Abstract
Higher plant species differ widely in their growth responses to ammonium (NH4+). However, the molecular genetic mechanisms underlying NH4+ sensitivity in plants remain unknown. Here, we report that mutations in the Arabidopsis gene encoding GDP-mannose pyrophosphorylase (GMPase) essential for synthesizing GDP-mannose confer hypersensitivity to NH4+. The in planta activities of WT and mutant GMPases all were inhibited by NH4+, but the magnitude of the inhibition was significantly larger in the mutant. Despite the involvement of GDP-mannose in both l-ascorbic acid (AsA) and N-glycoprotein biosynthesis, defective protein glycosylation in the roots, rather than decreased AsA content, was linked to the hypersensitivity of GMPase mutants to NH4+. We conclude that NH4+ inhibits GMPase activity and that the level of GMPase activity regulates Arabidopsis sensitivity to NH4+. Further analysis showed that defective N-glycosylation of proteins, unfolded protein response, and cell death in the roots are likely important downstream molecular events involved in the growth inhibition of Arabidopsis by NH4+.
Keywords: glycosylation, NH4+ toxicity, unfolded protein response, L-ascorbic acid
Ammonium (NH4+) is an essential ion in living cells. NH4+ and nitrate (NO3−) form the major sources of nitrogen nutrition for plants and microorganisms. Furthermore, NH4+ is also an indispensable intermediate in the biosynthesis of essential cellular components. However, NH4+ is toxic to cellular organisms when present in excess amounts (1). In fact, NH4+ sensitivity is a widespread phenomenon in animals, plants, and fungi, although the levels of sensitivity differ considerably among different species (1). Plants, being unable to escape from harmful environments, are especially prone to NH4+-induced growth inhibition. Owing to the application of large quantities of nitrogen fertilizers in intensive agriculture, high levels of NH4+ accumulation are becoming more common in many natural and agricultural soils (2). Consequently, NH4+ toxicity has been linked to plant species extinction and decline of forest under certain ecological conditions in recent years (3).
The molecular mechanisms underlying plant sensitivity to this ion are still unclear. Past studies have revealed important physiological changes (for example, acidification of external growth environment, disturbance in the acid/base balance, or excessive energy consumption in pumping the toxic level of NH4+ out of cells) accompanying NH4+ uptake and toxicity symptoms (1, 2). However, the genetic determinant of NH4+ sensitivity in plants remains unknown. Interestingly, recent biochemical investigations in animal cells have shown that NH4+ sensitivity is linked with reduced efficiencies in protein glycosylation and the correct processing and secretion of glycoproteins (4–6). But, again, the genetic trigger for these changes has not been determined. From the fact that different plant families, or different species in the same plant family, can differ considerably in their growth response to NH4+ (7), we deduce that NH4+ sensitivity is genetically controlled. Identification of a genetic determinant should provide valuable clues for understanding the molecular mechanisms of NH4+ sensitivity. To this end, we have taken a forward genetics approach by identifying an Arabidopsis thaliana mutant showing enhanced NH4+ sensitivity than WT control and have characterized the hypersensitive response of hsn1 and its allelic mutant vtc1 to NH4+.
We found that hsn1 was the result of a point mutation in the gene encoding GDP-mannose pyrophosphorylase (GMPase; EC 2.7.7.22), which has been established to be essential for synthesizing the vital cellular metabolite GDP-mannose in Arabidopsis (8). We provide molecular genetic and biochemical evidence on the inhibition of GMPase by NH4+ and describe the consequential downstream molecular events likely to be important for the inhibition of Arabidopsis growth by NH4+. The relevance of our finding on further studies of the mechanisms underlying NH4+ sensitivity in higher plants is discussed.
Results
Morphological and Physiological Characterization of hsn1 Mutant.
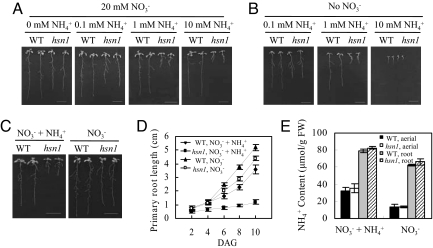
Previous studies have shown that Brassicaceae is one of the NH4+-sensitive plant families (1) and that the growth of the Col-0 ecotype of Arabidopsis, which is a member of Brassicaceae, is inhibited by physiological concentrations of NH4+ (9). When cultured on defined media with both NO3− and NH4+ as nitrogen sources, the growth of Arabidopsis, especially the elongation of its roots, is increasingly inhibited by rising concentrations of NH4+ (9). Based on these findings, we screened 20,000 ethylmethanesulfonate-mutagenized M2 seedlings on a half-strength Murashige–Skoog (MS) medium (with 20 mM NO3− and 10 mM NH4+ as nitrogen sources, hereafter referred as NO3− + NH4+ medium), and isolated a mutant exhibiting hypersensitivity to NH4+ (named as hsn1, hypersensitive to NH4+ 1). The hsn1 mutant showed dramatically reduced growth of both aerial and root organs in the media containing NH4+ irrespective of the presence or absence of NO3− as an additional nitrogen source (Fig. 1). Moreover, the level of growth retardation of hsn1, especially that of its root, increased substantially with rising NH4+ concentrations (Fig. 1 A and B), demonstrating that the growth of hsn1 (particularly its root) is more sensitive to NH4+ than that of WT Arabidopsis. A typical comparison of WT and hsn1 seedlings germinated on NO3− + NH4+ medium or a modified 1/2 MS medium with NO3− as the sole nitrogen source (henceforth named as NO3− medium) is shown in Fig. 1C. Quantitative analysis confirmed that the primary root of hsn1 was markedly shorter than that of WT control on both NO3− + NH4+ and NO3− medium, with the difference in primary root length between the 2 genotypes being much larger on NO3− + NH4+ medium (Fig. 1D). Consistent with previous findings (9), the primary root length of WT Arabidopsis seedlings was also significantly reduced on the medium containing NH4+ (i.e., NO3− + NH4+ medium; Fig. 1 C and D) compared with that attained on the medium with NO3− as the only nitrogen source (i.e., NO3− medium; Fig. 1 C and D).
