Abstract
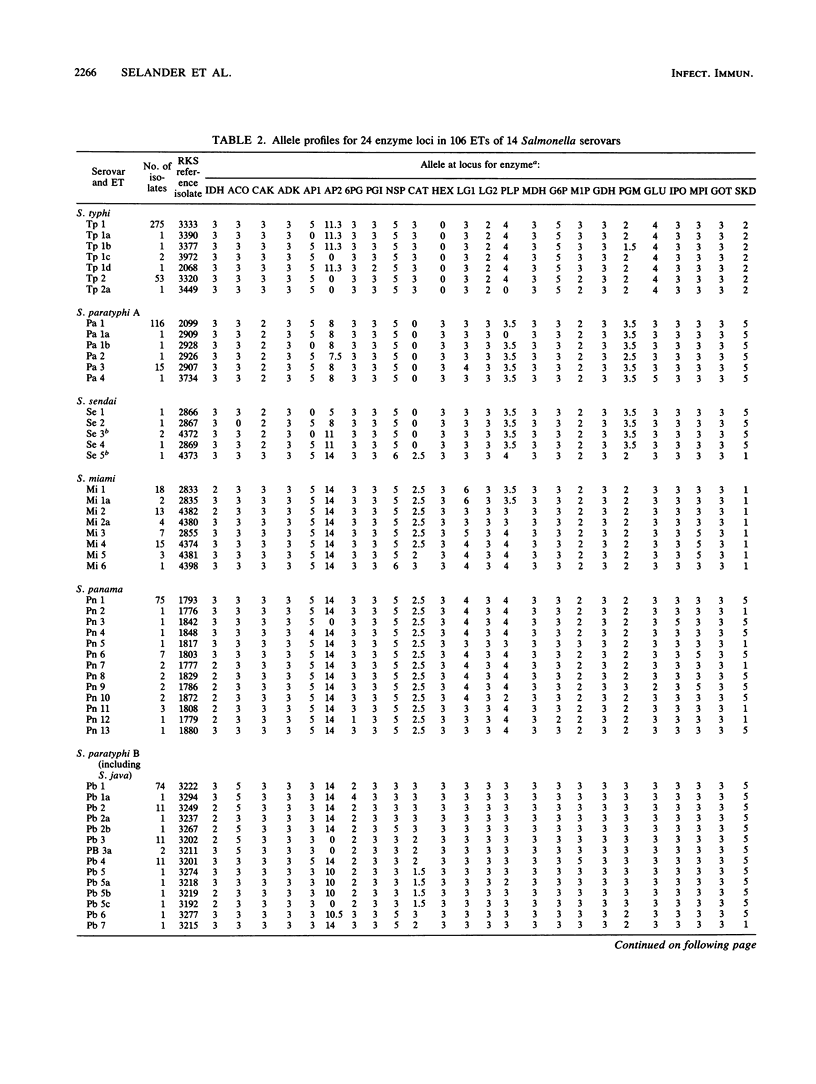
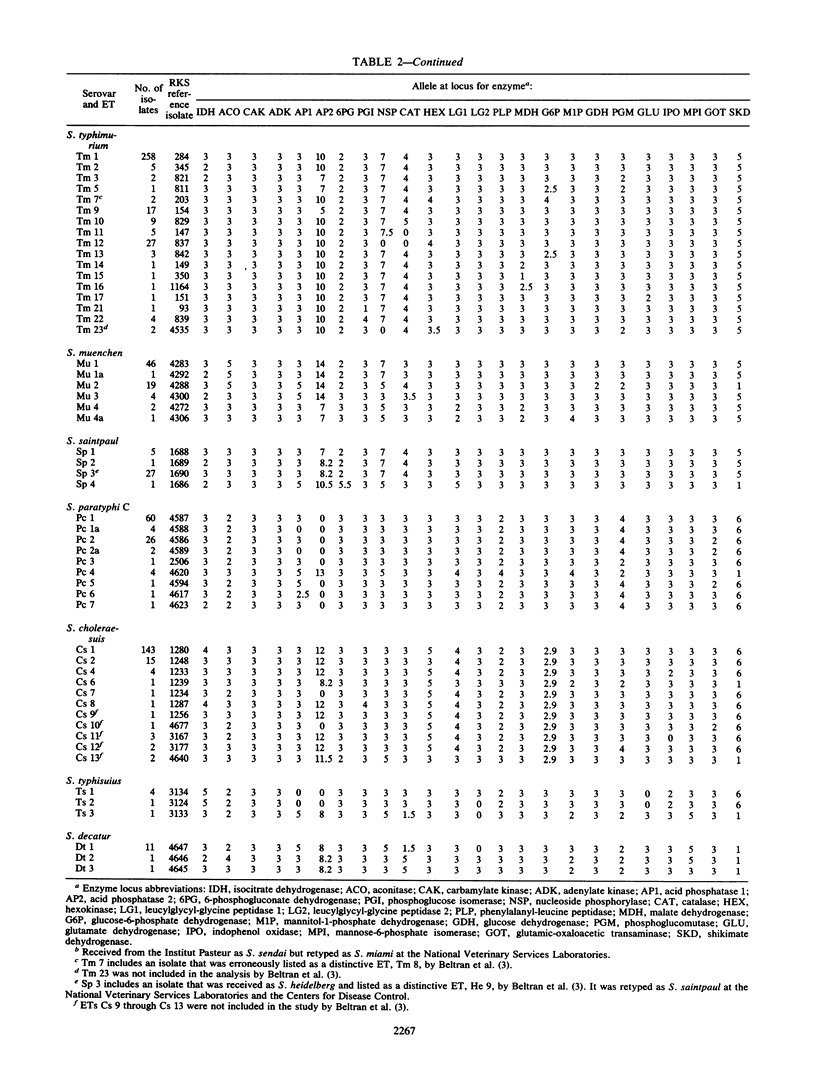
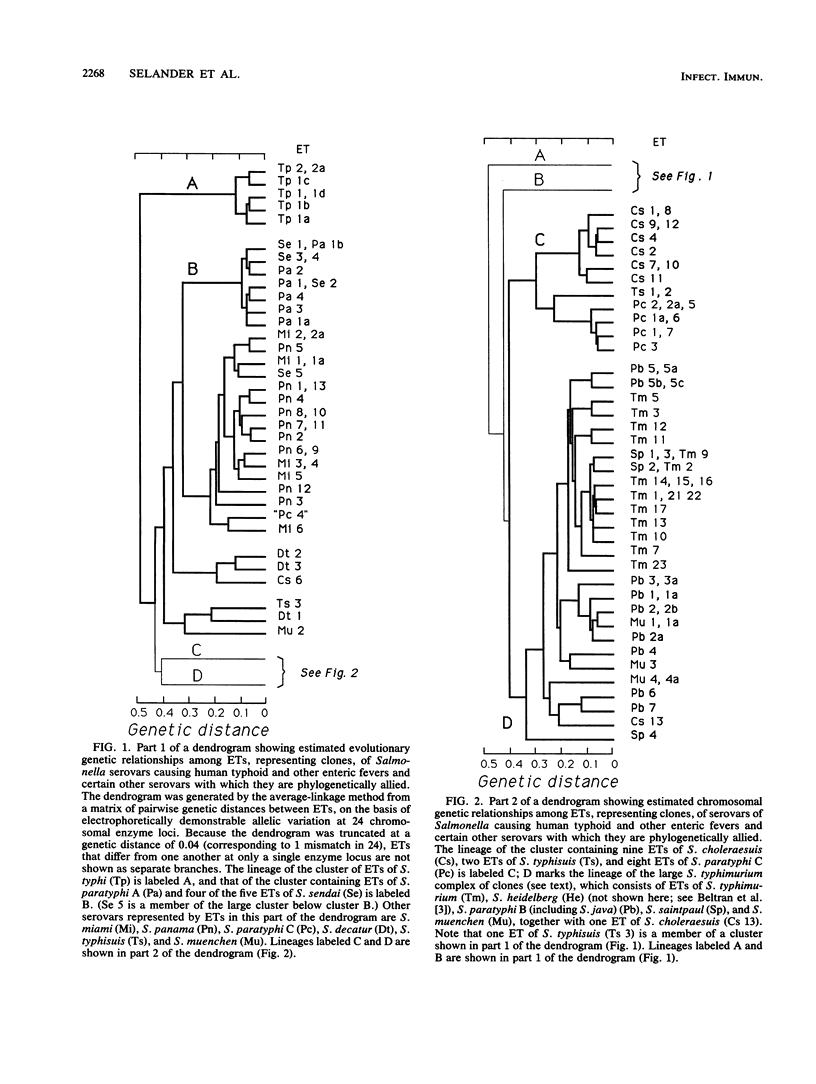
Multilocus enzyme electrophoresis was employed to measure chromosomal genotypic diversity and evolutionary relationships among 761 isolates of the serovars Salmonella typhi, S. paratyphi A, S. paratyphi B, S. paratyphi C, and S. sendai, which are human-adapted agents of enteric fever, and S. miami and S. java, which are serotypically similar to S. sendai and S. paratyphi B, respectively, but cause gastroenteritis in both humans and animals. To determine the phylogenetic positions of the clones of these forms within the context of the salmonellae of subspecies I, comparative data for 22 other common serovars were utilized. Except for S. paratyphi A and S. sendai, the analysis revealed no close phylogenetic relationships among clones of different human-adapted serovars, which implies convergence in host adaptation and virulence factors. Clones of S. miami are not allied with those of S. sendai or S. paratyphi A, being, instead, closely related to strains of S. panama. Clones of S. paratyphi B and S. java belong to a large phylogenetic complex that includes clones of S. typhimurium, S. heidelberg, S. saintpaul, and S. muenchen. Most strains of S. paratyphi B belong to a globally distributed clone that is highly polymorphic in biotype, bacteriophage type, and several other characters, whereas strains of S. java represent seven diverse lineages. The flagellar monophasic forms of S. java are genotypically more similar to clones of S. typhimurium than to other clones of S. java or S. paratyphi B. Clones of S. paratyphi C are related to those of S. choleraesuis. DNA probing with a segment of the viaB region specific for the Vi capsular antigen genes indicated that the frequent failure of isolates of S. paratyphi C to express Vi antigen is almost entirely attributable to regulatory processes rather than to an absence of the structural determinant genes themselves. Two clones of S. typhisuis are related to those of S. choleraesuis and S. paratyphi C, but a third clone is not. Although the clones of S. decatur and S. choleraesuis are serologically and biochemically similar, they are genotypically very distinct. Two clones of S. typhi were distinguished, one globally distributed and another apparently confined to Africa; both clones are distantly related to those of all other serovars studied.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altwegg M., Hickman-Brenner F. W., Farmer J. J., 3rd Ribosomal RNA gene restriction patterns provide increased sensitivity for typing Salmonella typhi strains. J Infect Dis. 1989 Jul;160(1):145–149. doi: 10.1093/infdis/160.1.145. [DOI] [PubMed] [Google Scholar]
