Abstract
Precise axon growth is required for making proper connections in development and after injury. One method of studying axon guidance and growth is through in vitro outgrowth assays that present controlled microenvironments. In this study, we applied circular statistical methods to evaluate directional neurite response. Visualization of data on a circular scale allows more accurate representation of the data, as neurite angles are inherently expressed on a circle. Here, the direction of neurite outgrowth from dorsal root ganglia derived neurons on different substrate types was quantitatively measured. Further, simulations of datasets with known circular parameters reflecting expected neurite angle distributions from different substrate types were also generated. Circular statistical methods were utilized and compared to linear statistical models widely used in the neuroscience literature. For small samples, Rao’s spacing test showed the smallest occurrence of Type I errors (false positives) when tested against simulated uniform distributions. V-test and Rayleigh’s test showed highest statistical power when tested against a unimodal distribution with known and unknown mean direction, respectively. For bimodal samples, Watson’s U2 test showed the highest statistical power. Overall, circular statistical uniformity tests showed higher statistical power than linear non-parametric tests, particularly for small samples (n=5). Circular analysis methods represent a useful tool for evaluation of directionality of neurite outgrowth with applications including: (1) assessment of neurite outgrowth potential; (2) determination of isotropy of cellular responses to single and multiple cues and (3) determination of the relative strengths of cues present in a complex environment.
Keywords: Axon guidance, Neurite alignment, Directional data, Simulation
Introduction
Axon guidance during development and after injury has been studied in traditional cell culture and in increasingly complex in vitro environments generated with tissue engineering and other biomedical engineering techniques. One approach has been to manipulate the cells’ local microenvironment and observe neurite outgrowth in microenvironments containing cues of interest. Studies of axon guidance often use in vitro neurite outgrowth assays (Ronn et al., 2000; Smit et al., 2003; Thompson and Buettner, 2006; Weaver et al., 2003) as models to elucidate the growth potential of neurons, the effects of the environment, and the mechanisms underlying the axon growth process.
Quantitative assessment of neurite outgrowth in these assays represents a critical step in gaining specific information on axon growth. Quantitative morphometric analyses depend heavily on microscopy techniques (Meijering et al., 2004; Mitchell et al., 2007) and automated (Karlon et al., 1998; Price et al., 2006; Weaver et al., 2003) or semi-automated (Bilsland et al., 1999; Hynds and Snow, 2002; Thompson and Buettner, 2006) image analysis tools which allow researchers to accurately assess neuronal and neurite growth. Parameters that provide information on neuronal response may include the area of the neuron or neurite (Abosch and Lagenaur, 1993), number of neurites (Abosch and Lagenaur, 1993; Le Roux and Reh, 1994), neurite orientation, neurite length (Abosch and Lagenaur, 1993) and path of migration. One widely used measure for the strength of a guidance cue is the direction of neurite outgrowth following some underlying directional stimulus (Alexander et al., 2006; Bruder et al., 2007; Deumens et al., 2004; Mahoney et al., 2005; Thompson and Buettner, 2006).
The geometry of neurite outgrowth is most meaningfully parameterized in a circular coordinate system centered on the cell and rotationally aligned to the stimulus applied. The distribution of neurite angles in culture can be described by circular statistical parameters, such as mean direction and length of the mean vector, in an analogous manner to linear statistical parameters mean and variance. For both linear and circular parameters, the mean refers to the expected value of a random variable. Length of mean vector and variance are both measures of the spread of the data, where the variance represents the average squared deviation from the mean, and length of mean vector is an inverse analogue of the variance. Circular variables have values that fall along a circle and hence have specific properties related to the cyclic nature of the circular scale. The application of these methods to neurite direction is analogous to the application of population biology measures to cellular function.
Statistical analysis of circular variables differs from analysis of linear variables as there are several properties of circular variables that need to be taken into account. Because circular variables are finite and closed when a circular data set comes back on itself (at 0° and 360°), the zero direction, the designation of magnitude, and the number and size of groups (in the case of grouped data) are arbitrary. In addition, the mean angle of orientation cannot be found by the simple summation of measured values and division by the sample size. The sums of circular variables must be taken either modulo 360° if the sample is circular, or taken modulo 180° if the sample is axial, i.e. where data occur as an undirected line as in the example of geological fractures (Tran, 2007). For axial data in the present study, there is symmetry about the y-axis hence there is no distinction between the north-south directions. Analysis for linear variables approximates randomness by using a Poisson distribution; this distribution does not translate to circular variables. In circular statistics, the null hypothesis describing a random distribution is taken to be a uniform distribution, where all directions may occur at equal probability, approximating randomness and reflecting the finite closure of a circle (Fisher, 1993).
Neurite outgrowth angles are generally simple distributions, requiring display of data and summary of a single random sample usually with single or bimodal groups. As such, a null hypothesis of uniformity and randomness is generally appropriate, with the objective to assess the uniformity of a given distribution of neurite angles cultured in different environmental conditions. When the comparison of two or more samples of neurites cultured in different conditions is of interest, regression analysis and statistical models may be useful for description and prediction of cell response. Circular statistical methods complement traditional linear statistical methods to describe and draw inferences about the population of neurons and neurites being studied (Batschelet, 1981; Fisher, 1993). We propose in this study that in many cases, circular statistical methods allow us to more robustly describe the complexity of neurite outgrowth phenomena.
In this work, we employed circular statistical models to evaluate directional growth in a variety of representative in vitro neurite outgrowth assays. Multiple statistical methods were used to evaluate in vitro neurite outgrowth ranging from Gaussian based models and nonparametric methods to hypothesis testing for circular samples. Here we report a comparison of circular and linear data presentation and statistical methods for evaluation of several types of neurite outgrowth patterns.
Methods
2.1 Substrate preparation
Three types of substrates were used to evaluate the use of circular statistical methods on directionality of neurite outgrowth: adsorbed uniform protein coating on glass, adsorbed protein stripes and adsorbed protein gradients. Uniform protein coating was performed by incubating protein solution for one hour on acid washed glass coverslips, washing twice with sterile water and air drying.
Micropatterned laminin (LN, 50µg/mL) and chondroitin sulfate proteoglycans (CSPG, 10µg/mL) stripes of 10 mm length, 50 µm width and 50 µm pitch were stamped onto glass coverslips via micro-contact printing techniques as described in Bruder et al. (Bruder et al., 2006). Briefly, grooved polydimethyl siloxane (PDMS) stamps fabricated using the method described in Goldner et al. (Goldner et al., 2006), were submerged in 10% sodium dodecyl sulfate in deionized water, rinsed in water, and incubated with 50µg/mL mouse LN in Hank’s balanced salt solution without calcium or magnesium (HBSS-CMF) for 1 h. Glass coverslips were plasma activated with a plasma cleaner/sterilizer (PDC-32 G, Medium RF level, 1min, in air), and incubated in contact with stamps overnight to achieve adsorbed alternating stripes of either LN or CSPG.
Protein gradients were generated with the use of a microfluidic gradient mixer, fabricated using soft lithography techniques in a modification of the method of Dertinger et al. (Dertinger et al., 2002), described in Li et al. (Li et al., 2008). Briefly, the gradient mixer pattern was designed in AutoCAD and transferred to a silicon wafer using photolithography. Using the silicon wafer as a template and PDMS as an elastomeric replica, soft lithography was used to fabricate the gradient mixer. The polymeric gradient mixer and a glass slide were irreversibly bonded by plasma activation of both surfaces for 1 min. Single cue gradients of LN or CSPG opposite bovine serum albumin (BSA, 3%, a neutral molecule for neurite guidance), were generated as substrates to evaluate neurite directionality. Protein solutions of LN or CSPG and BSA were pumped through the gradient mixer, at 0.2µL/min and allowed to interdiffuse and adsorb overnight. The glass substrates containing the adsorbed protein gradients were used as the substrates on which to culture dorsal root ganglia (DRG) neurons.
