Abstract
High-throughput, microarray-based chromatin immunoprecipitation (ChIP-chip) technology allows in vivo elucidation of transcriptional networks. However this complex is not yet readily accessible, in part because its many parameters have not been systematically evaluated and optimized. We address this gap by systematically assessing experimental-design parameters including antibody purity, dye-bias, array-batch, inter-day hybridization bias, amplification method and choice of hybridization control. The combined performance of these optimized parameters shows a 90% validation rate in ChIP-chip analysis of Myc genomic binding in HL60 cells using two different microarray platforms. Increased sensitivity and decreased noise in ChIP-chip assays will enable wider use of this methodology to accurately and affordably elucidate transcriptional networks.
INTRODUCTION
The combination of chromatin immunoprecipitation (ChIP) and high-throughput DNA microarray technology (chip) provides a powerful method (ChIP-chip) for mapping protein–DNA interactions in vivo. The ChIP-chip procedure was first used to study yeast transcription factors (1–4) and has recently been exploited for similar studies in mammalian cells (5–15). In the ChIP-chip procedure, genomic DNA is precipitated using specific and control antibodies, then labeled and hybridized to genomic microarrays. Several different array platforms have been used, including proximal promoter region arrays (11), CpG island arrays (12,15) and whole-genome tiling arrays (16,17). The technique is extremely versatile and can identify target genes whose regulatory regions are bound by transcription factors through both protein–DNA and protein–protein interactions. ChIP-chip can be applied to any cell or tissue, making it possible to decode gene regulatory networks in vivo. Certainly, the ability to profile bona fide target genes has been revolutionized by ChIP-chip. Unlike mRNA expression analysis, the genetic program specifically directed by the transcription factor can be distinguished from subsequent downstream regulatory events (18–22).
Although ChIP-chip has become the standard for discovering the genomic binding sites of transcriptional regulators there is wide variability in experimental design (23). This variability has complicated and delayed widespread application, and is a reflection of the large number of parameters that must be carefully optimized in ChIP-chip experiments. For example, the number of cells or amount of tissue used as starting material varies widely from one study to another (24,25). The protein–protein and/or protein–DNA cross-linking, chromatin sonication, as well as antibody sensitivity and purity characteristics can also vary significantly. In most studies, the enriched DNA recovered after the ChIP procedure is amplified. A variety of amplification methods have been developed, including ligation-mediated PCR (25,26), random primed PCR (27), T7 primed PCR (28) and Whole Genome Amplification (WGA) (29), and it is unclear which method is most appropriate for ChIP studies. Finally, when the amplified and labeled DNA is hybridized to a microarray a control sample must be selected and the effects of array batch and dye-swap status considered.
Experimental design parameters for mRNA expression arrays have been extensively evaluated by a number of groups over the past decade (30–36). As a result, the key factors are well understood and the assay has been optimized. It is possible, for example, to estimate the number of biological replicates required to sufficiently power a specific hypothesis-testing question (37). Despite this clear evidence that parameter optimization can greatly improve the quantity and quality of information retrieved from an array analysis, ChIP-chip design parameters have not yet been thoroughly and systematically investigated, and it cannot be assumed that parameters and processes would be the same for both mRNA and ChIP-chip arrays.
Here, we fill that gap by providing a comprehensive evaluation of experimental design parameters for ChIP-chip studies. Through a series of validation studies we address both the parameters previously investigated for mRNA expression studies as well as those specific to ChIP-chip experiments. We exploit a well-characterized system: the genomic binding of the Myc oncoprotein in HL60 cells, a human myelogenous leukemia cell line, combined with CpG island arrays (38). Many parameters for successful ChIP-chip studies were analyzed, including antibody purity, array batch variability, dye-bias, inter-day hybridization-variability, amplification procedure and hybridization control. In addition, we evaluated the combined effect of the optimized parameters by conducting a Myc ChIP-chip study using an alternative oligonucleotide array platform. Our results show a high rate of validation by real time Q-PCR. The raw data from this study, encompassing over 100 arrays has been deposited in the Gene Expression Omnibus (GEO) repository at NCBI. Our careful description of ChIP-chip experimental design is a key step towards enabling widespread use of this important technology for the rapid elucidation of global transcriptional regulatory networks.
MATERIALS AND METHODS
Antibody production and purification
The DNA fragment corresponding to the Myc 1-262 N-terminal domain polypeptide was cloned into pET15b vector (Novagen 69661-3) at 5′-NdeI and 3′-BamHI sites. His-c-Myc (1–262) fusion protein was purified under denatured conditions using Talon beads (BD Biosciences, Mississauga, ON, Canada). His-c-Myc (1–262) fusion protein was purified under denatured conditions using Talon beads (BD Biosciences) as follows, the cell pellet was homogenized in 40 ml of lysis buffer pH 8.0 (5 mM imidazole, 20 mM Tris, 500 mM NaCl, 10 µM ZnCl2, 6 M Guanidine hydrochloride, 0.1% Triton X-100, 1 mM β-mercaptoethanol) and sonicated three times for 3 min at duty cycle 30 and 30% output. The lysate was then centrifugated at 14 000 r.p.m. for 30 min at 4°C. Next, 4 ml of 50% Talon beads were washed with 50 ml of binding buffer pH 8.0 (lysis buffer without TX-100) and pelleted at 1800 r.p.m. for 5 min. The lysed supernatant was incubated with washed Talon beads with gentle swirling for 1 h at room temperature and then centrifugated at 1800 r.p.m. for 5 min at 4°C. The beads were washed once with binding buffer and once with washing buffer pH 8.0 (Binding buffer with 10 mM imidazole). The proteins were eluted by adding 1 ml of elution buffer pH 8.0 (binding buffer with 500 mM imidazole) to the beads and after centrifugation at 1800 r.p.m. for 5 min at 4°C the supernatant was collected. The protein concentration is checked and the elution step is repeated several times until no more or very low protein is detected. The purity of the protein sample is then checked by SDS–PAGE. The denatured protein sample was precipitated by adding 900 µl of 95% ethanol to 100 µl of protein sample, and then centrifugated at 14 000 r.p.m. for 10 min. The pellet was resuspended in 20 µl of SDS-loading buffer before loading. The purified protein was concentrated to 2.8 mg/ml before injecting into the rabbit. The purified protein was then dissolved in a volume of 500 µl of PBS per rabbit. Primary immunizations were done with 500 µg of purified protein per rabbit. First boost was done with 250 µg of purified protein. Two subsequent boosts were done with 50 µg of purified protein. The final bleed was then purified using Enchant IgG Purification Kit with Protein A, IgG Purification (Pall 5300-IGGPROA) as follows: the sample was diluted 1:1 with binding buffer and applied to the equilibrated Protein A affinity column. The column was washed once with the Protein A Binding Buffer and then eluted with the Protein A Elution Buffer to recover the bound IgG. Each microliter of purified fraction was then neutralized by adding 50 µl of 1 M Tris, pH 9.5.
Cell culture conditions
HL60 (ATCC) cells were maintained in α-MEM with 10% FBS.