- Barker R. M., Kearney G. M., Nicholson P., Blair A. L., Porter R. C., Crichton P. B. Types of Salmonella paratyphi B and their phylogenetic significance. J Med Microbiol. 1988 Aug;26(4):285–293. doi: 10.1099/00222615-26-4-285. [DOI] [PubMed] [Google Scholar]
- Beltran P., Musser J. M., Helmuth R., Farmer J. J., 3rd, Frerichs W. M., Wachsmuth I. K., Ferris K., McWhorter A. C., Wells J. G., Cravioto A. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin A. B., Parodos K., Lane D. J., Pace N. R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990 Feb 9;60(3):405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Brenner D. J., Ewing W. H., Falkow S. Molecular relationships among the Salmonelleae. J Bacteriol. 1973 Jul;115(1):307–315. doi: 10.1128/jb.115.1.307-315.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels E. M., Schneerson R., Egan W. M., Szu S. C., Robbins J. B. Characterization of the Salmonella paratyphi C Vi polysaccharide. Infect Immun. 1989 Oct;57(10):3159–3164. doi: 10.1128/iai.57.10.3159-3164.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman R., Levine M. M. Summary of an international workshop on typhoid fever. Rev Infect Dis. 1986 May-Jun;8(3):329–349. doi: 10.1093/clinids/8.3.329. [DOI] [PubMed] [Google Scholar]
- Everard C. O., Tota B., Bassett D., Ali C. Salmonella in wildlife from Trinidad and Grenada, W.I. J Wildl Dis. 1979 Apr;15(2):213–219. doi: 10.7589/0090-3558-15.2.213. [DOI] [PubMed] [Google Scholar]
- Frankel G., Newton S. M., Schoolnik G. K., Stocker B. A. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 1989 Oct;8(10):3149–3152. doi: 10.1002/j.1460-2075.1989.tb08468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein F. W., Chumpitaz J. C., Guevara J. M., Papadopoulou B., Acar J. F., Vieu J. F. Plasmid-mediated resistance to multiple antibiotics in Salmonella typhi. J Infect Dis. 1986 Feb;153(2):261–266. doi: 10.1093/infdis/153.2.261. [DOI] [PubMed] [Google Scholar]
- Gotuzzo E., Morris J. G., Jr, Benavente L., Wood P. K., Levine O., Black R. E., Levine M. M. Association between specific plasmids and relapse in typhoid fever. J Clin Microbiol. 1987 Sep;25(9):1779–1781. doi: 10.1128/jcm.25.9.1779-1781.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinée P. A., Jansen W. H., Maas H. M., Le Minor L., Beaud R. An unusual H antigen (Z66) in strains of Salmonella typhi. Ann Microbiol (Paris) 1981 May-Jun;132(3):331–334. [PubMed] [Google Scholar]
- Hartl D. L., Medhora M., Green L., Dykhuizen D. E. The evolution of DNA sequences in Escherichia coli. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):191–204. doi: 10.1098/rstb.1986.0001. [DOI] [PubMed] [Google Scholar]