2.2 Cell culture
DRG were dissected from the spinal columns of postnatal (P0-P4) rat pups and cleaned of axons, blood, and connective tissue. DRG were incubated in 0.05% trypsin-EDTA in HBSS-CMF at 37°C for 60 min and dissociated by trituration. Cells were plated onto substrates in Dulbecco’s modified eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 4 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin with 50 ng/ml nerve growth factor (NGF). Cells were seeded at a density of 100,000 cells/mL, on uniformly coated glass or micropatterned substrates and 12,500 cells/mL on gradient substrates. Phase contrast microscopy using a 10X objective was performed using a Nikon Eclipse TE2000-S, and images were captured with Hamamatsu-ORCA outputting to Openlab v.4.05 after 24 hours in culture.
2.3 Image analysis
To evaluate direction of neurite outgrowth on uniform substrates and micropatterned protein stripes, the angles of all neurites in at least 6 fields of view were measured as the angle between the vector from the cell body to the tip of the neurite and the vertical axis (0°, Fig. 1d), using the measure tool in OpenLab software on phase contrast images. To evaluate directional bias of neurites on gradient substrates, the angles (θ) of the longest neurites of all neurons adhered to the gradient channel were measured as described above.
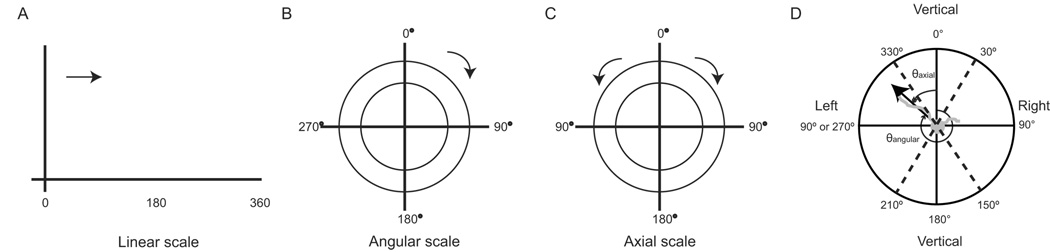
Figure 1. Presentation and measurement of neurite angles.
(a) Linear data fall in a straight line with values increasing along the axes, scale ranges between 0–360°. (b) Angular data is shown on a circular scale where 0–360° wraps around on a circle. Scale used in this study for neurite angles on gradient substrates. (c) Axial data is shown on a circular scale with an axis of symmetry about the y-axis such that the scale extends from 0–180°. Scale used in this study for neurite angles on striped substrates. Note that for circular histograms, concentric circles about the axes denote frequency of data. (d) Schematic shows groupings of neurite angles into categories “left”, “vertical” and “right” performed prior to plotting grouped bar graphs and performing statistical tests for categorical data. Angles are θangular on gradient substrates, θaxial on striped substrates.
2.4 Linear statistical analysis
Linear descriptive statistics such as mean and standard deviation were calculated by equations described in Table I. Conventional statistical tests were performed using SPSS 14. Linear probability density functions were tested against circular data as comparison (SPSS 14; listed in Table II). For the χ2 test, the angle data was grouped into three groups: neurites growing towards the left (210°–330°), right (30°–150°) and vertical (0°–30°, 150°–210°, 330°–360°). For the KS test, the angles of neurite outgrowth were grouped into 10° bins.
Table I. Equations of calculations of mean and standard deviation to determine preferred direction and spread of data.
Where θi = angular data for the ith observation as i= 1,…, n and n = number of observations, S= x-component of mean vector, C = y-component of mean vector. Variance calculations performed by Oriana, according to Zar (Zar, 1999).
| Statistic | Equation |
|---|---|
| CIRCULAR | |
| Vector components | |
| Mean direction(µc) | |
| Length of mean vector(R) | |
| Variance(Varc) | Varc = −2 ln R |
| Standard deviation(σc) | |
| LINEAR | |
| Mean direction(µ) | |
| Variance(Var) | |
| Standard deviation(σ) | |
Table II. Equations of test statistical parameters used in circular and linear tests.
Where θi = angular data for the ith observation as i= 1,…, n and n = number of observations. Tables providing critical values of each test statistic are available from Batschelet (Batschelet, 1981) or from circular statistical software programs such as Oriana. Refer to Table I for additional definitions of variables.
| Test | Data type | Null hypothesis | Alternate hypothesis | Formula for test statistic | Notes |
|---|---|---|---|---|---|
| CIRCULAR ONE-SAMPLE TESTS | |||||
| Rao’s Spacing | Continuous | Uniform | Any alternative | θ1 ≤ θ2 ≤ … ≤ θn | n ≥ 5 |
| Tn−1 = θn − θn−1 | where Ti is the arclength between consecutive angles | ||||
| Tn = 360 + θ1 − θn | |||||
| Kuiper’s | Continuous | Uniform | Any alternative | Vn = D+ + D− | where D is the deviation from (uniform) theoretical distribution |
| for small n, more powerful than χ2 test | |||||
| circular goodness-of-fit test corresponding to Kolmogorov-Smirnov | |||||
| Rayleigh’s | Continuous | Uniform | Unimodal | unimodal von Mises alternative | |
| Watson’s U2 | Continuous | Uniform | Unimodal | θ1 ≤ θ2 ≤ … ≤ θn | where F(θ) is the function of (uniform) theoretical distribution |
| suitable for unimodal and multimodal distributions | |||||
| V-test | Continuous | Uniform | Unimodal | external direction (µ0) specified a priori | |
| CIRCULAR MULTISAMPLE TESTS | |||||
| Mardia-Watson-Wheeler | Continuous | Two samples have identical distributions | Two samples have different distributions | data is ungrouped | |
| Watson’s U2 | Continuous | Two samples have same distributions | Two samples have different distributions | where N = n1 + n2 | |
| no ties between samples | |||||
| LINEAR TESTS | |||||
| χ2 | Grouped | Uniform | Non-uniform | where ei = expected value for ith term | |
| based on approximation, need large n | |||||
| suitable for both unimodal and multimodal distributions | |||||
| Kolmogorov-Smirnov | Grouped | Uniform | Non-uniform | Dmax = max(|di|) | where F̄i is the function of (uniform) theoretical distribution |
| where |di| = |Fi − F̄i| | more powerful than χ2 when n is small | ||||
| dependent on starting point, more sensitive near median than at tails | |||||
| Student’s t-test | Continuous | Two samples have same means | Two samples have different means | where x̄ is the sample mean and s is the sample standard deviation | |
| where | assuming equal variances | ||||
2.5 Circular data presentation
Circular data was plotted as a frequency distribution with neurites binned in ten degree bins, and “wrapped” around a circle. Circular histograms were plotted using circular statistical software package Oriana v2.02.
2.6 Circular Statistics
A common aim for analysis of directional data is to estimate the preferred direction and distribution of data. To describe circular distributions, measures have been developed such as circular mean angle, the length of mean vector (R) and particularly for the case of the von Mises (VM) distribution, commonly thought of as the circular analogue to the normal distribution, the concentration parameter (κ) (Batschelet, 1981; Fisher, 1993). κ̣ is a shape parameter (κ≥0) measuring the tendency of the data to cluster around the mean direction (µc). As κ approaches 0, the distribution converges to a uniform distribution, and as κ approaches infinity, the distribution tends to concentrate around the direction µc. These parameters take into account the periodicity in angular data by using trigonometric functions, and the equations describing the calculations are listed in Table I.
Six one-sample goodness-of-fit tests were compared in this paper to test against the null hypothesis of uniformity of neurite angle distributions: Rao’s spacing test, Kuiper’s test, Rayleigh’s test, Watson’s U2 test, chi-squared (χ2) test, and V-test, a modification of Rayleigh’s test. The χ2 goodness of fit tests are not strictly circular, but are generally accepted to be appropriate for circular variables under certain conditions (listed in Table 1, (Batschelet, 1981; Fisher, 1993; Zar, 1996)). Equations for each circular goodness-of-fit test are listed in Table II. Each type of test was developed for different data distributions. These tests differ in their alternate hypotheses, where Rao’s and Kuiper’s test for randomness in the sample against any alternative, and Rayleigh’s and Watson’s test against a unimodal alternative. The V-test tests against a specified mean direction and was only performed for alignment studies and computer simulations as these were the only cases in which an external direction was applied.
