Abstract
Prophenoloxidases (PPOs) are key enzymes of the melanization reaction, which is a prominent defense mechanism of arthropods. The mosquito Aedes aegypti has ten PPO genes in the genome, four of which (PPO1, PPO3, PPO5, and PPO8) were expressed in response to microbial infection. Cactus depletion resulted in transcriptional activation of these four genes, suggesting this up-regulation to be under the control of the Toll pathway. The silencing of Cactus also led to developmental arrest and death of the avian malaria parasite, Plasmodium gallinaceum. We discovered that RUNT-related transcription factor 4 (RUNX4), the orthologue of Drosophila Lozenge, bound to the RUNT binding motif in the promoter of mosquito PPO genes and stimulated the expression of Drosophila PPO-A1 and PPO3 in S2 cell line. The immune effects caused by Cactus depletion were eliminated by double knockdown of Cactus/RUNX4. These findings suggest that RUNX4 regulates PPO gene expression under the control of the Toll pathway and plays a critical role in restricting parasite development.
Keywords: immune pathways, prophenoloxidase, RUNX transcription factor
Mosquitoes are important vectors of numerous human diseases, most notably malaria and Dengue fever. Current research in mosquito immunity focuses on specific interactions between pathogens/parasites and their vectors to develop novel pest control strategies. Similar to Drosophila melanogaster, mosquitoes have two major immune signal transduction pathways—Toll and IMD—which lead to the activation of two NF-κκB transcription factors—REL1 and REL2, respectively. In addition, the melanization reaction serves as an important defense mechanism in the mosquito immune response (1). Melanotic encapsulation (the production and deposition of melanin pigments on the surface of pathogens or parasites) is a common phenomenon found in many arthropods, including flies and mosquitoes. This reaction requires the proteolytic activation of prophenoloxidase (PPO). After initiation of the reaction by pathogen pattern-recognition proteins, a serine protease (SP) cascade is sequentially activated to finally cleave PPO to attain its oxidase activity. The PPO activation cascade is negatively regulated by serpins (serine protease inhibitors), such as D. melanogaster Spn27A (2, 3), Anopheles gambiae Serpin-2 (4), and Manduca sexta Serpin-3 (5).
The D. melanogaster genome contains three PPO genes—PPO-A1, PPO-A3, and PPO3. Black cells (bc), which encode PPO-A1, have been used as a maturation-specific marker of hematopoiesis (6). RUNX are RUNT-related transcription factors containing a highly conserved RUNT domain. They are key regulators in multiple developmental processes, ranging from hematopoiesis to carcinogenesis. Recent reports have shown that a Drosophila RUNX [Lozenge (Lz)] and a GATA transcription factor (Serpent) are both necessary for hemocyte differentiation and proliferation. The crystal cell-specific expression of Drosophila PPOs is dependent on Lz (7); however, the effect of Lz on insect immunity has yet to be elucidated.
Genome analyses revealed an expansion of the PPO gene family in mosquitoes (8, 9). There are nine PPO genes in An. gambiae and ten in Ae. aegypti, but only one to three in flies and other insect species. In addition to melanization, PPOs are essential for wound healing and hardening of the egg chorion. The existence of multiple PPO genes in mosquitoes makes it difficult to elucidate the functions and regulation of each individual gene.
The melanization process in mosquitoes has drawn considerable attention since the discovery of an An. gambiae strain refractory to the simian malaria parasite, P. cynomolgi (10). The specific genetic traits of this strain lead to melanization of the ookinetes and prevention of parasites developing into the oocyst stage. In An. gambiae, the RNA interference (RNAi)-mediated knockdown of C-type lectin 4 (CTL4), CTL mannose binding lectin 2 (CTMA2), and Serpin-2 resulted in an increased resistance of the mosquito to the rodent malaria parasite Plasmodium berghei (4, 11). Both CTL4 and CTMA2 are hemolymph-circulating C-type lectins that, when depleted, arrest development of the rodent parasite and simultaneously enhance melanization (11). Serpin-2 is a mosquito orthologue of Drosophila Spn27A, which was proposed to control the melanization reaction by inhibiting a clip-domain serine protease (CLIP) that activates PPO. However, the role of melanization in defense against malaria parasites in mosquitoes remains controversial. The depletion of CTL4 and CTMA2 as well as Serpin-2 did not dramatically affect the parasite development in the natural vector–parasite system of An. gambiae and the human malaria parasite Plasmodium falciparum (12, 13). Melanization has been linked to the killing of malaria parasites in susceptible mosquitoes, although appears here only to be responsible for disposal of dead parasites in the refractory strain (14).