- Holmberg S. D., Wachsmuth I. K., Hickman-Brenner F. W., Cohen M. L. Comparison of plasmid profile analysis, phage typing, and antimicrobial susceptibility testing in characterizing Salmonella typhimurium isolates from outbreaks. J Clin Microbiol. 1984 Feb;19(2):100–104. doi: 10.1128/jcm.19.2.100-104.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegathesan M. Salmonella serotypes isolated from man in Malaysia over the 10-year period 1973-1982. J Hyg (Lond) 1984 Jun;92(3):395–399. doi: 10.1017/s0022172400064615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUFFMANN F. On the transduction of serological properties in the Salmonella group. Acta Pathol Microbiol Scand. 1953;33(4):409–420. doi: 10.1111/j.1699-0463.1953.tb01537.x. [DOI] [PubMed] [Google Scholar]
- KAUFFMANN F. Zur Differential diagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. Z Hyg Infektionskr. 1955;141(6):546–550. [PubMed] [Google Scholar]
- Kelterborn E. Uber die Häufigkeit des Vorkommens der Salmonella-Species. Eine Untersuchung an 1,5 Millionen Salmonella-Kulturen, die von 1934 bis 1975 in 109 Ländern isoliert worden waren. Zentralbl Bakteriol Orig A. 1979 Apr;243(2-3):289–307. [PubMed] [Google Scholar]
- LE MINOR L. Etude sur les Salmonella du groupe D possédant les antigènes flagellaires a: 1, 5. Ann Inst Pasteur (Paris) 1956 Nov;91(5):664–669. [PubMed] [Google Scholar]
- LE MINOR L. Variantes biochimiques de S. miami et S. sendai; étude de l'antigène a. Ann Inst Pasteur (Paris) 1955 Jan;88(1):76–83. [PubMed] [Google Scholar]
- Le Minor L., Beaud R., Laurent B., Monteil V. Etude des Salmonella possédant les facteurs antigéniques 6,7:c:1,5. Ann Inst Pasteur Microbiol. 1985 Sep-Oct;136B(2):225–234. doi: 10.1016/s0769-2609(85)80047-8. [DOI] [PubMed] [Google Scholar]
- Le Minor L., Popoff M. Y., Laurent B., Hermant D. Individualisation d'une septième sous-espèce de Salmonella: S. choleraesuis subsp. indica subsp. nov. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):211–217. [PubMed] [Google Scholar]
- Lehner A. F., Harvey S., Hill C. W. Mapping and spacer identification of rRNA operons of Salmonella typhimurium. J Bacteriol. 1984 Nov;160(2):682–686. doi: 10.1128/jb.160.2.682-686.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics. 1981 Sep;99(1):1–23. doi: 10.1093/genetics/99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher K. O., Morris J. G., Jr, Gotuzzo E., Ferreccio C., Ward L. R., Benavente L., Black R. E., Rowe B., Levine M. M. Molecular techniques in the study of Salmonella typhi in epidemiologic studies in endemic areas: comparison with Vi phage typing. Am J Trop Med Hyg. 1986 Jul;35(4):831–835. doi: 10.4269/ajtmh.1986.35.831. