflect intrinsic and extrinsic mechanisms that selectively target proteins to the axonal or somatodendritic compartments. Despite the central role of Nav channels in initiating and propagating neuronal electrical signals, the determinants of their discrete subcellular localizations in mammalian neurons are just beginning to be defined.
1. Axon initial segment determinants
The developmental expression and localization of Nav channels at the axon initial segment (Fig. 4C, D) has mainly been studied in retinal ganglion cells, cerebellar granule cells and hippocampal neurons (24). In all cases, during early development Nav1.2 is the predominant channel found at the axon initial segment. However, at later developmental stages Nav 1.6 is also detected at the axon initial segment together with Nav1.2. The reason for the difference in temporal expression of the two distinct Nav channels is unknown. Surprisingly, a recent study showed that the proximal axon initial segment of retinal ganglion cells and a subset of hippocampal CA3 neurons could also exhibit strong staining for Nav1.1 channels (Fig. 4D), which generally has a somatodendritic distribution (342). Targeting and sequestration of Nav channels at the axon initial segment involves a macromolecular complex of interacting proteins, including the cytoskeletal proteins ankyrin-G and βIV spectrin, the cell adhesion molecules neurofascin-186 (Nf-186) and neuron-glia related cell adhesion molecule NrCAM, and Navβ auxiliary subunits (137, 276, 386). The C-terminal cytoplasmic domain of the Nav1 α subunit appears to control the polarized localization of Nav channels in the axon (93). Appending the C-terminal cytoplasmic tail of Nav1.2 onto a reporter protein leads to its targeting to axons (93). Deletion analyses of the Nav1.2 C-terminus led to identification of a short sequence, CLDILFAFT that directs axonal targeting. This same sequence is also recognized by components of the endocytic pathway, suggesting that Nav1.2 is delivered to the surface of both axons and dendrites and retrieved by endocytosis from the soma and dendrites (93). Interestingly, this sequence is 100% conserved in all neuronal Nav channels (Nav1.1, Nav1.2, Nav1.3, and Nav1.6) that exhibit very different subcellular localizations in mammalian brain. As such, the contribution of this signal in the establishment and maintenance of the contrasting patterns of subcellular localization of different Nav isoforms in mammalian central neurons is unclear. However, the overall similarity of the distal C-termini of Nav1 α subunits is low, such that differences in the overall structure of this domain could impact the activity of the axonal targeting motif. A second Nav1 targeting determinant has been identified in the interdomain II-III linker that directs Nav1 localization to the axon initial segment (95, 170). This determinant mediates Nav1 interaction with the cytoskeletal scaffold ankyrin-G. However, it is important to note that this axon initial segment targeting motif is also conserved among all Nav1 channels. This suggests that, like many of the identified targeting signals described above, there exist additional determinants that modulate activity of this conserved signal. It is important to note that a similar signal targets Kv7.2 and Kv7.3 to axon initial segments (see above).
2. Node of Ranvier determinants
Nodes of Ranvier have molecular similarities with axon initial segment (e.g. high density of Nav channels, Kv channels, the cytoskeletal proteins ankyrin-G and βIV spectrin, the cell adhesion molecules Nf-186 and NrCAM). However, there exists a major distinction between these two domains: recruitment of proteins at nodes of Ranvier requires myelinating glia, but recruitment at the axon initial segment is neuronal autonomous (115). Although it is clear that myelinating oligodendrocytes are required for node of Ranvier formation and VDIC clustering, studies suggest that the Nav1 channel clustering can be induced by a soluble factor released by oligodendrocytes (146), whereas other studies suggest a requirement for intact myelin (24, 244, 246, 358). The cell adhesion molecule NF-186 is also required for the formation of CNS nodes, and mice with a neurofascin gene deletion, which appear to have intact myelination, fail to cluster Nav1 channels (as well as NrCAM, ankyrin-G and βIV spectrin) at the node (298). Clustering of Nav1 channels at axon initial segments, nodes of Ranvier and other sites in central neurons remains an exciting focus of much current research.
C. Cav Channels
Proper functioning and specific targeting of individual Cav channels into different subcellular compartments of neurons is crucial for the maintenance and physiological regulation of normal neuronal activity. As detailed above, different Cav channel subtypes exhibit different subcellular distributions in mammalian brain neurons, with Cav2.1 and Cav2.2 channels exhibiting predominantly axonal and presynaptic localization, and Cav1 and Cav3 channels mainly exhibiting somatodendritic expression.
Interaction with the Cavβ4 subunit has been suggested to promote the presynaptic targeting of Cav2.1 channels (28, 365). This coincides with the preferential presynaptic localization of Cavβ4 subunits in mammalian brain neurons (365). Coexpression of the Cavβ1b subunit in MDCK cells has also been shown to promote the targeting of Cav2.1 channels to apical membranes (28), suggesting a possible role for and Cav β1b in axonal targeting of Cav2.1 (75). The Cavβ subunits contain two conserved protein-protein interaction domains, a src Homology-3 (SH3) domain and a guanylate kinase (GK) domain, both of which are also found in channel clustering MAGUK scaffold proteins (108). These domains mediate the trafficking, targeting and voltage-dependent gating effects that Cavβ subunits exert on coexpressed Cav α1 subunits (92). A recent report suggests that the SH3 and GK domains can autonomously act to exert Cavβ subunit-like modulatory effects on Cav α1 trafficking and voltage-dependent gating (319). However, the precise mechanism underlying the Cavβ subunit-mediated targeting of Cavα1 subunits remains unclear.
Recent reports have strongly suggested the presence of a major intrinsic molecular determinant of presynaptic targeting in the Cav2.1 and Cav2.2 channels, namely a synaptic protein interaction site called the synprint site, which lies within the domain II - II cytoplasmic linker region (318). This domain II - III cytoplasmic linker region in Cav2.1 and Cav2.2 channels has been shown to assemble with synaptic protein such as syntaxin, SNAP-25, and synaptotagmin (135). Two Cav2.2 splice variants, which lack larger portions of the domain II - III cytoplasmic linker region and including the synprint motif exhibit axonal targeting but lack clustering in presynaptic terminals (211, 318). The synprint motif could not direct the axonal targeting of dendritic Cav1.2 when inserted into the Cav1.2 interdomain II - III cytoplasmic linker suggested that the synprint site is necessary but not sufficient to direct targeting of Cav channels to presynaptic (318). Another study using exogenous expression of Cav2.2 channels in cultured rat hippocampal neurons suggested that synaptic targeting of these channels is dependent on neuronal contacts and synapse formation (197). The cytoplasmic C-terminus of Cav2.2 channels also contains consensus motifs for binding to PDZ and SH3 domains that are both necessary and sufficient for Cavv2.2 axonal and synaptic targeting (197). The Cav2.2b splice variant lacks these motifs and does not exhibit axonal localization (197). The C-terminus of the Cav2.2a splice variant, which exhibits axonal and synaptic localization, contains these motifs and interacts with the adapter proteins Mint-1 and CASK, both of which are expressed in presynaptic nerve terminals. This suggests a model whereby interaction with Mint-1 and CASK promotes presynaptic recruitment of Cav2.2 channels in hippocampal neurons (197). However, another recent study suggested that deletion of the similar region in Cav2.1 does not affect presynaptic targeting, indicating possible differences in targeting of Cav2.1 and Cav2.2a channels to axon and presynaptic terminals (124). Overall, the precise molecular mechanisms underlying the axonal and presynaptic targeting of Cav2.1 and Cav2.2 channels through synprint-, Mint-1, CASK-mediate protein-protein interactions, as well as the involvement of these cytoplasmic C-terminal determinants are not completely defined.
Little is known of the molecular determinants and mechanisms underlying somatodendritic targeting of Cav1.2, Cav1.3 and Cav3 channels in mammalian neurons. One candidate is the auxiliary Cavβ2a subunit, which in polarized epithelial cells directs the targeting of Cav2.1 to basolateral membranes (28, 75). Cav1.3 colocalizes with the postsynaptic adapter protein Shank, and these proteins directly interact via a motif in the cytoplasmic C-terminus of Cav1.3 (383, 384). However, the role of Shank interaction with Cav1.3 on its dendritic targeting is not known. Cav1.2 is robustly expressed in the growth cone of developing neurons, yet upon neuronal maturation is targeted to the soma and dendrites, suggesting that specific targeting machinery in different neuronal compartments changes during neuronal differentiation (222). Presumably, differential interaction of Cav1.2 with as yet unidentified protein(s) leads to a switch between growth cone and somatodendritic targeting of Cav1.2. A recent report has suggested that calcium/calmodulin binding to the cytoplasmic C-terminus of Cav1.2 channels regulates the activity-dependent targeting of these channels to distal dendrites, however, the precise mechanism underlying this process is not known (348). Together, these preliminary studies on somatodendritic targeting of different Cav channels warrants further detailed investigation and elucidation of the underlying mechanism(s).
VII. CONCLUDING REMARKS
The intrinsic electrical properties, and rapid processing and transmission of synaptic signals in mammalian neurons depend largely on the abundance and function of a large repertoire of VDICs located at specific sites in neuronal somata, dendrites and axons. The remarkable molecular complexity of VDICs principal and auxiliary subunits leads to the generation of a remarkable diversity of VDIC function critical to fine control of neuronal electrical signaling. The selective placement of specific VDIC types at precise locations in mammalian neurons, and their dynamic regulation through local signaling pathways, allows for a complexity of neuronal function that underlies brain function. However, the molecular complexity of VDICs, the enormous cellular complexity of mammalian brain, and the extreme morphological complexity of neuronal form and poses a considerable challenge in understanding the specific role of individual VDICs in brain function, and how the specific and discrete subcellular compartmentalization of VDICs regulates signaling in mammalian neurons. Although molecular and biochemical studies have begun to define the cellular expression and subunit composition of native VDICs, defining the spatial localization of VDICs comprising specific subunit compositions remains an important challenge, for both fundamentally understanding neuronal function and validating specific VDICs as neurotherapeutic drug targets. The fundamental mechanisms that lead to the observed distributions in the neuronal membrane, and how these mechanisms are regulated during development, with aging, and in response to normal and pathological alterations in brain activity, have not yet been elucidated and remain a major topic of ongoing research.
ACKNOWLEDGMENTS
We thank Drs. Gary G. Matthews, Milena M. Menegola, Matthew N. Rasband and Kenneth J. Rhodes for numerous contributions to defining VDIC localization, and for sharing unpublished results. We also thank them and Drs. Edward C. Cooper and Bernardo Rudy for graciously granting permission to use published results herein.
GRANTS
The work from our laboratory that is discussed here was supported by NIH grants NS34383 and NS42225.
