Abstract
The program package ‘ClustScan’ (Cluster Scanner) is designed for rapid, semi-automatic, annotation of DNA sequences encoding modular biosynthetic enzymes including polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS) and hybrid (PKS/NRPS) enzymes. The program displays the predicted chemical structures of products as well as allowing export of the structures in a standard format for analyses with other programs. Recent advances in understanding of enzyme function are incorporated to make knowledge-based predictions about the stereochemistry of products. The program structure allows easy incorporation of additional knowledge about domain specificities and function. The results of analyses are presented to the user in a graphical interface, which also allows easy editing of the predictions to incorporate user experience. The versatility of this program package has been demonstrated by annotating biochemical pathways in microbial, invertebrate animal and metagenomic datasets. The speed and convenience of the package allows the annotation of all PKS and NRPS clusters in a complete Actinobacteria genome in 2–3 man hours. The open architecture of ClustScan allows easy integration with other programs, facilitating further analyses of results, which is useful for a broad range of researchers in the chemical and biological sciences.
INTRODUCTION
Bioprospecting for lead compounds from nature continues to be a corner stone in drug development. As well as isolating microorganisms from unique environments or biological diversity ‘hotspots’, approaches are also being developed to exploit the chemical diversity from > 98% of uncultivable microbes living in the natural environment. There is now unprecedented opportunity to access the natural diversity of small molecules made by such microbes by the isolation of metagenomic DNA and heterologous expression of biosynthetic pathways in a fermentable host. Discovery of novel biosynthetic gene clusters is the first goal of this culture-independent research that requires the application of molecular bioinformatics to identify DNA sequences of interest. We have developed an integrated set of computer programs for this task, which we call the ‘ClustScan’ (Cluster Scanner) program package.
Many important secondary metabolites in bacteria are synthesized on enzymes encoded by modular biosynthetic gene clusters: polyketide synthase (PKS) clusters, non-ribosomal peptide synthetase (NRPS) clusters, NRPS-independent siderophore (NIS) synthetase clusters or hybrid clusters (1–4). These secondary metabolites include polyketide antibiotics (e.g. erythromycin), immuno-suppressants (e.g. rapamycin) and antiparasitics (e.g. avermectin) as well as peptide antibiotics (e.g. vancomycin), immuno-suppressants (e.g. cyclosporin) and herbicides (e.g. bialaphos). Correlation of the chemical structures of the products with cluster DNA sequences shows that, in most cases, a defined series of catalytic domains that can be grouped into modules are responsible for each round of chain elongation. Thus, synthesis follows a co-linear principle in which the gene sequences of the individual modules determine the chemical outcomes of successive chain extension reactions. Large-scale DNA sequencing has revealed many gene clusters, whose products are not known (5–7). Predictions about the structures of the products based on the DNA sequences encoding enzyme modules can help decisions about which products may be interesting in the search for novel drugs. Modules are composed of domains that carry out the different reactions so that prediction of module specificity can be built up from that of domain specificity. In PKSs, each module usually contains an acyl carrier protein (ACP) domain and an acyl transferase (AT) domain, which is responsible for substrate selection and transferring the substrate to the ACP domain. For all modules except possibly the starter module (‘loading domain’) there is also a ketosynthase (KS) domain that performs condensation. Some AT domains select a malonyl–CoA substrate which results in a two carbon extension. However, other substrates can be used (e.g. methylmalonyl–CoA, ethylmalonyl–CoA, methoxy-malonyl–CoA) which result in the incorporation of more carbon atoms. However, the backbone chain is always extended by two carbon atoms and the other carbon atoms occur as side chains (e.g. methyl groups). Amino acid residues in AT domains that differ between malonyl–CoA-incorporating and methylmalonyl–CoA-incorporating have been identified from multiple alignments of AT sequences (8–12). There may be further reduction domains that carry out a sequential reduction of the introduced keto group: ketoreductase (KR) produces a hydroxyl group, which may be acted on by a dehydratase domain (DH) to produce a double bond that can be modified to a completely reduced product by an enoyl reductase (ER) domain. The stereochemistry of the addition step is also important. This can arise when the KR domain introduces a hydroxyl group and comparison of the sequences of KR domains introducing different stereochemistry identified specific residues correlated with this difference (13,14). A second source of differential stereochemistry is the incorporation of an extender unit with more than two carbon atoms resulting in a side chain with a choice of stereochemistry. At one time it was assumed that the KS domain was responsible for this choice. However, bioinformatic analyses could find no amino acid differences in the KS domain correlating with the stereochemical outcome and instead found correlations with the sequence of the KR domain (15). Studies of the 3D structure of KR domains provided mechanistic explanations of how the stereochemistry of the hydroxyl group and the α-carbon atom are controlled (16,17). The chirality of the α-carbon is lost if reduction of the hydroxyl group to a double bond on the β-carbon occurs, but this reduction may result in a new stereochemical choice between the cis- and trans-isomers that is probably determined by the DH domain carrying out the reduction. A new chirality may be created if full reduction occurs and is likely to be determined by the ER domain responsible for the final reduction step.
The annotation of the DNA sequence of a PKS cluster can be time-consuming because of the large number of domains and the necessity of integrating data from many sources. Several tools have been developed to assist this process. Identifying domains poses few problems as the sequences are well conserved. A much more difficult problem is predicting the activity and specificity of domains. The NRPS–PKS database (18, http://www.nii.res.in/nrps-pks.html), holds data on PKS and NRPS gene clusters including module and domain structure and chemical structures of the biosynthetic products. It allows users to input protein sequences to be used in BLAST (19) searches to identify domains and finds the closest sequences in the database. This allows prediction of whether an AT-domain uses malonyl–CoA or methylmalonyl–CoA as a substrate (i.e. whether a C2 or C3 unit is incorporated into the polyketide). The ASMPKS database (20, http://gate.smallsoft.co.kr:8008/%7Ehstae/asmpks/index.html) uses a similar methodology, but integrates it with a graphical display of the domains in genes so that modules can be easily recognized. It also allows the display of a predicted linear polyketide chain product for which the user has to select starter and extender units from lists. Minowa et al. (21) used an approach based on the creation of hidden Markov model profiles (22) to predict substrate specificity of AT domains. The company ECOPIA has also developed a software tool (23) DecipherIT™, which helps annotation of new gene clusters based on comparison with a database of known clusters. Although these approaches are useful, they do not make predictions about the stereochemistry of the products, which is extremely important for assessing their promise. As these analyses are essentially based on similarity to known clusters rather than identification of functional residues, they are less effective for clusters from novel organism groups. Another practical limitation is that they do not export information about chemical structures in a format that can be used by standard programs for further analyses.