Two multisample tests were performed in this study, to compare two datasets and determine whether their distributions are different: Mardia-Watson-Wheeler test and Watson’s U2 test. Equations for each multisample test are listed in Table II. Both tests determine whether the two samples differ significantly from each other in mean angle, angular variance or both measures. Neurite angles on LN and CSPG striped substrates, LN striped and gradient substrates, and LN and CSPG gradient substrates were compared against each other to test if neurite outgrowth directions differed significantly on these substrates. Mardia-Watson-Wheeler compares the resultant vector lengths (R), and Watson’s U2 test compares the deviation between the cumulative density functions of the two populations (Batschelet, 1981). All circular data analysis was performed using Oriana v2.02c. Student t-test was performed on the same datasets to compare circular methods to linear methods of comparing means between two samples.
2.7 Simulations of circular distributions
Simulations approximating various circular distributions were performed using a custom MATLAB program that generates circular random numbers from a specified distribution with input parameters. Algorithms for simulation of data types (uniform, VM, and bimodal (BM) distributions with corresponding probability density functions described in Table III) were taken from Fisher (Fisher, 1993). In each simulation, random numbers were generated (n=5, 30, 100), input parameters included mean (µc=0°), and for VM and BM simulations, concentration (κ=0.85, 1, 1.5, 3). Simulations of each distribution were run 100 times. Values for κ were chosen to generate distributions with varying dispersions (angular variances between 20–70°). Simulated uniform data were generated by transforming linear random numbers in the range of 0 to 1 into degrees by using the RAND function in MATLAB modulo 360. Random numbers from VM and BM distributions were generated according to Fisher (Fisher, 1993). BM distributions consisted of data drawn from two subsets of proportions p and (1-p), with parameters (µc1, κ1) and (µc2, κ2) corresponding to each subset. In this case, bimodal distributions were simulated as an equal mixture of two subpopulations with VM distribution (p1=p2=0.5, µc1=0°, µc2=90°, κ1=κ2=0.85,1, 1,5, 3).
Table III. Equations of probability density functions and parameters of statistical models used in simulation of neurite outgrowth.
| Theoretical distribution | Probability density function | n | Mean(µ)(°) | Concentration(κ) | Dispersion(°) |
|---|---|---|---|---|---|
| Uniform, U | 5,30,100 | 0 | 0 | ||
| von Mises, VM(µ, κ) | f(θ) = [2πI0(κ)]−1 exp[κ cos (θ − µ)], 0 ≤ θ ≤ ∞ | 5,30,100 | 0 | 0.85,1,1.5,3 | 180,120,60,20 |
| where | |||||
| Bimodal, VM(µ1, κ1), VM(µ2, κ2) | f(θ) = [2πI0(κ)]−1 exp[κ cos 2(θ − µ)], 0 ≤ θ ≤ ∞ | 5,30,100 | 0 | 0.85,1,1.5,3 | 180,120,60,20 |
Two linear and four circular goodness-of-fit tests were performed on each experimental condition similar to the experimental data described in section 2.5 to assess the probability density function that would best describe the data. The percentage of significant simulations (p<0.05) was determined for each experimental condition and Type I error was determined for each experimental condition in the simulation (Table VII).
Table VII. Comparison of circular and linear statistical tests for simulated data of known distributions.
Numbers reported here indicate the number of times out of 100 that the null hypothesis of uniformity was rejected as each simulated condition was run 100 times. Each one-sample uniformity test listed in Table II was run for each simulation. Ha indicates the alternate hypothesis used.
| Statistic Distribution Condition (µ,κ) | n | Type I error Uniform | Power VM (0,0.85) | Power VM (0,1) | Power VM (0,1.5) | Power VM (0,3) | Power BM(0, 90, 0.85, 0.85) | Power BM(0, 90, 1, 1) | Power BM(0, 90, 1.5, 1.5) | Power BM(0, 90, 3, 3) |
|---|---|---|---|---|---|---|---|---|---|---|
| CIRCULAR | ||||||||||
| Rao's Spacing | 5 | 8 | 15 | 19 | 36 | 65 | 13 | 22 | 41 | 65 |
| 30 | 6 | 39 | 45 | 83 | 100 | 6 | 14 | 17 | 85 | |
| 100 | 4 | 71 | 92 | 38 | 100 | 16 | 15 | 55 | 100 | |
| Kuiper's | 5 | 10 | 16 | 23 | 38 | 70 | 15 | 22 | 43 | 67 |
| 30 | 3 | 80 | 92 | 100 | 100 | 20 | 32 | 56 | 98 | |
| 100 | 3 | 100 | 100 | 100 | 100 | 59 | 82 | 97 | 100 | |
| Rayleigh | 5 | 10 | 17 | 24 | 34 | 75 | 13 | 22 | 46 | 79 |
| 30 | 3 | 85 | 92 | 100 | 100 | 21 | 29 | 56 | 98 | |
| 100 | 6 | 100 | 100 | 100 | 100 | 69 | 87 | 97 | 100 | |
| Watson's U2 | 5 | |||||||||
| 30 | 4 | 86 | 93 | 100 | 100 | 19 | 31 | 55 | 100 | |
| 100 | 6 | 100 | 100 | 100 | 100 | 67 | 88 | 98 | 100 | |
| V-test | 5 | 10 | 34 | 57 | 63 | 94 | 12 | 17 | 35 | 49 |
| 30 | 3 | 95 | 97 | 100 | 100 | 33 | 54 | 76 | 100 | |
| 100 | 4 | 100 | 100 | 100 | 100 | 85 | 93 | 98 | 100 | |
| LINEAR | ||||||||||
| Kolmogorov-Smirnov | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ha=normal | 30 | 0 | 6 | 4 | 33 | 93 | 0 | 0 | 1 | 0 |
| 100 | 2 | 99 | 99 | 100 | 100 | 7 | 0 | 47 | 97 | |
| Kolmogorov-Smirnov | 5 | 4 | 12 | 6 | 13 | 21 | 0 | 5 | 2 | 9 |
| Ha=uniform | 30 | 1 | 38 | 59 | 86 | 100 | 0 | 28 | 49 | 94 |
| 100 | 5 | 97 | 98 | 10 | 100 | 59 | 76 | 90 | 100 | |
Results
Here we describe the evaluation of directional neurite outgrowth using linear and circular statistics of two types of data: experimental data with unknown population parameters and simulated data with known (user defined) population parameters.
3.1 Experimental Results for Live Neurons
Experimental data of three different types of DRG neurite behavior were elicited from three differently micropatterned substrates and simulations of established uniform, unimodal and bimodal datasets were performed. Experimental neurite angle data were presented in circular histograms customary for circular data, and also in more conventional ways of presenting alignment, by bar graphs of neurites categorized in aligned and unaligned groups.
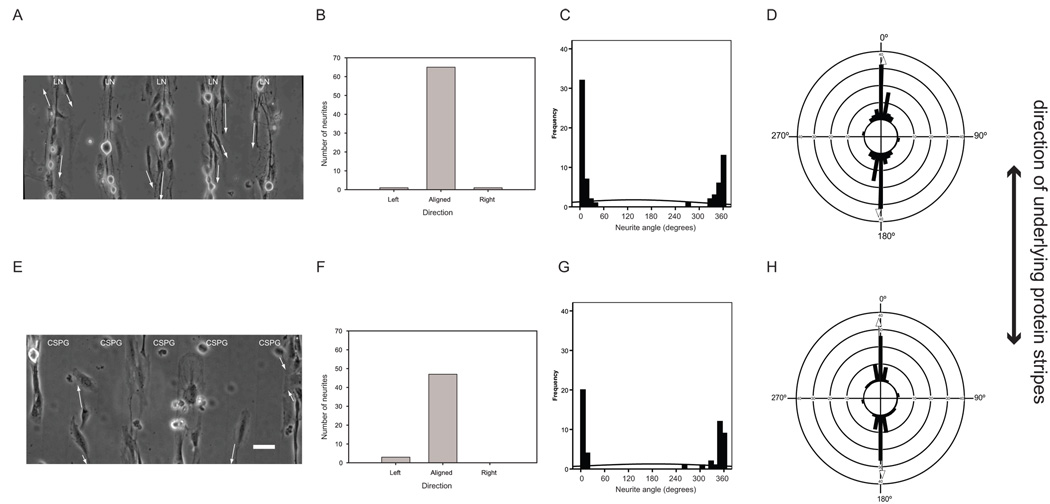
Phase contrast images of neurons on uniformly coated LN (Fig. 2a) showed neurite outgrowth in all directions. Mean neurite direction was not presented since as discussed previously, mean direction of a uniform distribution has no physical relevance in describing the data as the magnitude of the vector equals zero. All tests (circular and linear) showed no significant difference between the data and a uniform distribution (p>0.05; Table V). Graphically, we compared several ways of presenting the data: a grouped bar graph poorly reflected the uniformity of the dataset, where more neurites appeared to have grown in the “left” direction than in all others (Fig. 2b). A linear histogram of neurite data appeared to show four modal groups (0–80°, 90–170°, 190–290°, 300–360°) and revealed the poor fit of a normal distribution (Fig. 2c). Data plotted on a circular axis reflected the variability of the angular data and most clearly showed that angular data fell in even spacing around a circular scale (Fig. 2d).
