Abstract
The Leu33Pro polymorphism of the gene encoding β3 integrin (ITGB3) is associated with acute coronary syndromes and influences platelet aggregation. Three common promoter polymorphisms have also been identified. The aims of this study were to (1) investigate the influence of the ITGB3 −400C/A, −425A/C and −468G/A promoter polymorphisms on reporter gene expression and nuclear protein binding and (2) determine genotype and haplotype associations with platelet αIIbβ3 receptor density. Promoter haplotypes were introduced into an ITGB3 promoter-pGL3 construct by site directed mutagenesis and luciferase reporter gene expression analysed in HEL and HMEC-1 cells. Binding of nuclear proteins was assessed by electrophoretic mobility shift assay. The association of ITGB3 haplotype with platelet αIIbβ3 receptor density was determined in 223 subjects. Species conserved motifs were identified in the ITGB3 promoter in the vicinity of the 3 polymorphisms. The GAA, GCC, AAC, AAA and ACC constructs induced ~50% increased luciferase expression relative to the GAC construct in both cell types. Haplotype analysis including Leu33Pro indicated 5 common haplotypes; no associations between ITGB3 haplotypes and receptor density were found. However, the GCC-Pro33 haplotype was associated with significantly higher vWF activity (128.6 [112.1–145.1]%) compared with all other haplotypes (107.1 [101.2–113.0]%, p=0.02). In conclusion, the GCC-Pro33 haplotype was associated with increased vWF activity but not with platelet αIIbβ3 receptor density, which may indicate ITGB3 haplotype influences endothelial function.
Keywords: β3 integrins, polymorphisms, gene regulation
Introduction
Receptor-mediated interactions between leukocytes, endothelial cells and platelets and coagulation factors and extracellular matrix proteins at the sites of vascular injury are central to the pathogenesis of atherosclerosis and thrombosis [1]. β3 integrin is a component of the platelet-specific αIIbβ3 receptor and the more widely distributed αvβ3 receptor. Platelets play a key role in arterial thrombosis and the αIIbβ3 receptor is central to these processes as the mediator of platelet aggregation at the site of vascular injury. The αvβ3 receptor is variably expressed by a number of cell types including endothelial cells and vascular smooth muscle cells [2]. Increased endothelial cell and vascular smooth muscle cell expression of αvβ3 in coronary artery atherosclerotic plaques has been reported [3]. Consequently variants of the gene encoding β3 integrin (ITGB3) may influence the development of cardiovascular disease through modification of the function of αIIbβ3 and αvβ3.
Several polymorphisms in the ITGB3 gene have been identified including the Leu33Pro (PlA) variant and the −400C/A, −425A/C and −468G/A promoter polymorphisms [4]. Studies have demonstrated associations between the Pro33 allele (PlA2) and premature myocardial infarction (MI), ischemic stroke and restensosis following coronary angioplasty [5,6]. Pro33 is associated with a lower threshold for αIIbβ3 activation in response to ADP, and enhanced α-granule release, clot retraction and adhesion to fibrinogen [7,8]. Limited information is available regarding the functional effects of the −400C/A, −425A/C and −468G/A promoter polymorphisms. The aims of this study were to (i) investigate the influence of the −400C/A, −425A/C and −468G/A promoter polymorphisms on reporter gene expression and nuclear protein binding and (ii) determine genotype and haplotype associations with platelet αIIbβ3 receptor density.
Materials and Methods
Bioinformatics
Bioinformatics analyses were carried out to compare the 5’ region of the human (accession number AY706100), chimp (AY413572), mouse (AY413573) and chicken (X75348) ITGB3 genes to determine regions of conservation. The first 500 bp 5’ to the ATG of each species were analysed by FootPrinter (http://abstract.cs.washington.edu/~blanchem/FootPrinterWeb/FootPrinterInput2.pl) against the default phylogenetic tree to indicate potential regulatory elements [9]. The software produced short regions of sequences (up to 10 bp in length) that were highly conserved (maximum of 2 bp difference) between species.
Generation of Variant ITGB3Reporter Constructs
The ITGB3 promoter region was cut from a previously reported ITGB3-pGL2 vector [10] and ligated into pGL3 by digestion with 5 units of Kpn I and Nhe I, and ligation with T4 ligase. The wild type ITGB3 5’ region construct will be referred to as ITGB3-pGL3-GAC in relation to the more common alleles located at positions −468, −425 and −400. Haplotypes were introduced into the ITGB3-pGL3-GAC construct by site directed mutagenesis using the GeneEditor™ in-vitro Site Directed Mutagenesis system (Promega). Mutagenic oligonucleotides were designed with the relevant mismatch in the middle:
−468A: 5’-GGCATTCAACAGATGTTTG-3’;
−425C: 5’-GTGTGAATGAAT GACACTCGAGGTAGTGG-3’;
−400A: 5’-GTGAATGTGTACCAAGAATC-3’
.