In this paper, we describe a program that utilizes recent advances in understanding the function of KR domains to make knowledge-based predictions of activity and stereochemical specificity for hydroxyl groups and α-carbon atoms. This is combined with a fingerprint approach to predict specificity of AT domains and more conventional approaches for prediction of activity of DH and ER domains. The program predicts the chemical structures of products, which can be exported in a SMILES/SMARTS format for further analysis by standard Chemistry programs. The program is structured so that it can easily be updated to incorporate new knowledge about the specificity of domains. It has a convenient graphical interface that allows the rapid semi-automatic annotation of gene clusters encoding modular biosynthetic enzymes by non-expert users.
MATERIALS AND METHODS
GeneMark (24) (version 2.5; http://opal.biology.gatech.edu/GeneMark/) or Glimmer (25) (version 3.02; http://www.cbcb.umd.edu/software/glimmer/) were used to identify genes. HMMER (22) (version 2.3.2; http://hmmer.janelia.org/) was used for identification of protein domains. Profiles from Pfam (26) as well as specially constructed profiles were used. The gene prediction and protein domain prediction programs run on a Linux server and each user has a password to allow access to their own workspace. All user activities are performed via the Java client, which was written in Windows, MacIntosh and Linux versions.
To predict the specificity of AT or KR domains the amino acid sequence was aligned with an appropriate HMMER profile and the diagnostic amino acid residues extracted (Supplementary Data 1 Tables 2S and 3S). The diagnostic residues were compared to fingerprints corresponding to the different specificities (substrate specificity for AT; activity and stereochemistry for KR). The prediction of activity/inactivity of DH domains used a HMMER-profile based on active actinomycete domains. The prediction was based on the HMMER score. ER domains were detected using a profile based on a mixture of active and inactive domains.
To predict chemical structures, a table was constructed (see Supplementary Data 1 Table 4S) that contained different chemical building blocks written as isomeric SMILES (27). These were ordered on the basis of substrate and degree of reduction. In cases, where stereochemical prediction was not possible non-isomeric SMILES were used. Generic units as SMARTS (http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html) were also included for cases where prediction was not possible. The predictions were used to generate a description of the product in an XML format (http://www.w3.org/TR/xml/) organized in a hierarchical structure corresponding to module and domain architecture. This XML description was used to generate the chemical structure from the table of SMILES. This description was also used to generate a ring structure from the linear polyketide using a simple cyclization rule. The SMILES description can be drawn and displayed in ClustScan using Jmol v. 11.2.14, 2006 (Jmol; http://www.jmol.org/) or exported. Clustscan can be obtained by request from Novalis Ltd (novalis@novalis.hr).
RESULTS
The analyses of the DNA sequence data are carried out on a server and the results are cached so that each analysis only needs to be carried out once. This is important as the analysis of a whole Streptomyces genome may take several hours, but this can occur unsupervised overnight. The user accesses the results using a Java client that gives user-friendly presentation of the data. There is a password-protected workspace for each user on the server. The client allows the user to upload DNA sequences to the server and initiate analyses. The sequence is automatically translated in all six reading frames to allow HMMER (22) searches using a library of protein family profiles. The standard libraries contain PKS and/or NRPS domains, but it is possible to add other profiles if desired that makes the program package generic. These can be profiles from the Pfam (26) database or custom profiles created with the HMMER package (e.g. we have used profiles to find and annotate shikimic acid pathway genes; see Performance of ClustScan subsection). Independently of the search for protein patterns, the DNA can be analyzed to find probable coding regions using GeneMark (24) or Glimmer (25). GeneMark provides a library of models based on different bacteria and the appropriate model is chosen using a species related to the source of the DNA. Glimmer can construct a model for coding regions using long open reading frames (ORF) in the input sequence as training data. This is less effective for short input sequences. Also sequences with high G+C-content have long non-coding random ORFs, which may reduce the accuracy of coding sequence prediction. The program, therefore, also allows the user to create a model by supplying appropriate training data (e.g. the genome sequence of a related species) and the model can be stored by the user for future analyses.