Chromatin immunoprecipitation
Exponentially growing HL60 cells were cross-linked with 1% formaldehyde for 10 min at 37°C. The cross-linking reaction was quenched by addition of glycine to a final concentration of 0.125 M for 5 min, followed by two washes with phosphate-buffered saline (PBS). Cells were resuspended in cell lysis buffer (5 mM PIPES pH 8, 85 mM KCl, 0.5% [v/v] NP40, 1 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin) for 10 min on ice and then pelleted (5000 r.p.m., 5 min, 4°C). The pellet was resuspended in 1 ml of nuclei lysis buffer (50mM Tris–HCl pH 8.1, 10mM EDTA, 1% SDS, 1 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin) for 10 min on ice and then sonicated using 8 pulses (12–13 Watts, setting 10, 10 s per pulse, 45 s on ice between pulses) from a Model 60 Sonic Dismembrator (Fisher Scientific 15-338-53) to generate fragments between 600 bp and 1000 bp. Lysates were centrifuged for 10 min at 21 000g at 4°C. Supernatants were diluted into an equal volume with IP dilution buffer (0.01% SDS, 1.1% Triton-X100, 1.2 mN EDTA, 16.7 mM Tris–HCl pH 8.1, 0.2% Sarkosyl, 1 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin) and precleared for 30 min at 4°C with protein G-PLUS agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA, sc-2002). Prior to use, G-PLUS agarose beads were blocked with salmon sperm DNA at a final concentration of 50 µg/ml and rotated overnight at 4°C. Diluted and cleared extracts corresponding to 10 x 106 HL60 cells were incubated and rotated at 4°C for ∼12–16 h with each of the following antibodies: no-antibody control, 0.7 µg normal rabbit IgG (Santa Cruz Biotechnology sc-2027), 0.7 µg N262 (Santa Cruz Biotechnology sc-764), for the N262 home-made unpurified and purified antibodies, we determined empirically, by serial dilutions, the amount of antibody to be used. A 50 µl of salmon sperm DNA preblocked Protein G-PLUS agarose beads were added to each sample, incubated on a rotating platform at 4°C for 3 h. Each pellet was washed once with 1.4 ml of sonication buffer and then twice with 1.4 ml of high salt buffer (0.1% [v/v] SDS, 1% [v/v] Triton X-100, 1 mM EDTA, 50 mM HEPES, 500 mM NaCl, 0.1% [w/v] sodium deoxycholate) and then once with 1.4 ml LiCl Buffer (250 mM LiCl, 1% [v/v] NP-40, 1% [w/v] sodium deoxycholate, 1 mM EDTA, 1 mM Tris pH 8) and finally twice with 1.4 ml TE pH 8 (10 mM Tris pH8, 1 mM EDTA). For each wash, the pellets were mixed for 5 min at room temperature then pelleted (3000 r.p.m., 30 s, room temperature). After the last wash, the pellets were eluted in 300 µl of Elution buffer (1% [w/v] SDS, 10 mM Tris pH 8, 5 mM EDTA), incubated at 65°C for 15 min, and then pelleted (3000 r.p.m., 3 min, room temperature). Cross-links were reversed in the presence of 200 mM NaCl at 65°C over-night and samples were treated with RNase A (Sigma R5500). After ethanol precipitation, the samples were resuspended in 100 μl of TE (10 mM Tris, pH 7.5, 1 mM EDTA), 25 μl of 5× proteinase K buffer (1.25% SDS, 50 mM Tris, pH 7.5, 25 mM EDTA), and 1.5 μl of proteinase K (Roche 1413783) and incubated at 42°C for 2 h. DNA was extracted from each sample using phenol:chloroform:isoamyl alcohol (25:24:1), then precipitated with 1/10th volume of 3 M sodium acetate (pH 5.3), 5 μg of glycogen, and 2 volumes of ethanol at −20°C overnight. Pellets were collected by microcentrifugation and resuspended in 60 μl of H2O.
PCR
A total of 2 µl of the ChIP DNA was PCR amplified using promoter-specific primers. All PCR primers (synthesized by Sigma Genosys, Oakville, ON, Canada or Invitrogen, Burlington, ON, Canada) were resuspended in sterile RNase-/DNase-free water. In general, each 10 µl PCR reaction, set up at room temperature, was composed of 10 ng/µl of each primer (fwd/rev), 1× PCR buffer, 0.2 mM dNTP mix, sterile water, DNA template and 0.2 µl HotstarTaq DNA polymerase (Qiagen, Mississauga, ON, Canada, 203205); the HotstarTaq was activated by initiating the PCR program at 95°C for 15 min. Amplification of each PCR primer set was performed using multiple annealing temperatures along a 52–68°C gradient for 30 s. Primer sequences are provided in Supplementary Table 8. All PCR reactions were electrophoresed on 100 ml 1.2% agarose gels containing 4 µl of 10 mg/ml EtBr, and then visualized by UV florescence.
Real-time quantitative PCR
Real-time quantitative PCR (Q-PCR) amplification was conducted using the SYBR Green assay in the ABI PRISM 7900-HT (Applied Biosystems, Foster City, CA, USA). Each 12 µl quantitative PCR reaction was composed of the following: 2 µl of the DNA isolated in the ChIP, 1x PCR Buffer, 2.5 mM MgCl2, 0.17 mM dNTP mix, sterile water, 0.25× SYBR Green (Sigma S9430), 0.2 µl Rox reference dye (Invitrogen 12223-012) and 0.05 µl Platinum Taq DNA polymerase (Invitrogen 10966-034) and was performed in triplicate in a 384-well plate. The reactions began at 50°C for 2 min and then were activated at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Human male genomic DNA (Novagen 70572) was used as standard real-time Q-PCR was conducted in triplicates for each of the three independent biological replicates. The Q-PCR data were analyzed by calculating the Myc/IgG ratios for each target gene, and the Chr6 negative control. A symmetrical distribution was then obtained by log2-transformation of each individual ratio. Statistical testing involved comparison of the ratios for each target gene to the ratios of the Chr6 negative control using a two-tailed paired t-test. Primer sequences are provided in Supplementary Table 8. The primers were designed using Primer Express Software Version 2.0 (Applied Biosciences) with the following parameters: Tm range of 58–60°C, the primer length between 19 and 50 bp, optimal primer length of 20 bases, and amplicon length of 80–200 bp.
Random primer PCR amplification methods
The immunoprecipitated DNA was amplified as described elsewhere (www.microarrays.ca/support/PDFs/ChIP_Protocol_for_Microarray_Analysis_Beads_corr_01-12-06.pdf) with the following modifications: 15 µl of Round A are mixed with Round B1 Reaction Mix to a final volume of 100 µl [4 µl of 50 mM MgCl2, 10 µl of 10× PCR Buffer, 2 µl of 10mM dNTPs, 2.5 µl of 100 µM of Primer B (5′GTTTCCCAGTCACGATC3′), 1 µl of Taq DNA Polymerase (Promega, Madison, WI, USA, M1861)]. Amplification/Nucleotide Incorporation Program: (94°C for 30 s, 40°C for 30 s, 50°C for 30 s, 72°C for 1 min) × 20 cycles, 72°C for 2 min.
A total of 15 µl of Round B1 were mixed with Round B2 Reaction Mix to a final volume of 50 µl [2 µl of 50 mM MgCl2, 5 µl of 10× PCR Buffer, 0.5 µl of dNTP Mix B2 (25 mM of dATP, dCTP, dCTP, 12.5 mM dTTP), 1.5 µl of 2 mM aa-dUTP, 1 µl of 100 µM of Primer B (5′GTTTCCCAGTCACGATC3′), 1 µl of Taq DNA Polymerase (Promega M1861)]. Amplification/Nucleotide Incorporation Program: (92°C for 30 s, 40°C for 30 s, 50°C for 30 s, 72°C for 1 min) × 15 cycles, 72°C for 2 min. Round B2 amplified DNA was concentrated using Microcon columns (Millipore, Billerica, MA, USA, Microcon YM-30). Once the ChIP DNA was amplified to 3 µg, PCR reactions were performed with 10 ng amplified DNA and were identical to the reactions performed after the ChIP DNA was initially isolated. This PCR was used to determine whether specificity between the IgG or NoAb and antibody samples was maintained after the amplification.
Ligation-mediated PCR amplification
The DNA from each ChIP reaction was treated as described elsewhere (26). The PCR was performed using the following cycling protocol: 1 cycle of 55°C for 2 min, 72°C for 5 min, and 95°C for 2 min and the corresponding number of cycles for ligation-mediated PCR (LM-PCR) A and LM-PCR B (see below LM-PCR A and B cycle conditions) at 95°C for 30 s, 55°C for 30 s and 72°C for 1 min, ending with 4 min at 72°C and then an endless cooling at 4°C. After the PCR, the DNA was again isolated using the QIAquick PCR purification kit (Qiagen 28106). The concentration of the purified DNA was determined on the NanoDrop ND-1000 spectrophotometer. Often the 3 µg required for hybridization was not acquired after this first round of amplification, so another round of amplification would be performed, followed by purification and determination of the DNA concentration as previously described. LM-PCR A amplification: 20 cycles followed by QIAquick PCR purification, and then 15 cycles followed by QIAquick PCR purification. LM-PCR B amplification: 20 cycles followed by QIAquick PCR purification, repeated three rounds, sequentially. Once the ChIP DNA was amplified to 3 µg, PCR reactions of positive and negative control genes (Supplementary Table 8) were performed with 10 ng amplified DNA and were identical to the reactions performed after the ChIP DNA was initially isolated. This PCR was necessary to determine whether specificity between the IgG or NoAb and antibody samples was maintained after the amplification.