Recently, it was reported that activation of mosquito immunity by removing the NF-κB inhibitor Cactus blocked the proliferation of P. berghei (15). While the silencing of Cactus also increased the melanization of parasite ookinetes, the link between immune system activation by Cactus depletion and ookinete killing is not yet clear. In Ae. aegypti, the sequence analysis of mosquito PPOs, their gene expression profiles, and comparison between Cactus and Serpin-2 knockdown phenotypes against the avian malaria parasite suggest the presence of an immune-inducible killing mechanism of the parasite. Here, we show that four of the ten PPO genes are expressed in response to microbial infection and that transcription is regulated by a RUNT-related transcription (RUNX) factor and the Toll pathway. Furthermore, the RUNX4-mediated immune activation under regulation of the Toll pathway has a potential role that largely restricts the parasite development.
Results and Discussion
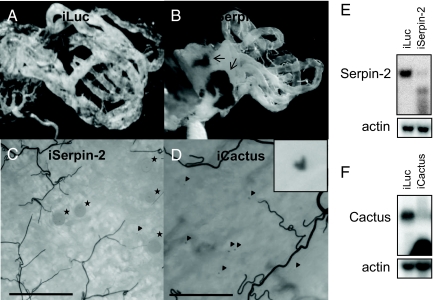
Using the laboratory model system of the parasite Plasmodium gallinaceum and the mosquito Ae. aegypti (16), the depletion of Serpin-2 was shown to trigger the melanization reaction, which resulted in formation of melanotic masses in the mosquito hemocoel (Fig. 1 A and B). However, this did not decrease the number of avian malaria parasite oocysts in the midgut (Fig. 1C). These results are in accord with studies that used An. gambiae and the human malaria parasite P. falciparum, but not the rodent malaria parasite P. berghei (4, 12). In contrast to Serpin-2 depletion, knockdown of Cactus expression appeared to have a broader effect on the immune system, which resulted in refractoriness of Ae. aegypti against the same avian malaria parasite. Developmental arrest and ookinete melanization occurred as a local reaction in the midgut (Fig. 1D). Since Cactus acts as a REL1 inhibitor, these results suggest that the Toll immune pathway has the ability to block development of the P. gallinaceum parasite into oocysts in Ae. aegypti.
Fig. 1.
The interaction between mosquito melanization and the development of the malaria parasites. The spontaneous melanotic masses were only found in the hemocoel cavities (internal tissue) of the mosquitoes with Serpin-2 depletion (B), but not in those with control luciferase dsRNA treatment (A) or Cactus depletion (not shown). However, the development of Plasmodium ookinetes to oocysts was not notably affected by the activation of melanization reaction by Serpin-2 depletion (C). In contrast, the proliferation of the malaria parasites to oocysts was almost not observed in mosquitoes with Cactus depletion by RNA interference; instead, the melanized ookinetes were found in mosquito midguts (D). The transcript knockdowns for Cactus and Serpin-2 were confirmed by means of Northern analysis (E and F). The black arrows, black arrowheads, and black stars indicate the melanotic masses, melanized ookinetes and fully developed oocysts, respectively. (Scale bars, 100 μm.) A and B were photographed using Leica DMR; C and D were photographed using Nikon DXM 1200.