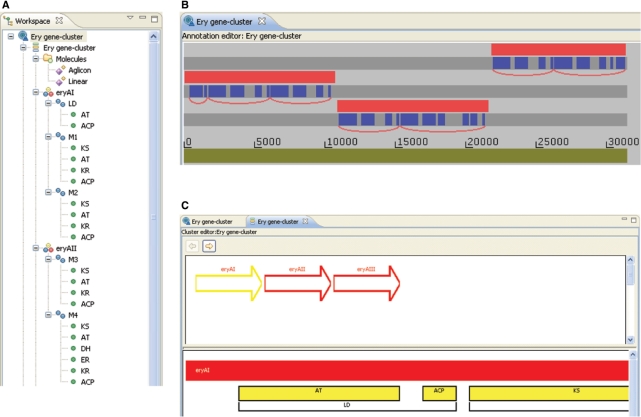
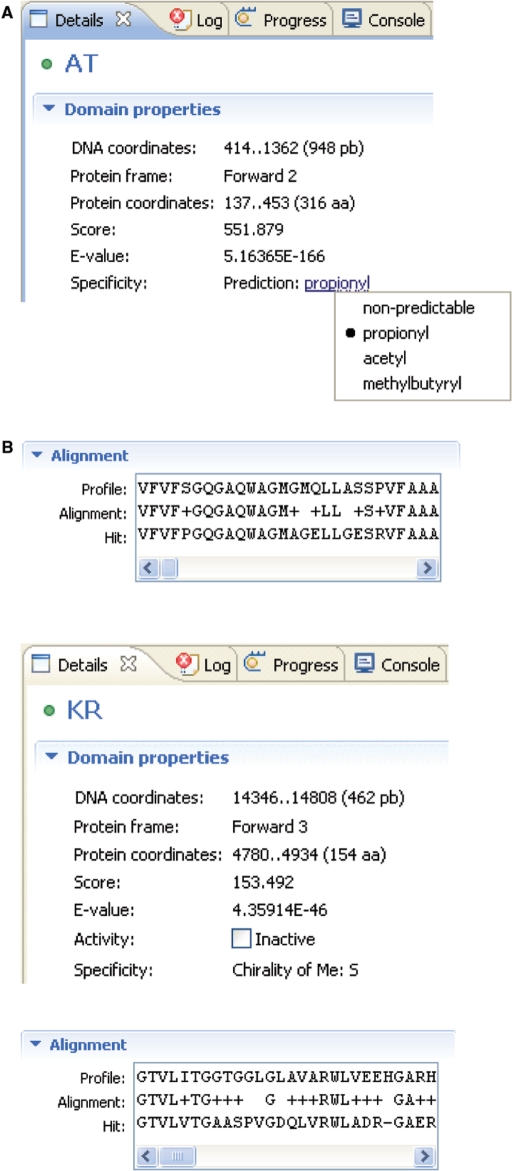
The results of the analyses are presented both as lists in the ‘workspace’ window (Figure 1A) and graphically in the ‘annotation’ editor window (Figure 1B). The workspace window shows the results in a tree format in which branches can be opened up or collapsed to show the genes and the protein domains. This is useful for obtaining an overview and it is possible to navigate through the thousands of genes present in a complete genome. The graphical ‘annotation’ editor window (Figure 1B) shows the positions of genes and protein domains on the six reading frames and can be viewed at different resolutions using a zoom function. It is possible using the mouse to displace genes and domains above and below the reading frames for better visualization of overlapping regions. It is usual to keep both the workspace window and the annotation editor window open and clicking on a feature in either, marks the corresponding feature in the other window. The protein domains are identified by HMMER analysis using a cut-off score that can be set to a stringent or relaxed value. This results in some putative protein domains, which may not be genuine. The user can choose to reject a protein domain so that it is removed from the analysis; the program tracks editing changes so that they can later be reversed if mistaken deletions occur. In many cases, the decision about the protein domain is taken on the basis of whether it occurs at an appropriate position with respect to other domains, which is easily seen in the graphical view of the annotation editor. To help the decision, the evidence for the identification of the protein domains can be viewed using the ‘details’ window (Figure 2). This shows the coordinates of the protein domain in the DNA and protein and the scores and E-values from the HMMER analysis. In addition, the alignment of the protein with the profile is shown. A prediction of the specificity of the protein domain is also shown. For AT domains (Figure 2A) this is the starter unit incorporated by the condensation reaction. For KR domains (Figure 2B) it is predicted whether the domain is active for reduction and, in addition, the stereochemistry of the hydroxyl group and the α-carbon atom are predicted. The predictions can be overridden if the user has extra information in conflict with the program's prediction. For instance, Figure 2A shows the (correct) prediction of propionyl as the starter unit for erythromycin. By clicking on the propionyl, a drop-down list is shown that enables selection of an alternative unit.
Figure 1.
(A) The workspace window gives an overview of the analysis in the form of collapsible trees. Detected genes and protein domains are shown. (B) The annotation editor window shows the location of genes (in red) and protein domains (in blue). In this case there are three genes on the three different forward open reading frames. The genes have been displaced from the reading frames by the user to allow better visualization of the domains. The annotation editor has been used for user definition of modules (shown as red curves below the open reading frames). (C) The cluster editor window. The user can define a set of contiguous genes as a cluster. The cluster editor window shows the genes in a cartoon form with an expanded view of the selected gene showing protein domains. Domains can be linked together to give modules. The modules are given identifying names and the program suggests a biosynthetic order that can be accepted or altered by the user.
Figure 2.
The details window allows the user to examine the evidence for assignment of protein domains. The HMMER scores and E-values as well as the alignment are displayed. The predictions of activity and specificity are also displayed and can be modified by the user. (A) The loading AT domain of the erythromycin cluster. The program makes the correct prediction of a propionyl starter unit. By clicking on this choice, a selection window has been opened that allows the user to override the automatic prediction and select an alternative choice. (B) The KR domain of module 3 of the erythromycin cluster.
On the basis of the results in the annotation editor, the user can define a gene cluster covering a region of adjacent genes. The annotation of the gene cluster is carried out using the ‘cluster’ editor window (Figure 1C), which shows the genes of the gene cluster in a simple cartoon form, hence the term semi-automatic. When a gene is selected, the protein domains are also shown. Modules can be assembled by marking protein domains and each module created is given a name. The program suggests a biosynthetic order of the genes of a cluster. For PKS clusters this is based on identifying a potential loading domain (i.e. typically a module containing only AT and ACP domains; Figure 1C) and looking for a thioesterase domain as identifying the last module. If there is ambiguity, it is assumed that the genes are used in the pathway in the same order as they occur in the DNA. This procedure identifies the correct biosynthetic order in most natural gene clusters. The user can alter the suggested order to incorporate any additional knowledge available.
The complete annotation by ClustScan can be stored as a file in an XML format so that it can be reimported into ClustScan. The hierarchical nature of XML makes it well suited for representing clusters in terms of genes, modules and protein domains. We developed an XML format that includes information about the biosynthetic order. Although the XML format is primarily designed for the internal use of ClustScan, it makes it easy for other applications to read or write ClustScan compatible files by adding an appropriate XML parser. In addition to the XML format, annotations can be exported as an EMBL or GenBank file for use in other applications or for submission to databases; this results in loss of information on biosynthetic order. In addition, the DNA or amino acid sequences of genes, domains or modules can be copied to the clipboard for further analyses with other programs.