Labeling reaction
A total of 3 µg of amplified DNA from the antibody sample, no antibody, or IgG sample were vacuum desiccated and resuspended in 2.5 µl of H2O (Sigma W4502). 4.5 µl of 0.1 M NaHCO3 at pH 9 was added. The DNA was resuspended by vortexing several times. 2 µl of Cy3 or Cy5 dye (Amersham, PA23001 and PA25001, respectively) were added to the sample. The mix was incubated for 1.5 h at room temperature in the dark. After incubation, 35 µl of 100 mM Na-Acetate pH5.2 was added and topped with H2O (Sigma W4502) to a total volume of 100 µl for each sample. The fluorescent-labeled probes were purified using QIAquick PCR Purification kit (Qiagen 28106). The DNA was eluted with 50 µl of EB buffer from the kit, heated to 65°C. The 100 µl combined ChIP and no antibody samples were vacuum desiccated using a SpeedVac for 60 min at maximum heat.
DNA microarray hybridization
The mixed DNA was resuspended in 5 µl of H2O and mixed with 85 µl hybridization mix [100 µl of DIG Easy Hyb solution (Roche 603 558), 5 µl of 10 mg/ml calf thymus DNA (Sigma D8661), 5 µl of 10 mg/ml yeast tRNA (Invitrogen 15401-029)]. The resulting solution was incubated at 65°C for 2 min, then cooled to room temperature and applied to a CpG island array slide (UHN Microarray Centre HCGI12K, design is available at www.microarrays.ca) and incubated overnight at 42°C in a hybridization chamber.
Microarray washing
After hybridization, the microarray slide was washed once with washing buffer 1 (1× SSC, 0.1% SDS) at 50°C for 5 min. This was followed by a wash with washing buffer 1 (room temperature for 5 min) and two washes with washing buffer 2 (0.2× SSC for 5 min) and one final wash with washing buffer 3 (0.1× SSC for 5 min at room temperature). The slide was then dried by spinning at 700 r.p.m. for 15 min. To preserve the Cy5 dye from ozone-dependent degradation, which was observed during the summertime when the hybridization occurred, the slides were then dipped in DyeSaver 2 (Genisphere Q500600) and allowed to air dry.
Microarray scanning
The microarray slides were scanned using a Gene Pix 4000B scanner (Axon Instruments) at multiple PMT voltages.
Validation using agilent promoter arrays
The Chromatin Immunoprecipitation of HL60 cells was performed as described above. In this case, the sonication was performed as follows, 11 pulses (setting high, 26 s per pulse, 26 s on ice between pulses) from a Model Diagenode BioRuptor Sonicator (Diagenode, BioRuptor 200, UCD-200 TM-EX) to generate fragments between 500 and 1000 bp. The immunoprecipitations were performed using 0.7 µg normal rabbit IgG (Santa Cruz Biotechnology sc-2027) or N262 home-made purified antibody as described above. The WGA was performed following a protocol described by O’Geen et al. (29). This method efficiently primes the DNA fragments to generate a library of DNA fragments with defined 3′ and 5′ termini. The library is then replicated using linear amplification in the initial steps followed by a limited round of geometric amplifications. Entire ChIP samples with an average size of 500–1000 bp of DNA fragments were used for the library generation and subsequent amplification. For the library preparation and the amplification (round 1), GenomePlex Complete WGA kit was used (Sigma WGA2) as follows. Library Preparation step: 2 µl of Library Preparation Buffer was added to the ChIP material that has been concentrated to 10 µl. One microliter of Library Stabilization Solution was added and heated at 95°C for 2 min in a thermal cycler and immediately cooled on ice. One microliter of Library Preparation Enzyme was added and incubated in a thermal cycler precooled to 16°C (16°C for 20 min, 24°C for 20 min, 37°C for 20 min, 75°C for 5 min). Amplification step (round 1): the following master mix was added to each sample prepared on the previous step, 7.5 µl of 10× Amplification Master Mix, 47.5 µl of nuclease-free H2O and 5 µl of WGA DNA Polymerase. Samples were incubated in a thermal cycler (95°C for 3 min, then 14 cycles of 94°C for 15 s, and 65°C for 5 min). Samples were then purified using QIAquick PCR Purification kit (Qiagen 28106). A 10 ng of each amplified sample was tested for specificity as described above under the PCR title. For the reamplification step (round 2), GenomePlex WGA amplification Kit was used (Sigma WGA3). A master mix of 7.5 µl of 10× Amplification Master Mix, 47.5 µl of nuclease-free H2O and 5 µl of WGA DNA Polymerase was added to 15 ng of purified amplification product previously diluted in nuclease-free H2O to 10 µl volume. Samples were then purified using QIAquick PCR Purification kit (Qiagen 28106). Amplified sample (10 ng) was tested for specificity as described above. WGA amplified DNA (4 µg) was hybridized to Agilent 2x244 promoter arrays at the UHN microarray center (Toronto, Canada). Based on our previous results a single batch of arrays was used with lot number 19 585. Arrays were labeled using the Agilent Genomic DNA Labelling Kit and hybridized using the aCGH Hybridization Kit following the ChIP-on-chip v10 protocol as follows: for each 1x244 promoter array, 2 µg of DNA was brought to 26 µl final volume. A 5 µl of Random Primers (supplied with Agilent Genomic DNA Labeling Kit PLUS) was added and the mix was incubated at 95°C for 3 min, then on ice for 5 min. The 31 µl were mixed with the Labeling Mix to a final volume of 50 µl (10 µl of 5× Buffer, 5 µl of 10× dNTP, 3 µl of 1 mM Cyanine 3-UTP or 1 mM Cyanine 5-dUTP, 1 µl of Exo-Klenow fragment) and incubated at 37°C for 2 h. The enzyme was then inactivated at 65°C for 10 min. The labeled Genomic DNA was cleaned with Microcon columns (Millipore, Microcon YM-30) and eluted with 80.5 µl of 1× TE. A 5 µg Cy5-labeled and 5 µg of Cy3-labeled DNAs were combined in a total volume of 158 µl. The 158 µl were mixed with 50 µl of 1 mg/ml of Human Cot-1 DNA, 52 µl of 10× Agilent Blocking Agent, 260 µl of 2× Agilent Hybridization Buffer. Samples were heated at 95°C for 3 min, and then incubated at 37°C for 30 min. A 490 µl of the sample were applied to the 1x244 promoter array assembled in a chamber and incubated in a rotisserie hybridization oven at 65°C and 20 r.p.m. for 40 h. After hybridization the microarray slide was washed with Oligo aCGH/ChIP-on-chip Wash Buffer for 5 min at room temperature, followed by a wash for 5 min at 31°C. QC metrics used were based on the ChIP-on-chip requirements and no arrays were removed or repeated.
CpG island microarray data preprocessing
Array images were first manually examined for image artifacts, then quantitated using GenePix Pro (v6.0.1.27). A scan that maximized the dynamic range without saturating high-intensity signals was selected and carried forward for analysis. This scan was quantified using the ‘circular’ segmentation algorithm, then preprocessed using the variance-stabilizing normalization algorithm (VSN). VSN was implemented in the R statistical environment (v2.4.1) in the BioConductor open-source project (39). Version 1.12.0 of the vsn package was used, with print-tip groups used as strata, and default parameterization except for using 1000 iterations for fitting (increased from the default of 10). All raw and processed have been made publicly available in the GEO repository at NCBI with IDs GSE8447, GSE8448, and GSE8449.