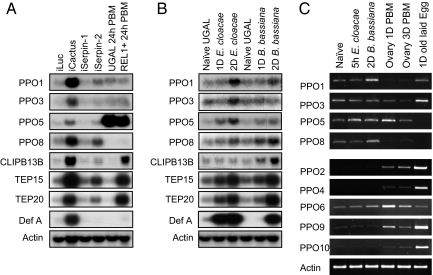
In the study of the Toll pathway, Cactus depletion resulted in enhanced expression of four PPO genes (PPO1, PPO3, PPO5, and PPO8) in Ae. aegypti (Fig. 2A). Therefore, we attempted to explore the connection between anti-parasitic response by Cactus depletion and transcriptional activation of PPO. When compared with Drosophila and other insects, mosquitoes have a significant expansion in PPO gene family. By amino acid sequence comparison, we found that some—but not all—mosquito PPOs have a typical cleavage site for activation (supporting information (SI) Fig. S1). This site, consisting of a decapeptide with NR*FG (*:scissile bond) and a Y/F residue (11th residue before *), is present in the majority of biochemically characterized PPOs from moths and beetles and in some PPOs from flies and mosquitoes. There are only a few exceptions: G to S in Drosophila PPO-A1 and Anopheles PPO7 and G to K in Tribolium beetle PPO2. The cleavage activation site of PPO is similar to the reactive site loop (RSL) sequence of Drosophila Spn27A and its orthologues. The RSL of a serpin is the site that interacts with its target protease to form a stable complex. The Spn27A-related serpins share the NK-FG (P2-P2′) consensus residues and Y/F at P11, which are very similar to the typical cleavage site for PPO activation. This suggests that the Spn27A subclass serpin could inhibit specific serine proteases, which proteolytically activate PPOs. Interestingly, six Aedes PPOs and four Anopheles PPOs do not have the typical cleavage site for PPO activation (Fig. S1). With few exceptions, the phylogenic relationship of insect PPOs generally supports the presence of a distinct group of mosquito PPOs with the atypical cleavage site (Fig. S2). The two unique characteristics of mosquito PPOs—mosquito-specific family expansion and the presence of PPOs with an atypical cleavage site—suggest the existence of another PPO activation mechanism distinct from the melanization reaction controlled by Serpin-2.
Fig. 2.
Indirect activation of four immune-inducible PPOs by the Toll pathway. Four immune-inducible PPO genes were activated by Cactus depletion but not by gain-of-function of REL1 in REL1+ transgenic mosquitoes (A), indicating that REL1 transcription factor is not the sole factor involved. CLIP-SPs, serpins, and TEPs were fully activated by both Cactus depletion and transgenic over-expression of REL1. Defensin A was also indirectly activated by the Toll pathway. Four PPO genes were immune inducible by bacterial (E. cloacae) and/or fungal (the spores of B. bassiana) infection (B). The expression profiles of nine of ten PPO genes were characterized using RT-PCR during immune response and embryonic development (C). Among the ten PPO genes, only transcripts of the PPO7 gene were not detected using RT-PCR. Those of PPO2, PPO4, PPO9, and PPO10 were present during ovary and egg development rather than in female mosquitoes with or without immune challenge. Using Northern analysis, immune induction of PPO3 was observed to have occurred in mosquitoes two days after E. cloacae infection (B); a time point was not used in the RT-PCR experiments. Aedes actin gene was used as a RNA loading control. Serpin-1 is a serpin with the most similarity to Serpin-2 and was used as a negative control. Naïve UGAL, UGAL mosquitoes without any treatment; 1D or 2D E. cloacae, UGAL mosquitoes 1 day or 2 days after septic injury with E. cloacae liquid culture; 1D or 2D B. bassiana, UGAL mosquitoes 1 day or 2 days after septic injury with B. bassiana. iLuc, iCactus, iSerpin-1 and iSerpin-2 indicate control (luciferase), Cactus, Serpin-1, and Serpin-2 dsRNA-treated mosquitoes, respectively. UGAL or REL1 + 24 h PBM, the wild-type UGAL, or the REL1+ transgenic mosquitoes 24 h after blood feeding.
The silencing of Cactus by RNAi resulted in up-regulation of a large group of genes, including four PPO, CLIP-B13B, TEP15, TEP20 (TEP15 and TEP20 are two mostly expressed TEP genes in the adult female mosquitoes; data not shown), and Defensin A genes (Fig. 2A). We also confirmed additional Toll-pathway-dependent activation of CLIPs, such as CLIP-B5, -B13B, -B15, -B24, -B29, -B35, and -B37, and serpins, including Serpin-2, Serpin-4, and Serpin-9 (data not shown). The knockdown of Serpin-2 resulted in weak activation of PPO, CLIP, and TEP genes (Fig. 2A). These results suggest a minor role of Serpin-2 in the suppression of the mosquito Toll pathway.