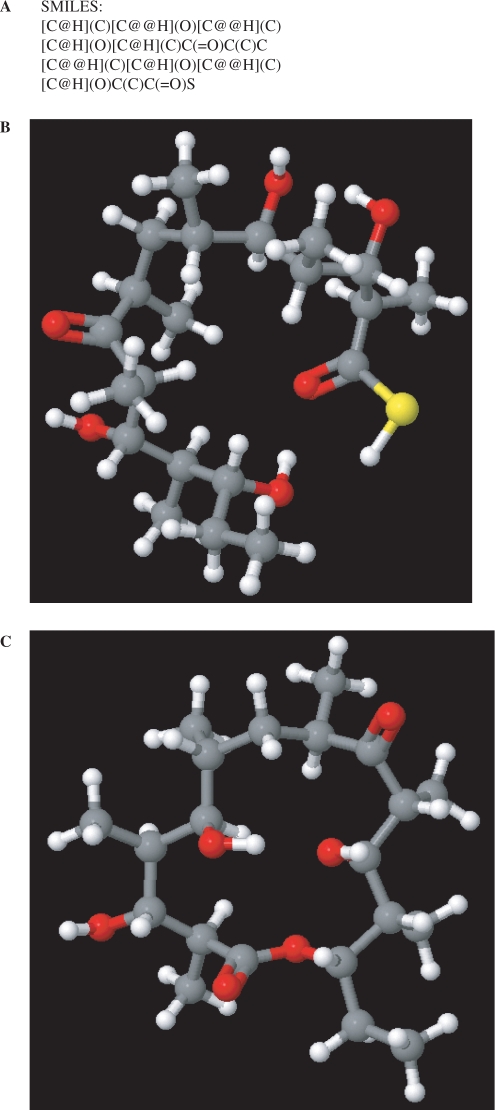
The prediction functions for the activity and specificity of protein domains are used to deduce module specificity and, thus, to predict the chemical structure of the linear polyketide chain product of the gene cluster. The structures are represented internally in the program as isomeric SMILES (27), which can be copied to the clipboard (Figure 3A) allowing export for use with standard chemical software. The user can define new module specificities and provide isomeric SMILES descriptions of the extender units. It is possible to edit the prediction of module specificity to allow incorporation of such novel extender units. The program allows the user to display the chemical structure of products in a 3D ‘molecules’ window (Figure 3B) in which the molecule can be rotated. The program can also produce a potential cyclic structure from a linear molecule (Figure 3C). It is assumed that the first hydroxyl or amino group introduced during synthesis reacts with the terminal extender unit.
Figure 3.
The molecules window. (A) The SMILES description for the linear backbone of erythromycin predicted from the DNA sequence of the cluster. The SMILES description can be copied to the clipboard for export. (B) The 3D structure of the predicted linear chain is shown. The mouse can be used to rotate the molecule. (C) The ring structure of the erythromycin aglycone as predicted using the cyclization function of the program.
The program is designed to allow easy incorporation of new knowledge. New or modified prediction of enzyme activity or specificity can be implemented without changing program structure. It is also possible for sophisticated users to write their own specific scripts to introduce specialized prediction functions.
Prediction of domain activities
The presence of any of the seven domains KS, AT, ER, DH, KR, ACP or TE is detected using the HMMER profiles. An extender module needs at least KS, AT and ACP. AT determines the substrate selection for the extension reaction. The three reduction domains (KR, DH and ER) may be absent or present as active or inactive domains. ClustScan predicts whether domains are active as well as predicting substrate specificity or stereochemical outcome when several outcomes are possible (see Supplementary Data 1 Table 1S).
The KR domain is the best characterized domain in terms of structural determination of differential activities. Active KR domains determine the chirality of the hydroxyl product and bioinformatic analysis identified amino acid residues involved in this choice (13,14). Bioinformatics also suggested that KR rather than KS determined the stereochemistry of β-carbon groups, when C3 or C4 units are incorporated (15). A comparison of 3D structures of two KR domains of different specificity gave more detailed information on amino acid residues involved in determining both hydroxyl and β-carbon stereochemistry (17). In ClustScan, alignment with a KR profile allows identification of all of these critical amino acids (the ‘fingerprint’) and, thus prediction of the product. The fingerprints used are shown in the Supplementary Data 1 Table 2S. There are six possible products (A1, A2, B1, B2, C1, C2), which correspond to three possible ketoreduction outcomes (either hydroxyl stereoisomer—A or B, or no reduction C) coupled with two β-carbon chiralities (called 1 and 2). The accuracy of prediction was tested using 49 KR domains for which the structure of the polyketide product provides information about activity and stereochemistry; if further active reduction domains are present, the product does not provide any information about the stereochemistry of the KR step. Ten of the KR domains processed 2-carbon extender units so that only hydroxyl stereochemistry was relevant: all 10 predictions were correct. Nine of the KR domains processing 3- or 4-carbon extender units were inactive: in eight cases the program predicted that the domains were inactive for reduction and also predicted the correct side chain stereochemistry. In one case the inactive KR domain was predicted as active. The other 30 KR domains processing 3- or 4-carbon extender units were active. In 25 cases the program predicted the correct stereochemistry. In one case, the program predicted the incorrect side chain stereochemistry (A1 instead of A2). In the other four cases, the alignment with the profile did not yield an amino acid fingerprint that fell into any of the groups: in these cases the program indicates that no prediction is possible. Thus, the KR prediction was correct in 88% of the cases, incorrect in 4% of the cases and the program was unable to provide a prediction in 8% of the cases.