Fig. 1.
Phenotypes of hsn1mutant. (A and B) Comparisons of seedling growth of WT Arabidopsis and hsn1 on modified MS media at 10 days after germination (DAG). In A, the nitrogen source included 20 mM NO3− with increasing concentrations of NH4+ (from 0 to 10 mM). In B, the nitrogen source was provided solely by ascending concentrations of NH4+ (from 0.1 to 10 mM). (Bars: 1 cm.) (C) A typical example of WT and hsn1 seedlings cultured on NO3− + NH4+ or NO3− medium. (Bars: 1 cm.) (D) Comparisons of postgerminative root growth between WT Arabidopsis and hsn1 on NO3− + NH4+ and NO3− medium. Statistically significant differences (P < 0.05) were found between the 4 data points at 8 or 10 DAG. (E) NH4+ contents (expressed as μmol/g of fresh weight) in the aerial and root tissues of WT Arabidopsis and hsn1 seedlings (10 DAG) grown on NO3− + NH4+ or NO3− medium. Significant differences (P < 0.01) were found between the NH4+ contents of aerial and root tissues for both WT Arabidopsis and hsn1 grown on either NO3− + NH4+ or NO3− medium.
The NH4+ content did not differ significantly between WT control and hsn1 in both aerial and root tissues on either NO3− + NH4+ or NO3− medium (Fig. 1E). For both WT Arabidopsis and hsn1 and on either NO3− + NH4+ or NO3− medium, the NH4+ content in the roots was significantly higher than that in the shoots (Fig. 1E), indicating that NH4+ uptake and translocation processes did not differ significantly between WT control and hsn1. The NH4+ contents in the aerial and root tissues of WT and hsn1 seedlings all were significantly higher on NO3− + NH4+ medium compared with their corresponding values on NO3− medium (Fig. 1E), reflecting a higher level of uptake and assimilation of the NH4+ present in the former medium by both genotypes.
Molecular and Biochemical Analysis of hsn1 and Its Allelic Mutant.
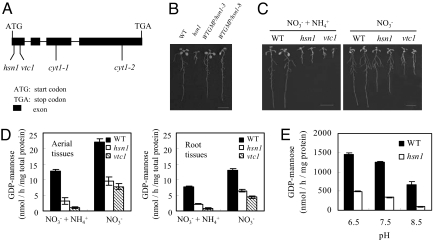
The hsn1 allele was isolated by using a positional cloning strategy and found to be caused by a point mutation in the gene (At2g39770) encoding GMPase (Fig. 2A). In addition to At2g39770, the Arabidopsis genome contains 2 more predicted genes (At3g55590, At4g30570) encoding putative GMPase (www.Arabidopsis.org). However, the 2 additional genes have been found not to complement the functional deficiency of At2g39770 (refs. 8 and 10). The mutation in hsn1 replaced a WT glycine residue (at position 11 of GMPase) by serine. The hsn1 phenotype on NO3− + NH4+ medium was fully complemented by transgenic expression of WT GMPase in the mutant background (Fig. 2B), confirming that a defective GMPase is responsible for the enhanced NH4+ sensitivity of hsn1.
Fig. 2.
Molecular characterization of hsn1. (A) A diagram illustrating the genomic coding sequence of Arabidopsis GMPase and its mutant alleles hsn1, vtc1, cyt1–1, and cyt1–2. (B) In the complementation experiments (n = 3), the NH4+-induced growth inhibition of hsn1 on NO3− + NH4+ medium was fully reversed by the expression of WT GMPase coding sequence in independent transgenic lines (WTGMP/hsn1–3, WTGMP/hsn1–8). (Bar: 1 cm.) (C) The vtc1 allele exhibited a similar tendency of growth inhibition as hsn1 on either NO3− + NH4+ (Left) or NO3− medium (Right), although the inhibition was much stronger for vtc1 than for hsn1. (Bars: 1 cm.) (D) Comparisons of in planta GMPase activity levels in the aerial and root tissues of 10 DAG seedlings of WT Arabidopsis and 2 GMPase mutants (hsn1, vtc1) cultured on NO3− + NH4+ or NO3− medium. GMPase activity was determined by monitoring the production of GDP-mannose (nmol/h per mg of total protein). Significant differences (P < 0.05) were found in GMPase activity levels among the 3 genotypes by either tissues types (aerial vs. root) or culture media (NO3− + NH4+ vs. NO3−). (E) Comparisons of the activities of recombinant WT GMPase and hsn1 enzymes under 3 pH regimes. Enzyme activity was determined as in D except that the purified recombinant proteins of WT GMPase and hsn1 were used for the assays. Significant differences (P < 0.01) were found in the activity levels between WT GMPase and hsn1 enzymes at all pH conditions and among the 3 activity levels exhibited by WT GMPase or hsn1.