To test for differential hybridization, we used general linear modeling, as implemented in the limma package (v2.9.13) (40) of the BioConductor open-source library (39) for the R statistical environment (v2.4.1). First, a spot-wise linear model was fit separately to each major design parameters: array batch, hybridization date and dye-status. No interaction terms were included. A Bayesian moderation of standard error was employed following model-fitting (40). A Gaussian kernel density was then fit to the effect estimates to compare the relative magnitudes of these design parameters. Next, a spot-wise linear model was fit comparing the hybridizations using a commercially available antibody to those using a locally produced one. The top 100 spots from each analysis were identified, and the pair-wise P-values plotted to identify trends in antibody specificity. A similar analysis was performed for purified and nonpurified locally produced antibody hybridizations. All model-fitting again employed the limma package (v2.9.13). To compare different amplification methods we fit separate linear models to the amplified versus nonamplified comparisons for each amplification method. The number of differentially bound spots between the two samples was calculated across the entire range of naïve P-value thresholds from 0 to 1, at 0.00 001 unit resolution. A similar analysis was used to investigate hybridization order, except that the models were fit simultaneously and the effect of each hybridization order was extracted separately using a contrast matrix. An identical tiling across P-values was used, in this case in the range 0 to 0.10 with a resolution of 0.0001. These analyses were performed in R (v2.4.1). All gene-lists presented are based on a P-value threshold of P < 0.05.
Agilent promoter microarray data preprocessing
Microarray data was scanned using the Agilent Feature Extraction Software (v9.5) and then loaded into the R statistical environment (v2.6.2) using the limma package (v2.12.0). Array data was preprocessed using variance-stabilizing normalization, as above, with 1000 iterations to generate highly robust results on these large arrays. Preprocessing employed the vsn package (v3.2.1) in the R statistical environment. The raw and preprocessed data were subjected to a series of quality control measures: all arrays were included in subsequent analyses. Significance-testing used t-tests to compare the antibody and control channels, followed by an Empirical Bayes’ moderation of standard error (40) and a false-discovery rate adjustment for multiple-testing (41). The raw and pre-processed data have been made available in the GEO repository with accession GSE11245.
Spot annotation
The published array annotations for the 12k UHN CpG island microarray were based on the UCSC build hg17 of the human genome (38). To update these results for the more polished hg18 build, we implemented a new annotation algorithm using a BioPerl (v1.5.1) based Perl (v5.8.8) script (42). First, the 3′ and 5′ sequence reads were overlapped using bl2seq (43) with a word-size of 7. In cases where a minimum 50 bp overlap was found the aggregated sequence was selected; otherwise the longer of the two sequence reads was chosen. Next, the sequences were aligned to the RepeatMasker filtered chromosomal sequence-reads of human genome build hg18 (44) using a locally compiled (gcc v4.1.1; AIX 5.2.0.0) NCBI BLASTN executable with a word-size of 7 (45). The best matching hit was selected as the most likely region of hybridization, and the ratio of the lengths of the top two hits was used as a measure of the likelihood of cross-hybridization. The Agilent default annotations were used for the oligonucleotide validation experiment.
RESULTS
Our analysis of ChIP-chip design parameters was conducted in the context of the genomic-binding of the well characterized Myc oncogene in an acute myelogenous leukemia cell line, HL60 (46). Optimization experiments utilized 12k CpG island microarrays which contain 12 192 genomic locations, representing 5411 distinct genomic loci (38). Validation experiments were performed using the Agilent human promoter whole genome array set.
Antibody selection, array batch, reciprocal labeling and inter-day processing
Our first series of experiments focused on assessing four key parameters of the ChIP-chip assay. Antibody selection was considered first because the immunoprecipitation step provides the starting material for the array analyses, and as such is a major determinant of the success of ChIP-chip studies. Second, we addressed the effect of array batch. This can be a major confounding variable because DNA spotted to a microarray slide may degrade or otherwise lose sensitivity, and even arrays printed in different print-runs from a single spotter can show variability. Third, we evaluated the bias introduced during the process of sample labeling. This bias refers to intensity differences between identical samples labeled with different dyes and can arise in several ways, including differential incorporation efficiencies for the two dyes, fluorophore degradation, as well as photo-bleaching rates or scanner-induced bias. Fourth, we determined whether day-to-day variations in atmospheric ozone, technical handling, scanner calibration or other, as yet unidentified, confounders affect the hybridization step.
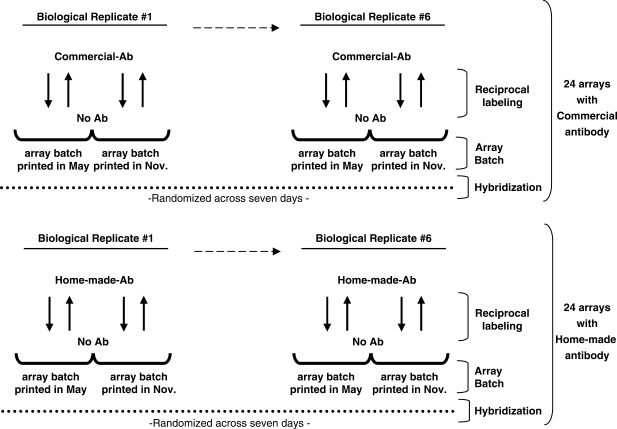
To evaluate the effect of these ChIP-chip parameters, we conducted a saturated, fully blocked experiment (Figure 1). First, 12 independent biological replicates of cross-linked HL60 cells were subjected to ChIP with polyclonal antibodies raised against the N-terminal 262 residues of the Myc oncoprotein. Six of the twelve replicates were ChIPed using a commercial antibody and the remaining six using a home-made antibody raised against the identical region. For each of these twelve replicate ChIP reaction we probed two batches of arrays separated in print-age by about 6 months. This allows us to evaluate the array batch effect. Each sample was hybridized twice to each batch with reciprocal labeling. Thus, each biological replicate was hybridized to four separate arrays, leading to 24 arrays for each antibody and 48 arrays in total. Finally, these 48 arrays were hybridized in batches over 7 days, allowing us to robustly estimate the effect of date-of-hybridization (Figure 1).
Figure 1.
Experimental design to evaluate antibody effect, array batch, reciprocal labeling and hybridization. Schematic representation of the experimental design in which six independent biological replicates of cross-linked HL60 cells were subjected to ChIP with either commercial or home-made antibodies raised against the N-terminal 262 residues of the Myc oncoprotein. Each sample was hybridized to individual arrays with its matched No Ab control. Dashed arrows indicate biological replicates #2 to #5, not included in the figure for simplicity. Black arrows indicate reciprocal labeling. Dotted lines indicate the period of seven days in which the 48 arrays were hybridized.
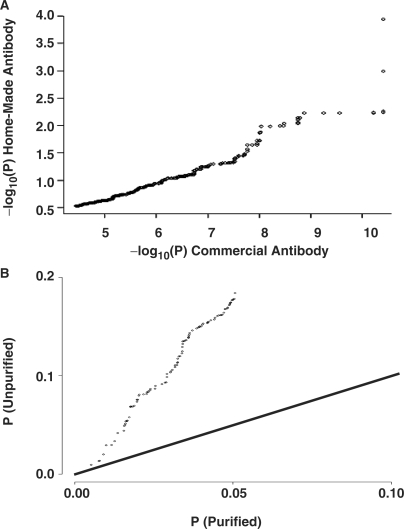
To quantify the importance of antibody selection, we compared two polyclonal antibodies raised against the same N-terminal region of the Myc protein. One antibody was purchased from a commercial vendor while the other was locally produced (Figure 1). Across the 48 arrays of our study, we observed a significant antibody effect. When the P-values of the top-ranked spots on the array are compared for the two antibodies, a linear trend is observed, suggesting specificity is comparable (Figure 2A). However, despite equivalent replicate numbers and full-blocking of other parameters, as described below, P-values are distinctly lower for the commercial antibody, suggesting that it results in better signal to noise characteristics. To increase the specific activity of our home-made antibody, we subjected it to a purification procedure to remove contaminating serum proteins, and compared purified and unpurified home-made antibody in a similar fashion (Figure 2B). A linear trend was again observed, with the purified antibody showing lower P-values than the unpurified. Note that the magnitude of P-values in Figure 2A and B are different because of the different replicate numbers involved (Figure 2A, n = 24; B, n = 4 for each antibody). These data suggest that antibody purity is a key factor influencing the specificity and sensitivity of ChIP-chip studies and may improve the performance of some antibodies.