An interesting result emerged when the expression profiles of the PPO genes were compared with other Toll-pathway-dependent immune genes in REL1+ transgenic mosquitoes with REL1 gain-of-function (17). We observed that the expression profiles of the PPO genes were not affected by transgenic over-expression of the REL1 transcriptional activator of the Toll pathway (Fig. 2A). However, expression of other Toll-pathway-dependent immune genes, including CLIP-B13B, TEP15, and TEP20, was substantially activated (Fig. 2A). This indicates that REL1 may not be the sole transcriptional activator of PPO genes and that other activator(s) might be involved in the regulation.
To elucidate the transcriptional regulation mechanism of PPO genes, we studied the expression profiles of the PPO genes after microbial infection in the mosquito Ae. aegypti. Transcriptions of PPO1, PPO3, PPO5, and PPO8 genes were induced after bacterial (Enterobacter cloacae) and fungal (Beauveria bassiana) infections in female mosquitoes (Fig. 2B). Interestingly, these four inducible genes belong to the group characterized as having mosquito-specific atypical cleavage sites (Figs. S1 and S2). Additionally, reverse-transcriptase PCR (RT-PCR) analysis of PPO6 showed a mild induction of transcription after fungal challenge (Fig. 2C), but the mRNA level was not high enough to be detected by Northern analyses. Also, the PPO6 gene has been shown to be generally expressed during larval and pupal development (18). PPO7, however, was not detected using either RT-PCR or Northern analyses. The transcripts of the remaining four PPO genes (PPO2, PPO4, PPO9, and PPO10), abundant during the embryonic stage, almost disappeared during the immune stage in female mosquitoes (Fig. 2C).
In Drosophila, a RUNX transcription factor, Lz, was reported to be a transcriptional activator of PPO genes during hemocyte differentiation and proliferation (7). All RUNX transcription factors share a highly conserved DNA-binding RUNT domain, which recognizes the consensus sequence TGYGGTY (19). The promoter sequences of the inducible PPO genes from Ae. aegypti contain at least one putative RUNT binding site (between −80 and −350) proceeding to the translation initiation codon ATG. Therefore, we suspected the role of RUNX factor(s) to be in the transcriptional activation of inducible PPO genes.
Although it has been predicted that the mosquito genome encodes four RUNT-related proteins (RUNX1–4), the annotation was incomplete with only the RUNT domain predicted. First, we characterized full-length transcripts of four RUNX genes by PCR-based cloning and RACE (Fig. S3). Mosquito RUNX genes are 1:1 orthologues to Drosophila RUNX genes when the RUNT domain was used for phylogenic analysis. Aedes RUNX4 is an orthologue of Drosophila Lz (Fig. S4).
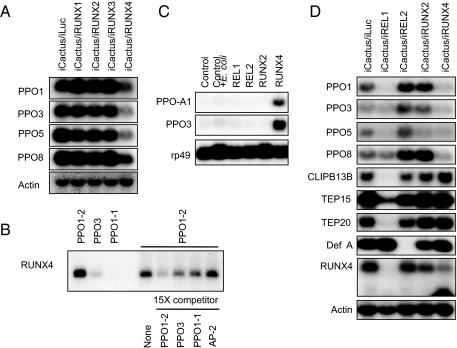
Among four mosquito RUNX genes, RUNX2 and RUNX4 were inducible by bacterial and fungal challenge, but transcripts of RUNX2 were most abundant in 1-day-old eggs (Fig. S5). This suggests a role for RUNX2 in embryonic development. The transcripts of RUNX4 were detected at the highest level in female mosquitoes 2 days after fungal infection (Fig. S5). The expression levels of RUNX1 and RUNX3 transcripts were not elevated by microbial infection (data not shown). Moreover, the double knockdown of Cactus and one of the four RUNX factors uncovered RUNX4 as a key transcriptional activator of the inducible PPO genes under the control of the Toll pathway, albeit the knockdown of RUNX2 slightly affected the transcriptional activation of the same genes (Fig. 3A).