The −400A and −468A variants were introduced using the manufacturer’s standard protocol. A modified protocol was utilised to generate the −425C variant; the mutagenic oligonucleotide was added at the alkaline denaturation stage, and the initial temperature for oligonucleotide annealing was increased to 83°C. Twenty colonies were picked for each mutagenesis reaction and successful mutagenesis confirmed by genotyping and sequencing. The sequence of the ITGB3 promoter (from the ATG codon to −2070) has been submitted to GenBank, accession number AY706100.
Cell Culture and Transient Transfections
Human erythroleukemic (HEL 92.1.7) cells (αIIbβ3 expressing representative of megakaryocytic cells) were grown in RPMI 1640 medium (Sigma), supplemented with 10% foetal calf serum (FCS) (First Link Ltd), 2 mM glutamine and antibiotic mix (10,000 U penicillin, 10 mg streptomycin and 25 µg amphotericin B per ml). Cells were transiently co-transfected by electroporation with the ITGB3-pGL3 constructs and Renilla Luciferase-CMV (pRL-CMV) vector (Promega), as the internal control, at a ratio of 50:1. For transfection, 1×107 cells per sample were pelleted and resuspended in 0.8 ml PBS, 10 µg of DNA was added and cells were electroporated at 500 µF, 0.4 kV (BioRad Gene Pulser). The electroporated cells were seeded in 25 ml of pre-warmed media and incubated at 37°C and 5% CO2 for 48 hours prior to reporter gene analysis.
Human Microvascular Endothelial cells (HMEC-1, αVβ3 expressing, representative of endothelial cells) were grown in MCDB 131 media (Invitrogen), supplemented with 10% FCS, antibiotic mix, 2mM glutamine, 0.001% epidermal growth factor and 1 µg/ml hydrocortisone. HMEC-1 cells were transiently transfected with Lipofectin (Invitrogen): 5×105 cells were plated on 60 mm plates 24 hours prior to transfection with 3µg DNA (100:1 ratio of ITGB3-pGL3 constructs to pRL-CMV) and 20 µl Lipofectin, according to manufacturer’s instructions, followed by incubation at 37°C and 5% CO2 for 24 hours prior to reporter gene analysis.
Luciferase Reporter Gene Expression Analysis
Luciferase reporter gene analysis was carried out using the Dual-Luciferase Reporter Assay system (Promega). Transfected HEL cells were pelleted by centrifugation, washed, and 1×107 cells lysed with 250 µl passive lysis buffer (PLB). Transfected HMEC-1 cells were washed, and lysed with 1 ml PLB. Renilla and firefly luciferase activities were determined in 20 µl of centrifuged cell lysate according to manufacturer’s instructions using a PHL luminometer (Mediators). Firefly luciferase values were normalised to Renilla luciferase activity. Eight independent sets of transfections were carried out, and results are expressed as the activity of each construct relative to the ITGB3-pGL3-GAC construct.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear proteins were extracted from 5×106 HEL and HMEC-1 cells according to the method described by Andrews & Faller [11]. Fluorescein-labelled sense oligonucleotides were synthesised with the polymorphic variant in the centre, as follows:
-
−468G/A: forward 5’- AGAGAAGGCATTCAG/ACAGATGTTTGCCAG -3’
reverse 5’- CTGGCAAACATCTGC/TTGAATGCCTTCTCT-3’
-
−425 A/C: forward 5’- GTGTGAATGAATGAA/CACTCGAGGTAGTGG-3’
reverse 5’- CCACTACCTCGAGTT/GTCATTCATTCACAC-3’
-
−400C/A: forward 5’- GTGGGTGAATGTGTC/ACAAGAATCCAGCG-3’
reverse 5’- CGCTGGATTCTTGG/TACACATTCACCCAC-3’
.
Sense and antisense oligonucleotides were annealed and gel purified, and 1.5 pmol used in each binding reaction. EMSA reactions were performed in binding buffer (20% glycerol, 20mM Tris, pH7.5, 100mM KCl, 1mM DTT) with 0.1 µg/µl poly dI/dC and 35 µg of nuclear protein extract. For competition assays, 1-, 10- and 50-fold molar excess of unlabelled specific or non-specific competitor (SP1: forward 5’- ATTCGATCGGGGCGGGGCGAGC-3’ and reverse 5’-GCTCGCCCCGCCCCGATCGAA-3’) were added. Reaction mixes were incubated at RT0 for 30 min and then electrophoresed on an 8% polyacrylamide gel with 5% stacking gel in 0.25x TBE (22.5 mM Tris-borate and 0.5 mM EDTA, pH 8.3) at 200V for 6 hours at 15°C. The gel was visualised using a FX Molecular Imager (BioRad).