Unlike the case of KR, structural information about AT domains is not sufficient to help in substrate prediction. The most common extender substrates are malonyl–CoA and methylmalonyl–CoA. Comparison by eye of alignments of AT domain sequences identified 13 amino acid residues, which differed significantly between domains incorporating the two substrates (8–12). The amino acid sequences of nine AT extender domains that incorporated ethylmalonyl–CoA were examined. It was found that the 13 amino acid residues had a common pattern that differed from those of the malonyl–CoA and methylmalonyl–CoA-specific AT domains. This information was used for prediction of specificity in the program. A further known extender substrate is methoxymalonyl–CoA and specific residues associated with choice of this substrate were identified in AT domains of the concanamycin A cluster (28). Eleven methoxymalonyl-incorporating AT domains were examined, but the 13 fingerprint residues used to characterize the other substrates did not show a conserved pattern. It was noticed that most had insertions with respect to the conserved alignment of all AT domains, which caused problems in identifying potential fingerprinting residues. After using a specific alignment for methoxymalonyl–CoA-incorporating AT domains, it was possible to use a modified form of the published pattern (28) to predict methoxymalonyl–CoA as a substrate.
The information about AT extender specificity was implemented in ClustScan. The amino acid sequence of the AT domain was aligned with a general AT-profile to identify the 13 diagnostic amino acid residues. These were compared to three fingerprints corresponding to the three substrates. If the amino acids did not fit any of the three fingerprints, the AT domain was aligned using a profile derived from the 11 methoxymalonyl–CoA AT domains. This alignment was used to test if one of the characterized insertions was present. If no match was found, the AT domain was assigned to an unknown substrate category.
In addition to AT domains in extender modules, there are often AT domains in loading domains. A set of AT domains that incorporate acetyl starters (nine domains), propionyl starters (eight domains) or methylbutyryl starters (three domains) were aligned with the general AT profile and the 13 diagnostic amino acid residues extracted. The fingerprints for acetyl and propionyl starters were identical to those for acetyl and propionyl extenders, respectively. The methylbutyryl starters showed a different pattern, which was also used to construct a specific fingerprint. This information was incorporated into ClustScan. When the user accepts the suggested biosynthetic order or defines a different order the loading domain is subjected to a special analysis. If an AT domain is present, the 13 diagnostic amino acids are extracted and tested for acetyl, propionyl or methylbutyryl fingerprints. All fingerprints for the specificity of AT starter and extender units are shown in Supplementary Data 1 Table 3S. A dataset of 196 known AT domains was analyzed with ClustScan (95 malonyl–CoA, 79 methylmalonyl–CoA, 9 ethylmalonyl–CoA and 13 methoxymalonyl–CoA. The remaining 25 were propionyl, acetyl, methylbutyryl and some unusual ones from loading domain ATs). This gave the correct prediction in 182 cases (93%), the wrong prediction in 9 cases (5%) and assignment to an unknown class in 5 cases (3%).
For DH domains, the prediction should distinguish active and inactive domains. As insufficient structural information was available to make predictions based on knowledge of function, it was decided to use a profile based on active domains to try to predict activity. The profile was built using the sequences of 57 active domains derived from actinomycetes. The profile was used to screen the active domains used in its construction as well as an additional 56 active and 46 inactive domains (159 total). All domains with a high score (>300) were active, whereas all with a low score (<200) were inactive. About 80% of the domains with intermediate scores were active, but the scores of inactive domains were distributed through the range. These results were used to define a prediction function with three outcomes: active (score >300), 80% probability of activity (scores between 200 and 300) and inactive (score < 200). This prediction function was tested on 159 domains (113 active and 46 inactive). Forty-six domains fell into the intermediate region (36 active, 10 inactive) with a prediction of 80% probability of activity. Sixty-seven domains were predicted to be active of which six were in fact inactive corresponding to a 9% false prediction rate. In contrast, the prediction of inactivity was less satisfactory: 43 DH domains were predicted to be inactive of which 16 were actually active. A closer examination of these false predictions showed that only 1 of the 16 was an actinomycete sequence, the other 15 being sequences from Gram-negative bacteria. When attention was confined to actinomycete sequences, 13 DH domains were predicted to be inactive of which only one was active.
Initially, a similar approach to that used for the DH domain was attempted with the ER domains. A profile was constructed using active actinomycete ER domains. However, it was found that better prediction was achieved with a profile based on a mixture of active and inactive domains. Sixty-six known ER domains were tested. In all cases the ER domain was detected. The HMMER score did not prove useful in distinguishing between active and inactive ER domains. However, there were only three cases of inactive ER domains in the presence of an active DH domain. There were four cases in which an ER domain was detected, but the DH domain was inactive. The program, therefore, predicts an active ER domain if a domain is found and there are active KR and DH domains present. This gives a false prediction in the 3/66 (5%) cases of an inactive ER domain with an active DH domain.
Performance of ClustScan
There are two main criteria for the usefulness of ClustScan: the accuracy of prediction and the speed and convenience of annotating large datasets. The accuracy of prediction was tested on two well-known gene clusters: the erythromycin gene cluster and the niddamycin gene cluster (GenBank accession numbers AY771999 and AF016585). For the erythromycin gene cluster, with one exception, all the protein domains of the six extender modules were accurately identified and the propionyl starter (Figure 2A) was also predicted. The only exception was that ClustScan was not able to predict the hydroxyl group stereochemistry of the KR domain of module 4; the prediction of the hydroxyl stereochemistry is flagged as unknown. This does not have an effect on the final prediction as an active DH domain forms a double bond. However, the active ER domain recreates a chiral center, which cannot be predicted with the current state of knowledge. This resulted in two possible structures, where the user can choose the correct chirality to obtain an accurate prediction of the chemistry of the linear backbone (Figure 3A and B). In this case, the cyclization was also predicted correctly (Figure 3C) (see also Supplementary Data 1 Figure 1S A and B). In the niddamycin gene cluster, the five genes, the loading domain and the seven extender modules containing 36 catalytically active domains were all correctly predicted with the exception that the substrate for module six was predicted as ethylmalonate instead of the correct methoxymalonate. The inactive KR in module 4 responsible for the β-carbon: S stereochemistry was predicted. The correct cyclization was also predicted (see Supplementary Data 1 Figure 2S A and B). The results with ClustScan were compared with those from the NRPS–PKS database prediction system (SEARCHPKS), which is the most popular current analysis tool for PKS clusters (see Supplementary Data 2 Figures 1–4). SEARCHPKS (http://www.nii.res.in/nrps-pks.html) requires protein sequences so the amino acid sequences of the genes were extracted with ClustScan and submitted. SEARCHPKS found two extra false positive ACP domains in the erythromycin cluster (Supplementary Data 2 Figure 1). The first at the end of the eryAI gene did not affect the prediction as it was an isolated ACP domain. The second occurred between the KS and AT domains of module five and resulted in the program predicting an additional module and making no prediction of the chemistry of the two modules generated. It is not possible to review the data behind the prediction or to manually reject the false positives. SEARCHPKS found all the other domains successfully, but does not attempt to make predictions of the activity or stereochemistry of the reduction domains. In particular, this results in the false prediction of an active KR domain in module 3 resulting in the prediction of a hydroxyl group rather than the correct keto group. The substrate choice of the loading domain was not predicted, but there was correct prediction of a C3 unit for five of the six extender modules; no prediction of substrate was possible for module 5. For niddamycin (Supplementary Data 2 Figure 2) there was also a false prediction of an additional ACP domain in module 2. This results in a false positive prediction of an additional module and an inability to predict the chemical structures associated with the two ‘modules’. In module 4, the KR domain was incorrectly predicted as active and the substrate for module 5 could not be predicted. Like ClustScan the wrong substrate for module 6 was predicted.