REFERENCES
- 1.Agnew WS, Miller JA, Ellisman MH, Rosenberg RL, Tomiko SA, Levinson SR. The voltage-regulated sodium channel from the electroplax of Electrophorus electricus. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 1):165–179. doi: 10.1101/sqb.1983.048.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Ahlijanian MK, Westenbroek RE, Catterall WA. Subunit structure and localization of dihydropyridine-sensitive calcium channels in mammalian brain, spinal cord, and retina. Neuron. 1990;4:819–832. doi: 10.1016/0896-6273(90)90135-3. [DOI] [PubMed] [Google Scholar]
- 3.Albensi BC, Ryujin KT, McIntosh JM, Naisbitt SR, Olivera BM, Fillous F. Localization of [125I]omega-conotoxin GVIA binding in human hippocampus and cerebellum. Neuroreport. 1993;4:1331–1334. doi: 10.1097/00001756-199309150-00011. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht B, Weber K, Pongs O. Characterization of a voltage-activated K-channel gene cluster on human chromosome 12p13. Receptors Channels. 1995;3:213–220. [PubMed] [Google Scholar]
- 5.An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 6.Anderson PA, Greenberg RM. Phylogeny of ion channels: clues to structure and function. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:17–28. doi: 10.1016/s1096-4959(01)00376-1. [DOI] [PubMed] [Google Scholar]
- 7.Antonucci DE, Lim ST, Vassanelli S, Trimmer JS. Dynamic localization and clustering of dendritic Kv2.1 voltage-dependent potassium channels in developing hippocampal neurons. Neuroscience. 2001;108:69–81. doi: 10.1016/s0306-4522(01)00476-6. [DOI] [PubMed] [Google Scholar]
- 8.Arikkath J, Campbell KP. Auxiliary subunits: essential components of the voltage-gated calcium channel complex. Curr Opin Neurobiol. 2003;13:298–307. doi: 10.1016/s0959-4388(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 9.Azimi-Zonooz A, Kawa CB, Dowell CD, Olivera BM. Autoradiographic localization of N-type VGCCs in gerbil hippocampus and failure of omega-conotoxin MVIIA to attenuate neuronal injury after transient cerebral ischemia. Brain Res. 2001;907:61–70. doi: 10.1016/s0006-8993(01)02471-4. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin TJ, Tsaur ML, Lopez GA, Jan YN, Jan LY. Characterization of a mammalian cDNA for an inactivating voltage-sensitive K+ channel. Neuron. 1991;7:471–483. doi: 10.1016/0896-6273(91)90299-f. [DOI] [PubMed] [Google Scholar]
- 11.Baranauskas G, Tkatch T, Nagata K, Yeh JZ, Surmeier DJ. Kv3.4 subunits enhance the repolarizing efficiency of Kv3.1 channels in fast-spiking neurons. Nat Neurosci. 2003;6:258–266. doi: 10.1038/nn1019. [DOI] [PubMed] [Google Scholar]
- 12.Baumann A, Krah-Jentgens I, Muller R, Muller-Holtkamp F, Seidel R, Kecskemethy N, Casal J, Ferrus A, Pongs O. Molecular organization of the maternal effect region of the Shaker complex of Drosophila: characterization of an I(A) channel transcript with homology to vertebrate Na channel. EMBO J. 1987;6:3419–3429. doi: 10.1002/j.1460-2075.1987.tb02665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bean BP. The action potential in mammalian central neurons. Nat Rev Neurosci. 2007;8:451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- 14.Beckers MC, Ernst E, Belcher S, Howe J, Levenson R, Gros P. A new sodium channel alpha-subunit gene (Scn9a) from Schwann cells maps to the Scn1a, Scn2a, Scn3a cluster of mouse chromosome 2. Genomics. 1996;36:202–205. doi: 10.1006/geno.1996.0447. [DOI] [PubMed] [Google Scholar]
- 15.Beckh S. Differential expression of sodium channel mRNAs in rat peripheral nervous system and innervated tissues. FEBS Lett. 1990;262:317–322. doi: 10.1016/0014-5793(90)80218-8. [DOI] [PubMed] [Google Scholar]
- 16.Beckh S, Noda M, Lubbert H, Numa S. Differential regulation of three sodium channel messenger RNAs in the rat central nervous system during development. EMBO J. 1989;8:3611–3616. doi: 10.1002/j.1460-2075.1989.tb08534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell DC, Butcher AJ, Berrow NS, Page KM, Brust PF, Nesterova A, Stauderman KA, Seabrook GR, Nurnberg B, Dolphin AC. Biophysical properties, pharmacology, and modulation of human, neuronal L-type (alpha(1D), Ca(V)1.3) voltage-dependent calcium currents. J Neurophysiol. 2001;85:816–827. doi: 10.1152/jn.2001.85.2.816. [DOI] [PubMed] [Google Scholar]
- 18.Bichet D, Cornet V, Geib S, Carlier E, Volsen S, Hoshi T, Mori Y, De Waard M. The I-II loop of the Ca2+ channel alpha1 subunit contains an endoplasmic reticulum retention signal antagonized by the beta subunit. Neuron. 2000;25:177–190. doi: 10.1016/s0896-6273(00)80881-8. [DOI] [PubMed] [Google Scholar]
- 19.Black JA, Yokoyama S, Higashida H, Ransom BR, Waxman SG. Sodium channel mRNAs I, II and III in the CNS: cell-specific expression. Brain Res Mol Brain Res. 1994;22:275–289. doi: 10.1016/0169-328x(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 20.Black JL., 3rd. The voltage-gated calcium channel gamma subunits: a review of the literature. J Bioenerg Biomembr. 2003;35:649–660. doi: 10.1023/b:jobb.0000008029.22650.c5. [DOI] [PubMed] [Google Scholar]
- 21.Blaine JT, Ribera AB. Heteromultimeric potassium channels formed by members of the Kv2 subfamily. J Neurosci. 1998;18:9585–9593. doi: 10.1523/JNEUROSCI.18-23-09585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobik M, Ellisman MH, Rudy B, Martone ME. Potassium channel subunit Kv3.2 and the water channel aquaporin-4 are selectively localized to cerebellar pinceau. Brain Res. 2004;1026:168–178. doi: 10.1016/j.brainres.2004.07.088. [DOI] [PubMed] [Google Scholar]
- 23.Bogdanov Y, Brice NL, Canti C, Page KM, Li M, Volsen SG, Dolphin AC. Acidic motif responsible for plasma membrane association of the voltage-dependent calcium channel beta1b subunit. Eur J Neurosci. 2000;12:894–902. doi: 10.1046/j.1460-9568.2000.00981.x. [DOI] [PubMed] [Google Scholar]
- 24.Boiko T, Rasband MN, Levinson SR, Caldwell JH, Mandel G, Trimmer JS, Matthews G. Compact myelin dictates the differential targeting of two sodium channel isoforms in the same axon. Neuron. 2001;30:91–104. doi: 10.1016/s0896-6273(01)00265-3. [DOI] [PubMed] [Google Scholar]
- 25.Boiko T, Van Wart A, Caldwell JH, Levinson SR, Trimmer JS, Matthews G. Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. J Neurosci. 2003;23:2306–2313. doi: 10.1523/JNEUROSCI.23-06-02306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boland LM, Jiang M, Lee SY, Fahrenkrug SC, Harnett MT, O’Grady SM. Functional properties of a brain-specific NH2-terminally spliced modulator of Kv4 channels. Am J Physiol Cell Physiol. 2003;285:C161–170. doi: 10.1152/ajpcell.00416.2002. [DOI] [PubMed] [Google Scholar]
- 27.Bosse E, Regulla S, Biel M, Ruth P, Meyer HE, Flockerzi V, Hofmann F. The cDNA and deduced amino acid sequence of the gamma subunit of the L-type calcium channel from rabbit skeletal muscle. FEBS Lett. 1990;267:153–156. doi: 10.1016/0014-5793(90)80312-7. [DOI] [PubMed] [Google Scholar]
- 28.Brice NL, Dolphin AC. Differential plasma membrane targeting of voltage-dependent calcium channel subunits expressed in a polarized epithelial cell line. J Physiol. 1999;515(Pt 3):685–694. doi: 10.1111/j.1469-7793.1999.685ab.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooke RE, Atkinson L, Batten TF, Deuchars SA, Deuchars J. Association of potassium channel Kv3.4 subunits with pre- and post-synaptic structures in brainstem and spinal cord. Neuroscience. 2004;126:1001–1010. doi: 10.1016/j.neuroscience.2004.03.051. [DOI] [PubMed] [Google Scholar]
- 30.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brysch W, Creutzfeldt OD, Luno K, Schlingensiepen R, Schlingensiepen KH. Regional and temporal expression of sodium channel messenger RNAs in the rat brain during development. Exp Brain Res. 1991;86:562–567. doi: 10.1007/BF00230529. [DOI] [PubMed] [Google Scholar]
- 32.Burkhalter A, Gonchar Y, Mellor RL, Nerbonne JM. Differential expression of I(A) channel subunits Kv4.2 and Kv4.3 in mouse visual cortical neurons and synapses. J Neurosci. 2006;26:12274–12282. doi: 10.1523/JNEUROSCI.2599-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldwell JH, Schaller KL, Lasher RS, Peles E, Levinson SR. Sodium channel Na(v)1.6 is localized at nodes of Ranvier, dendrites, and synapses. Proc Natl Acad Sci U S A. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caminos E, Garcia-Pino E, Martinez-Galan JR, Juiz JM. The potassium channel KCNQ5/Kv7.5 is localized in synaptic endings of auditory brainstem nuclei of the rat. J Comp Neurol. 2007;505:363–378. doi: 10.1002/cne.21497. [DOI] [PubMed] [Google Scholar]
- 35.Campomanes CR, Carroll KI, Manganas LN, Hershberger ME, Gong B, Antonucci DE, Rhodes KJ, Trimmer JS. Kv beta subunit oxidoreductase activity and Kv1 potassium channel trafficking. J Biol Chem. 2002;277:8298–8305. doi: 10.1074/jbc.M110276200. [DOI] [PubMed] [Google Scholar]
- 36.Candenas L, Seda M, Noheda P, Buschmann H, Cintado CG, Martin JD, Pinto FM. Molecular diversity of voltage-gated sodium channel alpha and beta subunit mRNAs in human tissues. Eur J Pharmacol. 2006;541:9–16. doi: 10.1016/j.ejphar.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 37.Cantrell AR, Catterall WA. Neuromodulation of Na+ channels: an unexpected form of cellular plasticity. Nat Rev Neurosci. 2001;2:397–407. doi: 10.1038/35077553. [DOI] [PubMed] [Google Scholar]
- 38.Castellano A, Chiara MD, Mellstrom B, Molina A, Monje F, Naranjo JR, Lopez-Barneo J. Identification and functional characterization of a K+ channel alpha-subunit with regulatory properties specific to brain. J Neurosci. 1997;17:4652–4661. doi: 10.1523/JNEUROSCI.17-12-04652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 40.Catterall WA. Structure and function of voltage-gated ion channels. Annu Rev Biochem. 1995;64:493–531. doi: 10.1146/annurev.bi.64.070195.002425. [DOI] [PubMed] [Google Scholar]
- 41.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 42.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 43.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 44.Catterall WA, Scheuer T, Thomsen W, Rossie S. Structure and modulation of voltage-gated ion channels. Ann N Y Acad Sci. 1991;625:174–180. doi: 10.1111/j.1749-6632.1991.tb33840.x. [DOI] [PubMed] [Google Scholar]
- 45.Chandy KG. Simplified gene nomenclature. Nature. 1991;352:26. doi: 10.1038/352026b0. [DOI] [PubMed] [Google Scholar]
- 46.Chandy KG, Gutman GA. Voltage-gated potassium channel genes. In: North RA, editor. Ligand- and Voltage-gated Ion Channels. CRC Press; Boca Raton: 1995. pp. 1–71. [Google Scholar]
- 47.Chandy KG, Williams CB, Spencer RH, Aguilar BA, Ghanshani S, Tempel BL, Gutman GA. A family of three mouse potassium channel genes with intronless coding regions. Science. 1990;247:973–975. doi: 10.1126/science.2305265. [DOI] [PubMed] [Google Scholar]
- 48.Chang SY, Zagha E, Kwon ES, Ozaita A, Bobik M, Martone ME, Ellisman MH, Heintz N, Rudy B. Distribution of Kv3.3 potassium channel subunits in distinct neuronal populations of mouse brain. J Comp Neurol. 2007;502:953–972. doi: 10.1002/cne.21353. [DOI] [PubMed] [Google Scholar]
- 49.Chen RS, Deng TC, Garcia T, Sellers ZM, Best PM. Calcium channel gamma subunits: a functionally diverse protein family. Cell Biochem Biophys. 2007;47:178–186. doi: 10.1007/s12013-007-0002-0. [DOI] [PubMed] [Google Scholar]
- 50.Chen X, Johnston D. Voltage-gated ion channels in dendrites of hippocampal pyramidal neurons. Pflugers Arch. 2006 doi: 10.1007/s00424-006-0097-y. [DOI] [PubMed] [Google Scholar]
- 51.Chien AJ, Gao T, Perez-Reyes E, Hosey MM. Membrane targeting of L-type calcium channels. Role of palmitoylation in the subcellular localization of the beta2a subunit. J Biol Chem. 1998;273:23590–23597. doi: 10.1074/jbc.273.36.23590. [DOI] [PubMed] [Google Scholar]
- 52.Chittajallu R, Chen Y, Wang H, Yuan X, Ghiani CA, Heckman T, McBain CJ, Gallo V. Regulation of Kv1 subunit expression in oligodendrocyte progenitor cells and their role in G1/S phase progression of the cell cycle. Proc Natl Acad Sci U S A. 2002;99:2350–2355. doi: 10.1073/pnas.042698399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow A, Erisir A, Farb C, Nadal MS, Ozaita A, Lau D, Welker E, Rudy B. K(+) channel expression distinguishes subpopulations of parvalbumin- and somatostatin-containing neocortical interneurons. J Neurosci. 1999;19:9332–9345. doi: 10.1523/JNEUROSCI.19-21-09332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu PJ, Rivera JF, Arnold DB. A role for Kif17 in transport of Kv4.2. J Biol Chem. 2006;281:365–373. doi: 10.1074/jbc.M508897200. [DOI] [PubMed] [Google Scholar]
- 55.Chung HJ, Jan YN, Jan LY. Polarized axonal surface expression of neuronal KCNQ channels is mediated by multiple signals in the KCNQ2 and KCNQ3 C-terminal domains. Proc Natl Acad Sci U S A. 2006;103:8870–8875. doi: 10.1073/pnas.0603376103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung YH, Joo KM, Kim MJ, Cha CI. Age-related changes in the distribution of Na(v)1.1 and Na(v)1.2 in rat cerebellum. Neuroreport. 2003;14:841–845. doi: 10.1097/00001756-200305060-00013. [DOI] [PubMed] [Google Scholar]
- 57.Coleman SK, Newcombe J, Pryke J, Dolly JO. Subunit composition of Kv1 channels in human CNS. J Neurochem. 1999;73:849–858. doi: 10.1046/j.1471-4159.1999.0730849.x. [DOI] [PubMed] [Google Scholar]
- 58.Coma M, Vicente R, Tsevi I, Grande M, Tamkun MM, Felipe A. Different Kv2.1/Kv9.3 heteromer expression during brain and lung post-natal development in the rat. J Physiol Biochem. 2002;58:195–203. doi: 10.1007/BF03179857. [DOI] [PubMed] [Google Scholar]