Genotyping of ITGB3 polymorphisms
The −468G/A polymorphism was genotyped using an Assays-by-DesignSM SNP genotyping assay (Applied Biosystems):
forward primer 5’- GGCAAGAAAAAAATTAGTGAATAATAAAGGACTGA-3’
reverse primer 5’- ACTCGAGGTAGTGGGTGAATGT-3’
probes 5’-AAGGCATTCAG/ACAGATG-3’
Probes were labelled with VIC™ for detecting the −468G allele and 6-FAM™ for detecting the −468A allele, and following PCR amplification, fluorescence detection was carried out using an ABI PRISM 7700 SDS (Applied Biosystems). The −425 A/C polymorphism was genotyped by PCR-RFLP with primers: forward 5’-CGAGGCTCTTCATGGACCTA-3’ and reverse 5’-CACATTCACCCACTACCTCGAAT-3’ and digestion with Apo I (the underlined base is a mismatch to introduce an Apo I site in the presence of the −425A allele). The −400 C/A polymorphism was genotyped by PCR-RFLP with primers: forward 5’-AGTGAATAATAAAGGACTGAACCG-3’ and reverse 5’-GCGCTCGCATCTCGTC-3’ and digestion with Rsa I. The Leu33Pro polymorphism was genotyped as previously described [12].
Subject recruitment and analysis of αIIbβ3 receptor expression
Subjects (n=233) were recruited by the Baylor College of Medicine Thrombosis Research Group for analysis of the relationship between ITGB3 polymorphisms and αIIbβ3 receptor expression. The study population comprised healthy subjects, recruited through local advertising, who were free from a history of cardiovascular disease and were not taking aspirin (n=138), as previously described [13] and subjects who had previously (>3 months) undergone coronary artery bypass surgery (CABG, n=85) who were taking aspirin. All subjects gave informed consent according to a protocol approved by the Baylor College of Medicine institutional review board. Platelet αIIbβ3 receptor expression was measured by flow cytometry using phycoerythrin-conjugated monoclonal antibody HIP8 (anti-CD41a; BD-Pharmingen) and expressed as mean fluorescent intensity, as previously described [14] Fibrinogen concentration was determined by the Clauss method and vWF activity was determined using a ristocetin cofactor assay, as previously described [14].
Statistical Analysis
Non-normally distributed variables were log-transformed to enable parametric analyses. Comparisons between two groups of continuous variables were carried out using Student’s Independent t-test and more than two groups by ANOVA. Data are presented as mean (geometric mean) with either SD (log SD) or 95% confidence intervals (CI). Age did not conform to normal distribution and is presented as median with 25th and 75th percentiles in descriptive statistics. Differences between categorical variables were ascertained using chi-square analysis. All statistical analyses were performed using SPSS for Windows version 12 (SPSS Inc).
Results
Bioinformatics Analysis of the ITGB3 Promoter
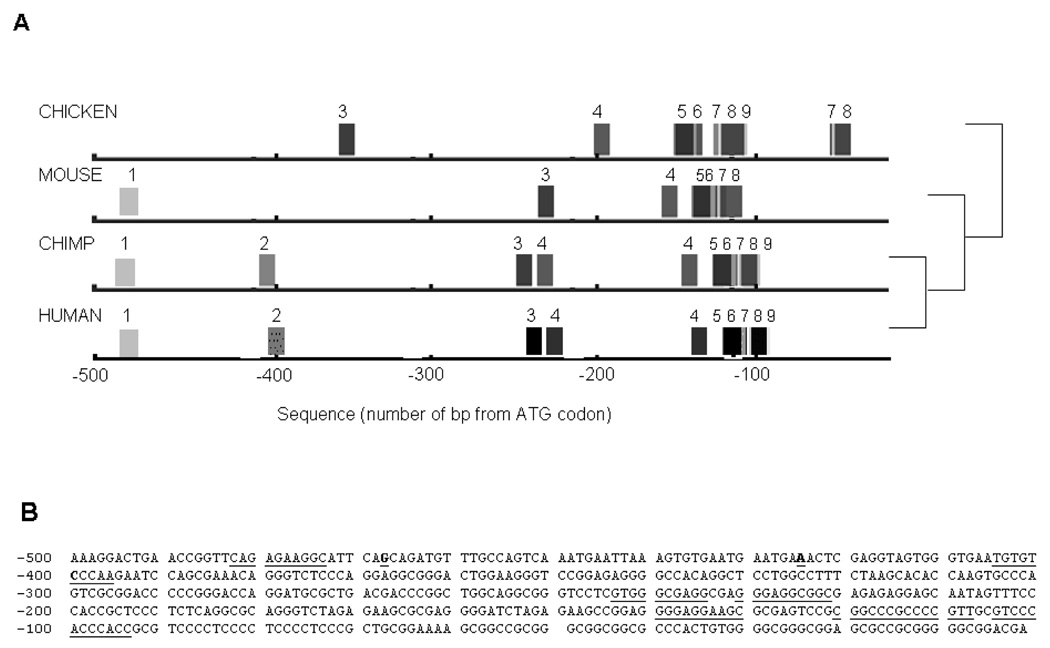
FootPrinter analysis (Fig. 1) indicated conserved motifs in the ITGB3 5’ region located in the region up to 500 bp from the ATG codon. The −400C/A polymorphism is within a motif conserved between human and chimp (TGTGTCCCAA) and the −425A/C and −468G/A polymorphisms are between conserved motifs. The evolutionary conservation of regions within the 5’ gene regulatory region indicated that these sequences might be functionally relevant. To determine if the promoter polymorphisms were functional, the luciferase reporter gene system was utilised together with analysis of nuclear protein binding.