Figure 2.
Measuring antibody effect. (A) Comparison of ChIP-chip results using a commercial and home-made polyclonal antibody raised against the same portion of Myc protein showed a similar linear trend and ordering by the two antibodies (P < 2.2 × 10−16; Wilcoxon Rank Test), however, they differed by orders of magnitude in sensitivity (axis scales). (B) Purification of the home-made antibody preparation improved sensitivity. The line represents equal sensitivity (y = x). Note that orders of magnitude on the axes differ between parts A and B because the replicate numbers are different (A, n = 24; B, n = 4).
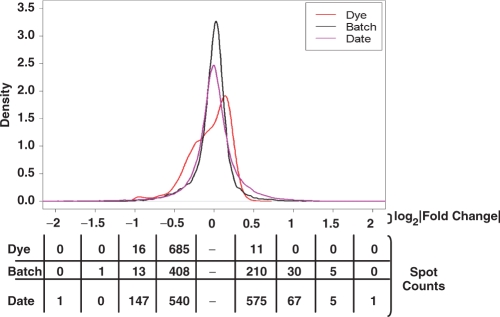
To evaluate the effect of array batch, dye bias and inter-day hybridization, we fit spot-wise linear models to the results of each parameter separately and plotted the Gaussian densities of the magnitudes of these effects (Figure 3). These Gaussian densities are essentially smoothed histograms. Their height (y-axis) reflects the number of spots on the arrays affected by each parameter at the given fold-change (x-axis). Each curve reflects a different source of variability (dye, batch and date). The numbers under the plot give the number of spots affected by each parameter at a given fold-change threshold. All three curves are unimodal, with a sharp peak at the origin. This shows that the majority of spots on the array are unaffected by each parameter. Of the spots that are affected, the magnitude is generally very modest, in the range of −0.5 to +0.5 log2 units. This corresponds to an effect of 1.4-fold. Overall, no >10% of spots on the array show large variability towards any of these parameters—suggesting that both the ChIP protocol and the CpG island arrays used are robust towards perturbations in any of these factors.
Figure 3.
Evaluating array batch effects, dye-bias and inter-day processing. To assess the effects of array batch, dye-swap, and hybridization date we fitted separate spot-wise linear models to the data from our study for each of these three parameters. Gaussian kernel densities were then plotted for the coefficients of the fits for each model. Each curve is unimodal, with a sharp peak around the origin, indicating that most spots on the array do not vary with these design parameters. The spot counts along the bottom of the figure further quantify the degree of variability at several thresholds (0.5 = 1.4-fold, 1.0 = 2.0-fold, 1.5 = 2.8-fold, 2.0 = 4.0-fold).
Despite this basal similarity there are differences across the three parameters. The curve of dye-bias (red) shows significant asymmetry, corresponding to a consistent Cy3 bias. While only 11 spots show Cy5/Cy3 ratios of 1.4-fold or higher, 685 spots show Cy3/Cy5 ratios of 1.4-fold or greater. This marked asymmetry highlights the importance of balancing experiments across the two dyes used in the study.
The effect of array batch shows a similar, but smaller asymmetry. Older arrays consistently show lower signal intensities than newer arrays, with 408 spots showing an old/new ratio of at least 1.4-fold, while only 210 spots show a new/old ratio of similar magnitude. This variability suggests that experiments should be conducted with arrays of a single batch, or experimental samples randomized across multiple batches.
Surprisingly, the effects of hybridization date affect almost 10% of the spots on the array, despite the same, experienced hands conducting all hybridizations in this series of experiments. This effect is symmetrical, with approximately equal numbers of spots showing date-dependent enrichment and depletion. The source of this bias is unknown. It is possible that atmospheric ozone may have differed from day-to-day. We and others have experienced Cy5 squelching due to ozone (47,48). However, in this case ozone is not likely to be the source of the bias, as we chose to conduct these experiments during times of consistent and low atmospheric ozone. Variations in the efficiencies of the labeling reaction and subsequent column-clean-up may account for this effect. It is also possible that variation in handling at the level of post-hybridization washes may contribute to this inter-day variability. Another possibility is that scanner calibration varied across the course of our experiment, but the biases described here are not temporally-dependent (data not shown). This suggests that if scanner-calibration is an issue, then it is a random drift rather than a progressive degradation.
As an alternative visualization of this data, we provide histograms of each factor as Supplementary Figure 1. In addition, the raw and normalized data are available in the GEO repository, while the linear model fits are available as Supplementary Tables 1–3.
Amplification method
Sufficient DNA for microarray hybridization cannot usually be obtained from a single ChIP reaction. The amount required can be achieved by pooling multiple ChIP reactions (12), but this approach is expensive and labor intensive. Further, when dealing with biologically limited samples, such as primary patient material, repeated ChIPs may not be possible. Indeed in such cases pooling multiple ChIPs may introduce intra-sample heterogeneity (49). To overcome this rate-limiting step, the precipitated DNA in a single ChIP reaction is usually amplified. A variety of PCR-based methods exists, including LM-PCR, WGA, random primed PCR (RP-PCR) and T7 primed PCR methods.
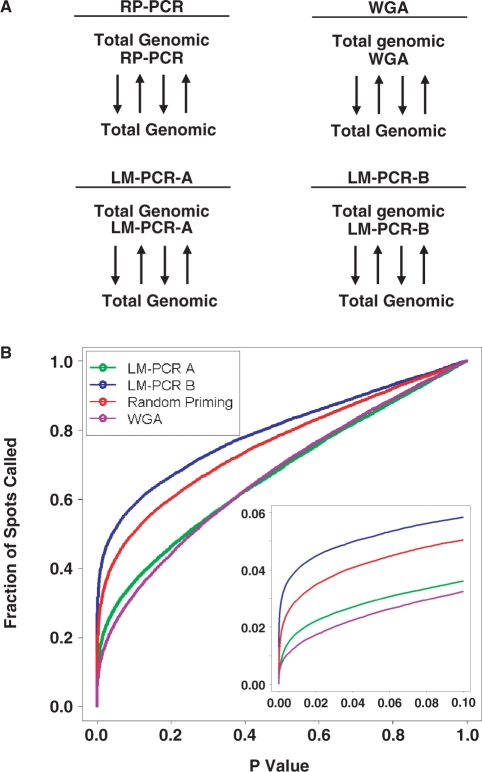
We develop an assay to evaluate the bias introduced by amplification protocols. For each amplification protocol we take a pool of total genomic DNA and amplify one aliquot. We then hybridize equal amounts of amplified and unamplified DNA on a microarray slide. This procedure is repeated to determine the number and nature of the spots showing amplification bias (Figure 4A). Using this method we tested the three most common amplification techniques employed in ChIP-chip experiments, RP-PCR, WGA and LM-PCR (Figure 4A). To consider parameter sensitivity for the LM-PCR method we evaluated protocols with low (LM-PCR A, 45 cycles) and high (LM-PCR B, 60 cycles) PCR cycle number.
Figure 4.
Comparing DNA amplification methods. To evaluate whether specific DNA amplification protocols introduce different degrees of bias, we surveyed the genome-wide effects of three amplification protocols. (A) Total genomic DNA was amplified with one of four different protocols: random priming (RP-PCR), WGA and two LM-PCR protocols (LM-PCR-A and LM-PCR-B). The resulting DNA was then hybridized to a CpG Island microarray against an equal amount of unamplified total genomic DNA. This procedure was repeated four times for each of the three amplification methods, and the raw data was pre-processed with VSN and model-based t-tests (see Materials and methods section). Black arrows indicate reciprocal labeling. (B) The fraction of spots on the array that are differentially amplified (fraction of spots called) is plotted against the P-value threshold for each amplification method. The y-axis gives the fraction of spots differentially amplified, thus a value of 0.1 indicates that 10% of the spots on the array have been affected by the amplification protocol. The y = x line reflects the fraction of spots that would be expected to be altered by amplification by chance alone. All four curves lie above this line, demonstrating the large effect of amplification. The curves for LM-PCR A and the WGA lie below that of the other two methods for all P-values, demonstrating that these two methods introduce the smallest amplification bias amongst the four protocols tested. The inset provides a zoom-in of the P-value range from 0 to 0.1, where thresholds for most experiments are selected.