GMPase is a highly conserved protein in eukaryotic organisms, with nearly 60% identity among Arabidopsis, human, and yeast orthologs and >85% identity between the orthologs from dicot and monocot plant species (data not shown). GMPase is necessary for synthesizing GDP-mannose, which is indispensable for proper N-glycosylation of proteins in eukaryotic cells (11). In higher plants, GDP-mannose is also an important intermediate for the biosynthesis of ascorbic acid (AsA) through the Smirnoff–Wheeler pathway (12). Previous studies have identified 2 types of mutations affecting the function of Arabidopsis GMPase. The vtc1 allele, causing a P22S change in GMPase protein (Fig. 2A), decreases GMPase activity and AsA content in Arabidopsis (8). Compared with WT controls, vtc1 seedlings are more susceptible to oxidation stress and their root length is constitutively shorter when grown with either single (NO3−) or dual (NO3− and NH4+) nitrogen sources (8, 13). Two cyt alleles (cyt1–1, cyt1–2; Fig. 2A), with a P89L substitution in the middle and an alteration of the C-terminal amino acid sequence of GMPase protein, respectively, display severe defects in N-glycosylation of proteins and are thus embryo-lethal (10). As anticipated, the root elongation of vtc1 was strongly inhibited on NO3− + NH4+ medium (Fig. 2C Left). Moreover, the degree of the inhibition displayed by vtc1 was more severe than that by hsn1 (Fig. 2C Left). Consistent with our earlier data (Fig. 1 C and D) and those published (13), the root lengths of hsn1 and vtc1 seedlings were both shorter than that of WT controls on NO3− medium (Fig. 2C Right).
The mutations in hsn1 and vtc1 did not decrease the transcript and protein accumulation levels of GMPase on either NO3− + NH4+ or NO3− medium [supporting information (SI) Fig. S1]. However, the in planta GMPase activity levels of hsn1 and vtc1 all were significantly lower than those of WT control in both the aerial and root tissues and on either NO3− + NH4+ or NO3− medium, although the decrease of GMPase activity in hsn1 was less severe than that in vtc1 (Fig. 2D). Further inspection of the data in Fig. 2D revealed 2 additional GMPase activity responses common to WT, hsn1, and vtc1 seedlings. First, in either the aerial or root tissues, GMPase activity levels in the 3 genotypes all were significantly lower when grown on NO3− + NH4+ medium compared with the corresponding values when NO3− medium used for the culture (Fig. 2D). Second, GMPase activity levels in the roots of the 3 genotypes were generally lower than those in the shoots irrespective of NO3− + NH4+ or NO3− medium used for the growth (Fig. 2D).
In addition to the experiments above, we investigated whether GMPase activity may be affected by alkaline pH, because one of the earliest cellular responses to NH4+ uptake is the alkalinization of the cytosol (reviewed in ref. 14). To this end, we compared the activities of recombinant WT GMPase and hsn1 proteins under 3 different pH regimes. At pH 7.5, which is within the range of cytosolic pH in unstressed plant cells (14), hsn1 enzyme activity was <30% of that WT GMPase (Fig. 2E). At pH 8.5, the activity of WT GMPase was reduced by >45% and that of hsn1 was reduced by >70% compared with the corresponding values at pH 7.5 (Fig. 2E). At pH 6.5, the activity of WT GMPase was enhanced by ≈15% and that of hsn1 was enhanced by ≈32% relative to the corresponding values at pH 7.5 (Fig. 2E). The data in Fig. 2E prompted us to test whether raising the rhizosphere pH would affect the in planta activity of GMPase in WT Arabidopsis and GMPase mutants. For this purpose, seedlings germinated on NO3− medium (pH 5.7) were transferred onto 3 different media, NO3− + NH4+ (pH 5.7), NO3− (pH 5.7), and NO3− (pH 6.7) and allowed to grow for 24 h, followed by measuring in planta GMPase activity. In agreement with the data depicted in Fig. 2D, GMPase activity levels in the shoots and roots were generally and significantly higher on NO3− medium (pH 5.7) than on NO3− + NH4+ medium (pH 5.7) (Fig. S2). However, raising the pH of NO3− medium from 5.7 to 6.7 caused considerable decreases of GMPase activity levels in both the shoots and roots and for either WT Arabidopsis or GMPase mutants (hsn1, vtc1) (Fig. S2). By contrast, GMPase protein level remained relatively stable in the shoots and roots of the 3 genotypes (Fig. S2).
Analysis of AsA Content and Protein N-Glycosylation in WT and Mutant Seedlings in Relation to NH4+ Sensitivity.