Figure 1. FootPrinter Analysis of Conserved Motifs in the 5’ Regions of the Human, Chimp, Mouse and Chicken ITGB3 Genes.
Five hundred base pairs of the 5’ region of the human, chimp, mouse and chicken ITGB3 genes underwent footprinting analysis to determine if there were any regions of conservation between the different species. Panel A: Identical motifs detected in each species’ sequence have the same number. The phylogenetic tree on the right hand side shows the evolutionary relationship between the different species. Panel B: Conserved motifs are underlined, several motifs between −90 and −140 overlapped. The positions of the −400C/A, −425A/C and −468G/A are in bold and underlined.
Influence of ITGB3 −468/−425/−400 Haplotypes on Luciferase Reporter Gene Expression and Nuclear Protein Binding Profile
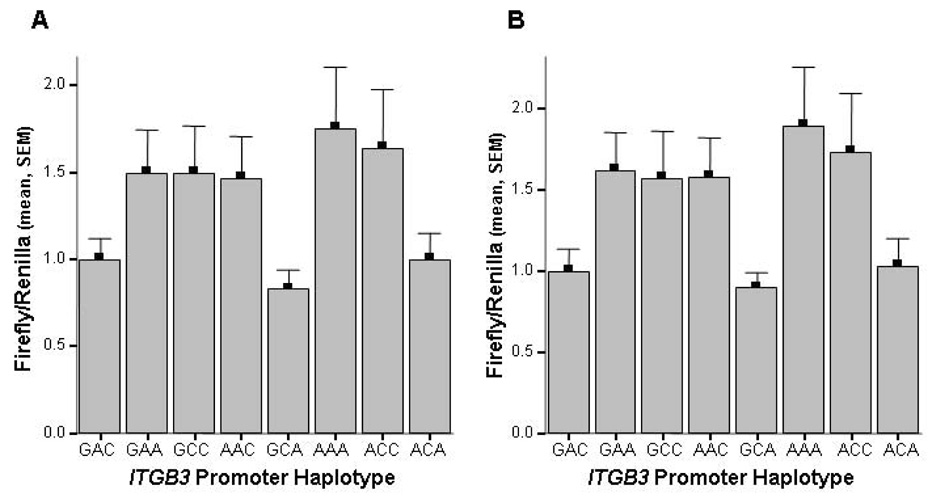
Luciferase reporter gene analyses indicated significant differences in reporter gene expression according to ITGB3 promoter haplotype in both HEL (p=0.035) and HMEC-1 cells (p=0.048), as shown in Figure 2. Similar luciferase expression patterns were obtained from the HEL and HMEC-1 cells. The GAA, GCC, AAC, AAA and ACC constructs induced ~50% increased luciferase expression relative to the GAC construct whereas the GCA and ACA constructs showed similar expression to GAC in both cell types. These results indicated that the three polymorphisms in the ITGB3 promoter may be functional and lead to a difference in nuclear protein binding. Each polymorphic variant was, therefore, analysed for differences in nuclear protein binding with HEL and HMEC-1 nuclear protein extracts. No difference in the nuclear protein binding profile was observed for the −400C/A, −425A/C and −468G/A allelic variants (data not shown).
Figure 2. Influence of the −468G/A, −425A/C and −400C/A polymorphisms in the 5’ gene regulatory region of ITGB3 on firefly reporter gene expression in HEL and HMEC-1 cells.
Firefly luciferase values were normalised to Renilla luciferase activity and expressed relative to the wild type GAC construct. Panel A: firefly/renilla luciferase values for HEL cells transiently transfected with each construct. ANOVA analysis, p=0.02. Panel B: firefly/renilla luciferase values for HMEC-1 cells transiently transfected with each construct. ANOVA analysis, p=0.048.
Association of ITGB3 polymorphisms and haplotype with platelet αIIbβ3 receptor density
The relationship between ITGB3 polymorphisms and αIIbβ3 receptor density was investigated in 223 subjects. The characteristics of the study population, including ethnicity, are presented in table 1. The mean receptor density (expressed as mean fluorescence intensity, MFI) was 136 (range: 81 to 192). There was no significant difference in mean receptor density between the different ethnic groups studied (Caucasian: 137 [SD 18]; African American: 134 [SD 19]; Mexican American: 135 [SD 16]; Asian: 143 [SD 27]; Other: 131 [SD 22], p=0.59). αIIbβ3 receptor density was significantly inversely correlated with BMI (r=−0.264, p<0.001) and fibrinogen (r=−0.155, p=0.02) but there was no correlation between αIIbβ3 receptor density and vWF activity (r=−0.019, p=0.78). There was no significant correlation between receptor density and age (r=−0.096, p=0.16) and no significant difference in αIIbβ3 receptor density between men and women, or between smokers and non-smokers (data not shown). There was no significant difference in receptor density between CABG patients taking aspirin and healthy subjects not taking aspirin, nor was there a significant difference in receptor density between those with and without a history of MI, or hypertension (data not shown).