For each amplification method, we considered the fraction of spots on the array that showed altered hybridization between amplified and unamplified samples as a function of the p-value threshold (Figure 4B). Methods with reduced amplification bias will show lower curves, especially at experimentally relevant lower P-value thresholds (Figure 4B, inset). The LM-PCR A and WGA amplification methods alter the hybridization of many fewer spots than either RP-PCR or LM-PCR B. These data demonstrate a practical and affordable method of evaluating a variety of amplification protocols using genomic DNA.
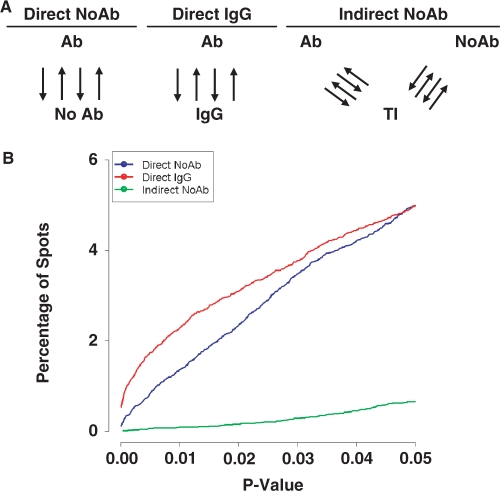
Hybridization control
A variety of different hybridization controls have been used for ChIP-chip studies. For example, some groups have employed a ‘no antibody’ control (12), while others have used total genomic DNA (11,50). To determine if there are significant differences between these hybridization controls, we designed a series of experiments to test each approach (Figure 5A). Specifically, we compared direct hybridization of ChIP samples against a mock treated control (Direct No Ab) and against an IgG antibody control (Direct IgG). We also considered the use of total input DNA as a ‘denominator’ by hybridizing ChIP and IgG samples separately against total-input DNA (Indirect No Ab), then comparing their intensities indirectly during statistical analysis by using a linear model. The results (Figure 5B) are surprisingly invariant from one control to the next, and at any given P-value the number of hits identified in each experiment is nearly identical. Nevertheless, the two direct designs show more sensitivity than the indirect approach in a threshold-independent manner.
Figure 5.
Comparing hybridization controls. A number of potential control samples can be used for ChIP-chip studies. (A) Three common approaches were compared, including two direct hybridization approaches (No Ab and IgG) and an indirect hybridization approach (No Ab). For each approach ChIP-chip studies of Myc in HL60 cells were performed. Black arrows indicate reciprocal labeling. (B) The sensitivity of each approach was plotted as the percentage of spots identified as differentially bound versus the P-value. While the three methods displayed similar performance, the two direct methods show higher sensitivity than the indirect method.
Validation studies
To evaluate the optimized parameters determined above, we conducted a new Myc ChIP-chip experiment with them. Thus, we used a purified antibody for the ChIP reaction, DNA was amplified by LM-PCR, we hybridized a single CpG island array batch from a recent print run, and IgG was used as the hybridization control. Dye swaps were randomized across hybridization dates, and the entire experiment was conducted at high replication (n = 13). By using this highly optimized design, we expected to both minimize bias and increase specificity. Following preprocessing and statistical analysis of the raw data we obtained a list of 534 Myc bound spots (Supplementary Table 4). This represents ∼9.9% of the 5411 distinct genomic loci represented on this array platform (38). Importantly, 35 of these 535 spots correspond to known Myc targets annotated in the Myc target gene database (51), providing strong support for the validity of this gene list.
To further generalize our results, we then performed a second validation experiment using a different microarray platform—Agilent genome-wide oligonucleotide promoter arrays. We again employed our optimized parameters, using a purified antibody, a single batch of arrays and IgG as a hybridization control. For this second validation, we used the WGA method to amplify the DNA. As these oligonucleotides are a relatively new platform we rigorously evaluated array quality and found no evidence of spatial or distributional artifacts (data not shown). Following preprocessing and statistical analysis we obtained a list of 53 453 genomic regions that bind c-Myc, corresponding to 10 196 unique genes or genomic features (Supplementary Tables 5 and 6). This represents 11.1% (53 453 of 481 909) of the unique genomic regions and 28.9% of the unique genes represented on the array, indicating wide-spread Myc binding. On average each unique gene was represented by 5.24 probes, giving a high-degree of internal replication within the array.
Comparison of the CpG island and promoter arrays is confounded by the lack of perfect correspondence between the genomic features represented on each array. For example, the CpG island arrays might interrogate a region downstream of the transcriptional start-site, while the promoter array interrogates one upstream of the start-site. Nevertheless, we compared the hits from the CpG island array that were within 10 kb of an annotated Entrez Gene transcriptional-start site to those from the promoter array. Fully, 81% (208/256) were confirmed on the promoter arrays at a 10% FDR. Some fraction of the unconfirmed cases are attributable to differences in the genomic regions interrogated, indicating that inter-platform variability is at most 19% in this study.
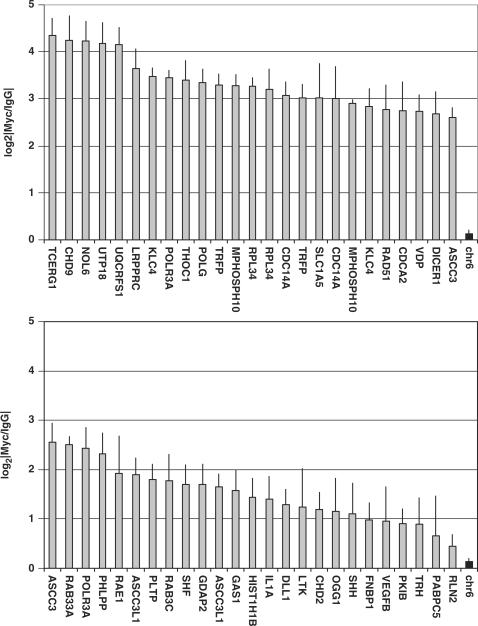
Surprised at the very large fraction of promoter regions that exhibited c-Myc binding in our genome-wide analysis, we sought to validate these results using gold-standard Q-PCR on independent biological replicates. We selected 50 random genomic loci from our gene-lists (Supplementary Tables 4–6) and primer pairs to interrogate them by Q-PCR. We used three Myc ChIP biological replicates in HL60 cells that were completely separate from any of the samples used in either the optimization or validation studies. As a negative control we employed a region of chromosome 6 that showed no evidence of Myc binding in the ENCODE study. Our Q-PCR results (Figure 6 and Supplementary Table 7) verify the extensive binding of c-Myc in HL60 cells, with an overall validation rate of 45/50. Further, the array and PCR results are highly correlated on a region-by-region basis, both in terms of statistical significance (Spearman's rho = 0.533, P = 7.80 × 10−5) and magnitude of enrichment (Spearman's rho = 0.598, P = 5.77 × 10−6). These data show that our optimized ChIP-chip experimental parameters allowed us to identify novel bona fide Myc target genes using two separate microarray platforms as well as quantitative PCR.
Figure 6.
Myc recruitment to regulatory regions of novel target genes in growing HL60 cells. ChIP assays with antibody to Myc and IgG control antibody from exponentially growing HL60 cells. DNA was Q-PCR amplified by using primers against 50 distinct genomic regions. A negative control region on chromosome six that showed no Myc binding in any cell-line of the ENCODE study was also included and is shown twice for reference purposes only. This region is shaded in black and is labeled ‘chr6’. Each column represents a specific region, identified by the HGNC gene-symbol. When two different regions of the same gene were tested, the same gene can be found twice in the figure (Supplementary Table 8). The y-axis gives the mean log2|Myc/IgG| values for each genomic region. Error bars represent SDs.
DISCUSSION
ChIP-chip is a relatively new technology that has quickly become a standard method for screening protein–DNA interactions in vivo. However, it is also a highly complex method, with many tunable parameters. We set out to systematically test a large subset of these experimental design parameters. Many previous studies of microarray reproducibility have employed spike-in or mixture designs (23,31,33,35), but it is unclear if spike-in or mixture experiments reproduce the underlying complexity of real biological samples. Accordingly, we chose to focus on a biologically well-characterized system: involving the Myc oncogene as a regulator of gene transcription in the HL60 human myeloid leukemia cells (12).