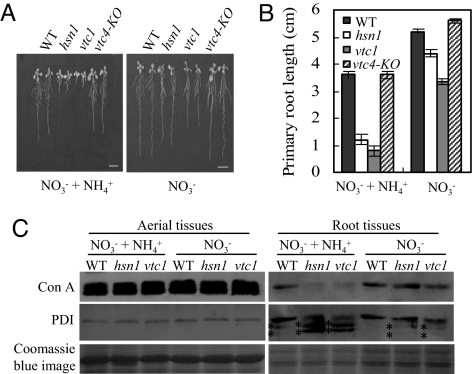
To find out whether decreased AsA content or defective N-glycosylation of proteins may be linked to the enhanced NH4+ sensitivity of hsn1 and vtc1, we first determined the total AsA content of hsn1 and compared the growth of WT Arabidopsis, hsn1, vtc1, and a knockout mutant (vtc4-KO) of the Arabidopsis VTC4 gene (15). VTC4 encodes l-Gal-1-P phosphatase devoted to AsA biosynthesis, and the total AsA content of vtc4-KO is <50% of that of WT Arabidopsis (15). As expected, the total AsA content in the aerial and root tissues of hsn1 was generally and substantially lower than those of WT control (Fig. S3). In contrast to the clear differences in the root length among WT, hsn1, and vtc1 seedlings on NO3− + NH4+ or NO3− medium, the growth of vtc4-KO seedlings was indistinguishable from that of WT controls on either of the 2 media (Fig. 3A). Quantitative measurements confirmed that the root length of vtc4-KO was highly similar to that of WT Arabidopsis and significantly longer than the one displayed by either hsn1 or vtc1 (Fig. 3B). Like WT control, the root length of vtc4-KO was much longer on NO3− medium than that on NO3− + NH4+ medium (Fig. 3).
Fig. 3.
Analysis of AsA and protein N-glycosylation in WT and mutant seedlings. (A) Comparisons of seedling growth of WT Arabidopsis, hsn1, vtc1, and vtc4-KO at 10 DAG. (Bars: 1 cm.) (B) Quantitative analysis of the root elongation of WT Arabidopsis, hsn1, vtc1, and vtc4-KO seedlings (10 DAG) cultured on NO3− + NH4+ or NO3− medium. Significant differences (P < 0.05 at least) were found between WT Arabidopsis and GMPase mutants (hsn1, vtc1) or among vtc4-KO and the 2 mutants but not between WT Arabidopsis and vtc4-KO. (C) Detection of defective N-glycosylation of proteins in WT and GMPase mutants. N-glycosylation of proteins was evaluated by using a Con A-peroxidase reagent (Top) or an antibody specific for PDI (Middle). The Coomassie blue-stained gel images (Bottom) illustrate equal loading of total protein samples during SDS/PAGE. Asterisks mark the underglycosylated PDI forms.
Two approaches were undertaken to investigate whether defective N-glycosylation of proteins may occur in hsn1 and vtc1 mutants on the medium containing NH4+. First, a peroxidase-conjugated Con A reagent, which binds to branches of oligomannose chains on N-glycoproteins (16), was used to compare the glycoproteins between WT and GMPase mutant (hsn1 and vtc1) tissues. This experiment revealed that the level of N-glycoproteins detected by Con A was drastically lower in the root tissues of hsn1 and vtc1 relative to that in WT Arabidopsis roots when grown on NO3− + NH4+ medium (Fig. 3C Right). However, no apparent reduction in the level of Con A-binding N-glycoproteins was observed in the root tissues of the 3 genotypes cultured on NO3− medium (Fig. 3C Right) or in the aerial tissues of the 3 genotypes irrespective of NO3− + NH4+ or NO3− medium used (Fig. 3C Left).
Second, the glycosylation status of protein disulfide isomerase (PDI), which is an abundant protein of the endoplasmic reticulum (ER) and posttranslationally modified with N-linked glycans under normal growth conditions (17), was examined by immunoblotting. PDI was resolved as a single protein band in the aerial tissues of WT control, hsn1, and vtc1 grown on either NO3− + NH4+ or NO3− medium (Fig. 3C Left). A single PDI band was also detected in the roots of WT Arabidopsis cultured on NO3− medium (Fig. 3C Right). By contrast, in the roots cultured on NO3− + NH4+ medium, 2 additional PDI bands, showing faster electrophoretic mobility and representing underglycosylated PDI forms (marked by asterisks in Fig. 3C Right), were detected for hsn1 and vtc1. Interestingly, the 2 underglycosylated PDI forms were also present, although in much smaller amounts, in the roots of WT Arabidopsis cultured on NO3− + NH4+ medium (Fig. 3C Right) or in the roots of hsn1 and vtc1 grown on NO3− medium (Fig. 3C Right).
Comparisons of Unfolded Protein Response (UPR) and Cell Death in the Roots of WT and Mutant Seedlings in the Presence of NH4+.
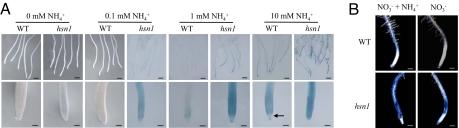
It is well known that N-glycosylation is an important form of posttranslational modification of proteins in eukaryotic cells (18). Consequently, the NH4+-induced decreases of GMPase activity and availability of GDP-mannose in WT and GMPase mutants may debilitate the processing, targeting, and functionality of many glycoproteins functioning in diverse cellular processes. Because delayed glycoprotein processing has been shown to trigger UPR in animal and yeast cells (19, 20), we tested whether UPR may occur in WT and GMPase mutants (hsn1 as a representative) in the presence of NH4+. This experiment was accomplished by monitoring the expression of BiP, an isoform of the HSP70 chaperone that is rapidly induced by N-glycosylation defects in the ER (21, 22). The biomarker BiP–β-glucuronidase (GUS) (21), which can be reliably followed by histochemical staining, was introduced into WT and hsn1 backgrounds. The marker-tagged lines were grown on a series of modified MS media with a constant level of NO3− (20 mM) but rising concentrations of NH4+. Compared with WT roots, in hsn1 roots, BiP–GUS expression (indicated by the blue signals in Fig. 4A) was observed at much lower NH4+ concentration (0.1 mM) and was substantially stronger at higher NH4+ concentrations (1–10 mM). Furthermore, at 10 mM NH4+, BiP–GUS expression was induced in the entire root tip region of hsn1 (Fig. 4A), whereas in WT Arabidopsis BiP–GUS signals were absent from the root apical meristematic region (Fig. 4A). Corresponding to the difference in BiP–GUS expression patterns, cell death, as revealed by the color precipitates after staining with Evans blue, was much more extensive in hsn1 roots than in WT controls in the presence of 10 mM NH4+ (in NO3− + NH4+ medium; Fig. 4B). A lower level of cell death, indicated by fewer Evans blue signals, was also observed in hsn1 roots in the absence of NH4+ (i.e., on NO3− medium; Fig. 4B).