Eight further well-characterized clusters were annotated. For the megalomycin, pimaricin and tylactone clusters the predicted module activities were in full agreement with the published results. For tylactone (Supplementary Data 2 Figure 3) SEARCHPKS found all the domains, but is unable to predict activity of reduction domains; thus, it predicts a chemistry based on an active KR in module 4, whereas ClustScan correctly identifies the domain as inactive. Also, the starter unit is not correctly predicted. The worst results for ClustScan were obtained with the rifamycin cluster, where the stereochemistry of three methyl groups could not be predicted and two of eight DH domains were falsely predicted as active. In comparison, SEARCHPKS falsely predicts five DH domains and one KR domain as active and does not attempt to predict the stereochemistry (see Supplementary Data 2 Figure 4). For the other four clusters (amphotericin, avermectin, nystatin and oleandomycin) there were fewer errors (data not shown). Six additional domains were identified, which were not present in the published annotations. Two were TE domains; as the presence of a TE domain does not directly affect the structure of the compound, it is likely that previous annotation work had not searched carefully for these domains. The other four new domains were all DH domains with significant deletions (a third to a half of the length). They are, thus, predicted as inactive by ClustScan. Although such partially deleted domains are not important for prediction of product structure, they are interesting for studies on the evolution of clusters.
A major problem with annotations in DNA database entries is that they are not uniform, but differ according to the person carrying out the annotation. ClustScan helps achieve a uniform annotation standard and we have reannotated published sequences to achieve a standard definition of domain boundaries and description of units. ClustScan has been used to annotate successfully more than 50 modular gene clusters from a variety of genomes and metagenomes; full details are available on request.
The speed and convenience of ClustScan were assessed using the genome sequences of Saccharopolyspora erythraea (7) which is 8.2 Mb in size. A graduate student was able to annotate the PKS and NRPS clusters in 2–3 h of work (the initial analysis using HMMER can take several hours of run time on the server, but this occurs unsupervised overnight). The ClustScan annotation identified genes, modules and protein domains and included prediction of activity, substrate specificity and stereochemical outcome for PKS domains. The published annotation (7) identified genes, modules and protein domains and, in addition, the AT domains are assigned to malonyl–CoA and methylmalonyl–CoA-incorporating classes. However the stereochemistry and activity of reduction domains are not annotated. The ClustScan annotation agreed with the published annotation and extended it with predictions of domain activity and stereochemistry of products. ClustScan has been used to annotate DNA sequences from a variety of bacterial species including cyanobacteria.
ClustScan is mainly designed for use with bacterial sequences. However, the more general utility of ClustScan program package was demonstrated by the analysis of lower eukaryote sequences, where intron prediction is often difficult. An example is provided by the slime mould Dictyostelium discoideum which has 45 PKS genes (29), which were annotated poorly by the standard annotation methods used in the genome project. Using ClustScan it was possible to use local HMMER profiles for the protein domains, which are effective in recognizing segments of the domains split by introns. When such an analysis is carried out, a PKS gene shows a characteristic signature with parts of protein domains in the correct order with gaps due to introns in between. The view in the annotation editor window allows easy recognition of genes and the coordinates of the domain segments help in detecting the intron boundaries.
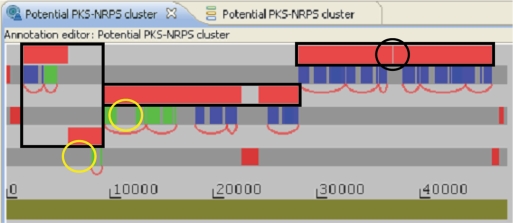
ClustScan is mainly designed for the annotation of gene clusters encoding modular biosynthetic enzymes, but it can also be used for annotating other genes by loading appropriate HMMER profiles. For instance, we have used seven profiles to find and annotate shikimic acid pathway genes in a marine organism (30). Recently there has been intensive activity with metagenomic sequences. The source organisms for sequences are not known, but they contain genomes from a number of culturable and non-culturable microorganisms. The contigs are often fairly small and the quality of the sequence is sometimes poor. These problems make an analysis using HMMER local profiles attractive. We used ClustScan to analyze a 200 kb DNA sequence (AACY020563593) from the J. Craig Venter Institute Global Ocean Sampling (GOS) Expedition metagenomic dataset (31). This revealed a potential PKS–NRPS hybrid gene cluster of about 50 kb in size (Figure 4). It starts with an NRPS loading module, followed by three PKS modules and seven NRPS modules and ends with an NRPS thioesterase domain. However, closer examination of the domain distribution between reading frames reveals several cases where domains forming a single module appear to be present in different neighboring genes. This is due to three apparent frameshifts and the anomalous occurrence of a stop codon, which probably arise due to sequencing errors. Thus, it seems likely that there are three genes rather than the seven genes indicated by both GenMark and Glimmer analysis. In the case of two of the potential PKS modules, no AT domains are recognized, but there are unassigned regions in the protein of appropriate sizes and locations for AT domains (Figure 4). Thus, the program allows rapid scanning of metagenomic datasets and makes it easy to identify potential sequencing errors and interesting features of clusters. With the growing importance of metagenomic data for drug discovery programs ClustScan helps to eliminate a major bottleneck in the analysis.