Fig. 3.
Direct activation of four immune-inducible PPO genes by RUNX4. Only RUNX4 depletion compromised the activation of these genes by Cactus depletion (A). Gel mobility-shift assay revealed the direct binding of RUNX4 to mosquito RUNT binding motifs (B). Two predicted motifs from PPO1 gene promoters, PPO1–1 and PPO1–2, and a motif from PPO3 were used for binding and competition assays. RUNX4 specifically activated the Drosophila PPO genes in Drosophila S2 cells (C). Drosophila PPO-A1 (bc) and PPO3 genes were activated by Aedes RUNX4 in Drosophila S2 cells. The activation of Drosophila PPO-A3 was not detected using this Northern analysis. The transcriptional activation of PPOs by Cactus depletion was compromised by both REL1 and RUNX4 (D). Defensin A was only partially activated by Cactus depletion (less than 10%, compared with full activation at 5 h after E. cloacae challenge; data not shown), and the activation by Cactus depletion was compromised only by REL2. This suggests that Cactus depletion partially activates the IMD pathway, which is independent of the REL1 transcription factor. CLIP-B13B and TEPs were shown to be regulated only by REL1, not by RUNX4. The transcript knockdown for each RUNX was confirmed by means of RT-PCR analysis (Fig. S8).
Gel mobility-shift assays revealed that the RUNX4 factor specifically bound to the RUNT binding motifs from the promoter sequences of immune-inducible PPO genes. The promoter region of the PPO1 gene contains two putative RUNT binding sites at approximately −350 and −280 from the translation initiation codon, ATG. The in vitro-translated RUNX4 factor bound directly to the −350 RUNT binding motif (PPO1–2), but not to the −280 motif (PPO1–1). Another RUNT binding motif (PPO3) at approximately −80 in the promoter of PPO3 gene also showed specific binding to the RUNX4 factor (Fig. 3B). In addition, mosquito RUNX4 strongly activated the expression of both Drosophila PPO-A1 and PPO3 genes in Drosophila S2 cell line (Fig. 3C), proving that RUNX4 binds to RUNT motifs and directly activates PPO genes.
After revealing the transcriptional activation of PPO1, PPO3, PPO5, and PPO8 genes by RUNX4, we established a relationship between the two immune pathways (Toll and IMD) and RUNX factors in the regulation of inducible PPO genes. RNAi complementation tests were performed between Cactus and the transcriptional activators REL1 and REL2, as well as with RUNX2 and RUNX4. The activation of CLIPs and TEPs by Cactus depletion was complementarily abolished by REL1 only; the activation of defensin A was eliminated only by REL2, a transcriptional activator of the IMD pathway (Fig. 3D) (20). Interestingly, the activation of PPO genes by Cactus depletion was dismissed by double knockdown of either Cactus/REL1 or Cactus/RUNX4 (Fig. 3D). Moreover, double knockdown of Cactus/REL1 completely eliminated the expression of the RUNX4 gene (Fig. 3D), which was shown to be specifically activated by B. bassiana infection using RT-PCR analysis (Fig. S5). Both REL1-dependent and fungal-specific activation of the RUNX4 gene clearly indicate that the Toll pathway regulates the expression of this gene in the mosquito. Thus, we propose that RUNX4 activates the PPO genes after microbial infection under the regulation of the Toll pathway.