Fig. 4.
Comparative analyses of UPR and cell death in the roots of WT Arabidopsis and hsn1 seedlings (10 DAG). (A) UPR in the roots (Upper) and root tips (Lower) was revealed by staining of the biomarker BiP–GUS that is frequently used in monitoring UPR in eukaryotic cells (21, 22). BiP–GUS was transferred into WT and hsn1 backgrounds through genetic crossing. The marker-tagged lines were cultured on modified MS media with a constant level of NO3− (20 mM) but rising concentrations of NH4+. The arrow indicates the root apical meristematic region. (Bars: Upper, 1 mm; Lower, 0.1 mm.) (B) WT Arabidopsis and hsn1 were grown on NO3− + NH4+ or NO3− medium. Cell death in the roots was revealed by the blue precipitates produced by histochemical staining with Evans blue. (Bars: 0.2 mm.)
Discussion
In this work, a forward genetic analysis of NH4+sensitivity was conducted with Arabidopsis as the model. We found that mutations in the gene encoding Arabidopsis GMPase, essential for synthesizing GDP-mannose, conferred hypersensitivity to NH4+ present in the growth medium, which provided a well-defined genetic system to study not only the mechanism underlying the function of GMPase, but also the downstream molecular events, in NH4+ sensitivity of Arabidopsis.
Our results reveal that strong correlations exist among the extents of decreased GMPase activity, the degrees of defective N-protein glycosylation in the roots, and the different magnitudes of growth retardation exhibited by WT, hsn1, and vtc1 seedlings in the presence of NH4+. From these correlations, we suggest that Arabidopsis GMPase activity is sensitive to inhibition by physiological concentrations of NH4+, which is further aggravated by mutations in GMPase protein (as in hsn1 and vtc1 seedlings). Consequently, the activity level of GMPase regulates Arabidopsis sensitivity to NH4+. From the findings that the in vitro GMPase activity was decreased by alkaline pH (Fig. 2E) and in planta GMPase activity was reduced by raising the pH of NO3− medium from 5.7 to 6.7 (Fig. S2), we hypothesize that cytosolic alkalinization associated with NH4+ uptake may play a role in the inhibition of GMPase activity by NH4+. However, further studies are needed to verify this hypothesis. Of the 2 consequences of decreased GMPase activity, defective protein glycosylation in the roots rather than lowered AsA content is responsible for the growth inhibition of WT, hsn1, and vtc1 seedlings by NH4+. This finding is compatible with the recent demonstration that oxidative stress does not contribute significantly to plant sensitivity to NH4+ (23).
In eukaryotic organisms, the accumulation of unfolded or misfolded proteins in the ER activates UPR signal-transduction pathways, which leads to increased protein folding capacity and cell death (to minimize the damage by the cells overaccumulating unfolded proteins; ref. 24). In agreement with previous reports (19, 20), this work revealed that defective N-glycosylation of proteins, observed in WT Arabidopsis and hsn1 roots grown in the presence of NH4+, rendered UPR and cell death. Importantly, UPR and cell death were more severe in hsn1 roots than in those of WT control in the presence of NH4+, which correlates tightly with the much larger decrease of GMPase activity in hsn1 roots and the enhanced inhibition of hsn1 root growth by NH4+. Thus, UPR and cell death are intensified in the Arabidopsis mutants with debilitated GMPase activity levels on the medium containing NH4+. In addition to this work, the association among disturbed protein N-glycosylation, augment of UPR and occurrence of cell death in the roots has been found in the Arabidopsis mutant with a defect in the STT3a gene encoding an essential subunit of the oligosaccharyltransferase complex (25). Interestingly, the stt3a-1 mutant displays increased sensitivity to salt/osmotic stress (25). Thus, it seems likely that the same paradigm (i.e., alteration of protein N-glycosylation, initiation of UPR, and occurrence of cell death) may be used to regulate the responses of Arabidopsis to different environmental stimuli (e.g., salt/osmotic stress, NH4+ toxicity).