The first experimental design parameter considered was the influence of different antibodies on the results of a ChIP-chip study. Intuitively, the nature of the antibody is a limiting step for ChIP-chip studies. To put a lower bound on the variability introduced by different antibodies we selected two polyclonal antibodies raised against the same portion of the Myc protein (the N-terminal 262 residues, N262) but derived from different sources. Critically, the two antibodies showed a broadly linear, well-ordered trend (P < 2.2 × 10−16; Wilcoxon Rank Test), but differed significantly in sensitivity (Figure 2A). We subjected the lower sensitivity antibody to a column purification procedure to remove contaminating serum proteins, and this purification procedure significantly improved performance (Figure 2B). Clearly, antibody selection is a critical element of ChIP-chip studies, and antibody-specific activity is an important parameter to consider prior to hybridization.
Next, we considered the effects of inter-batch variability of the CpG island microarrays. This array platform has been widely used by other groups (27,50,52), giving confidence in its reliability. We tested the variability between two batches of arrays and observed a consistent bias in signal-intensity, with the older array batch showing lower signal (Figure 3). This bias is highly reproducible, as we analyzed 24 arrays from each batch. The bias is symmetric, and therefore can lead to both false positives and false-negatives. Fortunately, straightforward methods exist to control this bias. First, whenever possible, arrays from a single batch should be used. Second, when multiple batches are used samples should be randomized across batches. Third, linear-modeling methods can be used to explicitly account for and remove batch variability. The first two methods are standard experimental design practices, while the third has been successfully used in the analysis of mRNA expression array data (53).
Third, we considered the effects of reciprocal labeling (dye-swapping) a single sample with the Cy3 and Cy5 dyes. This topic has been extensively studied from both theoretical (54,55), and empirical (56) perspectives. These studies found small, but consistent effects of reciprocal labeling. While reciprocal labeling has been shown to have less experimental value than true biological replicates, they appear to carry more information than simple repeated measures (56). Very moderate effects of reciprocal labeling were observed, affecting only about 0.1% of the spots on the array at a two-fold change level (Figure 3). There are many more spots affected by dye status at a less stringent 1.4-fold change threshold, however changes of this magnitude introduce a much smaller risk of altering the spot-wise statistical analyses. This indicates that, at least for ChIP-chip with our CpG island arrays, reciprocal labeling is not necessary and dye-bias is a negligible confounder. Nevertheless, it seems prudent to randomize samples across dye-status to account for unanticipated dye-bias in specific experimental scenarios, especially because of the profound asymmetry in this parameter.
The fourth experimental parameter considered was the effect of hybridization date. Because our experiment was fully blocked across all factors, we were able to directly estimate the effects of this parameter with a linear model. We were surprised to find that the day on which hybridization was performed had a greater influence on the final results than either the batch of arrays or the use of dye-swaps (Figure 3). The sources of this bias are unclear. The symmetrical distribution of the effect suggests that neither ozone-status nor scanner settings are a major factor, suggesting random drift within the scanner. Perhaps use of a mechanical hybridization and wash station would decrease inter-day variability, although this variability may also result from the labeling reaction or the subsequent column clean-up procedure. Control of this factor is straightforward: samples should be randomized across experimental day, so that biological replicates of a given condition are hybridized on different days.
The fifth experimental parameter considered was the method of DNA amplification. While the antibody-based ChIP protocol imparts specificity and sensitivity to ChIP-chip experiments, the quantity of DNA necessary for hybridization cannot usually be obtained in a single ChIP. Two basic strategies have evolved to cope with this issue: pooling of multiple ChIPs (12) and amplification of ChIP DNA (26). A variety of different PCR-based amplification methods have been developed and are in use by different groups. Assessing the performance of these methods has been challenging. To directly compare an amplified ChIP sample to an unamplified ChIP sample requires the pooling of a large number of ChIPs for each replicate, resulting in significant costs, time and labor. Further, care must be taken in the experimental design to ensure that the reduction in variance that results from pooling ChIP reactions does not confound the effects of the amplification procedure. As a result of these financial and technical factors, only one study has assessed the performance of amplification methods for ChIP-chip data (23), although several such studies do exist for mRNA experiments (57,58) where a much larger amount of starting material is readily available. To resolve this issue, we have developed a novel approach that hybridizes amplified against non-amplified genomic DNA (Figure 4A). We evaluated four amplification protocols: RP-PCR, WGA and two variations of LM-PCR. Because of its simplicity, this procedure could be readily repeated and an n = 4 was generated for each amplification procedure. The number of spots found to be differentially hybridized between the matched amplified and unamplified samples is a measure of amplification bias (Figure 4B). Interestingly, the performance of random-priming falls in between that of the two LM-PCR protocols, indicating that specific parameter choices within a protocol are important for determining overall performance. The WGA and low-cycle LM-PCR protocols showed similar performance, suggesting that one of these methods should be selected to minimize amplification bias. Our method of comparing amplification procedures provides a new way to measure variability, independent of antibody or biological question, at a reasonable cost.
The final experimental design parameter considered was the selection of a negative control for hybridization to the array. We considered three approaches (Figure 5A), including two direct and one indirect method. For each method we performed four Myc ChIP-chip experiments in HL60 cells. When we compared the sensitivity of these methods (Figure 5B) the two direct methods performed comparably, with no significant differences between the IgG and no-antibody controls. The indirect design, however, did show reduced sensitivity at any given P-value. Thus, direct designs, which use half as many arrays as indirect designs, are favored for both cost and sensitivity.
Having extensively optimized these six parameters, we set up two Myc ChIP-chip experiments to test our optimization in an integrated way. Our first experiment used extensive biological replication (n = 13) on CpG Island arrays, while our second used lesser replication (n = 5) on much larger whole-genome oligonucleotide promoter arrays. To validate the results of these two studies we employed Q-PCR using biologically independent samples. Of the 50 unique primer sets we analyzed, 45 were validated, for a success rate of 90%. This strong evidence that the vast majority of the 10 196 genomic locations found to be associated with Myc in HL60 cells are true positives. Importantly, we showed that the validation rate remained high in the bottom portion of our list. This suggests that the statistical analysis was conservative, and that additional hits might be uncovered by relaxing our P-value threshold from 0.05 to 0.10 or higher. This is particularly important because many published ChIP-chip studies use a P-value threshold of 0.001, 50 times more stringent than used here (11). Overall, our extensive validation confirms the success of our parameter optimizations.
Over the past decade, a series of studies from multiple groups characterized the major experimental design parameters for mRNA expression studies (33,56,59–61). A common theme of these studies is that it is necessary to use both empirical and statistical methodologies to optimize the experimental design. We have provided the first comprehensive overview of experimental design parameters for ChIP-chip data, testing both parameters related to all microarray experiments and those unique to ChIP-chip studies. We find good concordance of ChIP-chip and mRNA expression design characteristics. For example, in both cases, indirect hybridization designs appear to be more costly without providing a significant advantage over simpler direct designs. Similarly, we have shown that biological replicates are more important than dye-swaps, just as identified for mRNA expression arrays (56). We also demonstrated that antibody characteristics and amplification methods are major sources of variability in ChIP-chip studies. For example, using an antibody of maximal specific activity appears to be critical for increasing the sensitivity of ChIP-chip studies. In addition, we introduced a novel method for testing amplification bias in an antibody-independent manner. This study indicated that amplification procedures can significantly bias ChIP-chip results, and provides a simple, efficient method for benchmarking the bias of specific amplification protocols.
Our findings also raise a number of intriguing questions surrounding the antibody and amplification results. We demonstrate that antibody-specific baseline variability exists in ChIP-chip studies, and that while it can be mitigated it cannot be completely removed. It would be of great interest to replicate these studies with monoclonal antibodies, or polyclonal antibodies raised against different regions of a protein. In this manner, it might be possible to identify characteristics that predict which antibodies will be most sensitive or specific in a ChIP-chip study. Alternatively, this may not be a generalizable parameter and may be protein-specific. Similarly, we have presented a novel technique for studying amplification bias, and identified both a variant of LM-PCR and the WGA protocol as most favorable in this regard. Nevertheless, all amplification protocols tested here introduced a significant amount of bias. Our strategy for bias-identification should prove useful in the development of reduced-bias amplification protocols.