- 59.Cooper EC, Harrington E, Jan YN, Jan LY. M channel KCNQ2 subunits are localized to key sites for control of neuronal network oscillations and synchronization in mouse brain. J Neurosci. 2001;21:9529–9540. doi: 10.1523/JNEUROSCI.21-24-09529.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooper EC, Jan LY. M-channels: neurological diseases, neuromodulation, and drug development. Arch Neurol. 2003;60:496–500. doi: 10.1001/archneur.60.4.496. [DOI] [PubMed] [Google Scholar]
- 61.Cooper EC, Milroy A, Jan YN, Jan LY, Lowenstein DH. Presynaptic localization of Kv1.4-containing A-type potassium channels near excitatory synapses in the hippocampus. J Neurosci. 1998;18:965–974. doi: 10.1523/JNEUROSCI.18-03-00965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Craig PJ, Beattie RE, Folly EA, Banerjee MD, Reeves MB, Priestley JV, Carney SL, Sher E, Perez-Reyes E, Volsen SG. Distribution of the voltage-dependent calcium channel alpha1G subunit mRNA and protein throughout the mature rat brain. Eur J Neurosci. 1999;11:2949–2964. doi: 10.1046/j.1460-9568.1999.00711.x. [DOI] [PubMed] [Google Scholar]
- 63.Curtis BM, Catterall WA. Purification of the calcium antagonist receptor of the voltage-sensitive calcium channel from skeletal muscle transverse tubules. Biochemistry. 1984;23:2113–2118. doi: 10.1021/bi00305a001. [DOI] [PubMed] [Google Scholar]
- 64.Czirjak G, Toth ZE, Enyedi P. Characterization of the heteromeric potassium channel formed by kv2.1 and the retinal subunit kv8.2 in Xenopus oocytes. J Neurophysiol. 2007;98:1213–1222. doi: 10.1152/jn.00493.2007. [DOI] [PubMed] [Google Scholar]
- 65.Dallas ML, Atkinson L, Milligan CJ, Morris NP, Lewis DI, Deuchars SA, Deuchars J. Localization and function of the Kv3.1b subunit in the rat medulla oblongata: focus on the nucleus tractus solitarii. J Physiol. 2005;562:655–672. doi: 10.1113/jphysiol.2004.073338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davare MA, Avdonin V, Hall DD, Peden EM, Burette A, Weinberg RJ, Horne MC, Hoshi T, Hell JW. A beta2 adrenergic receptor signaling complex assembled with the Ca2+ channel Cav1.2. Science. 2001;293:98–101. doi: 10.1126/science.293.5527.98. [DOI] [PubMed] [Google Scholar]
- 67.Davies A, Hendrich J, Van Minh AT, Wratten J, Douglas L, Dolphin AC. Functional biology of the alpha(2)delta subunits of voltage-gated calcium channels. Trends Pharmacol Sci. 2007;28:220–228. doi: 10.1016/j.tips.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 68.Day NC, Shaw PJ, McCormack AL, Craig PJ, Smith W, Beattie R, Williams TL, Ellis SB, Ince PG, Harpold MM, Lodge D, Volsen SG. Distribution of alpha 1A, alpha 1B and alpha 1E voltage-dependent calcium channel subunits in the human hippocampus and parahippocampal gyrus. Neuroscience. 1996;71:1013–1024. doi: 10.1016/0306-4522(95)00514-5. [DOI] [PubMed] [Google Scholar]
- 69.Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat Rev Neurosci. 2005;6:850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 70.Devaux J, Alcaraz G, Grinspan J, Bennett V, Joho R, Crest M, Scherer SS. Kv3.1b is a novel component of CNS nodes. J Neurosci. 2003;23:4509–4518. doi: 10.1523/JNEUROSCI.23-11-04509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Devaux JJ, Kleopa KA, Cooper EC, Scherer SS. KCNQ2 is a nodal K+ channel. J Neurosci. 2004;24:1236–1244. doi: 10.1523/JNEUROSCI.4512-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dodson PD, Barker MC, Forsythe ID. Two heteromeric Kv1 potassium channels differentially regulate action potential firing. J Neurosci. 2002;22:6953–6961. doi: 10.1523/JNEUROSCI.22-16-06953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dodson PD, Billups B, Rusznak Z, Szucs G, Barker MC, Forsythe ID. Presynaptic rat Kv1.2 channels suppress synaptic terminal hyperexcitability following action potential invasion. J Physiol. 2003;550:27–33. doi: 10.1113/jphysiol.2003.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dodson PD, Forsythe ID. Presynaptic K+ channels: electrifying regulators of synaptic terminal excitability. Trends Neurosci. 2004;27:210–217. doi: 10.1016/j.tins.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 75.Dolphin AC. Beta subunits of voltage-gated calcium channels. J Bioenerg Biomembr. 2003;35:599–620. doi: 10.1023/b:jobb.0000008026.37790.5a. [DOI] [PubMed] [Google Scholar]
- 76.Dolphin AC, Wyatt CN, Richards J, Beattie RE, Craig P, Lee JH, Cribbs LL, Volsen SG, Perez-Reyes E. The effect of alpha2-delta and other accessory subunits on expression and properties of the calcium channel alpha1G. J Physiol. 1999;519(Pt 1):35–45. doi: 10.1111/j.1469-7793.1999.0035o.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dotti CG, Simons K. Polarized sorting of viral glycoproteins to the axon and dendrites of hippocampal neurons in culture. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- 78.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein- binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 79.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 80.Drewe JA, Verma S, Frech G, Joho RH. Distinct spatial and temporal expression patterns of K+ channel mRNAs from different subfamilies. J Neurosci. 1992;12:538–548. doi: 10.1523/JNEUROSCI.12-02-00538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Du J, Haak LL, Phillips-Tansey E, Russell JT, McBain CJ. Frequency-dependent regulation of rat hippocampal somato-dendritic excitability by the K+ channel subunit Kv2.1. J Physiol. 2000;522:19–31. doi: 10.1111/j.1469-7793.2000.t01-2-00019.xm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du J, Tao-Chang J-H, Zerfas P, McBain CJ. The K+ channel, Kv2.1, is apposed to astrocytic processes and is associated with inhibitory postsynaptic membranes in hippocampal and cortical principal neurons and inhibitory interneurons. Neuroscience. 1998;84:37–48. doi: 10.1016/s0306-4522(97)00519-8. [DOI] [PubMed] [Google Scholar]
- 83.Elezgarai I, Diez J, Puente N, Azkue JJ, Benitez R, Bilbao A, Knopfel T, Donate-Oliver F, Grandes P. Subcellular localization of the voltage-dependent potassium channel Kv3.1b in postnatal and adult rat medial nucleus of the trapezoid body. Neuroscience. 2003;118:889–898. doi: 10.1016/s0306-4522(03)00068-x. [DOI] [PubMed] [Google Scholar]
- 84.Elias GM, Nicoll RA. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell Biol. 2007;17:343–352. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Elliott EM, Malouf AT, Catterall WA. Role of calcium channel subtypes in calcium transients in hippocampal CA3 neurons. J Neurosci. 1995;15:6433–6444. doi: 10.1523/JNEUROSCI.15-10-06433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Engeland B, Neu A, Ludwig J, Roeper J, Pongs O. Cloning and functional expression of rat ether-a-go-go-like K+ channel genes. J Physiol. 1998;513(Pt 3):647–654. doi: 10.1111/j.1469-7793.1998.647ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ertel EA, Campbell KP, Harpold MM, Hofmann F, Mori Y, Perez-Reyes E, Schwartz A, Snutch TP, Tanabe T, Birnbaumer L, Tsien RW, Catterall WA. Nomenclature of voltage-gated calcium channels. Neuron. 2000;25:533–535. doi: 10.1016/s0896-6273(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 88.Felix JP, Bugianesi RM, Schmalhofer WA, Borris R, Goetz MA, Hensens OD, Bao JM, Kayser F, Parsons WH, Rupprecht K, Garcia ML, Kaczorowski GJ, Slaughter RS. Identification and biochemical characterization of a novel nortriterpene inhibitor of the human lymphocyte voltage-gated potassium channel, Kv1.3. Biochemistry. 1999;38:4922–4930. doi: 10.1021/bi982954w. [DOI] [PubMed] [Google Scholar]
- 89.Felts PA, Yokoyama S, Dib-Hajj S, Black JA, Waxman SG. Sodium channel alpha-subunit mRNAs I, II, III, NaG, Na6 and hNE (PN1): different expression patterns in developing rat nervous system. Brain Res Mol Brain Res. 1997;45:71–82. doi: 10.1016/s0169-328x(96)00241-0. [DOI] [PubMed] [Google Scholar]
- 90.Finnegan TF, Chen SR, Pan HL. Mu opioid receptor activation inhibits GABAergic inputs to basolateral amygdala neurons through Kv1.1/1.2 channels. J Neurophysiol. 2006;95:2032–2041. doi: 10.1152/jn.01004.2005. [DOI] [PubMed] [Google Scholar]
- 91.Frech G, VanDongen AMJ, Schuster G, Brown AM, Joho RH. A novel potassium channel with delayed rectifier properties isolated from rat brain by expression cloning. Nature. 1989;340:642–645. doi: 10.1038/340642a0. [DOI] [PubMed] [Google Scholar]
- 92.Gao T, Chien AJ, Hosey MM. Complexes of the alpha1C and beta subunits generate the necessary signal for membrane targeting of class C L-type calcium channels. J Biol Chem. 1999;274:2137–2144. doi: 10.1074/jbc.274.4.2137. [DOI] [PubMed] [Google Scholar]
- 93.Garrido JJ, Fernandes F, Giraud P, Mouret I, Pasqualini E, Fache MP, Jullien F, Dargent B. Identification of an axonal determinant in the C-terminus of the sodium channel Na(v)1.2. EMBO J. 2001;20:5950–5961. doi: 10.1093/emboj/20.21.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garrido JJ, Fernandes F, Moussif A, Fache MP, Giraud P, Dargent B. Dynamic compartmentalization of the voltage-gated sodium channels in axons. Biol Cell. 2003;95:437–445. doi: 10.1016/s0248-4900(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 95.Garrido JJ, Giraud P, Carlier E, Fernandes F, Moussif A, Fache MP, Debanne D, Dargent B. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 2003;300:2091–2094. doi: 10.1126/science.1085167. [DOI] [PubMed] [Google Scholar]
- 96.Geiger JRP, Jonas P. Dynamic control of presynaptic Ca2+ inflow by fast-inactivating K+ channels in hippocampal mossy fiber boutons. Neuron. 2000;28:927–939. doi: 10.1016/s0896-6273(00)00164-1. [DOI] [PubMed] [Google Scholar]
- 97.Goldin AL. Evolution of voltage-gated Na(+) channels. J Exp Biol. 2002;205:575–584. doi: 10.1242/jeb.205.5.575. [DOI] [PubMed] [Google Scholar]
- 98.Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA. Nomenclature of voltage-gated sodium channels. Neuron. 2001;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 99.Goldman-Wohl DS, Chan E, Baird D, Heintz N. Kv3.3b: a novel Shaw type potassium channel expressed in terminally differentiated cerebellar Purkinje cells and deep cerebellar nuclei. J Neurosci. 1994;14:511–522. doi: 10.1523/JNEUROSCI.14-02-00511.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gong B, Rhodes KJ, Bekele-Arcuri Z, Trimmer JS. Type I and type II Na(+) channel alpha-subunit polypeptides exhibit distinct spatial and temporal patterning, and association with auxiliary subunits in rat brain. J Comp Neurol. 1999;412:342–352. [PubMed] [Google Scholar]
- 101.Gu C, Jan YN, Jan LY. A conserved domain in axonal targeting of Kv1 (Shaker) voltage-gated potassium channels. Science. 2003;301:646–649. doi: 10.1126/science.1086998. [DOI] [PubMed] [Google Scholar]
- 102.Gu C, Zhou W, Puthenveedu MA, Xu M, Jan YN, Jan LY. The microtubule plus-end tracking protein EB1 is required for Kv1 voltage-gated K+ channel axonal targeting. Neuron. 2006;52:803–816. doi: 10.1016/j.neuron.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 103.Guan D, Lee JC, Higgs MH, Spain WJ, Foehring RC. Functional roles of Kv1 channels in neocortical pyramidal neurons. J Neurophysiol. 2007;97:1931–1940. doi: 10.1152/jn.00933.2006. [DOI] [PubMed] [Google Scholar]
- 104.Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K+ channel beta subunit. Cell. 1999;97:943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 105.Gulbis JM, Zhou M, Mann S, MacKinnon R. Structure of the cytoplasmic beta subunit-T1 assembly of voltage- dependent K+ channels. Science. 2000;289:123–127. doi: 10.1126/science.289.5476.123. [DOI] [PubMed] [Google Scholar]
- 106.Gurnett CA, De Waard M, Campbell KP. Dual function of the voltage-dependent Ca2+ channel alpha 2 delta subunit in current stimulation and subunit interaction. Neuron. 1996;16:431–440. doi: 10.1016/s0896-6273(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 107.Gutman GA, Chandy KG, Grissmer S, Lazdunski M, McKinnon D, Pardo LA, Robertson GA, Rudy B, Sanguinetti MC, Stuhmer W, Wang X. International Union of Pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol Rev. 2005;57:473–508. doi: 10.1124/pr.57.4.10. [DOI] [PubMed] [Google Scholar]
- 108.Hanlon MR, Berrow NS, Dolphin AC, Wallace BA. Modelling of a voltage-dependent Ca2+ channel beta subunit as a basis for understanding its functional properties. FEBS Lett. 1999;445:366–370. doi: 10.1016/s0014-5793(99)00156-8. [DOI] [PubMed] [Google Scholar]
- 109.Hanlon MR, Wallace BA. Structure and function of voltage-dependent ion channel regulatory beta subunits. Biochemistry. 2002;41:2886–2894. doi: 10.1021/bi0119565. [DOI] [PubMed] [Google Scholar]
- 110.Hansen HH, Waroux O, Seutin V, Jentsch TJ, Aznar S, Mikkelsen JD. Kv7 channels: interaction with dopaminergic and serotonergic neurotransmission in the CNS. J Physiol. 2008 doi: 10.1113/jphysiol.2007.149450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hanson JE, Smith Y. Subcellular distribution of high-voltage-activated calcium channel subtypes in rat globus pallidus neurons. J Comp Neurol. 2002;442:89–98. doi: 10.1002/cne.10075. [DOI] [PubMed] [Google Scholar]
- 112.Hartmann HA, Colom LV, Sutherland ML, Noebels JL. Selective localization of cardiac SCN5A sodium channels in limbic regions of rat brain. Nat Neurosci. 1999;2:593–595. doi: 10.1038/10147. [DOI] [PubMed] [Google Scholar]