Figure 2. Distribution of neurites after 24 hours in culture on uniformly coated LN substrates shows uniformity in neurite outgrowth angles.
(a) Phase contrast image of DRG neurons on uniform LN coated glass surface. Bar = 50µm. Arrows show the vectors that were used to evaluate the neurites. (b) Bar graph shows the corresponding grouped neurite outgrowth angle data. (c) Linear histogram shows corresponding distribution of neurite angles where each angle presented is the angle of the longest neurite per neuron. Normal curve is fitted to the linear histogram around the linear sample mean and standard deviation. (d) Circular histogram shows the corresponding distributions of neurite angles where each angle presented is the angle of the longest neurite per neuron.
Table V. Comparison of circular and linear goodness-of-fit statistical tests for all experimental conditions tested.
One sample uniformity tests developed for circular variables were performed for all experimental samples, including Rayleigh’s, Rao’s Spacing, Watson’s U2 and Kuiper’s test. Linear non-parametric one sample tests were also performed for all experimental samples including chi-squared (performed on data grouped in 3 bins) and KS tests (performed on ungrouped data). Significance levels taken to be p<0.05. Ha indicates the alternate hypothesis used.
| LN Uniform | LN Stripe | CSPG Stripe | LN Gradient | CSPG Gradient | |
|---|---|---|---|---|---|
| CIRCULAR | |||||
| Rao's Spacing | 0.50 > p > 0.10 | < 0.01 | < 0.01 | < 0.01 | < 0.05 |
| Kuiper's | > 0.15 | < 0.01 | < 0.01 | < 0.01 | < 0.01 |
| Rayleigh's | 0.386 | < 1E-12 | < 1E-12 | 8.18E-05 | 0.032 |
| Watson's U2 | 0.5 > p > 0.25 | < 0.005 | < 0.005 | < 0.005 | < 0.01 |
| LINEAR | |||||
| Chi squared ( 36 groups) | 0.317 | < 1E-12 | 1 | 0.04 | 0.089 |
| Chi squared (3 groups) | 0.097 | 0 | 0 | 0 | 0.006 |
| Kolmogorov-Smirnov (Ha=uniform) | 0.185 | 0 | 0 | 0 | 0.122 |
When permissive LN and inhibitory CSPG were presented on substrates, DRG neurons adhered to LN coated regions and avoided CSPG coated regions. DRG neurites extended and aligned to LN stripes (Fig. 3a) and to uncoated regions between CSPG stripes (Fig. 3e). Qualitative and quantitative analysis of the data showed a larger population and density of neurons and aligned neurites on LN striped substrates, as compared to CSPG striped substrates, but the mean neurite outgrowth direction (the parameter of interest) was similar for DRG neurons on both LN and CSPG striped substrates. Grouped bar graphs, with groups defined as left: 210–330°, right: 30–150°, and aligned: 330-30° and 150–210°) showed that data clustered around the vertical direction, which was set to correspond to alignment to the underlying pattern (Fig. 3b, f). Linear histograms split the data clusters, failed to recognize the relationship between 0–10° and 350–360°, and revealed a poor fit to a normal distribution around the linear mean of 180° (Fig. 3c, g). Data plotted on circular axes showed that the majority of the data fell within a 30° interval around the vertical 0° direction (Fig. 3d, h). This clustering around the vertical axis (0–180° axis) was reflected in all circular mean angles (µc=1.90° on LN and µc=178.61° on CSPG substrates). The linear mean angle of neurites on LN stripes showed the largest discrepancy from the graphed data, (µ=135.01°), most likely due to the high frequency of angles in the 0–10° bin (Figure 3c).
Figure 3. Distribution of neurites after 24 hours in culture on micropatterned LN or CSPG stripes shows clustered and directed neurite outgrowth angles.
Phase contrast images of DRG neurons on micropatterned LN (a) and CSPG (e) stripes respectively on glass surface. Bar = 50µm. Arrows show the vectors that were used to evaluate the neurites. (b, f) Bar graphs show the corresponding grouped neurite outgrowth angle data. (c, g) Linear histograms show corresponding distributions of neurite angles where each angle presented is the angle of the longest neurite per neuron. Normal curve is fitted to the linear histogram around the linear sample mean and standard deviation. Circular histograms show the corresponding distributions of neurite angles on LN (d) and CSPG (h) striped substrates, where each angle presented is the angle of the longest neurite per neuron. White arrows indicate mean neurite angles for directed distributions.
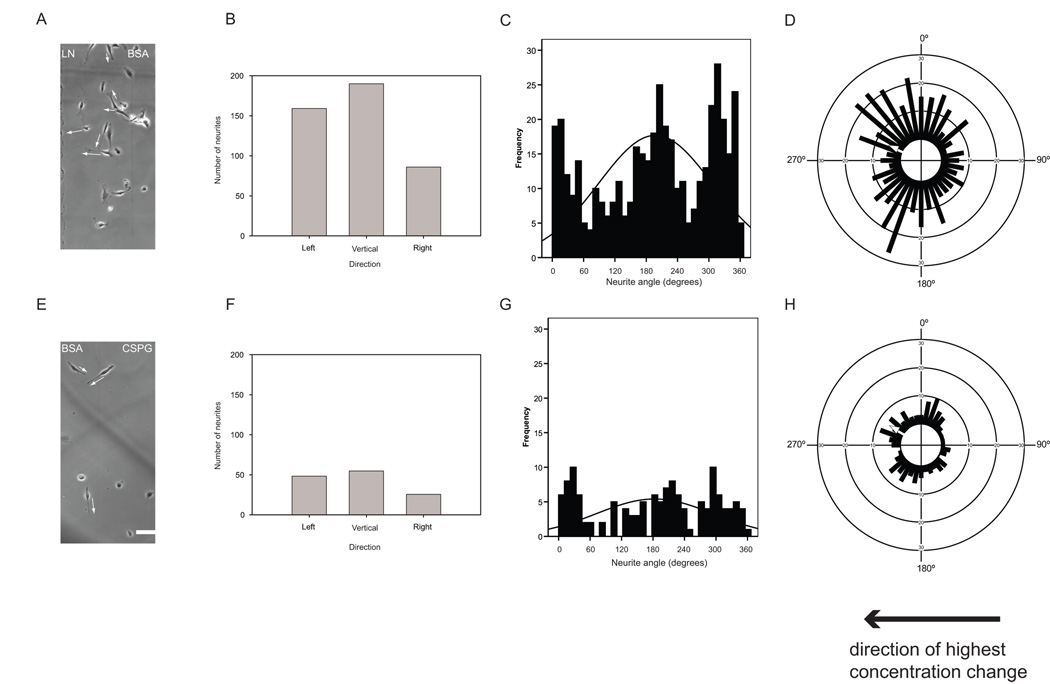
Analysis of neurite growth on gradient substrates illustrated how circular statistics can be used to analyze neurite outgrowth patterns that are more complex. Phase contrast images of DRGs plated on a LN gradient (Fig. 4a) and a CSPG gradient (Fig. 4e) showed an overall trend of neurite growth toward the permissive LN and away from the inhibitory CSPG, but the neurite response to the underlying substrate was not nearly as obvious as that of DRGs cultured on protein stripes in Fig. 3. Each method used to present the data graphically highlighted different features of the data. Grouped bar graphs (Figure 5b, f) showed the bias towards the “left” edge of the channel, which was the more permissive or the less inhibitory direction. Linear histograms appeared to show three modal groups (0–70°, 70–250°, 250–360°) for neurite growth on LN gradients (Fig. 4c). For DRG neurites on a CSPG gradient however, the neurite angle distribution was flattened and appeared more uniform (Fig 5g). Data plotted on circular axes showed a similar trend to the grouped bar graphs, where growth appeared to be towards the more permissive or less inhibitory regions (Figs. 5d, h). Similar to DRG response on protein stripes, the permissiveness of the substrate is reflected by the number of neurons and neurites present, whereas the directional guidance potential of the substrate is reflected by the angles at which the neurites extend. Circular uniformity tests determined that the neurite angles were directed on LN and on CSPG gradients (p < 0.05; Table V). The circular standard deviations of neurite angles were larger for neurites on gradients than on stripes, reflecting the complexity of the responses (Table IV).