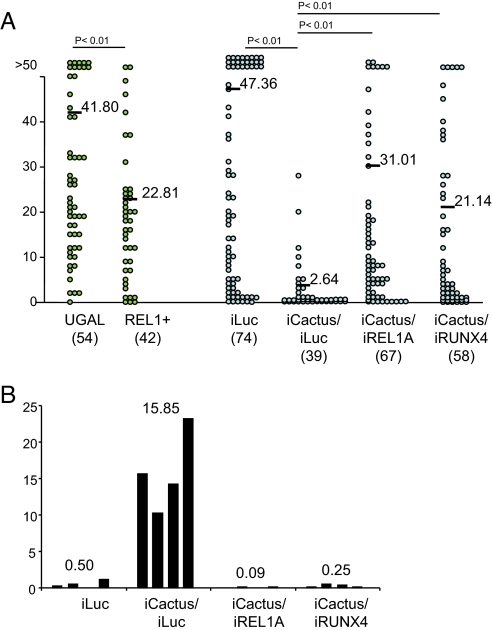
Next, we investigated the role of RUNX4 in the anti-parasitic response. Previous reports showed that development of malaria parasites was significantly hampered in mosquitoes in which immune systems had been activated before parasite infection (15, 21). In our system, we observed that boosting of mosquito immunity by Cactus depletion triggered killing and melanization of the avian malarial ookinetes (Figs. 1D and 4A). Double knockdown of Cactus/REL1 rescued the killing of parasite ookinetes by Cactus depletion (Fig. 4A). This implicates a role for the Toll pathway in the defense against the malaria parasites. However, only a partial restriction in Plasmodium development was seen in REL1+ transgenic mosquitoes with gain-of-function of Toll pathway when compared with the immune activation by Cactus depletion (Fig. 4A and Figs. S6–S9 for knockdown control). Moreover, transgenic over-expression of REL1 failed to activate the inducible PPO genes (Fig. 2A). These results indicate that other immune transcriptional factors are required for anti-parasitic activity of REL1. Indeed, the depletion of RUNX4 abolished immune effects caused by Cactus depletion. Cactus/RUNX4 double knockdown significantly compromised both the refractoriness of the mosquitoes against the parasite and ookinete melanization, albeit slightly less than with Cactus/REL1 double knockdown (Fig. 4 and S8 for the control of single REL1, REL2, and RUNX4 knockdown). With this comprehensive information, we propose that RUNX4 cooperates with REL1 in the anti-parasitic response.
Fig. 4.
Crosstalk between REL1 and RUNX4 transcription factors during the interaction with the avian malaria parasite. Cactus depletion restricts P. gallinaceum development and triggers ookinete melanization, both of which are compromised by both REL1 and RUNX4 (A and B). Mosquito midguts from each treated sample were dissected and the numbers of developed oocysts and the melanized ookinetes were scored eight days after blood feeding with P. gallinaceum-infected chicken. Data from oocysts were collected from three independent experiments in the comparison between UGAL and REL1+ transgenic mosquitoes (Fig. S6) and from four independent experiments with dsRNA-treated mosquitoes (Fig. S7) and pooled; resulting sample sizes are shown in brackets. (A) The number of fully developed oocysts in each midgut is shown as a circle. The mean number of parasite oocysts for each group is indicated with a black bar. (B) Mean values of melanized ookinetes for each independent experiment are indicated by black columns. The average of mean numbers from four experiments is shown on the top of columns.
In conclusion, our study has revealed that the expression of the four PPO genes is induced by microbial infection in the adult female Ae. aegypti mosquito. We also found that a transcription factor, RUNX4, is directly involved in the transcriptional activation of the PPO genes. Furthermore, RUNX4 might have a crucial role in the defense against the development of the avian malaria parasite P. gallinaceum in Ae. aegypti. This newly discovered immune mechanism contributes to our understanding of the immune interaction between the malaria parasite and its mosquito host at the molecular level.
Methods
Experimental Animals.
UGAL/Rockefeller, the wild-type strain of Ae. aegypti mosquitoes, and a transgenic mosquito strain, REL1+ (gain-of-function of REL1) (14), were maintained in laboratory culture. Adult mosquitoes were provided with water and a 10% sucrose solution. The avian malaria parasite Plasmodium gallinaceum was maintained in the laboratory by natural transmission between the Ae. aegypti UGAL/Rockefeller strain and chicks. Parasite oocyst numbers were determined in midguts dissected 8 days postmosquito infection and stained with 1% mercurochrome. Nikon E400 light microscopy was used to visualize and count oocysts.
Molecular Cloning of RUNX Factors.