Table 1.
Characteristics of study subjects
| Subjects (n=223) | |
|---|---|
| Age (years) | 42.3 (40.1–44.5) |
| Male | 112 (0.50) |
| BMI (kg/m2) | 26.6 (5.63) |
| Current smokers | 23 (0.10) |
| Ethnicity: | |
| Caucasian | 131 (0.59) |
| African American | 49 (0.22) |
| Mexican American | 28 (0.13) |
| Asian | 13 (0.06) |
| Other | 2 (0.01) |
| Fibrinogen (gL−1) | 3.39 (1.35) |
| vWF activity (%) | 110.6 (42.9) |
| CABG (%) | 85 (0.38) |
| Previous MI | 40 (0.18) |
| Hypertension | 59 (0.27) |
| Diabetes | 26 (0.12) |
| Aspirin* | 85 (0.38) |
Age presented as median (25th and 75th percentiles)
Other variables presented as mean or geometric mean (SD or antilog SD) or number (frequency)
CABG: coronary artery bypass graft
All subjects taking aspirin were CABG patients
The genotype distributions of the −400C/A, −425A/C, −468G/A and Leu33Pro polymorphisms conformed to Hardy Weinberg equilibrium. The −400C/A polymorphism was in significant linkage disequilibrium with the −425A/C (D’=−1.0, p=0.006) and −468G/A polymorphisms (D’=0.93, p<0.0001) but not with the Leu33Pro polymorphism. The −425A/C polymorphism was in significant linkage disequilibrium with the −468G/A (D’=−0.66, p=0.04) and the Leu33Pro (D’=0.67, p<0.0001) polymorphisms. There was no significant linkage disequilibrium between the −468G/A and Leu33Pro polymorphisms. Six promoter haplotypes were identified, as shown in table 2 (denoted A to F), with the 4 most frequent haplotypes accounting for 98% of the 446 chromosomes analysed. Inclusion of the Leu33Pro polymorphism in haplotype analysis gave rise to 11 different haplotypes, as shown in table 2 (denoted 1 to 11), with the 5 most common haplotypes accounting for 94% of the 446 chromosomes analysed.
Table 2.
ITGB3 haplotype frequencies of study subjects
| Promoter haplotye | Number of chromosomes (frequency) | Promoter and Leu33Pro haplotype | Number of chromosomes (frequency) | ||
|---|---|---|---|---|---|
| hapA: | GAC | 286 (0.64) | hap1: | GAC-Leu33 | 279 (0.62) |
| hap7: | GAC-Pro33 | 7 (0.02) | |||
| hapB: | AAA | 69 (0.15) | hap2: | AAA-Leu33 | 61 (0.13) |
| hap6: | AAA-Pro33 | 8 (0.2) | |||
| hapC: | GCC | 63 (0.14) | hap5: | GCC-Leu33 | 20 (0.04) |
| hap3: | GCC-Pro33 | 43 (0.10) | |||
| hapD: | AAC | 22 (0.05) | hap4: | AAC-Leu33 | 22 (0.05) |
| AAC-Pro33 | 0 | ||||
| hapE: | GAA | 4 (0.01) | hap8: | GAA-Leu33 | 3 (<0.01) |
| hap9: | GAA-Pro33 | 1 (<0.01) | |||
| hapF: | ACC | 2 (<0.01) | hap10: | ACC-Leu33 | 1 (<0.01) |
| hap11: | ACC-Pro33 | 1 (<0.01) | |||
Analysis of the relationship of individual SNPs with receptor density did not indicate any association of genotype with receptor density, as shown in table 3. The relationship of receptor density with promoter haplotype and haplotype including Leu33Pro was also analysed; there was no significant difference in receptor density according to ITGB3 haplotypes (data not shown).
Table 3.