Figure 4.
Annotation editor window showing the analysis of a potential PKS–NRPS hybrid cluster from a marine metagenomic sequence. The following coloring is used: genes (red), PKS protein domains (green) and NRPS protein domains (blue). Although seven genes are shown, the distribution of domains between genes suggest that sequencing errors have occurred. The three boxes indicate the positions of the probable genes. The first gene has one frameshift, the second gene has two frameshifts and the third gene has an anomalous stop codon (ringed in black) in it. The positions where two AT domains would be expected are also ringed (in yelow).
DISCUSSION
ClustScan is easy to use and allows rapid annotation of new gene clusters. This is very important for exploitation of the rapidly accumulating data from large-scale DNA sequencing projects. The facts that high-quality annotation with traditional methods is very time consuming and needs a high degree of experience have prevented full exploitation of the extensive DNA database to identify potentially interesting biosynthetic enzyme clusters. Although ClustScan is easy to use, it also allows the user to customize the result and override the automatic predictions. It is also designed to allow easy incorporation of new knowledge to improve predictive power. The server–client architecture means that such improvements as well as changes to reflect new versions of the standard analysis programs are implemented on the server and do not need changes in the client programs installed on users' computers. An important goal in the design of ClustScan was to give it an open architecture which would allow easy integration with other programs. The definition of an XML format for full gene cluster description allows interchange with other programs by simply adding an appropriate XML parser. The export of annotation as EMBL or GenBank formats and the export of chemical structures as SMILES (27) facilitate further analyses of results generated by ClustScan.
Knowledge about PKS protein domains is used to make predictions about chemical structure. In the case of the KR domain (14) there is detailed knowledge about protein structure and the role of the small number of amino acid residues that control reductase activity and stereospecificity. In the case of AT extender domains 13 amino acid residues that correlate with the choice between malonyl–CoA and methylmalonyl–CoA substrates were known (8–12). We found that these 13 amino acids could also be used to predict ethylmalonyl–CoA substrate. The incorporation of methoxymalonyl–CoA substrates was correlated with insertions. Initially, we tried to use a method similar to that of Minowa et al. (21) based on HMMER profiles of critical amino acids to predict AT specificity. However, this approach gave lower accuracy of prediction than the fingerprint method that we used subsequently. For both the KR and AT domains, the frequency of false prediction was low (4%). It was striking that good results were obtained for both Gram-negative sequences as well as for the majority of Gram-positive actinomycete sequences. This supports the idea that the diagnostic residues in AT domains are functional in substrate specificity rather than being evolutionary accidents. In contrast, the DH activity prediction, which was based on an actinomycete profile was only efficient for actinomycetes. In particular, many active Gram-negative DH domains were predicted to be inactive. This means that the profile mismatch is caused by the evolutionary distance. Although it would be possible in the short term to improve DH prediction using profiles for specific groups of organisms, the identification of important functional amino acid residues would give predictions less dependent on evolutionary distance. In contrast to other annotation programs (18,20,21,23,32), ClustScan predicts the stereochemistry of products. The dependence on functional residues in the KR and AT domains makes it especially valuable for novel gene clusters that are not closely related to known gene clusters. Such clusters are especially interesting in the search for novel drugs. We have not implemented specificity predictions for NRPS protein domains. However, there is some information available to allow partial prediction (32). When the prediction power is good enough it will be easy to add NRPS predictions to ClustScan and predict the chemical structure of products. We compared the performance of ClustScan to that of the SEARCHPKS prediction program of the NRPS–PKS database (18). This is less convenient to use as the genes must be identified and the deduced protein sequence input to the program. The output of predicted chemistry is not available in a standard chemical format. SEARCHPKS often predicts additional ACP domains that prevent accurate prediction of product chemistry. We also observed that BLAST (19) searches often gave problems in identifying ACP domains, whereas no problems were encountered with HMMER (22); this is probably because of the short length of ACP domains. The prediction of the specificity of extender AT domains is relatively good; this probably reflects the fact that AT specificity correlates well with phylogenetic trees (33) so that, in addition to critical functional amino acids, there are other amino acids that differ for evolutionary reasons. As the BLAST program does not weight residues according to conservation, it works best when differences at many residues correlate with activity. SEARCHPKS does not give good prediction of loading module specificities. It does not attempt to predict activity or stereochemistry of domains. As these predictions involve a small number of critical residues, they could not be effectively implemented using a BLAST-based approach. The ASMPKS database (20) could not be meaningfully compared to ClustScan as it's gene prediction for clusters with high G+C-content was very poor and it requires a DNA input. This is because it uses the Glimmer (25) program to predict genes and builds an HMM-model from input data. In ClustScan, we implemented the use of custom HMM-models to overcome this difficulty for subgenomic sequences. As the ASMPKS implements a similar approach to the NRPS–PKS database, it is likely that similar results would be found if this technical problem were overcome.
There are at least 15 known starters used by different modular PKSs. In many cases there is no AT domain in the loading domain. Acetyl, propionyl and methylbutyryl starters can be loaded by AT domains and it was found that they could be distinguished using diagnostic amino acid residues. It was striking that the acetyl and propionyl starter AT domains showed the same patterns as the malonyl–CoA and methylmalonyl–CoA extender domains. It is known that in some cases an acetyl starter is derived from decarboxylation of a malonyl–CoA substrate, but in other cases acetyl–CoA is the substrate (34). The fact that the commonest extenders' AT domains are closely related to starter AT domains suggests that it might be possible to evolve new PKS gene clusters from truncated clusters that have lost the starter module.
Most polyketides undergo cyclization. In ClustScan we have implemented a simple rule of cyclization by interaction of the first hydroxyl or amino group with the terminal group. This applies to many natural polyketides and raises the hope that a simple rule-based method can make correct predictions in many cases. Prediction of cyclization is important to obtain the full benefit of product prediction.