Our data also provide useful clues for explaining several interesting observations in this work. First, the root growth of WT Arabidopsis seedlings was inhibited on NO3− + NH4+ medium relative to that attained on NO3− medium. This inhibition correlates with a significant reduction of GMPase activity and the alteration of N-glycosylation of proteins in the WT roots cultured on NO3− + NH4+ medium, indicating a decrease of GMPase activity, and disturbance of protein N-glycosylation may at least be partly involved in the inhibition of WT Arabidopsis root growth by NH4+. Second, the roots of hsn1 and vtc1 seedlings were shorter than those of WT controls on NO3− medium (Fig. 2C). This phenomenon is accompanied by the substantially lower GMPase activity levels in the roots of the 2 mutants compared with that in the roots of WT control on NO3− medium (Fig. 2D Right). Moreover, on this medium, the roots of vtc1 seedlings were significantly shorter than those of hsn1 (Fig. 2C), which correlates with an even lower GMPase activity level in vtc1 roots (Fig. 2D Right). Together, these data indicate that the shorter root phenotype of hsn1 and vtc1 seedlings on NO3− medium is the result of constitutively decreased GMPase activity. Third, in either WT Arabidopsis or GMPase mutants, the roots were more pronouncedly inhibited by NH4+ than the shoots. Interestingly, on NO3− + NH4+ medium, the level of GMPase activity in the root tissues was generally lower than that in the aerial tissues (Fig. 2D). This result may be partly responsible for the enhanced susceptibility of root growth to NH4+. Because retarded root growth is a common and severe symptom of NH4+ sensitivity in higher plants (26, 27), it will be important to examine whether the inhibition of root growth by NH4+ in other plant species also involves a relatively lower level of GMPase activity in the root cells in future studies.
A further interesting observation in this work is that hsn1 and WT control did not differ significantly in the NH4+ content in either the shoot or root tissues on NO3− + NH4+ medium (Fig. 1E), indicating that NH4+ content does not play a major part in the hypersensitivity of hsn1 to NH4+ and in the enhanced inhibition of Arabidopsis roots by NH4+. In line with this observation, a previous study has shown that the transport process of NH4+ across the plasma membrane is an important step that distinguishes NH4+-sensitive plant species from those with NH4+ tolerance (2). Nevertheless, the NH4+ contents in the root and shoot tissues of the seedlings cultured on NO3− + NH4+ medium both are significantly higher than those of the seedlings cultured on NO3− medium. Whether these differences would contribute to the retarded Arabidopsis growth in the presence of NH4+ is a matter for future investigations.
In summary, the molecular genetic and biochemical investigations in this work have revealed that the NH4+ sensitivities of WT GMPase and its mutants result in profound changes in cellular activities and cell survival in Arabidopsis roots. Decreased GMPase activity and its consequences (defective protein N-glycosylation, activation of UPR, and cell death in the roots) are important molecular events in the NH4+ sensitivity of Arabidopsis. Although the responses of higher plants to NH4+ are complex and multiple mechanisms may be involved (1), the insights gained in this work may offer clues for future attempts to understand more completely the molecular genetic basis underlying the variations of NH4+ sensitivity among different plant families and species.
Materials and Methods
Plant Materials and Growth Conditions.
The plant materials used in this work included WT A. thaliana (Col-0 and Ler ecotypes) and genetic mutants derived from Col-0 ecotype. Seed germination and seedling growth were accomplished by using modified MS media, with the 2 most frequently used ones being NO3− + NH4+ medium (with 10 mM NH4+ and 20 mM NO3−, provided by KNO3 and NH4NO3, respectively, as nitrogen sources) and NO3− medium (with 20 mM NO3−, provided by KNO3, as sole nitrogen source). The above media all were supplemented with 0.5% (wt/vol) sucrose and 0.5 g/L Mes, adjusted to pH 5.7, and solidified with 1% agar-agar (Fisons). Arabidopsis growth took place in a growth cabinet (Percival Scientific), preset with a 16-h light/8-h dark photoperiod, 300 μmol·m−2·s−1 light intensity, and a constant temperature of 20 °C.
Chemical Reagents and Molecular Methods.
Unless otherwise stated, the chemicals used in this work were purchased from Sigma. The standard molecular and biochemical methods for nucleic acid manipulations, PCR amplifications, and protein investigations with SDS/PAGE and immunoblotting followed those described (28). The cloning experiments all were verified by DNA sequencing to exclude inadvertent introductions of sequence errors.
Ammonium Assay.
NH4+ content was determined colorimetrically at 410 nm after reaction with Nessler's reagent (29).
Map-Based Cloning.
A F2 population derived from a cross between hsn1 mutant and Ler ecotype was used for map-based cloning. Markers used in the initial mapping analysis were selected from The Arabidopsis Information Resource web site (www.Arabidopsis.org). Several new simple sequence length polymorphism (SSLP) markers were developed at the bottom of chromosome 2 based on differences between the Col-0 and Ler (available at www.Arabidopsis.org/Cereon/index.jsp) genomic sequences. The candidate gene was mapped between the 2 new SSLP markers H13 and H16. The primers for amplifying H13 were 5′-CCACCTCATTCCCGTAGTG-3′ and 5′-CCTCGGTA GATTCATGGTCAACTG-3′, and those for H16 were 5′-TGAACCGACAGTGTGACCTTT-3′ and 5′-AGTAAGAATCACGTTAATGAATGG-3′.
Complementation Assay.
A 4.2-kb genomic DNA fragment containing WT GMPase coding region and 2.7 kb of 5′ flanking sequence (upstream of the ATG start codon) was amplified by using the primers 5′-TGGTGGCGTGTATTTGTCAAAA-3′ and 5′-CAGCGTTTTGATTGATGCTTATTC-3′. The resulted fragment was blunt-ended and cloned into SmaI-digested pCAMBIA1300, creating the transformation construct for the complementation assay. Nine independent homozygous and genetically stable transgenic lines were developed based on hygromycine resistance. The 9 lines all complemented hsn1 phenotype although only data from 2 representatives (WTGMP/hsn1–3, WTGMP/hsn1–8) are presented.