The relationship of ITGB3 polymorphisms with platelet αIIbβ3 receptor density, plasma fibrinogen and plasma vWF activity
| −468 G/A | −425 A/C | −400 C/A | Leu33Pro | |||||
|---|---|---|---|---|---|---|---|---|
| mean (95% CI) | p | mean (95% CI) | p | mean (95% CI) | p | mean (95% CI) | p | |
| αIIbβ3 | GG: 136 (133, 139) | 0.12 | AA: 136 (133, 139) | 0.85 | CC: 137 (134, 140) | 0.62 | LL: 137 (134, 140) | 0.97 |
| (MFI) | GA: 139 (134, 143) | AC: 138 (133, 143) | CA: 237 (132, 142) | LP: 136 (131, 141) | ||||
| AA: 126 (119, 133) | CC: 138 (131, 144) | AA: 128 (120, 137) | PP: 138 (133, 143) | |||||
| Fibrinogen | GG: 3.36 (3.21, 3.52) | 0.62 | AA: 3.36 (3.21, 3.51) | 0.33 | CC: 3.40 (3.26, 3.55) | 0.92 | LL: 3.35 (3.20, 3.50) | 0.17 |
| (gL−1) | GA: 3.51 (3.24, 3.80) | AC: 3.60 (3.29, 3.94) | CA: 3.43 (3.14, 3.74) | LP: 3.68 (3.36, 4.02) | ||||
| AA: 3.40 (3.06, 3.78) | CC: 3.35 (2.60, 4.31) | AA: 3.58 (3.30, 3.90) | PP: 3.32 (2.67, 4.12) | |||||
| vWF | GG: 112.7 (105.1, 120.4) | 0.47 | AA: 105.4 (99.6, 111.2) | 0.01 | CC: 113.1 (106.0, 120.1) | 0.42 | LL: 106.8 (100.5, 113.2) | 0.06 |
| activity | GA: 108.3 (99.3, 117.3) | AC: 124.4 (110.0, 138.8) | CA: 105.3 (95.2, 115.3) | LP: 121.8 (109.6, 134.0) | ||||
| (%) | AA: 97.1 (76.7, 117.5) | CC: 139.7 (76.9, 202.5) | AA: 99.4 (70.2, 128.7) | PP: 131.6 (73.9, 189.3) |
Data presented as mean or geometric mean (95% confidence intervals)
MFI: Mean fluorescence intensity
Association of ITGB3 polymorphisms and haplotype with plasma fibrinogen and vWF activity
There was no significant difference in fibrinogen according to ITGB3 genotype, as shown in table 3, or haplotype (data not shown). There was no significant association of the −400C/A and −468G/A polymorphisms with vWF activity, however there was a significant association of the −425A/C polymorphism and a borderline association of Leu33Pro with vWF activity, as shown in table 3. Analysis of the relationship of promoter haplotypes and haplotypes including Leu33Pro with vWF activity did not indicate any significant associations (data not shown), however, the data was suggestive of potential differences in vWF activity, particularly in haplotype combinations including haplotype 3 (GCC-Pro33). To investigate this further we created a dummy variable including all subjects with haplotype combinations including haplotype 3 (n=43) compared with subjects homozygous for haplotype 1 (GACLeu33) and compared vWF activity: vWF was significantly higher in subjects possessing haplotype 3 (128.6 [112.1, 145.1]%) compared to subjects homozygous for haplotype 1 (106.3 [98.1, 114.5]%, p=0.02). vWF was also significantly higher in subjects possessing haplotype 3 when compared with all other subjects (107.1 [101.2, 113.0]%, p=0.02). vWF activity differed significantly by ethnicity, with African American subjects having significantly lower vWF (96.6 [SD 37.8]%) than Caucasian subjects (120.4 (SD 44.1)%, p=0.02 after Sheffe post hoc analysis). Furthermore, although ITGB3 haplotype did not differ significantly by ethnicity, haplotype 3 was less prevalent in African Americans (10.4%) than Caucasians (21.4%, p= 0.08). Therefore we analysed the relationship of haplotype 3 with vWF activity accounting for ethnicity and haplotype 3 remained significantly higher in those possessing haplotype 3 (p=0.03).
Discussion
Regions of a gene that are conserved between species are likely to be functionally significant and bioinformatics analysis of the region of ITGB3 promoter containing the −400C/A, −425A/C and −468G/A polymorphisms indicated a number of regions conserved between species, suggesting they may be important in regulation of ITGB3 expression. Luciferase reporter gene analyses in cells representative of megakaryocyte and endothelial cells indicated modest (~50%) increases in reporter gene expression for the GAA, GCC, AAC, AAA and ACC constructs relative to the GAC construct, supporting a potential role for the polymorphisms in modulating gene expression. In contrast, EMSA analyses did not indicate any clear difference in nuclear protein binding pattern between the alleles of each polymorphism. However, luciferase reporter gene and EMSA analyses do not take into account the native chromatin structure of the human ITGB3 gene and therefore the inconsistency of results between the 2 methods does not in itself negate an effect of the polymorphisms on ITGB3 expression in vivo. Therefore, to determine whether the differences in reporter gene expression translated into differences in αIIbβ3expression in platelets, genotype and haplotype associations with receptor expression were determined.