All of our analyses were performed studying the c-Myc transcription factor in the HL60 cell line. While this results in a consistent comparison across our many experiments, this also means that our results are formally restricted to a single biological system. However it is highly probable that our findings can be widely generalized. First, our validation experiment used a separate microarray platform with widely different feature sizes and again found a high confirmation rate by Q-PCR. Second, several of the parameters studied here are common to many microarray studies, including the choice of amplification method and the inter-batch and inter-day variability of microarrays and scanners. In particular, our analysis of amplification is relevant to studies of genomic variation using array-CGH and SNP-arrays. Third, the quality of antibody used in ChIP reactions will affect the quality of data retrieved by microarray, PCR and sequencing-based techniques. Thus we expect these findings to be broadly applicable to many fields.
As ChIP-chip studies continue to broaden in the scope and detail of the biological questions they probe, the application of optimized and validated experimental designs will be a significant advantage to the field.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Cancer Institute of Canada with funds from the Canadian Cancer Society (018298 to L.Z.P.); the Canadian Institutes of Health Research (MOP 67048 to L.Z.P.); the Canada Research Chairs Program (L.Z.P.); a Knudson Postdoctoral Fellowship (to R.P.); PreCarn Foundation scholarship (to P.C.B.); Natural Sciences and Engineering Research Council of Canada scholarship (to P.C.B.); Excellence In Radiation Research for the 21st Century Program fellowship (to P.C.B.); IBM (to I.J.); Terry Fox Foundation Fellowship through an award from the National Cancer Institute of Canada (to S.K.); Ontario Graduate Scholarships (to A.S. and A.P.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Penn Lab for helpful comments during the course of these experiments and in reviewing the article, and Mr Richard Lu for computer system support and administration.
REFERENCES
- 1.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 2.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 3.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 4.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 5.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horak CE, Mahajan MC, Luscombe NM, Gerstein M, Weissman SM, Snyder M. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc. Natl Acad. Sci. USA. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl Acad. Sci. USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, Farnham PJ, Huang TH, Penn LZ. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 13.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 14.Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat. Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 17.Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, Ijichi N, Ouchi Y, Shirahige K, ,,,,,,,,, Aburatani H, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26:4453–4463. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- 18.Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- 19.Kannan K, Kaminski N, Rechavi G, Jakob-Hirsch J, Amariglio N, Givol D. DNA microarray analysis of genes involved in p53 mediated apoptosis: activation of Apaf-1. Oncogene. 2001;20:3449–3455. doi: 10.1038/sj.onc.1204446. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- 21.Stanelle J, Stiewe T, Theseling CC, Peter M, Putzer BM. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002;30:1859–1867. doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberley MJ, Farnham PJ. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- 25.Ren B, Dynlacht BD. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 2004;376:304–315. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- 26.Oberley MJ, Tsao J, Yau P, Farnham PJ. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 27.Paris J, Virtanen C, Lu Z, Takahashi M. Identification of MEF2-regulated genes during muscle differentiation. Physiol. Genomics. 2004;20:143–151. doi: 10.1152/physiolgenomics.00149.2004. [DOI] [PubMed] [Google Scholar]
- 28.Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4:19. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat. Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 31.Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, Chen T, Herman D, Goodsaid FM, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat. Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 32.Shippy R, Fulmer-Smentek S, Jensen RV, Jones WD, Wolber PK, Johnson CD, Pine PS, Boysen C, Guo X, Chudin E, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat. Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 35.Tong W, Lucas AB, Shippy R, Fan X, Fang H, Hong H, Orr MS, Chu TM, Guo X, Collins PJ, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat. Biotechnol. 2006;24:1132–1139. doi: 10.1038/nbt1237. [DOI] [PubMed] [Google Scholar]
- 36.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 37.Wei C, Li J, Bumgarner RE. Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics. 2004;5:87. doi: 10.1186/1471-2164-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heisler LE, Torti D, Boutros PC, Watson J, Chan C, Winegarden N, Takahashi M, Yau P, Huang TH, Farnham PJ, et al. CpG Island microarray probe sequences derived from a physical library are representative of CpG Islands annotated on the human genome. Nucleic Acids Res. 2005;33:2952–2961. doi: 10.1093/nar/gki582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 41.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 44.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birnie GD. The HL60 cell line: a model system for studying human myeloid cell differentiation. Br. J. Cancer Suppl. 1988;9:41–45. [PMC free article] [PubMed] [Google Scholar]
- 47.Branham WS, Melvin CD, Han T, Desai VG, Moland CL, Scully AT, Fuscoe JC. Elimination of laboratory ozone leads to a dramatic improvement in the reproducibility of microarray gene expression measurements. BMC Biotechnol. 2007;7:8. doi: 10.1186/1472-6750-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fare TL, Coffey EM, Dai H, He YD, Kessler DA, Kilian KA, Koch JE, LeProust E, Marton MJ, Meyer MR, et al. Effects of atmospheric ozone on microarray data quality. Anal. Chem. 2003;75:4672–4675. doi: 10.1021/ac034241b. [DOI] [PubMed] [Google Scholar]
- 49.Bachtiary B, Boutros PC, Pintilie M, Shi W, Bastianutto C, Li JH, Schwock J, Zhang W, Penn LZ, Jurisica I, et al. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin. Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 50.Viganò MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, et al. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 2006;25:5105–5116. doi: 10.1038/sj.emboj.7601375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krieg AJ, Hammond EM, Giaccia AJ. Functional analysis of p53 binding under differential stresses. Mol. Cell Biol. 2006;26:7030–7045. doi: 10.1128/MCB.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semeralul MO, Boutros PC, Likhodi O, Okey AB, Van Tol HH, Wong AH. Microarray analysis of the developing cortex. J. Neurobiol. 2006;66:1646–1658. doi: 10.1002/neu.20302. [DOI] [PubMed] [Google Scholar]
- 54.Dobbin K, Shih JH, Simon R. Statistical design of reverse dye microarrays. Bioinformatics. 2003;19:803–810. doi: 10.1093/bioinformatics/btg076. [DOI] [PubMed] [Google Scholar]
- 55.Dobbin KK, Kawasaki ES, Petersen DW, Simon RM. Characterizing dye bias in microarray experiments. Bioinformatics. 2005;21:2430–2437. doi: 10.1093/bioinformatics/bti378. [DOI] [PubMed] [Google Scholar]
- 56.He YD, Dai H, Schadt EE, Cavet G, Edwards SW, Stepaniants SB, Duenwald S, Kleinhanz R, Jones AR, Shoemaker DD, et al. Microarray standard data set and figures of merit for comparing data processing methods and experiment designs. Bioinformatics. 2003;19:956–965. doi: 10.1093/bioinformatics/btg126. [DOI] [PubMed] [Google Scholar]
- 57.Dafforn A, Chen P, Deng G, Herrler M, Iglehart D, Koritala S, Lato S, Pillarisetty S, Purohit R, Wang M, et al. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques. 2004;37:854–857. doi: 10.2144/04375PF01. [DOI] [PubMed] [Google Scholar]
- 58.Marko NF, Frank B, Quackenbush J, Lee NH. A robust method for the amplification of RNA in the sense orientation. BMC Genomics. 2005;6:27. doi: 10.1186/1471-2164-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan W, Lin J, Le CT. How many replicates of arrays are required to detect gene expression changes in microarray experiments? A mixture model approach. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-research0022. research0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peixoto BR, Vencio RZ, Egidio CM, Mota-Vieira L, Verjovski-Almeida S, Reis EM. Evaluation of reference-based two-color methods for measurement of gene expression ratios using spotted cDNA microarrays. BMC Genomics. 2006;7:35. doi: 10.1186/1471-2164-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shih JH, Michalowska AM, Dobbin K, Ye Y, Qiu TH, Green JE. Effects of pooling mRNA in microarray class comparisons. Bioinformatics. 2004;20:3318–3325. doi: 10.1093/bioinformatics/bth391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.