GMPase Activity Assay.
GMPase activity in the supernatant was measured by monitoring the production of GDP-d-mannose from d-mannose-1-P and GTP. The extraction of total proteins and determination of protein concentrations were accomplished as detailed (30). The assay mixture contained 10 μg of total protein, 50 mM Tris·HCl (pH 7.5), 4 mM MgCl2, 1 mM GTP, and 1 mM d-mannose-1-P in a total volume of 50 μL. Incubations were done at 37 °C for 1 h. The reaction was stopped by boiling for 2 min followed by centrifugation to remove the precipitated proteins. Reaction products were analyzed by HPLC by using a C18 Bondapack column (Waters; 3.9 × 300 mm, 10-μm particle size) as described (31).
The above assay conditions were also used in the experiments comparing the activities of recombinant WT GMPase and hsn1 enzymes at 3 pH regimes (pH 6.5, 7.5, and 8.5). The cDNA coding sequences of WT GMPase and hsn1 were amplified by RT-PCR using the primers 5′-CTCATATGATGAAGGCACTCATTC-3′ and 5′-CTAAGCTTCATCACTATCTCTGG-3′ (the underlined nucleotides form the restriction sites for NdeI and HindIII, respectively), cleaved with NdeI and HindIII, and cloned into the bacterial expression vector pET30a. The resulted constructs were expressed in the bacterial cells, with the overexpressed proteins purified according to a published protocol (30). The purified recombinant protein (WT GMPase or hsn1 mutant, all in 100 ng) was used to initiate the enzyme assay. The different pH regimes in the enzyme reactions were created by using the Tris·HCl solutions with pH values adjusted to 6.5, 7.5, and 8.5.
N-Glycoprotein Analysis.
Peroxidase-conjugated Con A was purchased from Sigma. A mouse polyclonal antiserum against Arabidopsis PDI was developed as follows. The PDI cDNA was amplified by PCR using the primers 5′-CTCATATGACCGATACCATCAACAAACACG-3′ and 5′-CACTCGAGGTGGTT GTTTGGATCTTTGTC-3′ (the underlined nucleotides form the restriction sites for NdeI and XhoI, respectively). The bacterial expression and purification of Arabidopsis PDI were then performed as described above. The purified protein was used as an antigen to develop PDI-specific polyclonal antibodies in mouse (30).
Total protein samples were prepared as described above. Affinodetection of N-glycoproteins was carried out by using the Con A-peroxidase method (32). The PDI-specific antiserum described above was used to reveal the N-glycosylation status of PDI following a published procedure (33).
Detection of UPR in Arabidopsis Roots.
UPR was monitored by histochemical staining of the biomarker BiP–GUS that was rapidly activated by ER stress (21). The transgenic Arabidopsis line harboring BiP-GUS (GUS expression cassette directed by BiP2 gene promoter) was derived from a previous publication (21). By genetic crossing, the BiP–GUS marker was transferred into hsn1 background. Histochemical GUS staining was performed following a published protocol (34).
Cell Death Assay.
Cell death was examined by using Evans blue, a compound that selectively enters the dead cells as a result of their freely permeable plasma membranes (35). The root tips of 10 DAG seedlings were submerged in 20 mL of 1% (wt/vol) Evans blue water solution for 10 min, washed with distilled water for 2 h, and then photographed as above.
For additional methods see SI Text.
Supplementary Material
Acknowledgments.
We thank Prof. Patricia Conklin (State University of New York College, Cortland, NY) for the vtc4-KO (SAIL_843_G10) line; Dr. Nozomu Koizumi (Nara Institute of Science and Technology, Nara, Japan) for the transgenic line containing the biomarker BiP–GUS; and Prof. Dr. Paul Schulze-Lefert (Max-Planck-Institut für Züchtungsforschung, Cologne, Germany) for constructive suggestions for this work and the writing of the manuscript. This research was supported by National Basic Research and Development Program of China Grant 2005CB120901 and National Natural Science Foundation of China Grant 30521001.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806168105/DCSupplemental.