The extent of linkage disequilibrium between the promoter polymorphisms and the Leu33Pro polymorphism were similar to those reported by O’Haloran et al. [15]. Six of the 8 potential promoter haplotypes were identified in the present study and inclusion of the Leu33Pro polymorphism resulted in the detection of 11 haploytpes, in keeping with those of O’Halloran et al. [15]. The prevalence of the 3 most common haplotypes (GAC-Leu33, AAA-Leu33, GCC-Pro33) were approximately equivalent in the two study groups, with the majority of Pro33 alleles associated with the GCC promoter haplotype and the Leu33 allele associated predominantly with the GAC promoter haplotype. There was some discrepancy in the occurrence of the less frequent haplotypes, which likely reflects ethnic heterogeneity in the present study.
We found a wide range in platelet αIIbβ3 expression which was not explained by ITGB3 promoter and Leu33Pro genotype or haplotype. Previous reports of the association of Leu33Pro with receptor density have been inconsistent [7]; however, the results of the present study lend further support for a lack of association of ITGB3 promoter polymorphisms and Leu33Pro with receptor density in unstimulated platelets. These results are inconsistent with the luciferase reporter gene results in which the common GCC and AAA promoter haplotypes were associated with a 50% increase in luciferase reporter gene expression compared with the GAC haplotype. The influence of the promoter polymorphisms on αVβ3expression in human endothelial cells or smooth muscle cells cannot be inferred from the results of our study regarding αIIbβ3 receptor expression due to cell specific regulation of αIIbβ3 and αVβ3 receptor expression. Increased αIIbβ3 receptor expression is reported to arise from increased steady-state mRNA levels of both αIIb and β3 mRNA resulting from enhanced transcription [16]. In contrast, the expression of αVβ3 appears to be largely dependent upon regulation of β3 expression, both at the transcriptional and post-transcriptional level [17,18]. Therefore it cannot be excluded that the influence of the promoter polymorphisms on ITGB3 gene expression may have more of an influence on the ultimate expression of αVβ3than on the expression ofαIIbβ3.
We found an association of ITGB3 −425A/C and Leu33Pro polymorphisms and the GCC-Pro33 haplotype (hap3) with vWF activity. Since vWF is released from endothelial cell Weibel Palade bodies this may reflect an influence on endothelial cell function. Shear stress induces activation of endothelial cell αVβ3 [19] and it has been shown that shear stress also induces increased secretion of vWF from endothelial cell Weibel Palade bodies [20]. Therefore it is possible that shear stress elicits differential responses in endothelial cells expressing different ITGB3 haplotypes. Studies of the Leu33Pro polymorphism indicated differential effects on platelet function, with the Pro33 allele associated with enhanced outside-in signalling in platelets and increased α-granule secretion [21]. In addition, enhanced cell migration on vitronectin and osteopontin was observed in CHO cells expressing αVβ3 Pro33 [22], which may lend support for a functional effect of Leu33Pro in endothelial cells.
Conclusions
Bioinformatics and reporter gene analyses suggested that the ITGB3 −468 G/A, −425 A/C and −400 C/A polymorphisms may be functional. However, promoter haplotypes and promoter haplotypes in combination with the Leu33Pro polymorphism were not significantly associated with αIIbβ3 receptor density, despite wide inter-individual differences in receptor density. The GCC-Pro33 haplotype was, however, associated with enhanced vWF activity, which may indicate ITGB3 haplotype influences endothelial function. Further studies are required to determine whether ITGB3 haplotype is associated with enhanced αVβ3 expression or activity.
Acknowledgements
This study was funded through grants from the British Heart Foundation (PhD studentship FS/2001012, KEP) and the Heart, Lung and Blood Institute of the National Institutes of Health (HL65229, PB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jang Y, Lincoff AM, Plow EF, Topol EJ. Cell adhesion molecules in coronary artery disease. J Am Coll Cardiol. 1994;24:1591–1601. doi: 10.1016/0735-1097(94)90162-7. [DOI] [PubMed] [Google Scholar]
- 2.Wilhide CC, Jin Y, Guo Q, et al. The human integrin β3 gene is 63 kb and contains a 5′-UTR sequence regulating expression. Blood. 1997;90:3951–3961. [PubMed] [Google Scholar]
- 3.Hoshiga M, Alpers CE, Smith LL, Giachelli CM, Schwartz SM. αvβ3 integrin expression in normal and atherosclerotic artery. Circ Res. 1995;77:1129–1135. doi: 10.1161/01.res.77.6.1129. [DOI] [PubMed] [Google Scholar]