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Kimura M. Genetic variability and effective population size when local extinction and recolonization of subpopulations are frequent. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6710–6714. doi: 10.1073/pnas.77.11.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer L. W. Use of plasmid profiles in epidemiologic surveillance of disease outbreaks and in tracing the transmission of antibiotic resistance. Clin Microbiol Rev. 1988 Apr;1(2):228–243. doi: 10.1128/cmr.1.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray B. E., Levine M. M., Cordano A. M., D'Ottone K., Jayanetra P., Kopecko D., Pan-Urae R., Prenzel I. Survey of plasmids in Salmonella typhi from Chile and Thailand. J Infect Dis. 1985 Mar;151(3):551–555. doi: 10.1093/infdis/151.3.551. [DOI] [PubMed] [Google Scholar]
- Ochman H., Wilson A. C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26(1-2):74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Old D. C. Phylogeny of strains of Salmonella typhimurium. Microbiol Sci. 1984 Jun;1(3):69–72. [PubMed] [Google Scholar]
- Old D. C., Yakubu D. E., Senior B. W. Characterisation of a fimbrial, mannose-resistant and eluting haemagglutinin (MREHA) produced by strains of Salmonella of serotype Sendai. J Med Microbiol. 1989 Sep;30(1):59–68. doi: 10.1099/00222615-30-1-59. [DOI] [PubMed] [Google Scholar]
- Orskov F., Orskov I. From the national institutes of health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis. 1983 Aug;148(2):346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- Ou J. T., Baron L. S., Rubin F. A., Kopecko D. J. Specific insertion and deletion of insertion sequence 1-like DNA element causes the reversible expression of the virulence capsular antigen Vi of Citrobacter freundii in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4402–4405. doi: 10.1073/pnas.85.12.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M. W., Evins G. M., Heiba A. A., Plikaytis B. D., Farmer J. J., 3rd Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989 Feb;27(2):313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin F. A., Kopecko D. J., Noon K. F., Baron L. S. Development of a DNA probe to detect Salmonella typhi. J Clin Microbiol. 1985 Oct;22(4):600–605. doi: 10.1128/jcm.22.4.600-605.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Beltran P., Smith N. H., Barker R. M., Crichton P. B., Old D. C., Musser J. M., Whittam T. S. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect Immun. 1990 Jun;58(6):1891–1901. doi: 10.1128/iai.58.6.1891-1901.