References
- 1.Britto DT, Kronzucker HJ. NH4+ toxicity in higher plants: A critical review. J Plant Physiol. 2002;159:567–584. [Google Scholar]
- 2.Britto DT, Siddiqi MY, Glass AD, Kronzucker HJ. Futile transmembrane NH4+ cycling: A cellular hypothesis to explain ammonium toxicity in plants. Proc Natl Acad Sci USA. 2001;98:4255–4258. doi: 10.1073/pnas.061034698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Breemen N, Van Dijk HFG. Ecosystem effects of atmospheric deposition of nitrogen in The Netherlands. Environ Poll. 1988;54:249–274. doi: 10.1016/0269-7491(88)90115-7. [DOI] [PubMed] [Google Scholar]
- 4.Borys MC, Linzer DI, Papoutsakis ET. Culture pH affects expression rates and glycosylation of recombinant mouse placental lactogen proteins by Chinese hamster ovary (CHO) cells. Bio/Technology. 1993;11:720–724. doi: 10.1038/nbt0693-720. [DOI] [PubMed] [Google Scholar]
- 5.Yang M, Butler M. Effects of ammonia on CHO cell growth, erythropoietin production, and glycosylation. Biotechnol Bioeng. 2000;68:370–380. doi: 10.1002/(sici)1097-0290(20000520)68:4<370::aid-bit2>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 6.Gawlitzek M, Ryll T, Lofgren J, Sliwkowski MB. Ammonium alters N-glycan structures of recombinant TNFR-IgG: Degradative versus biosynthetic mechanisms. Biotechnol Bioeng. 2000;68:637–646. doi: 10.1002/(sici)1097-0290(20000620)68:6<637::aid-bit6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Falkengren-Grerup U. Interspecies differences in the preference of ammonium and nitrate in vascular plants. Oecologia. 1995;102:305–311. doi: 10.1007/BF00329797. [DOI] [PubMed] [Google Scholar]
- 8.Conklin PL, et al. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li BH, Shi WM. Effects of elevated NH4+ on Arabidopsis seedlings different in accessions. Acta Pedol Sinica. 2007;44:508–515. [Google Scholar]
- 10.Lukowitz W, et al. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA. 2001;98:2262–2267. doi: 10.1073/pnas.051625798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto H, Sakakibara A, Yamasaki M, Yoda K. Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J Biol Chem. 1997;272:16308–16314. doi: 10.1074/jbc.272.26.16308. [DOI] [PubMed] [Google Scholar]
- 12.Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- 13.Olmos E, Kiddle G, Pellny T, Kumar S, Foyer C. Modulation of plant morphology, root architecture, and cell structure by low vitamin C in Arabidopsis thaliana. J Exp Bot. 2006;57:1645–1655. doi: 10.1093/jxb/erl010. [DOI] [PubMed] [Google Scholar]
- 14.Britto DT, Kronzucker HJ. Nitrogen acquisition, PEP carboxylase, and cellular pH homeostasis: New views on old paradigms. Plant Cell Environ. 2005;28:1396–1409. [Google Scholar]
- 15.Conklin PL, et al. Arabidopsis thaliana VTC4 encodes l-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem. 2006;281:15662–15670. doi: 10.1074/jbc.M601409200. [DOI] [PubMed] [Google Scholar]
- 16.Keller R, Renz FS, Kossmann J. Antisense inhibition of the GDP-mannose pyrophosphorylase reduces the ascorbate content in transgenic plants leading to developmental changes during senescence. Plant J. 1999;19:131–141. doi: 10.1046/j.1365-313x.1999.00507.x. [DOI] [PubMed] [Google Scholar]
- 17.Shorrosh BS, Subramaniam J, Schubert KR, Dixon RA. Expression and localization of plant protein disulfide isomerase. Plant Physiol. 1993;103:719–726. doi: 10.1104/pp.103.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trombetta ES. The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology. 2003;13:77R–91R. doi: 10.1093/glycob/cwg075. [DOI] [PubMed] [Google Scholar]
- 19.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 20.Iwakoshi NN, et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 21.Oh DH, Kwon CS, Sano H, Chung WI, Koizumi N. Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem Biophys Res Commun. 2003;301:225–230. doi: 10.1016/s0006-291x(02)03019-x. [DOI] [PubMed] [Google Scholar]
- 22.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez-Valdivia MD, et al. Nitrogen nutrition and antioxidant metabolism in ammonium-tolerant and -sensitive plants. Physiol Plant. 2008;132:359–369. doi: 10.1111/j.1399-3054.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- 24.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 25.Koiwa H, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis OAM, Leidi EO, Lips SH. Effect of nitrogen source on growth response to salinity stress in maize and wheat. New Phytol. 1989;111:155–160. doi: 10.1111/j.1469-8137.1989.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 27.Guo S, Brück H, Sattelmacher B. Effects of supplied nitrogen form on growth and water uptake of French bean (Phaseolus vulgaris L.) plants. Plant Soil. 2002;239:267–275. [Google Scholar]
- 28.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 29.Leleu O, Vuylsteker C. Unusual regulatory nitrate reductase activity in cotyledons of Brassica napus seedlings: Enhancement of nitrate reductase activity by ammonium supply. J Exp Bot. 2004;55:815–823. doi: 10.1093/jxb/erh088. [DOI] [PubMed] [Google Scholar]
- 30.Qian W, et al. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J. 2007;49:399–413. doi: 10.1111/j.1365-313X.2006.02967.x. [DOI] [PubMed] [Google Scholar]
- 31.Albermann C, Distler J, Piepersberg W. Preparative synthesis of GDP-β-l-fucose by recombinant enzymes from enterobacterial sources. Glycobiology. 2000;10:875–881. doi: 10.1093/glycob/10.9.875. [DOI] [PubMed] [Google Scholar]
- 32.Faye L, Chrispeels MJ. Characterization of N-linked oligosaccharides by affinoblotting with concanavalin A-peroxidase and treatment of the blots with glycosidases. Anal Biochem. 1985;149:218–224. doi: 10.1016/0003-2697(85)90498-1. [DOI] [PubMed] [Google Scholar]
- 33.Lerouxel O, et al. Mutants in Defective Glycosylation, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 2005;42:455–468. doi: 10.1111/j.1365-313X.2005.02392.x. [DOI] [PubMed] [Google Scholar]
- 34.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaff DF, Okong'O-Ogola O. The use of nonpermeating pigments for testing the survival of cells. J Exp Bot. 1971;22:756–758. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.