- 113.Hartshorne RP, Catterall WA. Purification of the saxitoxin receptor of the sodium channel from rat brain. Proc Natl Acad Sci U S A. 1981;78:4620–4624. doi: 10.1073/pnas.78.7.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hattori S, Murakami F, Song WJ. Quantitative relationship between kv4.2 mRNA and a-type k+ current in rat striatal cholinergic interneurons during development. J Neurophysiol. 2003;90:175–183. doi: 10.1152/jn.00990.2002. [DOI] [PubMed] [Google Scholar]
- 115.Hedstrom KL, Rasband MN. Intrinsic and extrinsic determinants of ion channel localization in neurons. J Neurochem. 2006;98:1345–1352. doi: 10.1111/j.1471-4159.2006.04001.x. [DOI] [PubMed] [Google Scholar]
- 116.Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Helton TD, Horne WA. Alternative splicing of the beta 4 subunit has alpha1 subunit subtype-specific effects on Ca2+ channel gating. J Neurosci. 2002;22:1573–1582. doi: 10.1523/JNEUROSCI.22-05-01573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hidalgo P, Neely A. Multiplicity of protein interactions and functions of the voltage-gated calcium channel beta-subunit. Cell Calcium. 2007;42:389–396. doi: 10.1016/j.ceca.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 119.Hillman D, Chen S, Aung TT, Cherksey B, Sugimori M, Llinas RR. Localization of P-type calcium channels in the central nervous system. Proc Natl Acad Sci U S A. 1991;88:7076–7080. doi: 10.1073/pnas.88.16.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hiyama TY, Watanabe E, Ono K, Inenaga K, Tamkun MM, Yoshida S, Noda M. Na(x) channel involved in CNS sodium-level sensing. Nat Neurosci. 2002;5:511–512. doi: 10.1038/nn0602-856. [DOI] [PubMed] [Google Scholar]
- 121.Hofmann F, Biel M, Flockerzi V. Molecular basis for Ca2+ channel diversity. Annu Rev Neurosci. 1994;17:399–418. doi: 10.1146/annurev.ne.17.030194.002151. [DOI] [PubMed] [Google Scholar]
- 122.Holmqvist MH, Cao J, Hernandez-Pineda R, Jacobson MD, Carroll KI, Sung MA, Betty M, Ge P, Gilbride KJ, Brown ME, Jurman ME, Lawson D, Silos-Santiago I, Xie Y, Covarrubias M, Rhodes KJ, Distefano PS, An WF. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc Natl Acad Sci U S A. 2002;99:1035–1040. doi: 10.1073/pnas.022509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hopkins WF, Allen ML, Houamed KM, Tempel BL. Properties of voltage-gated K+ currents expressed in Xenopus oocytes by mKv1.1, mKv1.2 and their heteromultimers as revealed by mutagenesis of the dendrotoxin-binding site in mKv1.1. Pflugers Arch. 1994;428:382–390. doi: 10.1007/BF00724522. [DOI] [PubMed] [Google Scholar]
- 124.Hu Q, Saegusa H, Hayashi Y, Tanabe T. The carboxy-terminal tail region of human Cav2.1 (P/Q-type) channel is not an essential determinant for its subcellular localization in cultured neurones. Genes Cells. 2005;10:87–96. doi: 10.1111/j.1365-2443.2005.00820.x. [DOI] [PubMed] [Google Scholar]
- 125.Hugnot JP, Salinas M, Lesage F, Guillemare E, de Weille J, Heurteaux C, Mattei MG, Lazdunski M. Kv8.1, a new neuronal potassium channel subunit with specific inhibitory properties towards Shab and Shaw channels. EMBO J. 1996;15:3322–3331. [PMC free article] [PubMed] [Google Scholar]
- 126.Hwang PM, Cunningham AM, Peng YW, Snyder SH. CDRK and DRK1 K+ channels have contrasting localizations in sensory systems. Neuroscience. 1993;55:613–620. doi: 10.1016/0306-4522(93)90427-h. [DOI] [PubMed] [Google Scholar]
- 127.Hwang PM, Fotuhi M, Bredt DS, Cunningham AM, Snyder SH. Contrasting immunohistochemical localizations in rat brain of two novel K+ channels of the Shab subfamily. J Neurosci. 1993;13:1569–1576. doi: 10.1523/JNEUROSCI.13-04-01569.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Inda MC, DeFelipe J, Munoz A. Voltage-gated ion channels in the axon initial segment of human cortical pyramidal cells and their relationship with chandelier cells. Proc Natl Acad Sci U S A. 2006;103:2920–2925. doi: 10.1073/pnas.0511197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Isacoff EY, Jan YN, Jan LY. Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature. 1990;345:530–534. doi: 10.1038/345530a0. [DOI] [PubMed] [Google Scholar]
- 130.Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- 131.Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 132.Isom LL, Scheuer T, Brownstein AB, Ragsdale DS, Murphy BJ, Catterall WA. Functional co-expression of the beta 1 and type IIA alpha subunits of sodium channels in a mammalian cell line. J Biol Chem. 1995;270:3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- 133.Jan LY, Jan YN. Cloned potassium channels from eukaryotes and prokaryotes. Annu Rev Neurosci. 1997;20:91–123. doi: 10.1146/annurev.neuro.20.1.91. [DOI] [PubMed] [Google Scholar]
- 134.Jarnot M, Corbett AM. Immunolocalization of NaV1.2 channel subtypes in rat and cat brain and spinal cord with high affinity antibodies. Brain Res. 2006;1107:1–12. doi: 10.1016/j.brainres.2006.05.090. [DOI] [PubMed] [Google Scholar]
- 135.Jarvis SE, Zamponi GW. Masters or slaves? Vesicle release machinery and the regulation of presynaptic calcium channels. Cell Calcium. 2005;37:483–488. doi: 10.1016/j.ceca.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 136.Jay SD, Ellis SB, McCue AF, Williams ME, Vedvick TS, Harpold MM, Campbell KP. Primary structure of the gamma subunit of the DHP-sensitive calcium channel from skeletal muscle. Science. 1990;248:490–492. doi: 10.1126/science.2158672. [DOI] [PubMed] [Google Scholar]
- 137.Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–746. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jentsch TJ, Schroeder BC, Kubisch C, Friedrich T, Stein V. Pathophysiology of KCNQ channels: neonatal epilepsy and progressive deafness. Epilepsia. 2000;41:1068–1069. doi: 10.1111/j.1528-1157.2000.tb00302.x. [DOI] [PubMed] [Google Scholar]
- 139.Jerng HH, Pfaffinger PJ, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol Cell Neurosci. 2004;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 140.Jerng HH, Qian Y, Pfaffinger PJ. Modulation of Kv4.2 channel expression and gating by dipeptidyl peptidase 10 (DPP10) Biophys J. 2004;87:2380–2396. doi: 10.1529/biophysj.104.042358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnston D, Christie BR, Frick A, Gray R, Hoffman DA, Schexnayder LK, Watanabe S, Yuan LL. Active dendrites, potassium channels and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci. 2003;358:667–674. doi: 10.1098/rstb.2002.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kalman K, Nguyen A, Tseng-Crank J, Dukes ID, Chandy G, Hustad CM, Copeland NG, Jenkins NA, Mohrenweiser H, Brandriff B, Cahalan M, Gutman GA, Chandy KG. Genomic organization, chromosomal localization, tissue distribution, and biophysical characterization of a novel mammalian Shaker-related voltage-gated potassium channel, Kv1.7. J Biol Chem. 1998;273:5851–5857. doi: 10.1074/jbc.273.10.5851. [DOI] [PubMed] [Google Scholar]
- 144.Kamb A, Iverson LE, Tanouye MA. Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell. 1987;50:405–413. doi: 10.1016/0092-8674(87)90494-6. [DOI] [PubMed] [Google Scholar]
- 145.Kang MG, Chen CC, Felix R, Letts VA, Frankel WN, Mori Y, Campbell KP. Biochemical and biophysical evidence for gamma 2 subunit association with neuronal voltage-activated Ca2+ channels. J Biol Chem. 2001;276:32917–32924. doi: 10.1074/jbc.M100787200. [DOI] [PubMed] [Google Scholar]
- 146.Kaplan MR, Cho MH, Ullian EM, Isom LL, Levinson SR, Barres BA. Differential control of clustering of the sodium channels Na(v)1.2 and Na(v)1.6 at developing CNS nodes of Ranvier. Neuron. 2001;30:105–119. doi: 10.1016/s0896-6273(01)00266-5. [DOI] [PubMed] [Google Scholar]
- 147.Kavalali ET, Zhuo M, Bito H, Tsien RW. Dendritic Ca2+ channels characterized by recordings from isolated hippocampal dendritic segments. Neuron. 1997;18:651–663. doi: 10.1016/s0896-6273(00)80305-0. [DOI] [PubMed] [Google Scholar]
- 148.Kerr LM, Filloux F, Olivera BM, Jackson H, Wamsley JK. Autoradiographic localization of calcium channels with [125I]omega-conotoxin in rat brain. Eur J Pharmacol. 1988;146:181–183. doi: 10.1016/0014-2999(88)90501-8. [DOI] [PubMed] [Google Scholar]
- 149.Kerr LM, Yoshikami D. A venom peptide with a novel presynaptic blocking action. Nature. 1984;308:282–284. doi: 10.1038/308282a0. [DOI] [PubMed] [Google Scholar]
- 150.Khanna R, Roy L, Zhu X, Schlichter LC. K+ channels and the microglial respiratory burst. Am J Physiol Cell Physiol. 2001;280:C796–806. doi: 10.1152/ajpcell.2001.280.4.C796. [DOI] [PubMed] [Google Scholar]
- 151.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 152.Kim E, Sheng M. Differential K+ channel clustering activity of PSD-95 and SAP97, two related membrane-associated putative guanylate kinases. Neuropharmacology. 1996;35:993–1000. doi: 10.1016/0028-3908(96)00093-7. [DOI] [PubMed] [Google Scholar]
- 153.Kim J, Jung SC, Clemens AM, Petralia RS, Hoffman DA. Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron. 2007;54:933–947. doi: 10.1016/j.neuron.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kim J, Wei DS, Hoffman DA. Kv4 potassium channel subunits control action potential repolarization and frequency-dependent broadening in rat hippocampal CA1 pyramidal neurones. J Physiol. 2005;569:41–57. doi: 10.1113/jphysiol.2005.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim LA, Furst J, Butler MH, Xu S, Grigorieff N, Goldstein SA. Ito channels are octomeric complexes with four subunits of each Kv4.2 and K+ channel-interacting protein 2. J Biol Chem. 2004;279:5549–5554. doi: 10.1074/jbc.M311332200. [DOI] [PubMed] [Google Scholar]
- 156.Kim LA, Furst J, Gutierrez D, Butler MH, Xu S, Goldstein SA, Grigorieff N. Three-dimensional structure of I(to); Kv4.2-KChIP2 ion channels by electron microscopy at 21 Angstrom resolution. Neuron. 2004;41:513–519. doi: 10.1016/s0896-6273(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 157.Kitano J, Nishida M, Itsukaichi Y, Minami I, Ogawa M, Hirano T, Mori Y, Nakanishi S. Direct interaction and functional coupling between metabotropic glutamate receptor subtype 1 and voltage-sensitive Cav2.1 Ca2+ channel. J Biol Chem. 2003 doi: 10.1074/jbc.M303266200. [DOI] [PubMed] [Google Scholar]
- 158.Klugbauer N, Lacinova L, Marais E, Hobom M, Hofmann F. Molecular diversity of the calcium channel alpha2delta subunit. J Neurosci. 1999;19:684–691. doi: 10.1523/JNEUROSCI.19-02-00684.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koch RO, Wanner SG, Koschak A, Hanner M, Schwarzer C, Kaczorowski GJ, Slaughter RS, Garcia ML, Knaus HG. Complex subunit assembly of neuronal voltage-gated K+ channels. Basis for high-affinity toxin interactions and pharmacology. J Biol Chem. 1997;272:27577–27581. doi: 10.1074/jbc.272.44.27577. [DOI] [PubMed] [Google Scholar]
- 160.Kole MH, Letzkus JJ, Stuart GJ. Axon initial segment Kv1 channels control axonal action potential waveform and synaptic efficacy. Neuron. 2007;55:633–647. doi: 10.1016/j.neuron.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 161.Koopmann TT, Bezzina CR, Wilde AA. Voltage-gated sodium channels: action players with many faces. Ann Med. 2006;38:472–482. doi: 10.1080/07853890600969072. [DOI] [PubMed] [Google Scholar]
- 162.Krzemien DM, Schaller KL, Levinson SR, Caldwell JH. Immunolocalization of sodium channel isoform NaCh6 in the nervous system. J Comp Neurol. 2000;420:70–83. [PubMed] [Google Scholar]
- 163.Kues WA, Wunder F. Heterogeneous expression patterns of mammalian potassium channel genes in developing and adult rat brain. Eur J Neurosci. 1992;4:1296–1308. doi: 10.1111/j.1460-9568.1992.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 164.Lai HC, Jan LY. The distribution and targeting of neuronal voltage-gated ion channels. Nat Rev Neurosci. 2006;7:548–562. doi: 10.1038/nrn1938. [DOI] [PubMed] [Google Scholar]
- 165.Lambe EK, Aghajanian GK. The role of Kv1.2-containing potassium channels in serotonin-induced glutamate release from thalamocortical terminals in rat frontal cortex. J Neurosci. 2001;21:9955–9963. doi: 10.1523/JNEUROSCI.21-24-09955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lau D, Vega-Saenz de Miera EC, Contreras D, Ozaita A, Harvey M, Chow A, Noebels JL, Paylor R, Morgan JI, Leonard CS, Rudy B. Impaired fast-spiking, suppressed cortical inhibition, and increased susceptibility to seizures in mice lacking Kv3.2 K+ channel proteins. J Neurosci. 2000;20:9071–9085. doi: 10.1523/JNEUROSCI.20-24-09071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Laube G, Roper J, Pitt JC, Sewing S, Kistner U, Garner CC, Pongs O, Veh RW. Ultrastructural localization of Shaker-related potassium channel subunits and synapse-associated protein 90 to septate-like junctions in rat cerebellar Pinceaux. Brain Res Mol Brain Res. 1996;42:51–61. doi: 10.1016/s0169-328x(96)00120-9. [DOI] [PubMed] [Google Scholar]
- 168.Lauver A, Yuan LL, Jeromin A, Nadin BM, Rodriguez JJ, Davies HA, Stewart MG, Wu GY, Pfaffinger PJ. Manipulating Kv4.2 identifies a specific component of hippocampal pyramidal neuron A-current that depends upon Kv4.2 expression. J Neurochem. 2006;99:1207–1223. doi: 10.1111/j.1471-4159.2006.04185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Leicher T, Roeper J, Weber K, Wang X, Pongs O. Structural and functional characterization of human potassium channel subunit beta 1 (KCNA1B) Neuropharmacology. 1996;35:787–795. doi: 10.1016/0028-3908(96)00133-5. [DOI] [PubMed] [Google Scholar]
- 170.Lemaillet G, Walker B, Lambert S. Identification of a conserved ankyrin-binding motif in the family of sodium channel {alpha} subunits. J Biol Chem. 2003;278:27333–27339. doi: 10.1074/jbc.M303327200. [DOI] [PubMed] [Google Scholar]