Figure 4. Distribution of neurites after 24 hours in culture on micropatterned LN or CSPG gradients shows dispersed but directed neurite outgrowth angles.
Phase contrast images of DRG neurons on micropatterned LN (a) and CSPG (e) gradients respectively on glass surface. Bar = 50µm. Arrows show the vectors that were used to evaluate the neurites. (b, f) Bar graphs show the corresponding grouped neurite outgrowth angle data. (c, g) Linear histograms show corresponding distributions of neurite angles where each angle presented is the angle of the longest neurite per neuron. Normal curve is fitted to the linear histogram around the linear sample mean and standard deviation. Circular histograms show the corresponding distributions of neurite angles on LN (d) and CSPG (h) gradient substrates, where each angle presented is the angle of the longest neurite per neuron. White arrows indicate mean neurite angles for directed distributions.
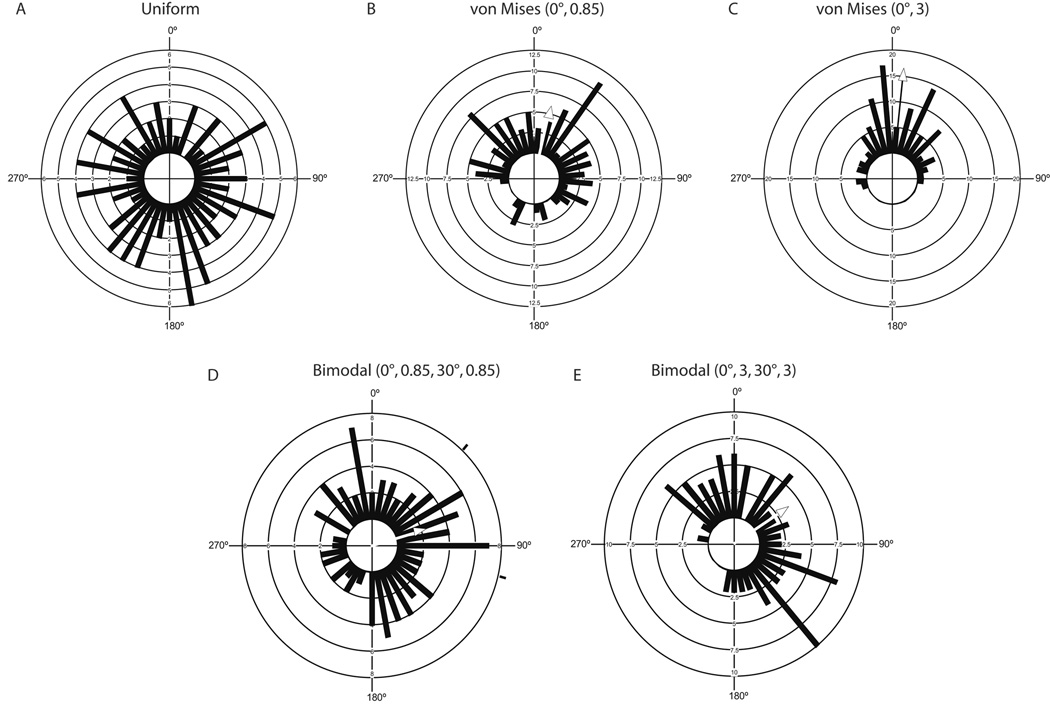
Figure 5. Representative circular histograms of simulated data generated by MATLAB algorithm.
Circular histograms of representative runs with n=100 angles generated from a (a) uniform circular distribution, (b) dispersed VM distribution (κ=0.85) with a mean of 0°, (c) tight VM distribution (κ =3) with a mean of 0°, (d) dispersed BM distribution (κ =0.85) with means 0° and 30° and (e) tight BM distribution (κ =3) with means 0° and 30°.
Table IV. Comparison of circular and linear descriptive statistics for all experimental conditions tested.
| LN Uniform | LN Stripe | CSPG Stripe | LN Gradient | CSPG Gradient | |
|---|---|---|---|---|---|
| n | 81 | 67 | 50 | 435 | 136 |
| CIRCULAR | |||||
| Mean angle (°) | n/a | 1.90 | 178.61 | 293.78 | 300.90 |
| Length of mean vector | n/a | 0.90 | 0.89 | 0.15 | 0.16 |
| Circular Standard Deviation (°) | 120.78 | 13.363 | 14.038 | 112.18 | 109.873 |
| Deviation from vertical axis | n/a | 1.9 | 1.39 | 66.22 | 59.1 |
| Corresponding figure | 3d | 4d | 4h | 5d | 5h |
| LINEAR | |||||
| Mean (°) | 188.55 | 135.01 | 182.93 | 197.58 | 183.09 |
| Standard deviation (°) | 105.43 | 166.43 | 173.01 | 109.04 | 111.47 |
| Deviation from vertical axis | 8.55 | 44.99 | 2.93 | 17.58 | 3.09 |
| Corresponding figure | 3c | 4c | 4g | 5c | 5g |
To compare the different available statistical tests for uniformity, varied protein micropatterns were presented in culture (uniform, striped, gradient) to direct neurite outgrowth towards different directions and yield corresponding different distributions of neurite angles. Comparison of mean neurite angles and deviations from the vertical axis calculated both with linear and with circular methods revealed differences in the abilities of linear and circular approaches to accurately reflect the complex distributions of neurites. The large linear standard deviations of neurite angles under all experimental conditions reflected how linear methods fail to account for data clustering around 0°. Deviation from the vertical axis 0° or 180°, on a circular or angular scale, (Fig. 1b,c; Table IV), showed that the circular mean angle avoided the convergence to 180°, providing a better estimate of mean angle.
For samples presenting stripes aligned to the vertical axis, perfect guidance would result in a µc of 0° or 180°. Linear analysis of the experimental data yielded means of 135.01° and 182.93° for LN and CSPG respectively, while circular statistical methods yielded means of 1.90° and 178.61° for striped substrates of LN and CSPG, respectively. For samples presenting concentration gradients, the direction of highest change in concentration is 270°. A larger dispersion would be expected as compared to the dispersion for neurite outgrowth on striped substrates. On gradient substrates, circular mean angles showed directed growth toward substrate regions that were more permissive and less inhibitory (293.89° on LN and 300.90° on CSPG). Corresponding linear mean angles converged towards 180°, showing little directionality (197.58° on LN and 183.09° on CSPG).
Several circular and linear goodness-of-fit tests were performed for all experimental data conditions with the null hypothesis of uniformity (Table V). For neurites cultured on uniform LN substrates, patterned LN and CSPG stripes, and LN gradients, circular and linear tests exhibited similar results. On uniform substrates, all circular and linear tests showed no significant difference from a uniform distribution. On striped substrates and LN gradients, all circular and linear tests showed significant difference from a uniform distribution. However, on CSPG gradients, a common linear test, KS, showed no significant difference from uniformity whereas Rayleigh’s, Watson’s, Rao’s and Kuiper’s circular tests showed significant difference from uniformity. Surprisingly, χ2 tests for grouped data with 3 groups were in better agreement with other circular tests than χ2 tests for grouped data with 36 groups.