- 4.Negrier C, Grenier C, Attali O, Dechavanne M, Vinciguerra C. Identification of new and known polymorphisms in glycoprotein IIb and IIIa genes by denaturing gradient gel electrophoresis. Platelets. 1998;9:374–380. doi: 10.1080/09537109876447. [DOI] [PubMed] [Google Scholar]
- 5.Weiss EJ, Bray PF, Tayback M, et al. A polymorphism of a platelet glycoprotein receptor as an inherited risk factor for coronary thrombosis. N Engl J Med. 1996;334:1090–1094. doi: 10.1056/NEJM199604253341703. [DOI] [PubMed] [Google Scholar]
- 6.Carter AM, Catto AJ, Bamford JM, Grant PJ. Platelet GPIIIa PlA and GPIb variable number tandem repeat (VNTR) polymorphisms and markers of platelet activation in acute stroke. Arteriosclerosis, Thrombosis & Vascular Biology. 1998;18:1124–1131. doi: 10.1161/01.atv.18.7.1124. [DOI] [PubMed] [Google Scholar]
- 7.Michelson AD, Furman MI, Goldschmidt-Clermont P, et al. Platelet GP IIIa Pl(A) polymorphisms display different sensitivities to agonists. Circulation. 2000;101:1013–1018. doi: 10.1161/01.cir.101.9.1013. [DOI] [PubMed] [Google Scholar]
- 8.Vijayan KV, Liu Y, Dong JF, Bray PF. Enhanced activation of mitogen-activated protein kinase and myosin light chain kinase by the Pro33 polymorphism of integrin β3. J Biol Chem. 2003;278:3860–3867. doi: 10.1074/jbc.M208680200. [DOI] [PubMed] [Google Scholar]
- 9.Blanchette M, Tompa M. FootPrinter: A program designed for phylogenetic footprinting. Nucleic Acids Res. 2003;31:3840–3842. doi: 10.1093/nar/gkg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y, Wilhide CC, Dang C, et al. Human integrin β3 gene expression: evidence for a megakaryocytic cell-specific cis-acting element. Blood. 1998;92:2777–2790. [PubMed] [Google Scholar]
- 11.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter AM, Ossei-Gerning N, Wilson IJ, Grant PJ. Association of the platelet Pl(A) polymorphism of glycoprotein IIb/IIIa and the fibrinogen Bβ448 polymorphism with myocardial infarction and extent of coronary artery disease. Circulation. 1997;96:1424–1431. doi: 10.1161/01.cir.96.5.1424. [DOI] [PubMed] [Google Scholar]
- 13.Yee DL, Sun CW, Bergeron AL, Dong JF, Bray PF. Aggregometry detects platelet hyperreactivity in healthy individuals. Blood. 2005;106:2723–2729. doi: 10.1182/blood-2005-03-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yee DL, Bergeron AL, Sun CW, Dong JF, Bray PF. Platelet hyperreactivity generalizes to multiple forms of stimulation. J Thromb Haemost. 2006;4:2043–2050. doi: 10.1111/j.1538-7836.2006.02089.x. [DOI] [PubMed] [Google Scholar]
- 15.O'Halloran AM, Curtin R, O'Connor F, et al. The impact of genetic variation in the region of the GPIIIa gene, on Pl expression bias and GPIIb/IIIa receptor density in platelets. Br J Haematol. 2006;132:494–502. doi: 10.1111/j.1365-2141.2005.05897.x. [DOI] [PubMed] [Google Scholar]
- 16.Zutter MM, Fong AM, Krigman HR, Santoro SA. Differential regulation of the α2β1 and αIIbβ3 integrin genes during megakaryocytic differentiation of pluripotential K562 cells. J Biol Chem. 1992;267:20233–20238. [PubMed] [Google Scholar]
- 17.Witzenbichler B, Kureishi Y, Luo Z, et al. Regulation of smooth muscle cell migration and integrin expression by the Gax transcription factor. J Clin Invest. 1999;104:1469–1480. doi: 10.1172/JCI7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X, Clark RA, Galanakis D, Tonnesen MG. Fibrin and collagen differentially regulate human dermal microvascular endothelial cell integrins: stabilization of αv/β3 mRNA by fibrin. J Invest Dermatol. 1999;113:913–919. doi: 10.1046/j.1523-1747.1999.00786.x. [DOI] [PubMed] [Google Scholar]
- 19.Tzima E, del Pozo MA, Shattil SJ, Chien S, Schwartz MA. Activation of integrins in endothelial cells by fluid shear stress mediates Rho-dependent cytoskeletal alignment. EMBO J. 2001;20:4639–4647. doi: 10.1093/emboj/20.17.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galbusera M, Zoja C, Donadelli R, et al. Fluid shear stress modulates von Willebrand factor release from human vascular endothelium. Blood. 1997;90:1558–1564. [PubMed] [Google Scholar]
- 21.Vijayan KV, Liu Y, Sun W, Ito M, Bray PF. The Pro33 isoform of integrin β3 enhances outside-in signaling in human platelets by regulating the activation of serine/threonine phosphatases. J Biol Chem. 2005;280:21756–21762. doi: 10.1074/jbc.M500872200. [DOI] [PubMed] [Google Scholar]
- 22.Sajid M, Vijayan KV, Souza S, Bray PF. PlA polymorphism of integrin β3 differentially modulates cellular migration on extracellular matrix proteins. Arterioscler Thromb Vasc Biol. 2002;22:1984–1989. doi: 10.1161/01.atv.0000043664.48689.7f. [DOI] [PubMed] [Google Scholar]