- 171.Levy-Mozziconacci A, Alcaraz G, Giraud P, Boudier JA, Caillol G, Couraud F, Autillo-Touati A. Expression of the mRNA for the beta 2 subunit of the voltage-dependent sodium channel in rat CNS. Eur J Neurosci. 1998;10:2757–2767. doi: 10.1046/j.1460-9568.1998.00283.x. [DOI] [PubMed] [Google Scholar]
- 172.Li W, Kaczmarek LK, Perney TM. Localization of two high-threshold potassium channel subunits in the rat central auditory system. J Comp Neurol. 2001;437:196–218. doi: 10.1002/cne.1279. [DOI] [PubMed] [Google Scholar]
- 173.Li Y, Um SY, McDonald TV. Voltage-gated potassium channels: regulation by accessory subunits. Neuroscientist. 2006;12:199–210. doi: 10.1177/1073858406287717. [DOI] [PubMed] [Google Scholar]
- 174.Lien CC, Jonas P. Kv3 potassium conductance is necessary and kinetically optimized for high-frequency action potential generation in hippocampal interneurons. J Neurosci. 2003;23:2058–2068. doi: 10.1523/JNEUROSCI.23-06-02058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lim ST, Antonucci DE, Scannevin RH, Trimmer JS. A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron. 2000;25:385–397. doi: 10.1016/s0896-6273(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 176.Liss B, Franz O, Sewing S, Bruns R, Neuhoff H, Roeper J. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J. 2001;20:5715–5724. doi: 10.1093/emboj/20.20.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 178.Llinas R, Sugimori M, Hillman DE, Cherksey B. Distribution and functional significance of the P-type, voltage-dependent Ca2+ channels in the mammalian central nervous system. Trends Neurosci. 1992;15:351–355. doi: 10.1016/0166-2236(92)90053-b. [DOI] [PubMed] [Google Scholar]
- 179.Llinas R, Sugimori M, Lin JW, Cherksey B. Blocking and isolation of a calcium channel from neurons in mammals and cephalopods utilizing a toxin fraction (FTX) from funnel-web spider poison. Proc Natl Acad Sci U S A. 1989;86:1689–1693. doi: 10.1073/pnas.86.5.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 181.Ludwig A, Flockerzi V, Hofmann F. Regional expression and cellular localization of the alpha1 and beta subunit of high voltage-activated calcium channels in rat brain. J Neurosci. 1997;17:1339–1349. doi: 10.1523/JNEUROSCI.17-04-01339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ludwig J, Terlau H, Wunder F, Bruggemann A, Pardo LA, Marquardt A, Stuhmer W, Pongs O. Functional expression of a rat homologue of the voltage gated either a go-go potassium channel reveals differences in selectivity and activation kinetics between the Drosophila channel and its mammalian counterpart. EMBO J. 1994;13:4451–4458. doi: 10.1002/j.1460-2075.1994.tb06767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ludwig J, Weseloh R, Karschin C, Liu Q, Netzer R, Engeland B, Stansfeld C, Pongs O. Cloning and functional expression of rat eag2, a new member of the ether-a-go-go family of potassium channels and comparison of its distribution with that of eag1. Mol Cell Neurosci. 2000;16:59–70. doi: 10.1006/mcne.2000.0851. [DOI] [PubMed] [Google Scholar]
- 184.Luneau CJ, Williams JB, Marshall J, Levitan ES, Oliva C, Smith JS, Antanavage J, Folander K, Stein RB, Swanson R, Kaczmarek LK, Buhrow SA. Alternative splicing contributes to K+ channel diversity in the mammalian central nervous system. Proc Natl Acad Sci U S A. 1991;88:3932–3936. doi: 10.1073/pnas.88.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Macica CM, von Hehn CA, Wang LY, Ho CS, Yokoyama S, Joho RH, Kaczmarek LK. Modulation of the kv3.1b potassium channel isoform adjusts the fidelity of the firing pattern of auditory neurons. J Neurosci. 2003;23:1133–1141. doi: 10.1523/JNEUROSCI.23-04-01133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.MacKinnon R. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature. 1991;350:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- 187.MacKinnon R. Potassium channels. FEBS Lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- 188.Magee JC, Carruth M. Dendritic voltage-gated ion channels regulate the action potential firing mode of hippocampal CA1 pyramidal neurons. J Neurophysiol. 1999;82:1895–1901. doi: 10.1152/jn.1999.82.4.1895. [DOI] [PubMed] [Google Scholar]
- 189.Magee JC, Christofi G, Miyakawa H, Christie B, Lasser-Ross N, Johnston D. Subthreshold synaptic activation of voltage-gated Ca2+ channels mediates a localized Ca2+ influx into the dendrites of hippocampal pyramidal neurons. J Neurophysiol. 1995;74:1335–1342. doi: 10.1152/jn.1995.74.3.1335. [DOI] [PubMed] [Google Scholar]
- 190.Maletic-Savatic M, Lenn NJ, Trimmer JS. Differential spatiotemporal expression of K+ channel polypeptides in rat hippocampal neurons developing in situ and in vitro. J Neurosci. 1995;15:3840–3851. doi: 10.1523/JNEUROSCI.15-05-03840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Malin SA, Nerbonne JM. Elimination of the fast transient in superior cervical ganglion neurons with expression of KV4.2W362F: molecular dissection of IA. J Neurosci. 2000;20:5191–5199. doi: 10.1523/JNEUROSCI.20-14-05191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Malin SA, Nerbonne JM. Molecular heterogeneity of the voltage-gated fast transient outward K+ current, I(Af), in mammalian neurons. J Neurosci. 2001;21:8004–8014. doi: 10.1523/JNEUROSCI.21-20-08004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Malo D, Schurr E, Dorfman J, Canfield V, Levenson R, Gros P. Three brain sodium channel alpha-subunit genes are clustered on the proximal segment of mouse chromosome 2. Genomics. 1991;10:666–672. doi: 10.1016/0888-7543(91)90450-s. [DOI] [PubMed] [Google Scholar]
- 194.Manganas LN, Trimmer JS. Subunit composition determines Kv1 potassium channel surface expression. J Biol Chem. 2000;275:29685–29693. doi: 10.1074/jbc.M005010200. [DOI] [PubMed] [Google Scholar]
- 195.Manganas LN, Wang Q, Scannevin RH, Antonucci DE, Rhodes KJ, Trimmer JS. Identification of a trafficking determinant localized to the Kv1 potassium channel pore. Proc Natl Acad Sci U S A. 2001;98:14055–14059. doi: 10.1073/pnas.241403898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Martina M, Yao GL, Bean BP. Properties and functional role of voltage-dependent potassium channels in dendrites of rat cerebellar Purkinje neurons. J Neurosci. 2003;23:5698–5707. doi: 10.1523/JNEUROSCI.23-13-05698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Maximov A, Bezprozvanny I. Synaptic targeting of N-type calcium channels in hippocampal neurons. J Neurosci. 2002;22:6939–6952. doi: 10.1523/JNEUROSCI.22-16-06939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.McCormack T, McCormack K. Shaker K+ channel beta subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1994;79:1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 199.McCormack T, Vega-Saenz de Miera EC, Rudy B. Molecular cloning of a member of a third class of Shaker-family K+ channel genes in mammals. Proc Natl Acad Sci U S A. 1991;88:4060. doi: 10.1073/pnas.88.9.4060-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McEwen DP, Meadows LS, Chen C, Thyagarajan V, Isom LL. Sodium channel beta1 subunit-mediated modulation of Nav1.2 currents and cell surface density is dependent on interactions with contactin and ankyrin. J Biol Chem. 2004;279:16044–16049. doi: 10.1074/jbc.M400856200. [DOI] [PubMed] [Google Scholar]
- 201.McKay BE, McRory JE, Molineux ML, Hamid J, Snutch TP, Zamponi GW, Turner RW. Ca(V)3 T-type calcium channel isoforms differentially distribute to somatic and dendritic compartments in rat central neurons. Eur J Neurosci. 2006;24:2581–2594. doi: 10.1111/j.1460-9568.2006.05136.x. [DOI] [PubMed] [Google Scholar]
- 202.McNamara NM, Averill S, Wilkin GP, Dolly JO, Priestley JV. Ultrastructural localization of a voltage-gated K+ channel alpha subunit (KV 1.2) in the rat cerebellum. Eur J Neurosci. 1996;8:688–699. doi: 10.1111/j.1460-9568.1996.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 203.McNamara NM, Muniz ZM, Wilkin GP, Dolly JO. Prominent location of a K+ channel containing the alpha subunit Kv 1.2 in the basket cell nerve terminals of rat cerebellum. Neuroscience. 1993;57:1039–1045. doi: 10.1016/0306-4522(93)90047-j. [DOI] [PubMed] [Google Scholar]
- 204.Meadows LS, Malhotra J, Loukas A, Thyagarajan V, Kazen-Gillespie KA, Koopman MC, Kriegler S, Isom LL, Ragsdale DS. Functional and biochemical analysis of a sodium channel beta1 subunit mutation responsible for generalized epilepsy with febrile seizures plus type 1. J Neurosci. 2002;22:10699–10709. doi: 10.1523/JNEUROSCI.22-24-10699.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Menegola M, Trimmer JS. Unanticipated region- and cell-specific downregulation of individual KChIP auxiliary subunit isotypes in Kv4.2 knock-out mouse brain. J Neurosci. 2006;26:12137–12142. doi: 10.1523/JNEUROSCI.2783-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Millar ID, Wang S, Brown PD, Barrand MA, Hladky SB. Kv1 and Kir2 potassium channels are expressed in rat brain endothelial cells. Pflugers Arch. 2007 doi: 10.1007/s00424-007-0377-1. [DOI] [PubMed] [Google Scholar]
- 207.Miller C. annus mirabilis of potassium channels. Science. 1990;1991;252:1092–1096. doi: 10.1126/science.252.5009.1092. [DOI] [PubMed] [Google Scholar]
- 208.Misonou H, Menegola M, Mohapatra DP, Guy LK, Park KS, Trimmer JS. Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J Neurosci. 2006;26:13505–13514. doi: 10.1523/JNEUROSCI.3970-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, Anderson AE, Trimmer JS. Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci. 2004;7:711–718. doi: 10.1038/nn1260. [DOI] [PubMed] [Google Scholar]
- 210.Miyake A, Mochizuki S, Yokoi H, Kohda M, Furuichi K. New ether-a-go-go K(+) channel family members localized in human telencephalon. J Biol Chem. 1999;274:25018–25025. doi: 10.1074/jbc.274.35.25018. [DOI] [PubMed] [Google Scholar]
- 211.Mochida S, Westenbroek RE, Yokoyama CT, Zhong H, Myers SJ, Scheuer T, Itoh K, Catterall WA. Requirement for the synaptic protein interaction site for reconstitution of synaptic transmission by P/Q-type calcium channels. Proc Natl Acad Sci U S A. 2003;100:2819–2824. doi: 10.1073/pnas.262787699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Monaghan MM, Trimmer JS, Rhodes KJ. Experimental localization of Kv1 family voltage-gated K+ channel alpha and beta subunits in rat hippocampal formation. J Neurosci. 2001;21:5973–5983. doi: 10.1523/JNEUROSCI.21-16-05973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Morgan K, Stevens EB, Shah B, Cox PJ, Dixon AK, Lee K, Pinnock RD, Hughes J, Richardson PJ, Mizuguchi K, Jackson AP. beta 3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci U S A. 2000;97:2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mouginot D, Bossu JL, Gahwiler BH. Low-threshold Ca2+ currents in dendritic recordings from Purkinje cells in rat cerebellar slice cultures. J Neurosci. 1997;17:160–170. doi: 10.1523/JNEUROSCI.17-01-00160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Muennich EA, Fyffe RE. Focal aggregation of voltage-gated, Kv2.1 subunit-containing, potassium channels at synaptic sites in rat spinal motoneurones. J Physiol. 2004;554:673–685. doi: 10.1113/jphysiol.2003.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nadal MS, Ozaita A, Amarillo Y, de Miera EV, Ma Y, Mo W, Goldberg EM, Misumi Y, Ikehara Y, Neubert TA, Rudy B. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron. 2003;37:449–461. doi: 10.1016/s0896-6273(02)01185-6. [DOI] [PubMed] [Google Scholar]
- 217.Nadal MS, Ozaita A, Amarillo Y, Vega-Saenz de Miera E, Ma Y, Mo W, Goldberg EM, Misumi Y, Ikehara Y, Neubert TA, Rudy B. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron. 2003;37:449–461. doi: 10.1016/s0896-6273(02)01185-6. [DOI] [PubMed] [Google Scholar]
- 218.Noda M, Ikeda T, Kayano T, Suzuki H, Takeshima H, Kurasaki M, Takahashi H, Numa S. Existence of distinct sodium channel messenger RNAs in rat brain. Nature. 1986;320:188–192. doi: 10.1038/320188a0. [DOI] [PubMed] [Google Scholar]
- 219.Noda M, Ikeda T, Suzuki H, Takeshima H, Takahashi T, Kuno M, Numa S. Expression of functional sodium channels from cloned cDNA. Nature. 1986;322:826–828. doi: 10.1038/322826a0. [DOI] [PubMed] [Google Scholar]
- 220.Noda M, Shimizu S, Tanabe T, Takai T, Kayano T, Ikeda T, Takahashi H, Nakayama H, Kanaoka Y, Minamino N, et al. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature. 1984;312:121–127. doi: 10.1038/312121a0. [DOI] [PubMed] [Google Scholar]
- 221.O’Connell KM, Rolig AS, Whitesell JD, Tamkun MM. Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J Neurosci. 2006;26:9609–9618. doi: 10.1523/JNEUROSCI.1825-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Obermair GJ, Szabo Z, Bourinet E, Flucher BE. Differential targeting of the L-type Ca2+ channel alpha 1C (CaV1.2) to synaptic and extrasynaptic compartments in hippocampal neurons. Eur J Neurosci. 2004;19:2109–2122. doi: 10.1111/j.0953-816X.2004.03272.x. [DOI] [PubMed] [Google Scholar]
- 223.Oh Y, Sashihara S, Waxman SG. In situ hybridization localization of the Na+ channel beta 1 subunit mRNA in rat CNS neurons. Neurosci Lett. 1994;176:119–122. doi: 10.1016/0304-3940(94)90885-0. [DOI] [PubMed] [Google Scholar]
- 224.Orlova EV, Papakosta M, Booy FP, van Heel M, Dolly JO. Voltage-gated K+ channel from mammalian brain: 3D structure at 18A of the complete (alpha)4(beta)4 complex. J Mol Biol. 2003;326:1005–1012. doi: 10.1016/s0022-2836(02)00708-8. [DOI] [PubMed] [Google Scholar]