Multisample analysis showed no difference in neurite angles between LN striped and CSPG striped samples. It also showed no difference in angles between LN gradient and CSPG gradient samples. Both Mardia-Watson-Wheeler and Watson’s U2 test showed no significant differences with p-values greater than 0.49 (Table VI). Student t-test was performed as a comparison and also found neurite angles to be not significantly different on these substrates at p<0.05 significance level, but p-values were much lower (p<0.1). Further examination of data shows similar mean angles on LN and CSPG striped substrates and on LN and CSPG gradient substrates (Table IV). Multisample analysis with Mardia-Watson-Wheeler and Watson’s U2 test showed that neurites on striped LN and LN gradient substrates were significantly different from each other. Comparison with Student t-test shows that linear methods also show significant difference between neurite angles on LN stripe and gradient substrates (Table VI). Further examination of data shows differences in both mean angles and circular standard deviation (Table IV). For CSPG striped versus CSPG gradient substrates, Student t-test comparison shows no significant difference in neurite growth. However, circular analysis does show significant differences in neurite angles (Table IV). Further examination of the data shows that this distinction is due to the convergence of the linear mean towards 180° (Table IV).
Table VI. Comparison of circular and linear multisample tests for all experimental conditions tested.
Circular multisample tests Mardia-Watson-Wheeler and Watson’s U2 test were performed to determine differences between neurite outgrowth on pairs of substrate types. Linear comparisons were performed using Student t-tests. Significance levels taken to be p<0.05.
| CIRCULAR | LINEAR | |||||
|---|---|---|---|---|---|---|
| Substrate | Mardia-Watson | Wheeler | Watson's | U2 | Student t | |
| W | p | U2 | p | t | p | |
| LN versus CSPG stripe | 1.409 | 0.494 | 0.079 | 0.5 > p > 0.2 | −1.515 | 0.066 |
| LN versus CSPG gradient | 0.056 | > 0.5 | 0.021 | 0.989 | 1.345 | 0.090 |
| LN stripe versus gradient | 33.867 | 0.000 | 1.385 | < 0.001 | −4.033 | 0.000 |
| CSPG stripe versus gradient | 9.479 | 0.009 | 0.721 | < 0.001 | −0.007 | 0.497 |
3.2 Simulation Results
Simulations of circular data were performed to compare the results of circular and linear goodness-of-fit tests for one sample statistical analysis of known distributions. Simulations of neurite angles were drawn from established circular statistical models such as uniform distribution to model the experimental condition of applying no directional cue, VM distribution to model the condition of applying one unidirectional cue and BM distribution to model the condition of applying two cues from two different directions. Because the sample size of neurite angles obtained from experimental data is variable based on the permissiveness of the substrate, the sample size in simulated data was varied from n=5 to n=100 to cover a range of sample sizes used in neurite outgrowth analysis. Simulated circular data was only presented in circular histograms.
For uniform distributions, all directions between 0° and 360° are equally likely to occur, and µc is undefined as R=0. VM distribution is the most commonly used model for unimodal samples of circular data. As κ approaches 0, the distribution converges to a uniform distribution, and as κ approaches infinity, the distribution tends to concentrate around the direction µc. Sample means of VM based data approached the population means (user-defined µc=0). Circular means were better approximated for simulated samples with tighter distributions, µc=11° where κ=0.85 and µc=5° where κ=3 (Fig. 5b, c). Circular histograms of BM data highlight the difficulty in graphically assessing multimodality in data (Fig. 5d, e). The means calculated for a BM distribution of (µ1=0°, µ2=90°) were 79° and 61°. For BM distributions, histograms do not show a clear distinction between the two subpopulations of data.
Table VII shows the number of statistically significant (non-uniform) simulations out of 100 trials, from each type of uniformity test listed in rows (Rayleigh, Rao’s spacing, Watson’s U2, Kuiper’s and V-test). Watson’s U2 test was unavailable for n=5 samples as the test’s assumptions require n=10 for analysis using this method. Simulated data is listed in columns, with the type of distribution (uniform, VM and BM) described by its parameters (µ, κ).
For uniform distributions, the number presented in Table VII when taken as a percentage corresponds to the Type I error of the given test. Type I error occurs when the null hypothesis (in this case, distribution is uniform) is rejected by the test when the null hypothesis is in fact true. Hence this column records the number of times the uniformity test incorrectly finds the distribution directed. A lower Type I error value indicates a better test for this type of data, as the test is wrong less often. As expected, for all tests, as n increases, Type I error decreases. Linear goodness-of-fit tests showed low Type I error for uniform distributions, indicating that nonparametric tests against a null hypothesis of uniformity such as KS tests are effective for determining uniformity. For circular tests, where n is small, Rao’s spacing test has the lowest Type I error, which is consistent with other studies comparing one sample statistical tests (Bergin, 1991). For VM and BM distributions, the number of significant trials out of 100 corresponded to the power of the statistical test of interest. The expectation was that the tests would find the data significantly different from the null hypothesis of uniformity. Power is defined as the probability that the null hypothesis will be rejected if it is false, or the probability that the test will not produce a Type II error or false negative. In this case, power is the probability that the test rejects uniformity (or indicates directedness) if the distribution is known to be unimodal or bimodal. As expected, increasing sample size corresponded to increasing power of each test.
For VM and BM distributions with large sample sizes (approaching n=100) KS tests found VM and BM distributions to be significantly different from a Gaussian distribution. Overall circular tests performed similarly for VM distributions, although Rao’s Spacing test was less powerful than other circular statistical one-sample tests. V-test had the highest power for VM datasets with small sample size and higher dispersion. For simulated data, the hypothesized mean direction was known as it was user defined. It is important to note that this hypothesized direction (θ0=0°) must be assigned in advance of experimentation and if the null hypothesis is not rejected by the V-test, it is unknown whether the population is distributed uniformly or whether the distribution has a mean direction other than θ0. For one-sample data with an unknown external directional component, the Rayleigh test is usually recommended for unimodal data (Batschelet, 1981; Fisher, 1993). Both the Rayleigh test and Kuiper’s test yielded similar power levels for unimodal VM distributions, even for distributions with relatively high angular dispersion (corresponding to a lower κ value). The power of all circular statistical tests were higher than the corresponding linear KS test against uniformity for simulated VM and BM distributions, except in the case where n=100. However, the power of the same test against a null hypothesis of a normal distribution is very low, such that most simulations showed that a KS test did not find the simulated uniform distribution to be significantly different from a normal distribution.
As expected, one-sample uniformity tests exhibited low power when tested on BM distributions, as the cluster of data around two peaks began to resemble uniform data when both distributions had low concentration values. V-test showed the highest power when testing BM data against a null hypothesis. When the BM distribution was relatively clustered (at the highest concentration value tested κ=3), all tests showed higher power in detecting non-uniformity.
Discussion
The present study demonstrates that circular statistical methods may be used to analyze biological data containing directional biases and anisotropy, particularly to quantify neurite outgrowth. To analyze the direct effects of the underlying substratum of the culture surface on neuronal outgrowth, we plated and cultured neurons at low density for 24 hours and imaged the cultures for analysis. In the absence of directional cues, on uniform substrates, neurites were randomly oriented. The neurons on substrates with patterned protein stripes were highly aligned to the underlying stripe geometry. The neurons on substrates with graded anisotropy in protein concentration were directed towards the more permissive regions of the substrate.
We have shown that circular statistical methods show sufficient power and are a better model than linear statistical methods to analyze directional neurite outgrowth on micropatterned substrates. Circular histograms and categorization allow easy visualization of data clustering, as demonstrated by histograms of neurite angles grouped around the direction of alignment. Circular data presentation avoids observational and truncation biases which occur in linear statistical analyses, for more accurate characterization of data. Circular statistical tests are more sensitive for smaller sample sizes, as shown by higher power of circular tests for simulations containing low n’s. Further, they are more sensitive to complex distributions, as shown by the performance of multisample tests in comparing neurite angles on CSPG stripes versus CSPG gradients. The approximate linearity of a small arc, in the case when data is clustered, is sometimes used to justify the application of linear models to simplify data analysis; however, different degrees of dispersion depending on the variability of the data can strongly affect the validity of this assumption. If there is any appreciable variability of circular data, it has been noted that the average of the dataset is better described by a resultant vector rather than the arithmetic mean (Fisher, 1993).