The Runt binding domains from Drosophila RUNX factors were used as query sequences to search AaegL1.1, June 2006 (Vectorbase) by TBLASTX. Four Runt-domain-containing proteins were screened out from the genome, RUNX1 AAEL006160 (EAT42277.1), RUNX2 AAEL006167 (EAT42279.1), RUNX3 AAEL007036 (EAT41317.1), and RUNX4 AAEL007040 (EAT41314.1). Although automated annotations predicted only the Runt binding domain of each RUNX gene, the prediction was improved using other gene prediction programs (BGF: Beijing gene finder, Genewise, and NCBI blast 2.2.14). Finally, the full-length cDNA sequences of four Ae. aegypti RUNX factors were obtained by a combination of PCR-based cloning and both end RACE experiments. Rapid Amplification of cDNA Ends (RACE) was performed using the SMART RACE cDNA Amplification Kit (Clontech). Total RNAs from mosquitoes were reverse-transcribed to first-strand cDNA with adapters and then applied to PCR with RACE primers; RT-PCR for conventional cloning was done using a two-step method. Reverse transcription was carried out using an Omniscript reverse-transcriptase kit (Qiagen) with oligo(dT) primers and PCR was performed using Platinum High Fidelity Supermix (Invitrogen). The Ae. aegypti sequences reported in this paper have been deposited in the GenBank database, with the following accession numbers: EU604099 (RUNX1), EU604100 (RUNX2), EU604101 (RUNX3), and EU604102 (RUNX4). Primers used for molecular cloning are indicated in materials and methods of SI Methods.
Gene Expression Knockdowns.
For gene silencing, double-stranded RNA (dsRNA) was synthesized with T7 RNA polymerase. The T7-phage promoter sequences into both sense and antisense sequences of target genes were incorporated to generate template cDNAs containing T7 tag. RT-PCR was performed using the Titan one-step RT-PCR kit (Roche) with samples of 0.2 μg total RNA as templates to generate 400-bp to 1-kb gene-specific cDNA fragments. Synthesis of dsRNA was accomplished by simultaneous transcription of both strands of template DNA using the MEGAscript kit (Ambion). The luciferase gene was used to generate control iLuc dsRNA. A Picospritzer II (General Valve) was used to introduce corresponding dsRNA into the thorax of CO2-anesthetized mosquito females, at 1–2 days posteclosion. The knockdown of a specific transcript was confirmed by Northern analysis for Cactus and Serpin-2 (Fig. 1 E and F) or RT-PCR for RUNX genes (Fig. S9). Primers used for amplification are indicated in SI Methods.
Septic Injury, RNA Preparation and Northern Analysis.
Septic injuries were performed by pricking mosquitoes in the rear part of the abdomen with an acupuncture needle (0.20 × 25 mm) dipped into either Enterobacter cloacae bacterial culture (stationary phase of bacteria in LB broth; OD approximately 2.0) or a fungal spore suspension (approximately 5 × 107 viable spores/ml) of B. bassiana strain GHA. The viable spore number was calculated by spreading the suspension onto Sabouraud dextrose Emmons agar plates. Total RNA was prepared using TRIzol (GIBCO/BRL); 5 μg of total RNA from each sample was separated on a formaldehyde gel, blotted, and hybridized with the corresponding 32P-labeled DNA probe.
Electrophoretic Gel Mobility-Shift Assay (EMSA).
RUNX4 protein was synthesized by a coupled in vitro transcription–translation (TNT) system (Promega). The cDNA clone encoding the full length of RUNX4 was subcloned into pcDNA3.1/Zeo (+) (Invitrogen). Sense and antisense oligonucleotides for each motif were synthesized for three predicted RUNT binding motifs from two mosquito PPO gene promoters. The sense oligonucleotide sequences are:
PPO1–1, 5′-ATCGTGCAGTTGTGGTTTGGAATATGT-3′
PPO1–2, 5′-ATCACGGTTGTGCGGTCCCACAGATTT-3′
PPO3, 5′-TTGTGGAGATTGCGGTTTCCGTCCATG-3′.