The ability to rapidly acquire knowledge of new gene clusters from their DNA sequence has a variety of implications in the search for pharmacologically relevant compounds. The identification of novel gene clusters with interesting and unusual product chemistry will direct the choice of targets for lead discovery. Another application of the new sequences is to use them to construct new polyketides based on known modules in silico; i.e. use them as input for a program such as the Biogenerator program (35). ClustScan will help eliminate the bottleneck posed by the annotation of DNA sequences and allow the full utilization of the rapidly increasing DNA sequence data. Studies on the evolution of secondary metabolite clusters (36) can reveal biological constraints on the structures that can be attained; such studies are greatly assisted by the ability to rapidly and accurately annotate new clusters.
We used a top-down approach based on HMM models to annotate gene clusters encoding modular biosynthetic enzymes. We showed that by choice of appropriate profiles, ClustScan could also be used for annotating other primary and secondary metabolic pathways in a variety of microbial and invertebrate organisms. It seems likely that extensions of this approach could be useful for more general annotation tasks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Ministry of Science, Education and Sports, Republic of Croatia (grant 058-0000000-3475 to D.H.); the German Academic Exchange Service (DAAD) and the Ministry of Science, Education and Sports, Republic of Croatia cooperation grant (to D.H. and J.C.); the Leverhulme Trust; Japanese Bio-Industry Association; The School of Pharmacy, University of London (to P.F.L.).
Conflict of interest statement. J.S. declares a financial interest related to this work with regard to the potential future commercial marketing of the software and databases.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Janko Diminic and Vedran Rodic for programming and Professor Arnold Demain for helpful advice and encouragement. J.S. declares a financial interest related to this work with regard to the potential future commercial marketing of the software and databases.
REFERENCES
- 1.Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 2.Finking R, Marahiel MA. Biosynthesis of non-ribosomal peptides. Ann. Rev. Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 3.Hranueli D, Cullum J, Basrak B, Goldstein P, Long PF. Plasticity of the Streptomyces genome - evolution and engineering of new antibiotics. Curr. Med. Chem. 2005;12:1697–1704. doi: 10.2174/0929867054367176. [DOI] [PubMed] [Google Scholar]
- 4.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 5.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 7.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 8.Haydock SF, Aparicio JF, Molnar I, Schwecke T, Khaw LE, König A, Marsden AF, Galloway IS, Staunton J, Leadlay PF. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA: acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 1995;374:246–248. doi: 10.1016/0014-5793(95)01119-y. [DOI] [PubMed] [Google Scholar]
- 9.Lau J, Fu H, Cane DE, Khosla C. Dissecting the role of acyltransferase domains of modular polyketide synthases in the choice and stereochemical fate of extender units. Biochemistry. 1999;38:1643–1651. doi: 10.1021/bi9820311. [DOI] [PubMed] [Google Scholar]
- 10.Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, McDaniel R. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry. 2001;25:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- 11.Del Vecchio F, Petkovic H, Kendrew SG, Low L, Wilkinson B, Lill R, Cortes J, Rudd BAM, Staunton J, Leadlay PF. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 2003;30:489–494. doi: 10.1007/s10295-003-0062-0. [DOI] [PubMed] [Google Scholar]
- 12.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003;328:335–363. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 13.Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 14.Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 15.Starcevic A, Cullum J, Jaspars M, Hranueli D, Long PF. Predicting the nature and timing of epimerisation on a modular polyketide synthase. Chembiochem. 2007;8:28–31. doi: 10.1002/cbic.200600399. [DOI] [PubMed] [Google Scholar]
- 16.Castonguay R, He W, Chen AY, Khosla C, Cane DE. Stereospecificity of ketoreductase domains of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc. 2007;129:13758–13769. doi: 10.1021/ja0753290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Yadav G, Gokhale RS, Mohanty D. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 2003;31:3654–3658. doi: 10.1093/nar/gkg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Tae H, Kong EB, Park K. ASMPKS: an analysis system for modular polyketide synthases. BMC Bioinformatics. 2007;8:327. doi: 10.1186/1471-2105-8-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minowa Y, Araki M, Kanehisa M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- 22.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 23.Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat. Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 24.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–454. doi: 10.1093/nar/gki487. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 28.Haydock SF, Appleyard AN, Mironenko T, Lester J, Scott N, Leadlay PF. Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiology. 2005;151:3161–3169. doi: 10.1099/mic.0.28194-0. [DOI] [PubMed] [Google Scholar]
- 29.Zucko J, Skunca N, Curk T, Zupan B, Long PF, Cullum J, Kessin R, Hranueli D. Polyketide synthase genes and the natural products potential of Dictyostelium discoideum. Bioinformatics. 2007;23:2543–2549. doi: 10.1093/bioinformatics/btm381. [DOI] [PubMed] [Google Scholar]
- 30.Starcevic A, Akthar S, Dunlap WC, Shick JM, Hranueli D, Cullum J, Long PF. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc. Natl Acad. Sci. USA. 2008;105:2533–2537. doi: 10.1073/pnas.0707388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenke-Kodama H, Börner T, Dittmann E. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLoS Comput. Biol. 2006;2:e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long PF, Wilkinson CJ, Bisang CP, Cortes J, Dunster N, Oliynyk M, McCormick E, McArthur H, Mendez C, Salas JA, et al. Engineering specificity of starter unit selection by the erythromycin-producing polyketide synthase. Mol. Microbiol. 2002;43:1215–1225. doi: 10.1046/j.1365-2958.2002.02815.x. [DOI] [PubMed] [Google Scholar]
- 35.Zotchev SB, Stepanchikova AV, Sergeyko AP, Sobolev BN, Filimonov DA, Poroikov VV. Rational design of macrolides by virtual screening of combinatorial libraries generated through in silico manipulation of polyketide synthases. J. Med. Chem. 2006;49:2077–2087. doi: 10.1021/jm051035i. [DOI] [PubMed] [Google Scholar]
- 36.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl Acad. Sci. USA. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.