Evaluation of neurite outgrowth in vitro have included qualitative scoring systems with grouped categorical quantification (Dertinger et al., 2002; Sonigra et al., 1999; Sorensen et al., 2007), and measurement of neurite characteristics such as neurite length, area, number and branching patterns (Kim et al., 2006; Mann et al., 1998; Recknor et al., 2004). A number of studies initially demonstrated the ability of patterned striped substrates to support directional neurite outgrowth in vitro (Clark et al., 1993; Gomez and Letourneau, 1994) and the ability of concentration gradients to direct neuronal growth up concentration gradients (Walsh et al., 2005) by using traditional linear statistical methods. Categorization of circular data prior to using linear statistical methods has been a common approach to analyzing directional data. After the categorization of angular data into bins for plotting histograms (generally in bins of 10–20°), or into categories of “aligned” versus “unaligned,” linear statistical tests can be applied to the groups of categorical data. ANOVAs are commonly used to test the differences between the degrees of neurite alignment over different substrates or experimental conditions (Macias et al., 2000; Sorensen et al., 2007). It is important to note that ANOVAs assume a Gaussian model for the distribution of angles which may not be accurate, depending on the population. Previous studies of neurite growth and orientation have used nonparametric statistical tests such as the χ2 test (Biran et al., 2003; Dertinger et al., 2002; Manwaring et al., 2004; Smeal et al., 2005) and the Kolmogorov-Smirnov (KS) test (Ming et al., 2001; Thompson and Buettner, 2006; Yuan et al., 2003). A rational for using nonparametric tests is that circular data of neurite outgrowth angles are rarely expected to approach a normal distribution (in a linear presentation) or a VM distribution (in a circular presentation). The χ2 and KS tests were also performed in this study and shown in some cases to perform differently from circular tests. One limitation of linear statistical methods for the application of directional neurite outgrowth in culture systems is that there may be an overemphasis on the tails of the linear scale, in this case 0° and 180° or 360°. This overemphasis can result in artificially inflating the calculated variance of the data.
Other studies that analyze cellular phenomenon, particularly of cell migration, have recognized the need to present data in a nonlinear fashion in order to most appropriately visualize and analyze movement data which is usually highly complex, with cell trajectories tracing a relatively noisy path. If the nature of the path in response to a directional stimulus is of interest, the migration angle is usually an important aspect of analysis. Recent studies have utilized circular visualizations for data presentation, most commonly using Rose diagrams, where the frequency of migration angles are plotted around a circular axis with the area of each bar corresponding to frequency (Frevert et al., 2006; Papakonstanti et al., 2007; Saadi et al., 2006). In neuroscience literature, examples of visualization of angular data using circular methods have included neuronal migration (Ward et al., 2003), neurite outgrowth (Tailby et al., 2005), response to magnetic stimulation (Macias et al., 2000), and mitochondrial organization in axonal transport (Miller and Sheetz, 2004).
In this report, we demonstrated the utility of a circular analysis approach for evaluating a wide range of neurite outgrowth phenomena. Comparison of six, one-sample uniformity or goodness-of-fit tests on simulated samples of three distributions (uniform, unimodal VM, BM) revealed relative strengths: Rao’s spacing test performed best for small samples, V-test and Rayleigh’s test performed best for unimodal samples and Watson’s U2 performed best for bimodal samples. Comparison of circular tests versus linear tests on experimental angular data of neurites grown on three types of micropatterned protein substrates (uniform, striped and gradient) demonstrated the strengths of the circular approaches. Out of the linear goodness-of-fit tests performed, only the χ2 test grouped into 3 bins gave results that were significantly different from a uniform distribution. In contrast, all circular uniformity tests were consistent in determining significance from a uniform distribution for all samples tested.
Conclusions
In conclusion, we have applied a statistical method for graphically representing and analyzing directional data pertaining to neurite growth that can be used to investigate neuronal cultures and their interactions with their microenvironment in vitro. Despite recent advances, current approaches to nerve repair fall short of restoring complete function, and in vitro systems that have been developed to more systematically study parameters affecting neurite growth have become more specific and quantitative. The techniques described here are useful in identifying directional neurite outgrowth patterns on in vitro platforms, allowing us to better evaluate neurite growth trajectories that exhibit circular geometry. Statistical methods such as uniformity tests provide a formal means to test hypotheses relating to neuronal responses to complex microenvironments, and circular histograms provide a visualization tool to investigate neuronal processes exhibiting circular geometries. This approach offers a more informative and rigorous way to probe the mechanisms of neurite outgrowth and guidance.
Acknowledgements
The authors thank Elizabeth Deweerd for assistance with alignment and gradient experiments, and Michael Sherback for assistance with MATLAB programming and for helpful discussion of the manuscript. This work was funded by R21 EB004506-01, R01 EB005722-01, an NSF CAREER grant to DHK and a Robert and Susan Kaplan Fellowship to GNL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abosch A, Lagenaur C. Sensitivity of neurite outgrowth to microfilament disruption varies with adhesion molecule substrate. Journal of Neurobiology. 1993;24:344–355. doi: 10.1002/neu.480240307. [DOI] [PubMed] [Google Scholar]
- Alexander JK, Fuss B, Colello RJ. Electric field-induced astrocyte alignment directs neurite outgrowth. Neuron Glia Biology. 2006;2:93. doi: 10.1017/S1740925X0600010X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschelet E. Circular statistics in biology. London: Academic Press; 1981. [Google Scholar]
- Bergin TM. A Comparison Of Goodness-Of-Fit Tests For Analysis Of Nest Orientation In Western Kingbirds (Tyrannus-Verticalis) Condor. 1991;93:164–171. [Google Scholar]
- Bilsland J, Rigby M, Young L, Harper S. A rapid method for semi-quantitative analysis of neurite outgrowth from chick DRG explants using image analysis. Journal of neuroscience methods. 1999;92:75–85. doi: 10.1016/s0165-0270(99)00099-0. [DOI] [PubMed] [Google Scholar]
- Biran R, Noble MD, Tresco PA. Directed nerve outgrowth is enhanced by engineered glial substrates. Experimental Neurology. 2003;184:141–152. doi: 10.1016/s0014-4886(03)00253-x. [DOI] [PubMed] [Google Scholar]
- Bruder JM, Lee AP, Hoffman-Kim D. Biomimetic materials replicating Schwann cell topography enhance neuronal adhesion and neurite alignment in vitro. J Biomater Sci Polym Ed. 2007;18:967–982. doi: 10.1163/156856207781494412. [DOI] [PubMed] [Google Scholar]
- Bruder JM, Monu NC, Harrison MW, Hoffman-Kim D. Fabrication of Polymeric Replicas of Cell Surfaces with Nanoscale Resolution. Langmuir. 2006;22:8266–8270. doi: 10.1021/la0608563. [DOI] [PubMed] [Google Scholar]
- Clark P, Britland S, Connolly P. Growth cone guidance and neuron morphology on micropatterned laminin surfaces. J Cell Sci. 1993;105:203–212. doi: 10.1242/jcs.105.1.203. [DOI] [PubMed] [Google Scholar]