- 225.Osorio N, Alcaraz G, Padilla F, Couraud F, Delmas P, Crest M. Differential targeting and functional specialization of sodium channels in cultured cerebellar granule cells. J Physiol. 2005;569:801–816. doi: 10.1113/jphysiol.2005.097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Osten P, Stern-Bach Y. Learning from stargazin: the mouse, the phenotype and the unexpected. Curr Opin Neurobiol. 2006;16:275–280. doi: 10.1016/j.conb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 227.Ottschytsch N, Raes A, Van Hoorick D, Snyders DJ. Obligatory heterotetramerization of three previously uncharacterized Kv channel alpha-subunits identified in the human genome. Proc Natl Acad Sci U S A. 2002;99:7986–7991. doi: 10.1073/pnas.122617999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ozaita A, Martone ME, Ellisman MH, Rudy B. Differential subcellular localization of the two alternatively spliced isoforms of the Kv3.1 potassium channel subunit in brain. J Neurophysiol. 2002;88:394–408. doi: 10.1152/jn.2002.88.1.394. [DOI] [PubMed] [Google Scholar]
- 229.Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, Bennett V, Scherer SS, Cooper EC. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pannasch U, Farber K, Nolte C, Blonski M, Yan Chiu S, Messing A, Kettenmann H. The potassium channels Kv1.5 and Kv1.3 modulate distinct functions of microglia. Mol Cell Neurosci. 2006;33:401–411. doi: 10.1016/j.mcn.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 231.Papa M, Boscia F, Canitano A, Castaldo P, Sellitti S, Annunziato L, Taglialatela M. Expression pattern of the ether-a-gogo-related (ERG) K+ channel-encoding genes ERG1, ERG2, and ERG3 in the adult rat central nervous system. J Comp Neurol. 2003;466:119–135. doi: 10.1002/cne.10886. [DOI] [PubMed] [Google Scholar]
- 232.Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 233.Park KS, Yang JW, Seikel E, Trimmer JS. Potassium channel phosphorylation in excitable cells: providing dynamic functional variability to a diverse family of ion channels. Physiology (Bethesda) 2008;23:49–57. doi: 10.1152/physiol.00031.2007. [DOI] [PubMed] [Google Scholar]
- 234.Patel AJ, Lazdunski M, Honore E. Kv2.1/Kv9.3, a novel ATP-dependent delayed-rectifier K+ channel in oxygen-sensitive pulmonary artery myocytes. EMBO J. 1997;16:6615–6625. doi: 10.1093/emboj/16.22.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Perney TM, Kaczmarek LK. Localization of a high threshold potassium channel in the rat cochlear nucleus. J Comp Neurol. 1997;386:178–202. [PubMed] [Google Scholar]
- 236.Perney TM, Marshall J, Martin KA, Hockfield S, Kaczmarek LK. Expression of the mRNAs for the Kv3.1 potassium channel gene in the adult and developing rat brain. J Neurophysiol. 1992;68:756–766. doi: 10.1152/jn.1992.68.3.756. [DOI] [PubMed] [Google Scholar]
- 237.Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, Furley AJ, Peles E. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ponce A, Vega-Saenz de Miera E, Kentros C, Moreno H, Thornhill B, Rudy B. K+ channel subunit isoforms with divergent carboxy-terminal sequences carry distinct membrane targeting signals. J Membr Biol. 1997;159:149–159. doi: 10.1007/s002329900278. [DOI] [PubMed] [Google Scholar]
- 239.Pongs O. Voltage-gated potassium channels: from hyperexcitability to excitement. FEBS Lett. 1999;452:31–35. doi: 10.1016/s0014-5793(99)00535-9. [DOI] [PubMed] [Google Scholar]
- 240.Pongs O, Leicher T, Berger M, Roeper J, Bahring R, Wray D, Giese KP, Silva AJ, Storm JF. Functional and molecular aspects of voltage-gated K+ channel beta subunits. Ann N Y Acad Sci. 1999;868:344–355. doi: 10.1111/j.1749-6632.1999.tb11296.x. [DOI] [PubMed] [Google Scholar]
- 241.Qu Y, Isom LL, Westenbroek RE, Rogers JC, Tanada TN, McCormick KA, Scheuer T, Catterall WA. Modulation of cardiac Na+ channel expression in Xenopus oocytes by beta 1 subunits. J Biol Chem. 1995;270:25696–25701. doi: 10.1074/jbc.270.43.25696. [DOI] [PubMed] [Google Scholar]
- 242.Raman IM, Sprunger LK, Meisler MH, Bean BP. Altered subthreshold sodium currents and disrupted firing patterns in Purkinje neurons of Scn8a mutant mice. Neuron. 1997;19:881–891. doi: 10.1016/s0896-6273(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 243.Rasband MN. It’s “juxta” potassium channel! J Neurosci Res. 2004;76:749–757. doi: 10.1002/jnr.20073. [DOI] [PubMed] [Google Scholar]
- 244.Rasband MN, Kagawa T, Park EW, Ikenaka K, Trimmer JS. Dysregulation of axonal sodium channel isoforms after adult-onset chronic demyelination. J Neurosci Res. 2003;73:465–470. doi: 10.1002/jnr.10675. [DOI] [PubMed] [Google Scholar]
- 245.Rasband MN, Park EW, Zhen D, Arbuckle MI, Poliak S, Peles E, Grant SG, Trimmer JS. Clustering of neuronal potassium channels is independent of their interaction with PSD-95. J Cell Biol. 2002;159:663–672. doi: 10.1083/jcb.200206024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rasband MN, Peles E, Trimmer JS, Levinson SR, Lux SE, Shrager P. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J Neurosci. 1999;19:7516–7528. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rasband MN, Trimmer JS. Developmental clustering of ion channels at and near the node of Ranvier. Dev Biol. 2001;236:5–16. doi: 10.1006/dbio.2001.0326. [DOI] [PubMed] [Google Scholar]
- 248.Rasband MN, Trimmer JS, Peles E, Levinson SR, Shrager P. K+ channel distribution and clustering in developing and hypomyelinated axons of the optic nerve. J Neurocytol. 1999;28:319–331. doi: 10.1023/a:1007057512576. [DOI] [PubMed] [Google Scholar]
- 249.Rasband MN, Trimmer JS, Schwarz TL, Levinson SR, Ellisman MH, Schachner M, Shrager P. Potassium channel distribution, clustering, and function in remyelinating rat axons. J Neurosci. 1998;18:36–47. doi: 10.1523/JNEUROSCI.18-01-00036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rasmussen HB, Frokjaer-Jensen C, Jensen CS, Jensen HS, Jorgensen NK, Misonou H, Trimmer JS, Olesen SP, Schmitt N. Requirement of subunit co-assembly and ankyrin-G for M-channel localization at the axon initial segment. J Cell Sci. 2007;120:953–963. doi: 10.1242/jcs.03396. [DOI] [PubMed] [Google Scholar]
- 251.Ratcliffe CF, Qu Y, McCormick KA, Tibbs VC, Dixon JE, Scheuer T, Catterall WA. A sodium channel signaling complex: modulation by associated receptor protein tyrosine phosphatase beta. Nat Neurosci. 2000;3:437–444. doi: 10.1038/74805. [DOI] [PubMed] [Google Scholar]
- 252.Ratcliffe CF, Westenbroek RE, Curtis R, Catterall WA. Sodium channel beta1 and beta3 subunits associate with neurofascin through their extracellular immunoglobulin-like domain. J Cell Biol. 2001;154:427–434. doi: 10.1083/jcb.200102086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Restituito S, Thompson RM, Eliet J, Raike RS, Riedl M, Charnet P, Gomez CM. The polyglutamine expansion in spinocerebellar ataxia type 6 causes a beta subunit-specific enhanced activation of P/Q-type calcium channels in Xenopus oocytes. J Neurosci. 2000;20:6394–6403. doi: 10.1523/JNEUROSCI.20-17-06394.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 255.Rettig J, Sheng ZH, Kim DK, Hodson CD, Snutch TP, Catterall WA. Isoform-specific interaction of the alpha1A subunits of brain Ca2+ channels with the presynaptic proteins syntaxin and SNAP-25. Proc Natl Acad Sci U S A. 1996;93:7363–7368. doi: 10.1073/pnas.93.14.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rettig J, Wunder F, Stocker M, Lichtinghagen R, Mastiaux F, Beckh S, Kues W, Pedarzani P, Schröter KH, Ruppersberg JP, Veh R, Pongs O. Characterization of a Shaw-related potassium channel family in rat brain. Embo J. 1992;11:2473–2486. doi: 10.1002/j.1460-2075.1992.tb05312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, Burke SL, Strassle BW, Buchwalder L, Menegola M, Cao J, An WF, Trimmer JS. KChIPs and Kv4 alpha subunits as integral components of A-type potassium channels in mammalian brain. J Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Rhodes KJ, Keilbaugh SA, Barrezueta NX, Lopez KL, Trimmer JS. Association and colocalization of K+ channel alpha- and beta-subunit polypeptides in rat brain. J Neurosci. 1995;15:5360–5371. doi: 10.1523/JNEUROSCI.15-07-05360.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rhodes KJ, Monaghan MM, Barrezueta NX, Nawoschik S, Bekele-Arcuri Z, Matos MF, Nakahira K, Schechter LE, Trimmer JS. Voltage-gated K+ channel beta subunits: expression and distribution of Kv beta 1 and Kv beta 2 in adult rat brain. J Neurosci. 1996;16:4846–4860. doi: 10.1523/JNEUROSCI.16-16-04846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rhodes KJ, Strassle BW, Monaghan MM, Bekele-Arcuri Z, Matos MF, Trimmer JS. Association and colocalization of the Kvbeta1 and Kvbeta2 beta-subunits with Kv1 alpha-subunits in mammalian brain K+ channel complexes. J Neurosci. 1997;17:8246–8258. doi: 10.1523/JNEUROSCI.17-21-08246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rhodes KJ, Trimmer JS. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J Neurosci. 2006;26:8017–8020. doi: 10.1523/JNEUROSCI.2728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rhodes KJ, Trimmer JS. Antibody-based validation of CNS ion channel drug targets. J Gen Physiol. 2008 doi: 10.1085/jgp.200709926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Richards MW, Butcher AJ, Dolphin AC. Ca2+ channel beta-subunits: structural insights AID our understanding. Trends Pharmacol Sci. 2004;25:626–632. doi: 10.1016/j.tips.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 264.Rivera J, Chu PJ, Lewis TL, Jr., Arnold DB. The role of Kif5B in axonal localization of Kv1 K(+) channels. Eur J Neurosci. 2007;25:136–146. doi: 10.1111/j.1460-9568.2006.05277.x. [DOI] [PubMed] [Google Scholar]
- 265.Rivera JF, Ahmad S, Quick MW, Liman ER, Arnold DB. An evolutionarily conserved dileucine motif in Shal K+ channels mediates dendritic targeting. Nat Neurosci. 2003;6:243–250. doi: 10.1038/nn1020. [DOI] [PubMed] [Google Scholar]
- 266.Roberds SL, Tamkun MM. Cloning and tissue-specific expression of five voltage-gated potassium channel cDNAs expressed in rat heart. Proc Natl Acad Sci U S A. 1991;88:1798–1802. doi: 10.1073/pnas.88.5.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Roche JP, Westenbroek R, Sorom AJ, Hille B, Mackie K, Shapiro MS. Antibodies and a cysteine-modifying reagent show correspondence of M current in neurons to KCNQ2 and KCNQ3 K+ channels. Br J Pharmacol. 2002;137:1173–1186. doi: 10.1038/sj.bjp.0704989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Roosild TP, Le KT, Choe S. Cytoplasmic gatekeepers of K+-channel flux: a structural perspective. Trends Biochem Sci. 2004;29:39–45. doi: 10.1016/j.tibs.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 269.Rudy B, Chow A, Lau D, Amarillo Y, Ozaita A, Saganich M, Moreno H, Nadal MS, Hernandez-Pineda R, Hernandez-Cruz A, Erisir A, Leonard C, Vega-Saenz de Miera E. Contributions of Kv3 channels to neuronal excitability. Ann NY Acad Sci. 1999;868:304–343. doi: 10.1111/j.1749-6632.1999.tb11295.x. [DOI] [PubMed] [Google Scholar]
- 270.Rudy B, Sen K, Vega-Saenz de Miera E, Lau D, Ried T, Ward DC. Cloning of a human cDNA expressing a high voltage-activating, TEA-sensitive, type-A K+ channel which maps to chromosome 1 band p21. J Neurosci Res. 1991;29:401–412. doi: 10.1002/jnr.490290316. [DOI] [PubMed] [Google Scholar]
- 271.Ruppersberg JP, Schroter KH, Sakmann B, Stocker M, Sewing S, Pongs O. Heteromultimeric channels formed by rat brain potassium-channel proteins. Nature. 1990;345:535–537. doi: 10.1038/345535a0. [DOI] [PubMed] [Google Scholar]
- 272.Saganich MJ, Machado E, Rudy B. Differential expression of genes encoding subthreshold-operating voltage-gated K+ channels in brain. J Neurosci. 2001;21:4609–4624. doi: 10.1523/JNEUROSCI.21-13-04609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Saganich MJ, Vega-Saenz de Miera E, Nadal MS, Baker H, Coetzee WA, Rudy B. Cloning of components of a novel subthreshold-activating K(+) channel with a unique pattern of expression in the cerebral cortex. J Neurosci. 1999;19:10789–10802. doi: 10.1523/JNEUROSCI.19-24-10789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Salinas M, Duprat F, Heurteaux C, Hugnot JP, Lazdunski M. New modulatory alpha subunits for mammalian Shab K+ channels. J Biol Chem. 1997;272:24371–24379. doi: 10.1074/jbc.272.39.24371. [DOI] [PubMed] [Google Scholar]
- 275.Salkoff L, Baker K, Butler A, Covarrubias M, Pak MD, Wei A. An essential ‘set’ of K+ channels conserved in flies, mice and humans. Trends Neurosci. 1992;15:161–166. doi: 10.1016/0166-2236(92)90165-5. [DOI] [PubMed] [Google Scholar]
- 276.Salzer JL. Polarized domains of myelinated axons. Neuron. 2003;40:297–318. doi: 10.1016/s0896-6273(03)00628-7. [DOI] [PubMed] [Google Scholar]
- 277.Sano Y, Mochizuki S, Miyake A, Kitada C, Inamura K, Yokoi H, Nozawa K, Matsushime H, Furuichi K. Molecular cloning and characterization of Kv6.3, a novel modulatory subunit for voltage-gated K(+) channel Kv2.1. FEBS Lett. 2002;512:230–234. doi: 10.1016/s0014-5793(02)02267-6. [DOI] [PubMed] [Google Scholar]
- 278.Saper CB. An open letter to our readers on the use of antibodies. J Comp Neurol. 2005;493:477–478. doi: 10.1002/cne.20839. [DOI] [PubMed] [Google Scholar]
- 279.Saper CB, Sawchenko PE. Magic peptides, magic antibodies: guidelines for appropriate controls for immunohistochemistry. J Comp Neurol. 2003;465:161–163. doi: 10.1002/cne.10858. [DOI] [PubMed] [Google Scholar]
- 280.Sashihara S, Oh Y, Black JA, Waxman SG. Na+ channel beta 1 subunit mRNA expression in developing rat central nervous system. Brain Res Mol Brain Res. 1995;34:239–250. doi: 10.1016/0169-328x(95)00168-r. [DOI] [PubMed] [Google Scholar]