The annealed deoxyoligonucleotide of each RUNT motif was purified from 15% TBE Criterion Precast Gel (Bio-Rad). Double-stranded oligonucleotides were labeled with γ-32P ATP. EMSA was performed using a gel-shift assay system (Promega). The DNA–protein complex was separated on 5% TBE Criterion Precast Gel (Bio-Rad) and visualized by means of autoradiography. For competition assay, 15-fold cold motif or non-specific control AP-2 motif was incubated with RUNX4 for 10 min and then further incubated with labeled PPO1–2 motif for 20 min.
Transfection Assay in Drosophila S2 Cell Line.
Coding region sequences of Ae. aegypti REL1 (REL1-A isoform), REL2 (REL2 Rel-type isoform), RUNX2, and RUNX4 were amplified by means of PCR, inserted into pAC5.1/V5/HisA vector (Invitrogen) and used to transfect Drosophila S2 cells. Cells (2 × 106 per mL) were distributed on 35-mm plates in Drosophila Schneider's medium (GIBCO) supplemented with 5% FBS (GIBCO) and 1X antibiotic–antimycotic (GIBCO). The cells were transfected with 1 μg of each plasmid construct and incubated for 6 h in serum-free Schneider's medium, which was then removed and replaced with complete medium. The cells were then incubated for 48 h, followed by bacterial challenge using heat-inactivated E. cloacae and then incubated for an additional 5 h. Total RNA was isolated from harvested cells using the TRIzol method (Invitrogen), and samples were then subjected to Northern analysis.
Computational Analysis.
All sequences retrieved from NCBI or Ensembl were aligned using ClustalX2.0 (Blosum 30 matrix, a gap penalty of 10, and an extension gap penalty of 0.1). A phylogenetic tree was constructed using the neighbor-joining method and displayed using Treeview. Plasmodium data collected from three or four independent experiments were analyzed using the Kolmogorov-Smirnov goodness of fit test and pooled. Sample sizes with brackets are shown. Statistically significant differences between samples were evaluated using the Mann-Whitney U test (Graphpad 5.0) and P values are marked on the graphics.
Supplementary Material
Acknowledgments.
We thank Dr. H. Jiang for helpful suggestions in preparing the manuscript. This work was supported by NIH/NIAID Grant R01AI05942.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition footnote: The Aedes aegypti sequences reported in this paper have been deposited in the GenBank database, with the following accession numbers: EU604099 (RUNX1), EU604100 (RUNX2), EU604101 (RUNX3), EU604102 (RUNX4).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804658105/DCSupplemental.
References
- 1.Cerenius L, Lee BK, Söderhäll K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 2.De Gregorio E, et al. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 3.Ligoxygakis P, et al. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J Biol Chem. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
- 6.Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- 7.Ferjoux G, Auge B, Boyer K, Haenlin M, Waltzer L. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol. 2007;305:726–734. doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 9.Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins FH, et al. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 11.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 12.Cohuet A, et al. Anopheles and Plasmodium: from laboratory models to natural systems in the field. EMBO Rep. 2006;7:1285–1289. doi: 10.1038/sj.embor.7400831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel K, et al. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc Natl Acad Sci USA. 2006;103:16858–16863. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volz J, Muller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8:1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 15.Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25:677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Slater LB. Malarial birds: Modeling infectious human disease in animals. Bull Hist Med. 2005;79:261–294. doi: 10.1353/bhm.2005.0092. [DOI] [PubMed] [Google Scholar]
- 17.Bian G, Shin SW, Cheon HM, Kokoza V, Raikhel AS. Transgenic alteration of Toll immune pathway in the female mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2005;102:13568–13573. doi: 10.1073/pnas.0502815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JS, Ruyl Kim S, Christensen BM, Li J. Purification and primary structural characterization of prophenoloxidases from Aedes aegypti larvae. Insect Biochem Mol Biol. 2005;35:1269–1283. doi: 10.1016/j.ibmb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 19.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 20.Shin SW, Kokoza V, Lobkov I, Raikhel AS. Relish-mediated immune deficiency in the transgenic mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100:2616–2621. doi: 10.1073/pnas.0537347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowenberger CA, et al. Mosquito-Plasmodium interactions in response to immune activation of the vector. Exp Parasitol. 1999;91:59–69. doi: 10.1006/expr.1999.4350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.