- Dertinger SK, Jiang X, Li Z, Murthy VN, Whitesides GM. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc Natl Acad Sci U S A. 2002;99:12542–12547. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deumens R, Koopmans GC, den Bakker CGJ, Maquet V, Blacher S, Honig WMM, Jerome R, Pirard JP, Steinbusch HWM, Joosten EAJ. Alignment of glial cells stimulates directional neurite growth of CNS neurons in vitro. Neuroscience. 2004;125:591. doi: 10.1016/j.neuroscience.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Fisher NI. Statistical Analysis of Circular Data. Cambridge, U.K: Cambridge University Press; 1993. [Google Scholar]
- Frevert CW, Boggy G, Keenan TM, Folch A. Measurement of cell migration in response to an evolving radial chemokine gradient triggered by a microvalve. Lab Chip. 2006;6:849–856. doi: 10.1039/b515560f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldner JS, Bruder JM, Li G, Gazzola D, Hoffman-Kim D. Neurite bridging across micropatterned grooves. Biomaterials. 2006;27:460–472. doi: 10.1016/j.biomaterials.2005.06.035. [DOI] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Filopodia initiate choices made by sensory neuron growth cones at laminin/fibronectin borders in vitro. J. Neurosci. 1994;14:5959–5972. doi: 10.1523/JNEUROSCI.14-10-05959.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynds DL, Snow DM. A semi-automated image analysis method to quantify neurite preference/axon guidance on a patterned substratum. Journal of Neuroscience Methods. 2002;121:53. doi: 10.1016/s0165-0270(02)00231-5. [DOI] [PubMed] [Google Scholar]
- Karlon WJ, Covell JW, McCulloch AD, Hunter JJ, Omens JH. Automated measurement of myofiber disarray in transgenic mice with ventricular expression of ras. Anat Rec. 1998;252:612–625. doi: 10.1002/(SICI)1097-0185(199812)252:4<612::AID-AR12>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Kim IA, Park SA, Kim YJ, Kim SH, Shin HJ, Lee YJ, Kang SG, Shin JW. Effects of mechanical stimuli and microfiber-based substrate on neurite outgrowth and guidance. J Biosci Bioeng. 2006;101:120–126. doi: 10.1263/jbb.101.120. [DOI] [PubMed] [Google Scholar]
- Le Roux PD, Reh TA. Regional differences in glial-derived factors that promote dendritic outgrowth from mouse cortical neurons in vitro. J. Neurosci. 1994;14:4639–4655. doi: 10.1523/JNEUROSCI.14-08-04639.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu J, Hoffman-Kim D. Multi-Molecular Gradients of Permissive and Inhibitory Cues Direct Neurite Outgrowth. Annals of Biomedical Engineering. 2008;36:889–904. doi: 10.1007/s10439-008-9486-z. [DOI] [PubMed] [Google Scholar]
- Macias MY, Battocletti JH, Sutton CH, Pintar FA, Maiman DJ. Directed and enhanced neurite growth with pulsed magnetic field stimulation. Bioelectromagnetics. 2000;21:272–286. [PubMed] [Google Scholar]
- Mahoney MJ, Chen RR, Tan J, Saltzman WM. The influence of microchannels on neurite growth and architecture. Biomaterials. 2005;26:771–778. doi: 10.1016/j.biomaterials.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Mann F, Zhukareva V, Pimenta A, Levitt P, Bolz J. Membrane-Associated Molecules Guide Limbic and Nonlimbic Thalamocortical Projections. J. Neurosci. 1998;18:9409–9419. doi: 10.1523/JNEUROSCI.18-22-09409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwaring ME, Walsh JF, Tresco PA. Contact guidance induced organization of extracellular matrix. Biomaterials. 2004;25:3631–3638. doi: 10.1016/j.biomaterials.2003.10.043. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Miller KE, Sheetz MP. Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117:2791–2804. doi: 10.1242/jcs.01130. [DOI] [PubMed] [Google Scholar]
- Ming G-l, Henley J, Tessier-Lavigne M, Song H-j, Poo M-m. Electrical Activity Modulates Growth Cone Guidance by Diffusible Factors. Neuron. 2001;29:441–452. doi: 10.1016/s0896-6273(01)00217-3. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Hanson JC, Quets-Nguyen AT, Bergeron M, Smith RC. A quantitative method for analysis of in vitro neurite outgrowth. Journal of neuroscience methods. 2007;164:350–362. doi: 10.1016/j.jneumeth.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Papakonstanti EA, Ridley AJ, Vanhaesebroeck B. The p110delta isoform of PI 3-kinase negatively controls RhoA and PTEN. EMBO J. 2007;26:3050–3061. doi: 10.1038/sj.emboj.7601763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RD, Oe T, Yamaji T, Matsuoka N. A simple, flexible, nonfluorescent system for the automated screening of neurite outgrowth. J Biomol Screen. 2006;11:155–164. doi: 10.1177/1087057105283344. [DOI] [PubMed] [Google Scholar]
- Recknor JB, Recknor JC, Sakaguchi DS, Mallapragada SK. Oriented astroglial cell growth on micropatterned polystyrene substrates. Biomaterials. 2004;25:2753–2767. doi: 10.1016/j.biomaterials.2003.11.045. [DOI] [PubMed] [Google Scholar]
- Ronn LC, Ralets I, Hartz BP, Bech M, Berezin A, Berezin V, Moller A, Bock E. A simple procedure for quantification of neurite outgrowth based on stereological principles. Journal of neuroscience methods. 2000;100:25–32. doi: 10.1016/s0165-0270(00)00228-4. [DOI] [PubMed] [Google Scholar]
- Saadi W, Wang S-J, Lin F, Jeon N. A parallel-gradient microfluidic chamber for quantitative analysis of breast cancer cell chemotaxis. Biomedical Microdevices. 2006;8:109–118. doi: 10.1007/s10544-006-7706-6. [DOI] [PubMed] [Google Scholar]
- Smeal RM, Rabbitt R, Biran R, Tresco PA. Substrate Curvature Influences the Direction of Nerve Outgrowth. Annals of Biomedical Engineering. 2005;33:376. doi: 10.1007/s10439-005-1740-z. [DOI] [PubMed] [Google Scholar]
- Smit M, Leng J, Klemke RL. Assay for neurite outgrowth quantification. BioTechniques. 2003;35:254–256. doi: 10.2144/03352bm01. [DOI] [PubMed] [Google Scholar]
- Sonigra RJ, Brighton PC, Jacoby J, Hall S, Wigley CB. Adult rat olfactory nerve ensheathing cells are effective promoters of adult central nervous system neurite outgrowth in coculture. Glia. 1999;25:256–269. doi: 10.1002/(sici)1098-1136(19990201)25:3<256::aid-glia6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Sorensen A, Alekseeva T, Katechia K, Robertson M, Riehle MO, Barnett SC. Long-term neurite orientation on astrocyte monolayers aligned by microtopography. Biomaterials. 2007;28:5498–5508. doi: 10.1016/j.biomaterials.2007.08.034. [DOI] [PubMed] [Google Scholar]
- Tailby C, Wright LL, Metha AB, Calford MB. Activity-dependent maintenance and growth of dendrites in adult cortex. Proc Natl Acad Sci U S A. 2005;102:4631–4636. doi: 10.1073/pnas.0402747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Buettner HM. Neurite outgrowth is directed by schwann cell alignment in the absence of other guidance cues. Ann Biomed Eng. 2006;34:161–168. doi: 10.1007/s10439-005-9013-4. [DOI] [PubMed] [Google Scholar]
- Tran NH. Fracture orientation characterization: Minimizing statistical modelling errors. Computational Statistics & Data Analysis. 2007;51:3187–3196. [Google Scholar]
- Walsh JF, Manwaring ME, Tresco PA. Directional Neurite Outgrowth Is Enhanced by Engineered Meningeal Cell-Coated Substrates. Tissue Engineering. 2005;11:1085–1094. doi: 10.1089/ten.2005.11.1085. [DOI] [PubMed] [Google Scholar]
- Ward M, McCann C, DeWulf M, Wu JY, Rao Y. Distinguishing between directional guidance and motility regulation in neuronal migration. J Neurosci. 2003;23:5170–5177. doi: 10.1523/JNEUROSCI.23-12-05170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CM, Pinezich JD, Lindquist WB, Vazquez ME. An algorithm for neurite outgrowth reconstruction. Journal of neuroscience methods. 2003;124:197–205. doi: 10.1016/s0165-0270(03)00017-7. [DOI] [PubMed] [Google Scholar]
- Yuan X-b, Jin M, Xu X, Song Y-q, Wu C-p, Poo M-m, Duan S. Signalling and crosstalk of Rho GTPases in mediating axon guidance. Nat Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice-Hall; 1996. [Google Scholar]