- 281.Scannevin RH, Murakoshi H, Rhodes KJ, Trimmer JS. Identification of a cytoplasmic domain important in the polarized expression and clustering of the Kv2.1 K+ channel. J Cell Biol. 1996;135:1619–1632. doi: 10.1083/jcb.135.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Schaller KL, Caldwell JH. Developmental and regional expression of sodium channel isoform NaCh6 in the rat central nervous system. J Comp Neurol. 2000;420:84–97. [PubMed] [Google Scholar]
- 283.Schaller KL, Caldwell JH. Expression and distribution of voltage-gated sodium channels in the cerebellum. Cerebellum. 2003;2:2–9. doi: 10.1080/14734220309424. [DOI] [PubMed] [Google Scholar]
- 284.Schaller KL, Krzemien DM, Yarowsky PJ, Krueger BK, Caldwell JH. A novel, abundant sodium channel expressed in neurons and glia. J Neurosci. 1995;15:3231–3242. doi: 10.1523/JNEUROSCI.15-05-03231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schroeder BC, Hechenberger M, Weinreich F, Kubisch C, Jentsch TJ. KCNQ5, a novel potassium channel broadly expressed in brain, mediates M-type currents. J Biol Chem. 2000;275:24089–24095. doi: 10.1074/jbc.M003245200. [DOI] [PubMed] [Google Scholar]
- 286.Schroeder BC, Kubisch C, Stein V, Jentsch TJ. Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature. 1998;396:687–690. doi: 10.1038/25367. [DOI] [PubMed] [Google Scholar]
- 287.Schulte U, Thumfart JO, Klocker N, Sailer CA, Bildl W, Biniossek M, Dehn D, Deller T, Eble S, Abbass K, Wangler T, Knaus HG, Fakler B. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron. 2006;49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 288.Scott VE, Muniz ZM, Sewing S, Lichtinghagen R, Parcej DN, Pongs O, Dolly JO. Antibodies specific for distinct Kv subunits unveil a heterooligomeric basis for subtypes of alpha-dendrotoxin-sensitive K+ channels in bovine brain. Biochem. 1994;33:1617–1623. doi: 10.1021/bi00173a001. [DOI] [PubMed] [Google Scholar]
- 289.Sekirnjak C, Martone ME, Weiser M, Deerinck T, Bueno E, Rudy B, Ellisman M. Subcellular localization of the K+ channel subunit Kv3.1b in selected rat CNS neurons. Brain Res. 1997;766:173–187. doi: 10.1016/s0006-8993(97)00527-1. [DOI] [PubMed] [Google Scholar]
- 290.Serodio P, Rudy B. Differential expression of Kv4 K+ channel subunits mediating subthreshold transient K+ (A-type) currents in rat brain. J Neurophysiol. 1998;79:1081–1091. doi: 10.1152/jn.1998.79.2.1081. [DOI] [PubMed] [Google Scholar]
- 291.Sewing S, Roeper J, Pongs O. Kv beta 1 subunit binding specific for shaker-related potassium channel alpha subunits. Neuron. 1996;16:455–463. doi: 10.1016/s0896-6273(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 292.Shah BS, Stevens EB, Pinnock RD, Dixon AK, Lee K. Developmental expression of the novel voltage-gated sodium channel auxiliary subunit beta3, in rat CNS. J Physiol. 2001;534:763–776. doi: 10.1111/j.1469-7793.2001.t01-1-00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Shamotienko OG, Parcej DN, Dolly JO. Subunit combinations defined for K+ channel Kv1 subtypes in synaptic membranes from bovine brain. Biochemistry. 1997;36:8195–8201. doi: 10.1021/bi970237g. [DOI] [PubMed] [Google Scholar]
- 294.Sharp AH, Black JL, 3rd, Dubel SJ, Sundarraj S, Shen JP, Yunker AM, Copeland TD, McEnery MW. Biochemical and anatomical evidence for specialized voltage-dependent calcium channel gamma isoform expression in the epileptic and ataxic mouse, stargazer. Neuroscience. 2001;105:599–617. doi: 10.1016/s0306-4522(01)00220-2. [DOI] [PubMed] [Google Scholar]
- 295.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 296.Sheng M, Tsaur ML, Jan YN, Jan LY. Contrasting subcellular localization of the Kv1.2 K+ channel subunit in different neurons of rat brain. J Neurosci. 1994;14:2408–2417. doi: 10.1523/JNEUROSCI.14-04-02408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sheng M, Tsaur ML, Jan YN, Jan LY. Subcellular segregation of two A-type K+ channel proteins in rat central neurons. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- 298.Sherman DL, Tait S, Melrose S, Johnson R, Zonta B, Court FA, Macklin WB, Meek S, Smith AJ, Cottrell DF, Brophy PJ. Neurofascins are required to establish axonal domains for saltatory conduction. Neuron. 2005;48:737–742. doi: 10.1016/j.neuron.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 299.Shi G, Nakahira K, Hammond S, Rhodes KJ, Schechter LE, Trimmer JS. Beta subunits promote K+ channel surface expression through effects early in biosynthesis. Neuron. 1996;16:843–852. doi: 10.1016/s0896-6273(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 300.Shi W, Wang HS, Pan Z, Wymore RS, Cohen IS, McKinnon D, Dixon JE. Cloning of a mammalian elk potassium channel gene and EAG mRNA distribution in rat sympathetic ganglia. J Physiol. 1998;511(Pt 3):675–682. doi: 10.1111/j.1469-7793.1998.675bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Shibasaki K, Nakahira K, Trimmer JS, Shibata R, Akita M, Watanabe S, Ikenaka K. Mossy fibre contact triggers the targeting of Kv4.2 potassium channels to dendrites and synapses in developing cerebellar granule neurons. J Neurochem. 2004;89:897–907. doi: 10.1111/j.1471-4159.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- 302.Shibata R, Misonou H, Campomanes CR, Anderson AE, Schrader LA, Doliveira LC, Carroll KI, Sweatt JD, Rhodes KJ, Trimmer JS. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J Biol Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- 303.Shibata R, Wakazono Y, Nakahira K, Trimmer JS, Ikenaka K. Expression of Kv3.1 and Kv4.2 genes in developing cerebellar granule cells. Dev Neurosci. 1999;21:87–93. doi: 10.1159/000017370. [DOI] [PubMed] [Google Scholar]
- 304.Shu Y, Yu Y, Yang J, McCormick DA. Selective control of cortical axonal spikes by a slowly inactivating K+ current. Proc Natl Acad Sci U S A. 2007;104:11453–11458. doi: 10.1073/pnas.0702041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Snutch TP, Sutton KG, Zamponi GW. Voltage-dependent calcium channels--beyond dihydropyridine antagonists. Curr Opin Pharmacol. 2001;1:11–16. doi: 10.1016/s1471-4892(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 306.Sokolov S, Weiss RG, Timin EN, Hering S. Modulation of slow inactivation in class A Ca2+ channels by beta-subunits. J Physiol. 2000;527(Pt 3):445–454. doi: 10.1111/j.1469-7793.2000.t01-1-00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sokolova O, Kolmakova-Partensky L, Grigorieff N. Three-dimensional structure of a voltage-gated potassium channel at 2.5 nm resolution. Structure. 2001;9:215–220. doi: 10.1016/s0969-2126(01)00578-0. [DOI] [PubMed] [Google Scholar]
- 308.Soler-Llavina GJ, Chang TH, Swartz KJ. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker K(v) channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 309.Song P, Yang Y, Barnes-Davies M, Bhattacharjee A, Hamann M, Forsythe ID, Oliver DL, Kaczmarek LK. Acoustic environment determines phosphorylation state of the Kv3.1 potassium channel in auditory neurons. Nat Neurosci. 2005;8:1335–1342. doi: 10.1038/nn1533. [DOI] [PubMed] [Google Scholar]
- 310.Song WJ, Tkatch T, Baranauskas G, Ichinohe N, Kitai ST, Surmeier DJ. Somatodendritic depolarization-activated potassium currents in rat neostriatal cholinergic interneurons are predominantly of the A type and attributable to coexpression of Kv4.2 and Kv4.1 subunits. J Neurosci. 1998;18:3124–3137. doi: 10.1523/JNEUROSCI.18-09-03124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Srinivasan J, Schachner M, Catterall WA. Interaction of voltage-gated sodium channels with the extracellular matrix molecules tenascin-C and tenascin-R. Proc Natl Acad Sci U S A. 1998;95:15753–15757. doi: 10.1073/pnas.95.26.15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Stocker M, Kerschensteiner D. Cloning and tissue distribution of two new potassium channel alpha-subunits from rat brain. Biochem Biophys Res Commun. 1998;248:927–934. doi: 10.1006/bbrc.1998.9072. [DOI] [PubMed] [Google Scholar]
- 313.Storm JF. K(+) channels and their distribution in large cortical pyramidal neurones. J Physiol. 2000;525:565–566. doi: 10.1111/j.1469-7793.2000.t01-1-00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Strassle BW, Menegola M, Rhodes KJ, Trimmer JS. Light and electron microscopic analysis of KChIP and Kv4 localization in rat cerebellar granule cells. J Comp Neurol. 2005;484:144–155. doi: 10.1002/cne.20443. [DOI] [PubMed] [Google Scholar]
- 315.Strong M, Chandy KG, Gutman GA. Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol Biol Evol. 1993;10:221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 316.Stuhmer W, Ruppersberg JP, Schroter KH, Sakmann B, Stocker M, Giese KP, Perschke A, Baumann A, Pongs O. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. Embo J. 1989;8:3235–3244. doi: 10.1002/j.1460-2075.1989.tb08483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Swartz KJ. Towards a structural view of gating in potassium channels. Nat Rev Neurosci. 2004;5:905–916. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- 318.Szabo Z, Obermair GJ, Cooper CB, Zamponi GW, Flucher BE. Role of the synprint site in presynaptic targeting of the calcium channel CaV2.2 in hippocampal neurons. Eur J Neurosci. 2006;24:709–718. doi: 10.1111/j.1460-9568.2006.04947.x. [DOI] [PubMed] [Google Scholar]
- 319.Takahashi SX, Miriyala J, Tay LH, Yue DT, Colecraft HM. A CaVbeta SH3/guanylate kinase domain interaction regulates multiple properties of voltage-gated Ca2+ channels. J Gen Physiol. 2005;126:365–377. doi: 10.1085/jgp.200509354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Takemura M, Kiyama H, Fukui H, Tohyama M, Wada H. Autoradiographic visualization in rat brain of receptors for omega-conotoxin GVIA, a newly discovered calcium antagonist. Brain Res. 1988;451:386–389. doi: 10.1016/0006-8993(88)90790-1. [DOI] [PubMed] [Google Scholar]
- 321.Takemura M, Kiyama H, Fukui H, Tohyama M, Wada H. Distribution of the omega-conotoxin receptor in rat brain. An autoradiographic mapping. Neuroscience. 1989;32:405–416. doi: 10.1016/0306-4522(89)90089-4. [DOI] [PubMed] [Google Scholar]
- 322.Takimoto K, Yang EK, Conforti L. Palmitoylation of KChIP splicing variants is required for efficient cell surface expression of Kv4.3 channels. J Biol Chem. 2002;277:26904–26911. doi: 10.1074/jbc.M203651200. [DOI] [PubMed] [Google Scholar]
- 323.Talley EM, Cribbs LL, Lee JH, Daud A, Perez-Reyes E, Bayliss DA. Differential distribution of three members of a gene family encoding low voltage-activated (T-type) calcium channels. J Neurosci. 1999;19:1895–1911. doi: 10.1523/JNEUROSCI.19-06-01895.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Tamkun MM, O’Connell KM, Rolig AS. A cytoskeletal-based perimeter fence selectively corrals a sub-population of cell surface Kv2.1 channels. J Cell Sci. 2007;120:2413–2423. doi: 10.1242/jcs.007351. [DOI] [PubMed] [Google Scholar]
- 325.Tanabe T, Takeshima H, Mikami A, Flockerzi V, Takahashi H, Kangawa K, Kojima M, Matsuo H, Hirose T, Numa S. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature. 1987;328:313–318. doi: 10.1038/328313a0. [DOI] [PubMed] [Google Scholar]
- 326.Tempel BL, Jan YN, Jan LY. Cloning of a probable potassium channel gene from mouse brain. Nature. 1988;332:837–839. doi: 10.1038/332837a0. [DOI] [PubMed] [Google Scholar]
- 327.Tiffany AM, Manganas LN, Kim E, Hsueh YP, Sheng M, Trimmer JS. PSD-95 and SAP97 exhibit distinct mechanisms for regulating K(+) channel surface expression and clustering. J Cell Biol. 2000;148:147–158. doi: 10.1083/jcb.148.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Tinel N, Lauritzen I, Chouabe C, Lazdunski M, Borsotto M. The KCNQ2 potassium channel: splice variants, functional and developmental expression. Brain localization and comparison with KCNQ3. FEBS Lett. 1998;438:171–176. doi: 10.1016/s0014-5793(98)01296-4. [DOI] [PubMed] [Google Scholar]
- 329.Tippens AL, Pare JF, Langwieser N, Moosmang S, Milner TA, Smith Y, Lee A. Ultrastructural evidence for pre- and postsynaptic localization of Cav1.2 L-type Ca2+ channels in the rat hippocampus. J Comp Neurol. 2008;506:569–583. doi: 10.1002/cne.21567. [DOI] [PubMed] [Google Scholar]
- 330.Tkatch T, Baranauskas G, Surmeier DJ. Kv4.2 mRNA abundance and A-type K(+) current amplitude are linearly related in basal ganglia and basal forebrain neurons. J Neurosci. 2000;20:579–588. doi: 10.1523/JNEUROSCI.20-02-00579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Torres YP, Morera FJ, Carvacho I, Latorre R. A marriage of convenience: beta-subunits and voltage-dependent K+ channels. J Biol Chem. 2007;282:24485–24489. doi: 10.1074/jbc.R700022200. [DOI] [PubMed] [Google Scholar]
- 332.Trimmer JS. Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc Natl Acad Sci U S A. 1991;88:10764–10768. doi: 10.1073/pnas.88.23.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Trimmer JS. Regulation of ion channel expression by cytoplasmic subunits. Curr Opin Neurobiol. 1998;8:370–374. doi: 10.1016/s0959-4388(98)80063-9. [DOI] [PubMed] [Google Scholar]
- 334.Trimmer JS, Cooperman SS, Tomiko SA, Zhou JY, Crean SM, Boyle MB, Kallen RG, Sheng ZH, Barchi RL, Sigworth FJ, et al. Primary structure and functional expression of a mammalian skeletal muscle sodium channel. Neuron. 1989;3:33–49. doi: 10.1016/0896-6273(89)90113-x. [DOI] [PubMed] [Google Scholar]
- 335.Trimmer JS, Rhodes KJ. Localization of voltage-gated ion channels in mammalian brain. Annu Rev Physiol. 2004;66:477–519. doi: 10.1146/annurev.physiol.66.032102.113328. [DOI] [PubMed] [Google Scholar]