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Beltran P., Selander R. K. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J Bacteriol. 1990 May;172(5):2209–2216. doi: 10.1128/jb.172.5.2209-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Selander R. K. Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):603–609. doi: 10.1128/jb.172.2.603-609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellings N. J., Johnson E. M., Baron L. S. Genetic basis of Vi antigen expression in Salmonella paratyphi C. J Bacteriol. 1977 Jul;131(1):57–62. doi: 10.1128/jb.131.1.57-62.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellings N. J., Johnson E. M., Kopecko D. J., Collins H. H., Baron L. S. Genetic regulation of variable Vi antigen expression in a strain of Citrobacter freundii. J Bacteriol. 1981 Feb;145(2):1010–1017. doi: 10.1128/jb.145.2.1010-1017.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Tamura K., Sakazaki R., Kuramochi S., Yoshizaki E. [On propriety of distinguishing Salmonella java from Salmonella paratyphi-B]. Kansenshogaku Zasshi. 1982 Nov;56(11):1025–1031. doi: 10.11150/kansenshogakuzasshi1970.56.1025. [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Brose E. C. Characterization of incompatibility group HI1 plasmids from Salmonella typhi by restriction endonuclease digestion and hybridization of DNA probes for Tn3, Tn9, and Tn10. Can J Microbiol. 1985 Aug;31(8):721–729. doi: 10.1139/m85-136. [DOI] [PubMed] [Google Scholar]
- Tompkins L. S., Troup N., Labigne-Roussel A., Cohen M. L. Cloned, random chromosomal sequences as probes to identify Salmonella species. J Infect Dis. 1986 Jul;154(1):156–162. doi: 10.1093/infdis/154.1.156. [DOI] [PubMed] [Google Scholar]
- Vieu J. F., Binette H., Leherissey M. Absence de l'antigène H:z66 chez 2355 souches de Salmonella typhi provenant de Madagascar et de plusieurs pays d'Afrique tropicale. Bull Soc Pathol Exot Filiales. 1986;79(1):22–26. [PubMed] [Google Scholar]
- Vieu J. F., Binette H., Leherissey M. Salmonella paratyphi B d-tartrate positif (var. java): lysotypie de 1200 souches isolées en France (1975-1985). Zentralbl Bakteriol Mikrobiol Hyg A. 1988 May;268(3):424–432. doi: 10.1016/s0176-6724(88)80027-0. [DOI] [PubMed] [Google Scholar]
- Vieu J. F., Lehérissey M. Recherche de l'antigène H:z66 chez 1,000 souches de Salmonella typhi provenant des Antilles, d'Amérique Centrale et d'Amérique du Sud. Bull Soc Pathol Exot Filiales. 1988;81(2):198–201. [PubMed] [Google Scholar]
- Wachsmuth K. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis. 1986 Sep-Oct;8(5):682–692. doi: 10.1093/clinids/8.5.682. [DOI] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]