- 336.Tseng TT, McMahon AM, Johnson VT, Mangubat EZ, Zahm RJ, Pacold ME, Jakobsson E. Sodium channel auxiliary subunits. J Mol Microbiol Biotechnol. 2007;12:249–262. doi: 10.1159/000099646. [DOI] [PubMed] [Google Scholar]
- 337.Usowicz MM, Sugimori M, Cherksey B, Llinas R. P-type calcium channels in the somata and dendrites of adult cerebellar Purkinje cells. Neuron. 1992;9:1185–1199. doi: 10.1016/0896-6273(92)90076-p. [DOI] [PubMed] [Google Scholar]
- 338.Vabnick I, Trimmer JS, Schwarz TL, Levinson SR, Risal D, Shrager P. Dynamic potassium channel distributions during axonal development prevent aberrant firing patterns. J Neurosci. 1999;19:747–758. doi: 10.1523/JNEUROSCI.19-02-00747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Vacher H, Misonou H, Trimmer JS. Determinants of voltage-gated potassium channel distribution in mammalian neurons. In: Bean AJ, editor. Protein Trafficking in Neurons. Elsevier Academic Press; London: 2006. pp. 244–270. [Google Scholar]
- 340.Vacher H, Trimmer JS. Shaker family Kv1 voltage-gated potassium channels in mammalian brain neurons. In: Gribkoff VK, Kaczmarek LK, editors. Structure, Function and Modulation of Neuronal Voltage Gated Ion Channels. Hoboken, NJ: John Wiley: in press. [Google Scholar]
- 341.Van Hoorick D, Raes A, Keysers W, Mayeur E, Snyders DJ. Differential modulation of Kv4 kinetics by KCHIP1 splice variants. Mol Cell Neurosci. 2003;24:357–366. doi: 10.1016/s1044-7431(03)00174-x. [DOI] [PubMed] [Google Scholar]
- 342.Van Wart A, Trimmer JS, Matthews G. Polarized distribution of ion channels within microdomains of the axon initial segment. J Comp Neurol. 2007;500:339–352. doi: 10.1002/cne.21173. [DOI] [PubMed] [Google Scholar]
- 343.Vega-Saenz de Miera EC, Rudy B, Sugimori M, Llinas R. Molecular characterization of the sodium channel subunits expressed in mammalian cerebellar Purkinje cells. Proc Natl Acad Sci U S A. 1997;94:7059–7064. doi: 10.1073/pnas.94.13.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Veh RW, Lichtinghagen R, Sewing S, Wunder F, Grumbach IM, Pongs O. Immunohistochemical localization of five members of the Kv1 channel subunits: contrasting subcellular locations and neuron-specific co- localizations in rat brain. Eur J Neurosci. 1995;7:2189–2205. doi: 10.1111/j.1460-9568.1995.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 345.Verma-Kurvari S, Border B, Joho RH. Regional and cellular expression patterns of four K+ channel mRNAs in the adult rat brain. Brain Res Mol Brain Res. 1997;46:54–62. doi: 10.1016/s0169-328x(96)00271-9. [DOI] [PubMed] [Google Scholar]
- 346.Wang H, Kunkel DD, Martin TM, Schwartzkroin PA, Tempel BL. Heteromultimeric K+ channels in terminal and juxtaparanodal regions of neurons. Nature. 1993;365:75–79. doi: 10.1038/365075a0. [DOI] [PubMed] [Google Scholar]
- 347.Wang H, Kunkel DD, Schwartzkroin PA, Tempel BL. Localization of Kv1.1 and Kv1.2, two K channel proteins, to synaptic terminals, somata, and dendrites in the mouse brain. J Neurosci. 1994;14:4588–4599. doi: 10.1523/JNEUROSCI.14-08-04588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Wang HG, George MS, Kim J, Wang C, Pitt GS. Ca2+/calmodulin regulates trafficking of Ca(V)1.2 Ca2+ channels in cultured hippocampal neurons. J Neurosci. 2007;27:9086–9093. doi: 10.1523/JNEUROSCI.1720-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wang HS, Pan Z, Shi W, Brown BS, Wymore RS, Cohen IS, Dixon JE, McKinnon D. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science. 1998;282:1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- 350.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 351.Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci U S A. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Weiser M, Bueno E, Sekirnjak C, Martone ME, Baker H, Hillman D, Chen S, Thornhill W, Ellisman M, Rudy B. The potassium channel subunit KV3.1b is localized to somatic and axonal membranes of specific populations of CNS neurons. Journal of Neuroscience. 1995;15:4298–4314. doi: 10.1523/JNEUROSCI.15-06-04298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Weiser M, Vega-Saenz de Miera E, Kentros C, Moreno H, Franzen L, Hillman D, Baker H, Rudy B. Differential expression of Shaw-related K+ channels in the rat central nervous system. Journal of Neuroscience. 1994;14:949–972. doi: 10.1523/JNEUROSCI.14-03-00949.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Weng J, Cao Y, Moss N, Zhou M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281:15194–15200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Westenbroek RE, Ahlijanian MK, Catterall WA. Clustering of L-type Ca2+ channels at the base of major dendrites in hippocampal pyramidal neurons. Nature. 1990;347:281–284. doi: 10.1038/347281a0. [DOI] [PubMed] [Google Scholar]
- 356.Westenbroek RE, Hell JW, Warner C, Dubel SJ, Snutch TP, Catterall WA. Biochemical properties and subcellular distribution of an N-type calcium channel alpha 1 subunit. Neuron. 1992;9:1099–1115. doi: 10.1016/0896-6273(92)90069-p. [DOI] [PubMed] [Google Scholar]
- 357.Westenbroek RE, Merrick DK, Catterall WA. Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron. 1989;3:695–704. doi: 10.1016/0896-6273(89)90238-9. [DOI] [PubMed] [Google Scholar]
- 358.Westenbroek RE, Noebels JL, Catterall WA. Elevated expression of type II Na+ channels in hypomyelinated axons of shiverer mouse brain. J Neurosci. 1992;12:2259–2267. doi: 10.1523/JNEUROSCI.12-06-02259.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Westenbroek RE, Sakurai T, Elliott EM, Hell JW, Starr TV, Snutch TP, Catterall WA. Immunochemical identification and subcellular distribution of the alpha 1A subunits of brain calcium channels. J Neurosci. 1995;15:6403–6418. doi: 10.1523/JNEUROSCI.15-10-06403.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Whitaker WR, Clare JJ, Powell AJ, Chen YH, Faull RL, Emson PC. Distribution of voltage-gated sodium channel alpha-subunit and beta-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol. 2000;422:123–139. doi: 10.1002/(sici)1096-9861(20000619)422:1<123::aid-cne8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 361.Whitaker WR, Faull RL, Waldvogel HJ, Plumpton CJ, Emson PC, Clare JJ. Comparative distribution of voltage-gated sodium channel proteins in human brain. Brain Res Mol Brain Res. 2001;88:37–53. doi: 10.1016/s0169-328x(00)00289-8. [DOI] [PubMed] [Google Scholar]
- 362.Williams ME, Brust PF, Feldman DH, Patthi S, Simerson S, Maroufi A, McCue AF, Velicelebi G, Ellis SB, Harpold MM. Structure and functional expression of an omega-conotoxin-sensitive human N-type calcium channel. Science. 1992;257:389–395. doi: 10.1126/science.1321501. [DOI] [PubMed] [Google Scholar]
- 363.Wimmers S, Bauer CK, Schwarz JR. Biophysical properties of heteromultimeric erg K+ channels. Pflugers Arch. 2002;445:423–430. doi: 10.1007/s00424-002-0936-4. [DOI] [PubMed] [Google Scholar]
- 364.Wimmers S, Wulfsen I, Bauer CK, Schwarz JR. Erg1, erg2 and erg3 K channel subunits are able to form heteromultimers. Pflugers Arch. 2001;441:450–455. doi: 10.1007/s004240000467. [DOI] [PubMed] [Google Scholar]
- 365.Wittemann S, Mark MD, Rettig J, Herlitze S. Synaptic localization and presynaptic function of calcium channel beta 4-subunits in cultured hippocampal neurons. J Biol Chem. 2000;275:37807–37814. doi: 10.1074/jbc.M004653200. [DOI] [PubMed] [Google Scholar]
- 366.Wong W, Newell EW, Jugloff DG, Jones OT, Schlichter LC. Cell surface targeting and clustering interactions between heterologously expressed PSD-95 and the Shal voltage-gated potassium channel, Kv4.2. J Biol Chem. 2002;277:20423–20430. doi: 10.1074/jbc.M109412200. [DOI] [PubMed] [Google Scholar]
- 367.Wu H, Cowing JA, Michaelides M, Wilkie SE, Jeffery G, Jenkins SA, Mester V, Bird AC, Robson AG, Holder GE, Moore AT, Hunt DM, Webster AR. Mutations in the gene KCNV2 encoding a voltage-gated potassium channel subunit cause “cone dystrophy with supernormal rod electroretinogram” in humans. Am J Hum Genet. 2006;79:574–579. doi: 10.1086/507568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Wu L, Nishiyama K, Hollyfield JG, Wang Q. Localization of Nav1.5 sodium channel protein in the mouse brain. Neuroreport. 2002;13:2547–2551. doi: 10.1097/01.wnr.0000052322.62862.a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Xu M, Cao R, Xiao R, Zhu MX, Gu C. The axon-dendrite targeting of Kv3 (Shaw) channels is determined by a targeting motif that associates with the T1 domain and ankyrin G. J Neurosci. 2007;27:14158–14170. doi: 10.1523/JNEUROSCI.3675-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Xu R, Thomas EA, Gazina EV, Richards KL, Quick M, Wallace RH, Harkin LA, Heron SE, Berkovic SF, Scheffer IE, Mulley JC, Petrou S. Generalized epilepsy with febrile seizures plus-associated sodium channel beta1 subunit mutations severely reduce beta subunit-mediated modulation of sodium channel function. Neuroscience. 2007;148:164–174. doi: 10.1016/j.neuroscience.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 371.Yan L, Figueroa DJ, Austin CP, Liu Y, Bugianesi RM, Slaughter RS, Kaczorowski GJ, Kohler MG. Expression of voltage-gated potassium channels in human and rhesus pancreatic islets. Diabetes. 2004;53:597–607. doi: 10.2337/diabetes.53.3.597. [DOI] [PubMed] [Google Scholar]
- 372.Yokoyama CT, Westenbroek RE, Hell JW, Soong TW, Snutch TP, Catterall WA. Biochemical properties and subcellular distribution of the neuronal class E calcium channel alpha 1 subunit. J Neurosci. 1995;15:6419–6432. doi: 10.1523/JNEUROSCI.15-10-06419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Yokoyama S, Imoto K, Kawamura T, Higashida H, Iwabe N, Miyata T, Numa S. Potassium channels from NG108-15 neuroblastoma-glioma hybrid cells. Primary structure and functional expression from cDNAs. FEBS Lett. 1989;259:37–42. doi: 10.1016/0014-5793(89)81488-7. [DOI] [PubMed] [Google Scholar]
- 374.Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 375.Yu FH, Westenbroek RE, Silos-Santiago I, McCormick KA, Lawson D, Ge P, Ferriera H, Lilly J, DiStefano PS, Catterall WA, Scheuer T, Curtis R. Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J Neurosci. 2003;23:7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol Rev. 2005;57:387–395. doi: 10.1124/pr.57.4.13. [DOI] [PubMed] [Google Scholar]
- 377.Yu W, Xu J, Li M. NAB domain is essential for the subunit assembly of both alpha-alpha and alpha-beta complexes of shaker-like potassium channels. Neuron. 1996;16:441–453. doi: 10.1016/s0896-6273(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 378.Yuan LL, Chen X. Diversity of potassium channels in neuronal dendrites. Prog Neurobiol. 2006;78:374–389. doi: 10.1016/j.pneurobio.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 379.Yuan W, Burkhalter A, Nerbonne JM. Functional role of the fast transient outward K+ current IA in pyramidal neurons in (rat) primary visual cortex. J Neurosci. 2005;25:9185–9194. doi: 10.1523/JNEUROSCI.2858-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Yunker AM, Sharp AH, Sundarraj S, Ranganathan V, Copeland TD, McEnery MW. Immunological characterization of T-type voltage-dependent calcium channel Ca(V)3.1 (alpha1G) and Ca(V)3.3 (alpha1I) isoforms reveal differences in their localization, expression, and neural development. Neuroscience. 2003;117:321–335. doi: 10.1016/s0306-4522(02)00936-3. [DOI] [PubMed] [Google Scholar]
- 381.Yus-Najera E, Munoz A, Salvador N, Jensen BS, Rasmussen HB, Defelipe J, Villarroel A. Localization of KCNQ5 in the normal and epileptic human temporal neocortex and hippocampal formation. Neuroscience. 2003;120:353–364. doi: 10.1016/s0306-4522(03)00321-x. [DOI] [PubMed] [Google Scholar]
- 382.Zagha E, Ozaita A, Chang SY, Nadal MS, Lin U, Saganich MJ, McCormack T, Akinsanya KO, Qi SY, Rudy B. DPP10 modulates Kv4-mediated A-type potassium channels. J Biol Chem. 2005;280:18853–18861. doi: 10.1074/jbc.M410613200. [DOI] [PubMed] [Google Scholar]
- 383.Zhang H, Fu Y, Altier C, Platzer J, Surmeier DJ, Bezprozvanny I. Ca1.2 and CaV1.3 neuronal L-type calcium channels: differential targeting and signaling to pCREB. Eur J Neurosci. 2006;23:2297–2310. doi: 10.1111/j.1460-9568.2006.04734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zhang H, Maximov A, Fu Y, Xu F, Tang TS, Tkatch T, Surmeier DJ, Bezprozvanny I. Association of Cav1.3 L-type calcium channels with Shank. J Neurosci. 2005;25:1037–1049. doi: 10.1523/JNEUROSCI.4554-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Zhao P, Waxman SG, Hains BC. Sodium channel expression in the ventral posterolateral nucleus of the thalamus after peripheral nerve injury. Mol Pain. 2006;2:27. doi: 10.1186/1744-8069-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–1304. doi: 10.1083/jcb.143.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Zhu XR, Netzer R, Bohlke K, Liu Q, Pongs O. Structural and functional characterization of Kv6.2 a new gamma-subunit of voltage-gated potassium channel. Receptors Channels. 1999;6:337–350. [PubMed] [Google Scholar]
- 388.Zito K, Fetter RD, Goodman CS, Isacoff EY. Synaptic clustering of Fascilin II and Shaker: essential targeting sequences and role of Dlg. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]
- 389.Zou A, Lin Z, Humble M, Creech CD, Wagoner PK, Krafte D, Jegla TJ, Wickenden AD. Distribution and functional properties of human KCNH8 (Elk1) potassium channels. Am J Physiol Cell Physiol. 2003;285:C1356–1366. doi: 10.1152/ajpcell.00179.2003. [DOI] [PubMed] [Google Scholar]